Introduction
Oral squamous cell carcinoma (OSCC) accounts for
>90% of oral and maxillofacial cancers and for 3% of all
malignant types of tumour (1).
Tissue biopsies and their histological analysis are conventionally
the gold standard in diagnosing OSCC and determining its type and
stage based on which to make clinical management decisions;
however, limitations such as invasive and difficult to perform
sampling methods, as well as and time-consuming analysis, are
primary challenges (2,3). With the rapid development of molecular
biological technology, previous studies have focused on early
detection and sensitive noninvasive biomarkers for OSCC diagnosis
(1,4).
Exosomes are microvesicles with a lipid bilayer
membrane; their size ranges from 30 to 150 nm and their density is
1.13–1.19 g/ml of sucrose. Exosomes can be identified using the
specific membrane marker ALG-2 interacting protein X (ALIX) and
tumor susceptibility 101 proteins (TSG101). Studies have indicated
that exosomes may be abundantly released by different cell types
into physiological fluids, such as the peripheral blood, urine and
breast milk (5). Depending on the
type, biogenesis and physiological condition of cells, exosomes
contain a wide range of proteins, lipids and nucleic acids,
reflecting the characteristics of parental cells (6). In exosomes, nucleic acids [including
microRNAs (miRNAs/miRs) and long non-coding RNAs (lncRNAs)] are
protected from rapid degradation by a lipid bilayer membrane to
maintain their stability; they may be transported into target cells
to remain stable and may be regulated over long distances (7). Therefore, exosomes are regarded as an
entirely new type of cell-to-cell communication; their cellular
messengers trigger a variety of physiological and pathological
responses and achieve cross-talk to adjacent or distant cells
(8).
lncRNAs are a class of ncRNAs with >200
nucleotides. Although lncRNAs have been initially considered
‘transcriptional noise’, they have been regarded as emerging
regulators of biological functions and described to have an
essential role in gene expression at multiple levels, such as
chromatin remodelling, X chromosome silencing and transcriptional
activation; they also participate in cell growth, differentiation,
metabolism and oncogenesis (9).
Studies have also indicated that aberrant lncRNA expression is
associated with numerous cancer types, e.g., colorectal cancer,
breast cancer and liver cancer (10). Therefore, differential expression of
exosomal RNA (exo-RNA), including miRNAs and lncRNAs, may exist in
various cancer types (11,12).
A considerable amount of studies has focused on the
aberrant expression of exo-miRNAs in OSCC (13–15), but
only a few studies have explored exo-lncRNAs (16,17).
Thus, the present study aimed to investigate exo-lncRNA expression
profiles and their detection potential for OSCC through a
bioinformatics analysis.
Materials and methods
Cell isolation and culture
Human oral epithelial cells (HOECs) were isolated
and used in accordance with the ethical standards established in
the Declaration of Helsinki. The present study was approved by the
ethics committee of the School/Hospital of Stomatology at Lanzhou
University (Lanzhou, China). HOECs were isolated and cultured as
previously described (18–20). Normal oral mucosa was sampled and
pooled between March and June 2018 from 10 patients (4 females and
6 males) who had surgery for cleft lip reconstruction (age range,
0.3–1 years; mean age, 0.78 years) and did not have any tumours.
Informed consent forms were signed by the patients' parents prior
to their participation in the study. The specimens were washed
using sterile PBS containing penicillin and streptomycin. After the
connective tissue was removed, the specimens were cut into small 2
mm × 1 mm pieces and incubated in Dispase II (Merck Millipore) at
4°C for 18 h. The epithelial layer was mechanically separated,
digested with 0.25% trypsin/0.02% EDTA (Thermo Fisher Scientific,
Inc.) at 37°C for 7 min and vigorously mixed. The mixture was
collected in a tube and centrifuged at 100 × g for 7 min at 4°C.
The supernatant was carefully removed, and the cells were
resuspended in an EpiGROTM Human Epidermal Keratinocyte Expansion
Medium (Merck Millipore). The cell line CAL-27 was obtained from
the American Type Culture Collection and cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal
bovine serum (FBS; both Gibco; Thermo Fisher Scientific, Inc.).
Once the cells grew in a stable manner, they were washed with PBS
and the CAL-27 medium was replaced with DMEM supplemented with 10%
(v/v) exosome-depleted FBS (System Biosciences). All of the
supernatants were collected and maintained for 2 days at 4°C for
further analysis.
Exosome isolation
As described in a previous study by our group, the
general approach of isolating exosomes from the cell culture
supernatant was based on differential ultracentrifugation. Once the
volume of the collected supernatant reached 100 ml, all the
following operations were performed at 4°C: The supernatant was
first centrifuged at 300 × g for 10 min, then at 2,000 × g for 10
min and finally at 10,000 × g for 30 min to discard the pellets
(cells, dead cells and cell debris). The supernatant was then
ultracentrifuged at 100,000 × g for 70 min at 4°C to produce a
sediment and concentrate the exosomes. The pellet was resuspended
in PBS and collected in a centrifuge tube. The sample was
centrifuged at 100,000 × g for 70 min at 4°C and the supernatant
was carefully removed by using a pipette. The pellet after
centrifugation was washed with PBS, whilst the pellet containing
the exosomes was resuspended in 100 µl PBS and maintained at −80°C
for further analysis.
After the particles were isolated from CAL-27 and
HOECs, the morphological characteristics and phenotype of the
particles were identified using the procedures described below.
Western blot analysis
The particles were thawed and treated with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) and protease inhibitor cocktail (Beyotime Institute
of Biotechnology) in accordance with the manufacturer's protocols.
Subsequently, 2X SDS-PAGE loading buffer (Sigma-Aldrich; Merck
KGaA) was added, followed by heating to 95°C for 5 min. The
proteins were separated using 10% SDS-PAGE and transferred onto
nitrocellulose (NC) membranes (Beyotime Institute of
Biotechnology). The NC membranes were blocked with 5% skimmed milk
powder at room temperature for 1.5 h, washed thrice with TBS-Tween
(0.1% Tween-20), by using an orbital shaker and incubated with
primary antibodies at 4°C overnight. The following primary
antibodies were used: Rabbit monoclonal anti-ALIX (1:1,000
dilution; cat. no. F-2609; System Biosciences) and goat monoclonal
anti-TSG 101 (1:1,000 dilution; cat. no. K-1711; System
Biosciences). Subsequently, the NC membranes were incubated with
horseradish peroxidase-conjugated secondary goat anti-mouse
antibody (1:5,000 dilution; System Biosciences) and rabbit
anti-mouse antibody (1:5,000 dilution; System Biosciences) at room
temperature for 1 h. The membranes were visualized using Immobilon
Western Chemiluminescent HRP Substrate (cat. no. WBKLS0100; EMD
Millipore) on a Bio-Rad gel imaging system (Gel Doc™
XR+; Bio-Rad Laboratories, Inc.).
