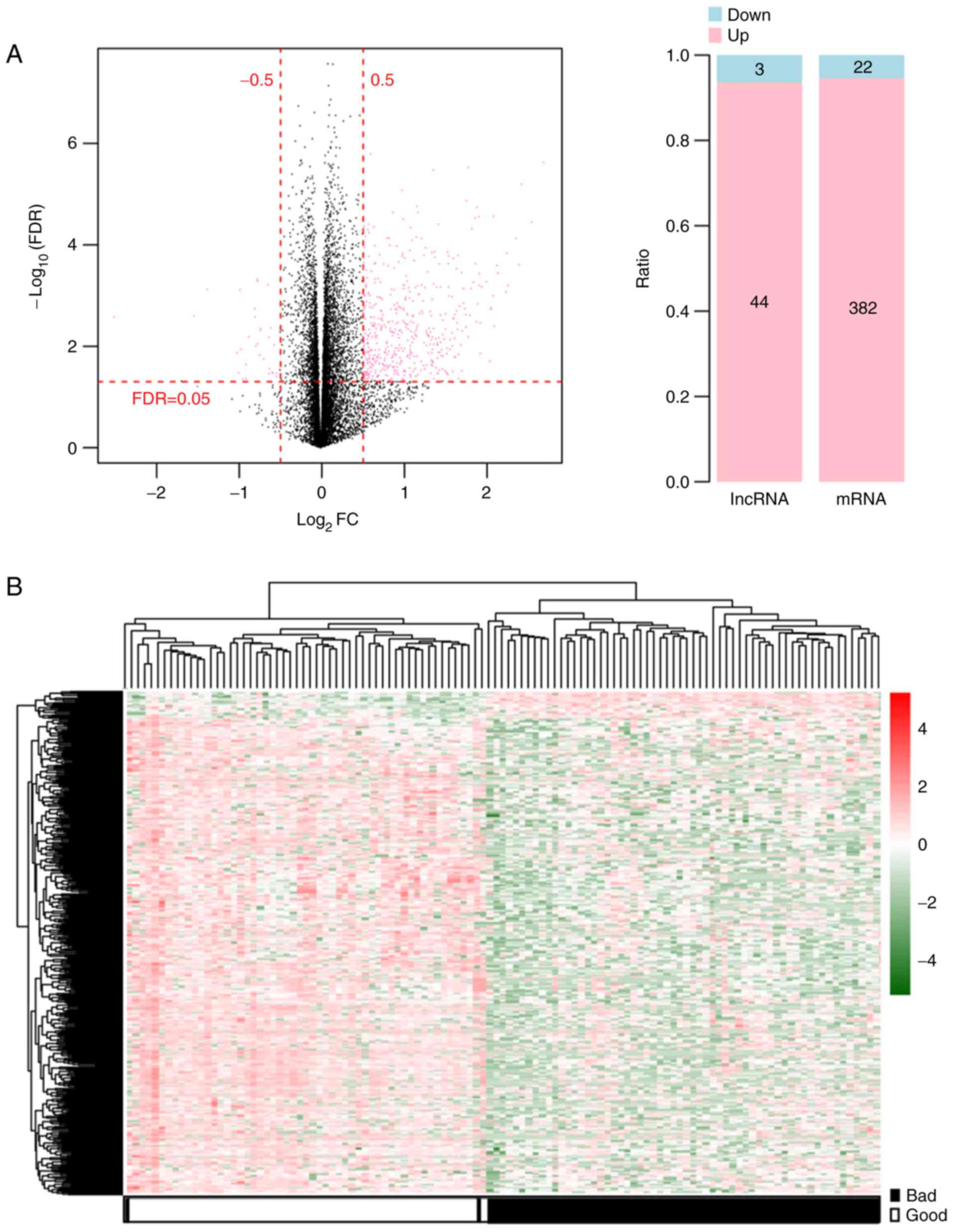