Transmission electron microscopy
Isolated particles were resuspended in PBS and 20 µl
of the mixture was loaded onto Formvar-coated 200-mesh copper
electron microscopy grids at room temperature for 10 min. Excess
liquid was drained by touching the grid edge against a piece of
filter paper. Counterstaining was performed by floating the grid on
a drop of 3% phosphotungstic acid solution (pH 6.8) at room
temperature for 5 min. Excess liquid was drained and the grid was
dried for several minutes at room temperature. Microphotographs
were obtained using an HT7700 transmission electron microscope
(Hitachi).
Particle size analysis
The particles isolated from 100 ml collected
supernatant were thoroughly resuspended in 1 ml PBS. The size
distribution of the particles was determined using a Zetasizer Nano
ZS (Malvern) in accordance with the manufacturer's protocols.
High-throughput lncRNA sequencing
Following the manufacturer's protocol, the total RNA
of exosomes was extracted using the Total Exosome RNA and Protein
Isolation kit (Invitrogen; Thermo Fisher Scientific, Inc.). The
total RNA of exosomes was used for lncRNA library preparation and
sequencing, which was performed at RiboBio Co., Ltd., where the
total RNAs from CAL-27-exo and HOEC-exo were reverse-transcribed
and their ends were repaired and amplified through PCR. The PCR
products were sequenced using the Illumina HiSeq 2500 (Illumina,
Inc.). Raw data were initially filtered in terms of low-quality
reads, ribosomal RNA contamination and reads containing >10%
uncertain nucleotides. Subsequently, clean reads were generated and
mapped with the reference genome by using TopHat 2 for the
subsequent analysis (21).
Bioinformatics analysis
After the sequencing data were obtained, Auddics was
used as a classic procedure to identify differential RNA
expression. The target genes of lncRNAs were predicted using
GENCODE (https://www.gencodegenes.org/), the European Molecular
Biology Laboratory-European Bioinformatics Institute (https://www.ebi.ac.uk/), lncRNAdb (http://www.lncrnadb.org/) and the gene database from
the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) according to their
cis or trans mechanism to systematically uncover the
functions of lncRNAs. Their comprehensive functions were analysed
using Gene Oncology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (http://www.genome.jp/). Furthermore,
the |log2 (fold-change)|>2, significance level
(P<0.05) and cancer-associated pathways of lncRNA-targeted genes
were set to select the candidate lncRNAs and evaluate their
diagnostic potential.
Relative expression levels of the
selected lncRNAs
The relative expression of the selected lncRNAs was
assessed using reverse transcription-quantitative (RT-q)PCR to
further validate the data from the high-throughput lncRNA
sequencing. The total RNAs were extracted from the cells and
exosomes using an RNeasy Mini kit (Qiagen GmbH) and the Total
Exosome RNA and Protein Isolation kit (Invitrogen; Thermo Fisher
Scientific, Inc.), respectively. The RNAs were treated with
RNase-free DNase I (Takara Bio, Inc.) to remove any DNA
contamination and eluted in 25 µl RNase-free ultrapure water. The
relative lncRNA expression was determined using PrimeScript™ RT
Master Mix (Perfect Real Time; Takara Bio, Inc.) and SYBR-Green™
Premix Ex Taq™ II (Tli RNaseH Plus; Takara Bio, Inc.) on a 7500
sequence detector system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). PCR was performed in a mixture (20 µl)
containing 2 µl of complementary DNA template, 10 µl 2X SYBR-Green
PCR Mix, 0.4 µl Rox II and 0.8 µl each of sense and antisense
primers. RT-qPCR was performed in triplicate for each sample. GAPDH
was used as the control and the specificity of the PCR products was
estimated from the melting curve. The following primer sequences
used for qPCR were synthesized by Tsingke Biological Technology
Co., Ltd.: NR_026892.1 forward, 5′-GGTCTACCAGTTGCACAGATT-3′ and
reverse, 5′-CAGAGAAAGAAGGTGGGAGTTAG-3′; NR_036586.1 forward,
5′-CCAACATGGGCTCTCAATACA-3′ and reverse,
5′-CACCATACCTGGCACATACAA-3′; NR_126435.1 forward,
5′-GTCTGACATCCAGAGCCAATAC-3′ and reverse,
5′-AGGCCTAACCATGTTTCCTTAC-3′; and GAPDH forward,
5′-GGTGAAGGTCGGAGTCAACGG-3′ and reverse,
5′-GAGGTCAATGAAGGGGTCATTG-3′. The relative expression of each
lncRNA was calculated using the 2−ΔΔCq method (22).
Statistical analysis
Data were presented as the mean ± SD (n=3). GraphPad
Prism version 6.0 (GraphPad Software, Inc.) was used for all of the
calculations. An unpaired Student's t-test was applied to examine
the differences in lncRNA expression obtained via RT-qPCR.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Exosomes from CAL-27 and HOEC culture
supernatant
The particles isolated from the supernatant of
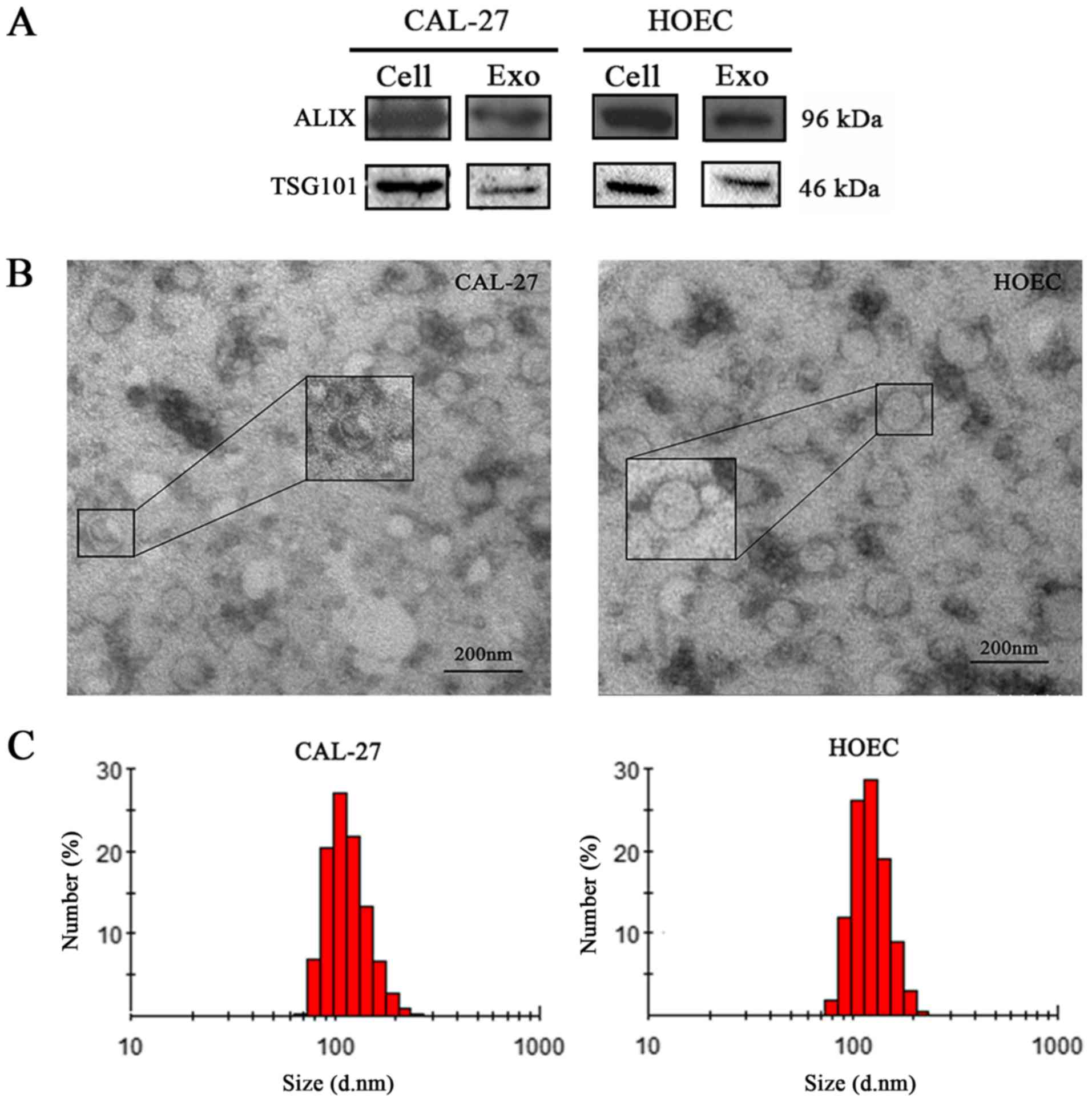
CAL-27 and HOEC were verified by detecting the expression of ALIX
and TSG101, which are known markers of exosomes (23) (Fig.
1A). The exosomes had a round or oval shape and a membrane
structure (Fig. 1B). The size of
most particles ranged from 30 to 150 nm and the cumulative
percentages of the particle diameter interval for CAL-27 in the
ranges of 0–30, 30–150 and >150 nm were 0, 89.6 and 10.4%,
respectively, and those for HOEC were 0, 87.8 and 12.4%,
respectively (Fig. 1C). These
observations confirmed that the particles isolated from the
supernatant were exosomes.
ncRNA and lncRNA expression in
exosomes
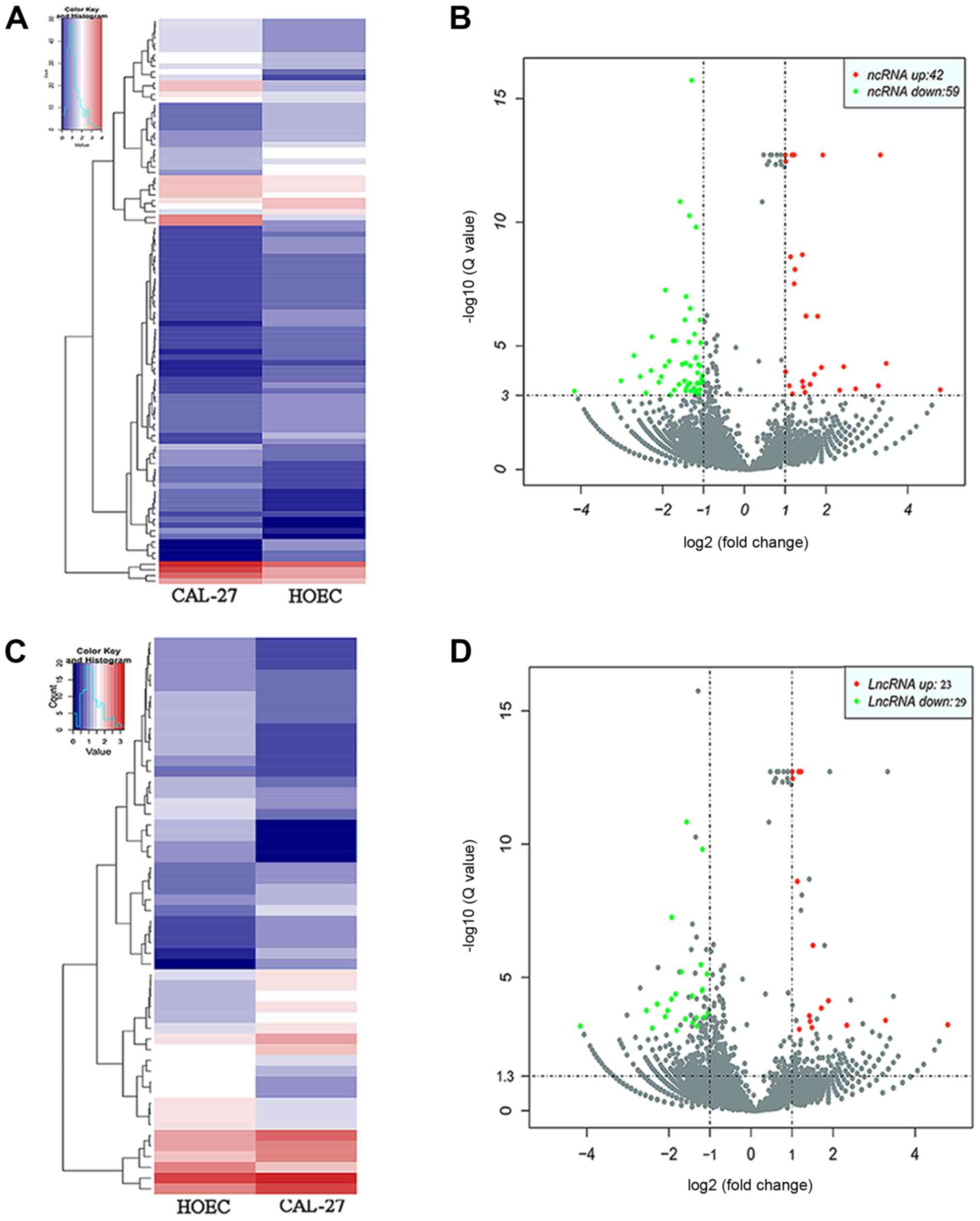
The sequencing data identified 28,437 ncRNAs in
CAL-27 and 29,254 ncRNAs in HOECs. By using the parameters
difference multiple (|log2 (fold-change)|>1), false
discovery rate (FDR)<0.001 and P<0.05, a total of 101
differentially expressed ncRNAs between CAL-27-exo and HOEC-exo
were identified. Amongst them, 42 ncRNAs were upregulated, whereas
59 ncRNAs were downregulated (Fig. 2A
and B). Of the 101 ncRNAs, 52 differentially expressed lncRNAs
were identified with 23 upregulated lncRNAs and 29 downregulated
lncRNAs (Fig. 2C and D).
Functional enrichment analysis of the
target genes of differentially expressed lncRNAs
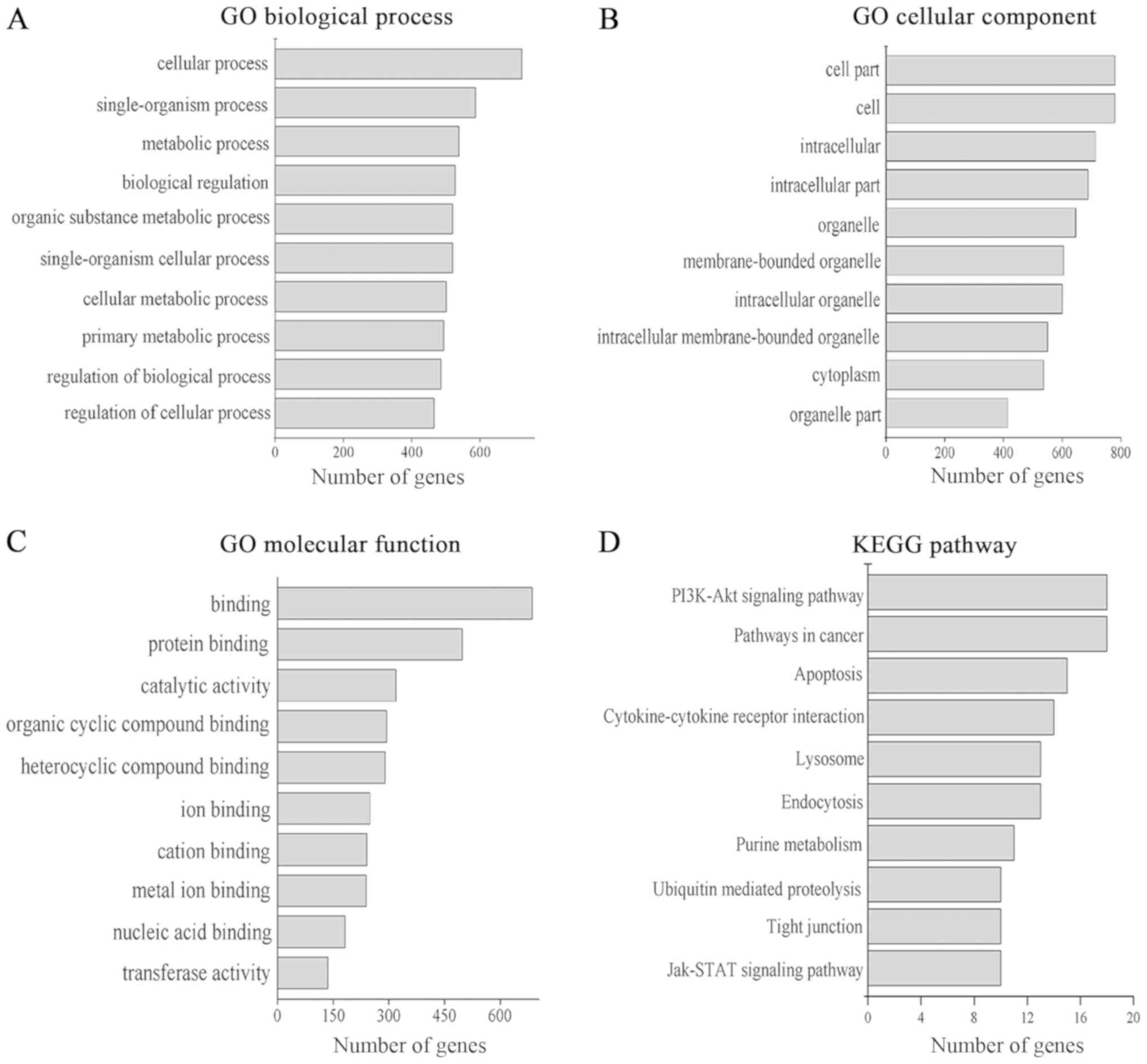
GO annotation in the categories biological process,
cellular component and molecular function was performed. A total of
1,549 GO terms in the biological process category (P<0.05), 227
GO terms in the cellular component category (P<0.05) and 320 GO
terms in the molecular function category (P<0.05) were
identified. The top 10 GO terms of each category are presented in
Fig. 3A-C, respectively; ‘cell’ and
‘cell part’ were the most prominent terms in the cellular component
category, while terms in the molecular function and biological
process categories were mainly associated with cellular processes,
binding functions and catalytic activity. KEGG pathway analysis
identified 50 pathways (P<0.05). The top 10 KEGG pathways are
presented in Fig. 3D. The target
genes of lncRNAs were mainly associated with the regulation of the
‘PI3K/Akt signaling pathway’, ‘Jak/STAT signaling pathway’,
‘pathways in cancer’ and ‘apoptosis’. These data confirmed that
exosomal lncRNAs may be involved in cell metabolism and oncogenesis
in general.
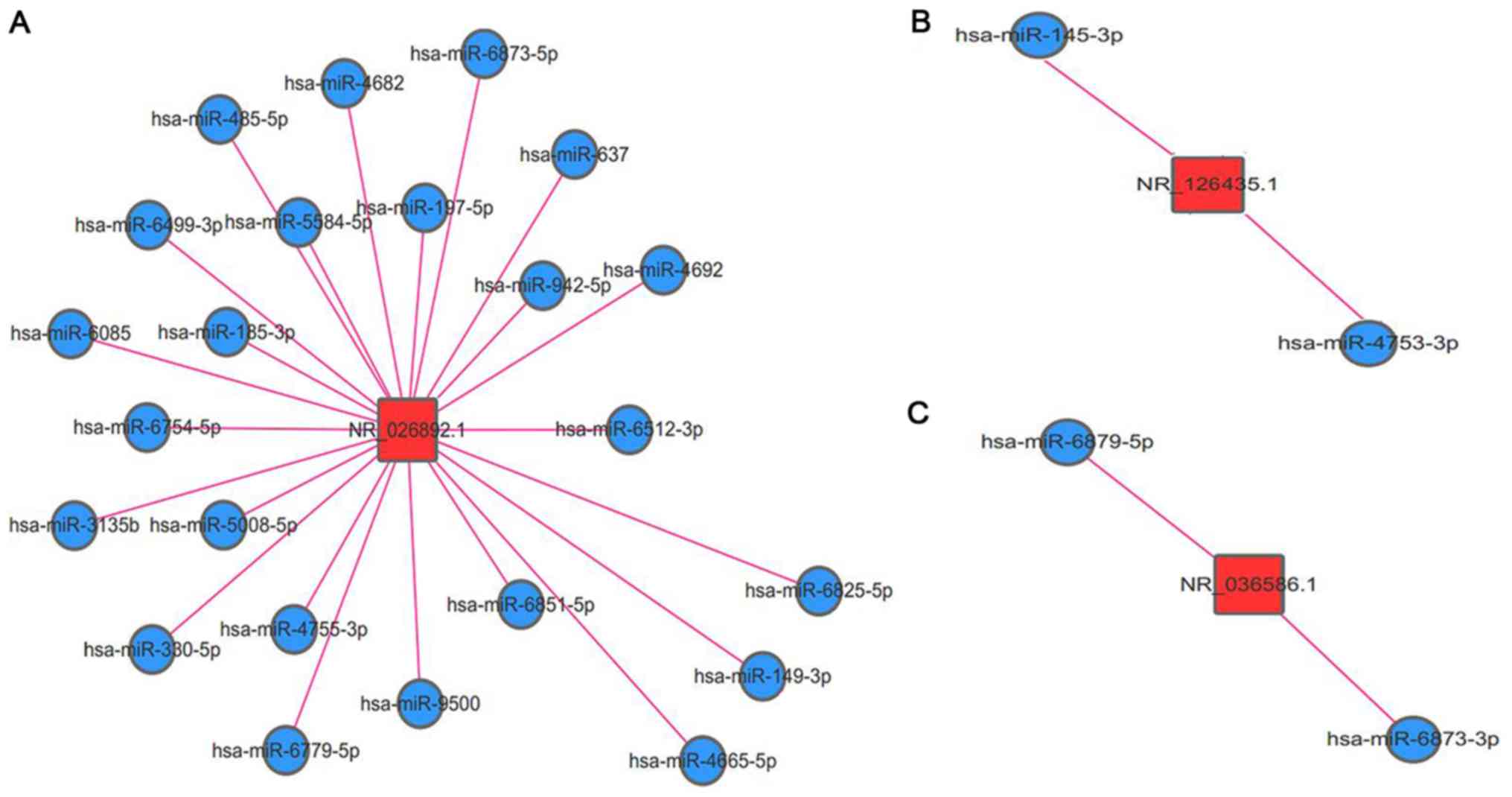
Selection and functional prediction of
candidate lncRNAs
After 52 differentially expressed lncRNAs were
screened, the expression levels of 12 lncRNAs were identified to
match |log2 (fold-change)|>2, FDR<0.001 and
P<0.05. These 12 lncRNAs were further filtered if their target
genes were involved in tumour-associated pathways and 3 lncRNAs
(i.e., NR-026892.1, NR-126435.1 and NR-036586.1) were ultimately
selected as the potential biomarkers of exo-lncRNAs. The
lncRNA-miRNA interactions were also predicted to elucidate the
functions of NR-026892.1, NR-126435.1 and NR-036586.1 (Fig. 4A-C, respectively). The network
revealed that NR-026892.1 interacted with 23 miRNAs, whilst
NR-126435.1 and NR-036586.1 interacted with 2 miRNAs each.
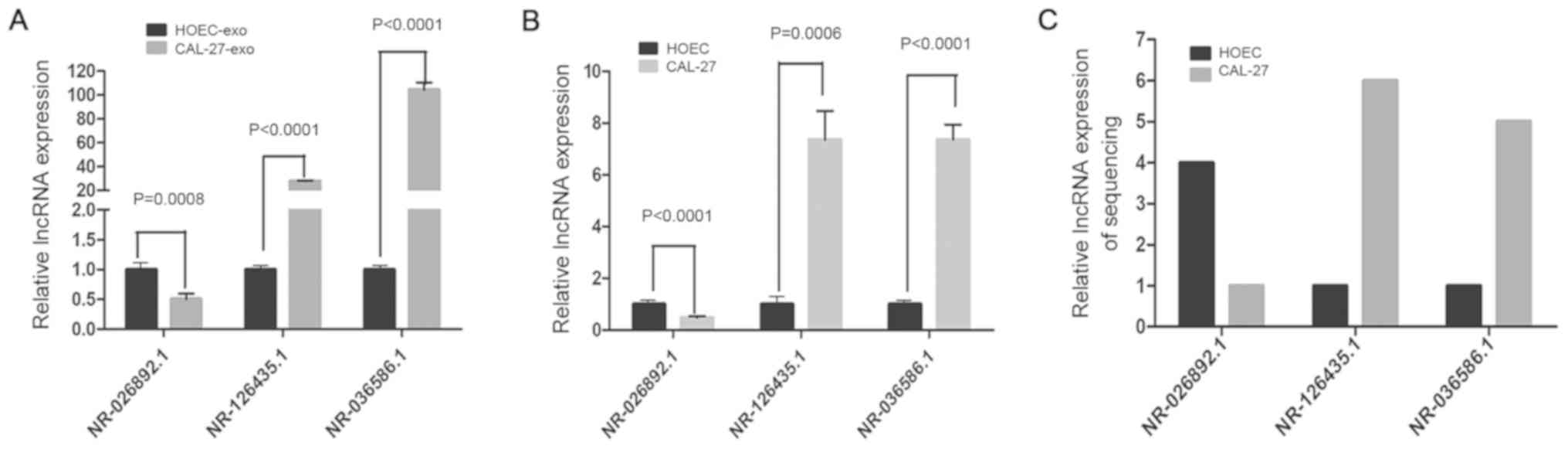
Relative expression levels of the
three candidate lncRNAs in cells and exosomes
RT-qPCR analysis confirmed that the expression of
the selected lncRNAs in CAL-27-exo was significantly different from
that in HOEC-exo (P<0.001), and there were also significant
differences between CAL-27 and HOECs (P<0.001). The relative
expression levels of NR-026892.1 in CAL-27-exo and CAL-27 were
approximately half of those in HOEC-exo and HOECs, respectively
(1.002±0.04328 vs. 0.4784±0.02827 in the cell group and
1.002±0.0451 vs. 0.5022±0.05389 in the exosome group), and the
differences were significant (Fig. 5A
and B). Conversely, the relative expression levels of
NR-126435.1 and NR-036586.1 were markedly higher in CAL-27-exo and
CAL-27 than in HOEC-exo and HOECs, respectively (Fig. 5A-C). For NR-126435.1, the
overexpression was more marked in CAL-27-exo vs. HOEC-exo than in
CAL-27 vs. HOEC (1.001±0.02648 vs. 27.77±0.3024 in the exosome
group and 1.009±0.09347 vs. 7.358±0.6427 in the cell group).
NR-036586.1 was upregulated by nearly 100-fold in the exosomes of
CAL-27 vs. HOEC and nearly 7-fold at the cellular level
(104.4±3.544 vs. 1.001±0.03821in the exosome group and 7.104±0.4076
vs. 1.001±0.033 in the cell group). The RT-qPCR results were
consistent with the sequencing data, which suggested that the
results of the exosomal lncRNA sequencing were credible.
Discussion
Despite advances in treatment, the 5-year survival
rate of patients with OSCC remains <50% due to the lack of early
diagnosis. To improve this situation, more sensitive and accurate
biomarkers should be developed for the early detection of OSCC.
According to The Cancer Genome Atlas, cancer initiation and
progression are likely a result of the interaction of environmental
factors and genetic alterations (24,25).
Evidence has demonstrated that lncRNAs modulate diverse processes
in tumour progression, metastasis and suppression (26). Hence, a tumour may be detected in its
early stage based on the evaluation of lncRNA expression levels in
tissues. However, this procedure is limited due to the invasiveness
of tissue collection. Of note, exosomal lncRNAs have a differential
abundance in exosomes that may reflect the characteristics of
parental cells (5), distinguishing
certain exosomal lncRNAs as potential cancer markers.
Tumour exosomes (TEXs) have been extensively studied
as novel biomarkers for the early detection and prognosis of
cancer, as they may exist stably in the circulatory system.
Signaling molecules, including RNAs and proteins, from exosomes may
be targeted to cells; these cells may then produce various factors
that are able to promote tumour proliferation, invasion and
migration (27). TEX-lncRNA has an
important utility in early cancer detection (28,29).
Various OSCC cell lines, including OSC-4, SCC1,
SCC2, SCC4, SCC9, CAL-27, UM1 and UM2, have been established.
Continuously established OSCC cell lines have become important
research tools to gain a better understanding of the pathogenesis
of this disease and search for efficient therapeutic strategies.
Amongst the cell lines mentioned above, CAL-27 is frequently used
and regarded as a representative cell line in the field of OSCC.
Jiang et al (30) selected
CAL-27 to build OSCC models for in vitro and in vivo
studies. While the in vitro studies have revealed satisfactory
results, in vivo studies have demonstrated abnormal growth
patterns of CAL-27 ×enografts, with vesicles slowly growing at the
surface and deeper areas of tumours. Jiang et al (30) concluded that certain established cell
lines are not suitable for building OSCC models for in vitro
and in vivo studies, as their growth patterns differ from
one another. Therefore, CAL-27 cells were chosen to be used in the
present study.
In the present study, KEGG analysis suggested that
lncRNA-associated target genes were mostly enriched to the PI3K/Akt
signalling pathway, which is a classic signalling pathway of
cancer. Previous studies suggested that this pathway may be
activated by numerous types of cellular stimuli or toxic insults
and may regulate fundamental cellular functions, including
transcription, translation, proliferation, growth and survival
(31–35). Once activated, Akt may control key
cellular processes by phosphorylating substrates involved in
proliferation, metastasis and invasion. Therefore, differentially
expressed lncRNAs may be involved in this cancer-associated
pathway.
The function of lncRNAs is similar to that of
miRNAs, which are able to regulate the expression of neighboring
target genes (36,37). The present study indicated that
certain lncRNAs are significantly associated with their target
genes and that those lncRNAs exert their function through their
predicted target genes.
lncRNAs and target genes may interact in cis
and trans forms. The target genes obtained by using the
cis model must meet two criteria: i) Genes should be located
in the range of 10 kb upstream and downstream of three
significantly and differentially expressed exo-lncRNAs; ii) a
significant co-expression trend should be observed in three
exo-lncRNAs. In the trans model, target genes acquire the
sequences of candidate exo-lncRNAs and differentially express mRNAs
first. These sequences are then screened by consecutively using
blast (e<1×10−5) and RNAplex (G<20) (38,39).
NR_126435.1, one of the selected lncRNAs, is located
on chromosome 4 and its target genes are associated with fibroblast
growth factor 5 (FGF5) and von Hippel-Lindau tumour suppressor
(VHL). FGF5, identified as an oncogene, belongs to the FGF family.
FGF family members are able to promote mitogenesis and cell
survival, which are involved in various biological processes,
including embryonic development, cell growth, angiogenesis, tissue
repair, tumour growth and progression. FGF5 expression is markedly
upregulated in patients with cancer (40,41). VHL
is able to form a multimeric complex that functions in the
ubiquitination and degradation of hypoxia-inducible-factor (HIF).
Exosomes derived from hypoxic OSCCs promote the migration and
invasion of normoxic OSCCs via HIF-1α and HIF-2α (13,42). The
interaction between NR_126435.1 and its target genes, e.g., FGF5
and VHL, implied that NR_126435.1 may participate in oncogenesis.
NR_126435.1 was overexpressed in CAL-27 and CAL-27-exo (fold-change
>6, P=0.0006). NR-026892.1 is located on chromosome 4 and its
target genes are associated with multiple coagulation factor
deficiency 2 (MCFD2), which is a marker of testicular germ cell
tumours (43). Studies have
demonstrated that MCFD2 promotes OSCC metastasis by regulating the
expression levels of lectin, mannose binding 1 and galectin 3
binding protein. Hence, MCFD2 may be a promising novel therapeutic
target in patients with metastatic OSCC (44). In the present study, the network of
lncRNA-miRNA interactions revealed that NR-026892.1 interacted with
23 miRNAs. These miRNAs also have a role in numerous types of
cancers. For instance, miR-485-5p is a potential tumour suppressor
in different cancer types and its overexpression may inhibit SCC25
proliferation by PAK1 (45).
miR-9500 is able to reduce the expression levels of Akt1 and affect
tumourigenesis and metastasis (46).
miR-637 acts as a tumour suppressor in pancreatic cancer cells
(47). miR-149-3p targets Akt1 and
induces cell apoptosis (48). In
previous studies, Akt1, as a serine/threonine-protein kinase, was
demonstrated to have anti-proliferative and anti-migratory effects
and functioned as an oncogene in multiple types of cancer (46). These results suggested that
NR-026892.1 may participate in cancer metastasis. NR_036586.1 is
located in chromosome 2 and its target gene is associated with
period circadian regulator 2 (PER2), whose gene polymorphisms may
be closely linked to certain types of cancer. Downregulation of
PER2, which is a promising predictor of OSCC, may contribute to the
tumourigenesis and development of OSCC; PER2 may also regulate its
anti-carcinogenic biological function by interacting with several
signalling pathways, including the P53/cyclin dependent kinase
inhibitor 2A, PIK3CA/AKT and caspase 8 pathways (49). Therefore, similar to NR_126435.1,
NR_036586.1 might be prone to tumourigenesis.
In conclusion, to the best of our knowledge, the
present study was the first to report that TEX-lncRNA may
contribute to the early detection of OSCC based on bioinformatics
analysis outcomes. NR-026892.1, NR-126435.1 and NR-036586.1 may be
explored as future detection tools through rigorous verification
and mechanistic investigation. Further studies are warranted to
conclusively determine whether the three candidate lncRNAs may be
considered useful OSCC biomarkers. Therefore, in further studies by
our group, exosomes will be isolated from pathological samples of
OSCC and adjacent tissues and from salivary/blood specimens of
patients with oral premalignant lesions and OSCC in the near
future.
Acknowledgements
Not applicable.
Funding
The present study was supported, in part, by the
National Natural Science Foundation of China (grant no. 31572522),
Gansu Natural Science Foundation (grant no. 17JR5RA217) and the
Fundamental Research Funds for the Central Universities (grant
nos.lzujbky-2017-it44, lzujbky-2018-it44 and lzukqky-2019-t01).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JJ and ZH analyzed and interpreted the data
regarding the exosomes and bioinformatics, and were major
contributors in writing the manuscript. XLu, SW, MJ and XLi
collected the clinical samples, performed the experiments and
collected the experimental data. ZL and XH designed the
experiments, reviewed and edited the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Human oral epithelial cells were isolated and used
in accordance with the ethical standards established in the
Declaration of Helsinki. The present study was approved by the
Ethics Committee of the School/Hospital of Stomatology at Lanzhou
University (Lanzhou, China) and informed consent forms were signed
by the patients' parents prior to their participation in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shpitzer T, Hamzany Y, Bahar G, Feinmesser
R, Savulescu D, Borovoi I, Gavish M and Nagler RM: Salivary
analysis of oral cancer biomarkers. Br J Cancer. 101:1194–1198.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharma A, Kim JW and Paeng JY: Clinical
analysis of neck node metastasis in oral cavity cancer. J Korean
Assoc Oral Maxillofac Surg. 44:282–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McNamara KK, Martin BD, Evans EW and
Kalmar JR: The role of direct visual fluorescent examination
(VELscope) in routine screening for potentially malignant oral
mucosal lesions. Oral Surg Oral Med Oral Pathol Oral Radiol.
114:636–643. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Colombo M, Raposo G and Théry C:
Biogenesis, secretion, and intercellular interactions of exosomes
and other extracellular vesicles. Annu Rev Cell Dev Biol.
30:255–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles. 4:270662015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z:
miRNA in plasma exosome is stable under different storage
conditions. Molecules. 19:1568–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Théry C: Exosomes: Secreted vesicles and
intercellular communications. F1000 Biol Rep. 3:152011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rafiee A, Riazi-Rad F, Havaskary M and
Nuri F: Long noncoding RNAs: Regulation, function and cancer.
Biotechnol Genet Eng Rev. 34:153–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan Q, Yang L, Zhang X, Peng X, Wei S, Su
D, Zhai Z, Hua X and Li H: The emerging role of exosome derived non
coding RNAs in cancer biology. Cancer Lett. 414:107–115. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F, Li L, Piontek K, Sakaguchi M and
Selaru FM: Exosome miR 335 as a novel therapeutic strategy in
hepatocellular carcinoma. Hepatology. 67:940–954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Li C, Wang S, Wang Z, Jiang J, Wang
W, Li X, Chen J, Liu K, Li C, et al: Exosomes derived from hypoxic
oral squamous cell carcinoma cells deliver miR 21 to normoxic cells
to elicit a prometastatic phenotype. Cancer Res. 76:1770–1780.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He
J, Peng JY, Chen QY, Mo HY, Jun-Cui, et al: Exosomal miR 24 3p
impedes T cell function by targeting FGF11 and serves as a
potential prognostic biomarker for nasopharyngeal carcinoma. J.
Pathol. 240:329–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu T, Chen G, Sun D, Lei M, Li Y, Zhou C,
Li X, Xue W, Wang H, Liu C and Xu J: Exosomes containing miR 21
transfer the characteristic of cisplatin resistance by targeting
PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim
Biophys Sin (Shanghai). 49:808–816. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gomes CC, de Sousa SF, Calin GA and Gomez
RS: The emerging role of long noncoding RNAs in oral cancer. Oral
Surg Oral Med Oral Pathol Oral Radiol. 123:235–241. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin N, Jin N, Bu W, Li X, Liu L, Wang Z,
Tong J and Li D: Long non-coding RNA TIRY promotes tumor metastasis
by enhancing epithelial-to-mesenchymal transition in oral cancer.
Exp Biol Med (Maywood). 245:585–596. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oda D and Wastson E: Human Oral Epithelial
Cell Culture I. Improved Conditions for Reproducible Culture in
Serum Free Medium. In Vitro Cell Dev Biol. 26:589–595. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sdek P, Zhang ZY, Cao J, Pan HY, Chen WT
and Zheng JW: Alteration of cell cycle regulatory proteins in human
oral epithelial cells immortalized by HPV16 E6 and E7. Int J Oral
Maxillofac Surg. 35:653–657. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moffatt-Jauregui CE, Robinson B, de Moya
AV, Brockman RD, Roman AV, Cash MN, Culp DJ and Lamont RJ:
Establishment and characterization of a telomerase immortalized
human gingival epithelial cell line. J Periodontal Res. 48:713–721.
2013.PubMed/NCBI
|
|
21
|
Brueffer C and Saal LH:
TopHat-Recondition: A post-processor for TopHat unmapped reads. BMC
Bioinformatics. 17:1992016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Théry C: Exosomes secreted vesicles and
intercellular communications. F1000 Biol Rep. 3:1–8. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee H, Palm J, Grimes SM and Ji HP: The
Cancer Genome Atlas Clinical Explorer: A Web and Mobile Interface
for Identifying Clinical-Genomic Driver Associations. Genome Med.
7:1122015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guttman M and Rinn JL: Modular regulatory
principles of large non coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J. Control Release. 219:278–294. 2015.
View Article : Google Scholar
|
|
28
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao R and Zhang Y, Zhang X, Yang Y, Zheng
X, Li X, Liu Y and Zhang Y: Exosomal long noncoding RNA HOTTIP as
potential novel diagnostic and prognostic biomarker test for
gastric cancer. Mol Cancer. 17(68)2018.
|
|
30
|
Jiang L, Ji N, Zhou Y, Li J, Liu X, Wang
Z, Chen Q and Zeng X: CAL 27 is an oral adenosquamous carcinoma
cell line. Oral Oncol. 45:e204–e207. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu Q, Lin X, Ding L, Zeng Y, Pang D,
Ouyang N, Xiang Y and Yao H: ARHGAP42 promotes cell migration and
invasion involving PI3K/Akt signaling pathway in nasopharyngeal
carcinoma. Cancer Med. 7:3862–3874. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhong C, Chen Y, Tao B, Peng L, Peng T,
Yang X, Xia X and Chen L: LIM and SH3 protein 1 regulates cell
growth and chemosensitivity of human glioblastoma via the PI3K/AKT
pathway. BMC Cancer. 18:7222018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martins F, de Sousa SC, Dos Santos E, Woo
SB and Gallottini M: PI3K AKT mTOR pathway proteins are differently
expressed in oral carcinogenesis, J. Oral Pathol Med. 45:746–752.
2016. View Article : Google Scholar
|
|
34
|
Hou T, Zhou L, Wang L, Kazobinka G, Chen
Y, Zhang X and Chen Z: Leupaxin promotes bladder cancer
proliferation, metastasis, and angiogenesis through the PI3K/AKT
pathway. Cell Physiol Biochem. 47:2250–2260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sukawa Y, Yamamoto H, Nosho K, Ito M,
Igarashi H, Naito T, Mitsuhashi K, Matsunaga Y, Takahashi T, Mikami
M, et al: HER2 expression and PI3K Akt pathway alterations in
gastric cance. Digestion. 89:12–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang G, Lan Y, Xie A, Shi J, Zhao H, Xu
L, Zhu S, Luo T, Zhao T, Xiao Y and Li X: Comprehensive analysis of
lncRNA chromatin interactions reveals lncRNA functions dependent on
binding diverse regulatory elements. J Biochem. 294:15613–15622.
2019.
|
|
38
|
Kopp F and Mendell JT: Functional
Classification and Experimental Dissection of Long Noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Petruk S, Sedkov Y, Riley KM, Hodgson J,
Schweisguth F, Hirose S, Jaynes JB, Brock HW and Mazo A:
Transcription of bxd noncoding RNAs promoted by trithorax represses
Ubx in cis by transcriptional interference. Cell. 127:1209–1221.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang Y, Wang H and Yang Y: Expression of
fibroblast growth factor 5 (FGF5) and its influence on survival of
breast cancer patients. Med Sci Monit. 24:3524–3530. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu D, Zhang C, Li X, Zhang H, Pang Q and
Wan A: MicroRNA-567 inhibits cell proliferation, migration and
invasion by targeting FGF5 in osteosarcoma. EXCLI J.
17:102–112. 2018.PubMed/NCBI
|
|
42
|
Balamurugan K: HIF 1 at the crossroads of
hypoxia, inflammation, and cancer. Int. J Cancer. 138:1058–1066.
2016.
|
|
43
|
Gashaw I, Dushaj O, Behr R, Biermann K,
Brehm R, Rübben H, Grobholz R, Schmid KW, Bergmann M and
Winterhager E: Novel germ cell markers characterize testicular
seminoma and fetal testis. Mol Hum Reprod. 13:721–727. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fukamachi M, Kasamatsu A, Endo-Sakamoto Y,
Fushimi K, Kasama H, Iyoda M, Minakawa Y, Shiiba M, Tanzawa H and
Uzawa K: Multiple coagulation factor deficiency protein 2 as a
crucial component in metastasis of human oral cancer. Exp Cell Res.
368:119–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin XJ, He CL, Sun T, Duan XJ, Sun Y and
Xiong SJ: hsa miR 485 5p reverses epithelial to mesenchymal
transition and promotes cisplatin induced cell death by targeting
PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med.
40:83–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yoo JK, Jung HY, Lee JM, Yi H, Oh SH, Ko
HY, Yoo H, Kim HR, Song H, Kim S and Kim JK: The novel miR 9500
regulates the proliferation and migration of human lung cancer
cells by targeting Akt1. Cell Death Differ. 21:1150–1159. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu RL, He W, Tang J, Guo W, Zhuang P, Wang
CQ, Fu WM and Zhang JF: Primate specific miRNA 637 inhibited
tumorigenesis in human pancreatic ductal adenocarcinoma cells by
suppressing Akt1 expression. Exp Cell Res. 363:310–314. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Si L, Xu L, Yin L, Qi Y, Han X, Xu Y, Zhao
Y, Liu K and Peng J: Potent effects of dioscin against pancreatic
cancer via miR 149 3P mediated inhibition of the Akt1 signalling
pathway. Br J Pharmacol. 174:553–568. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xiong H, Yang Y, Yang K, Zhao D, Tang H
and Ran X: Loss of the clock gene PER2 is associated with cancer
development and altered expression of important tumor-related genes
in oral cancer. Int J Oncol. 52:279–287. 2018.PubMed/NCBI
|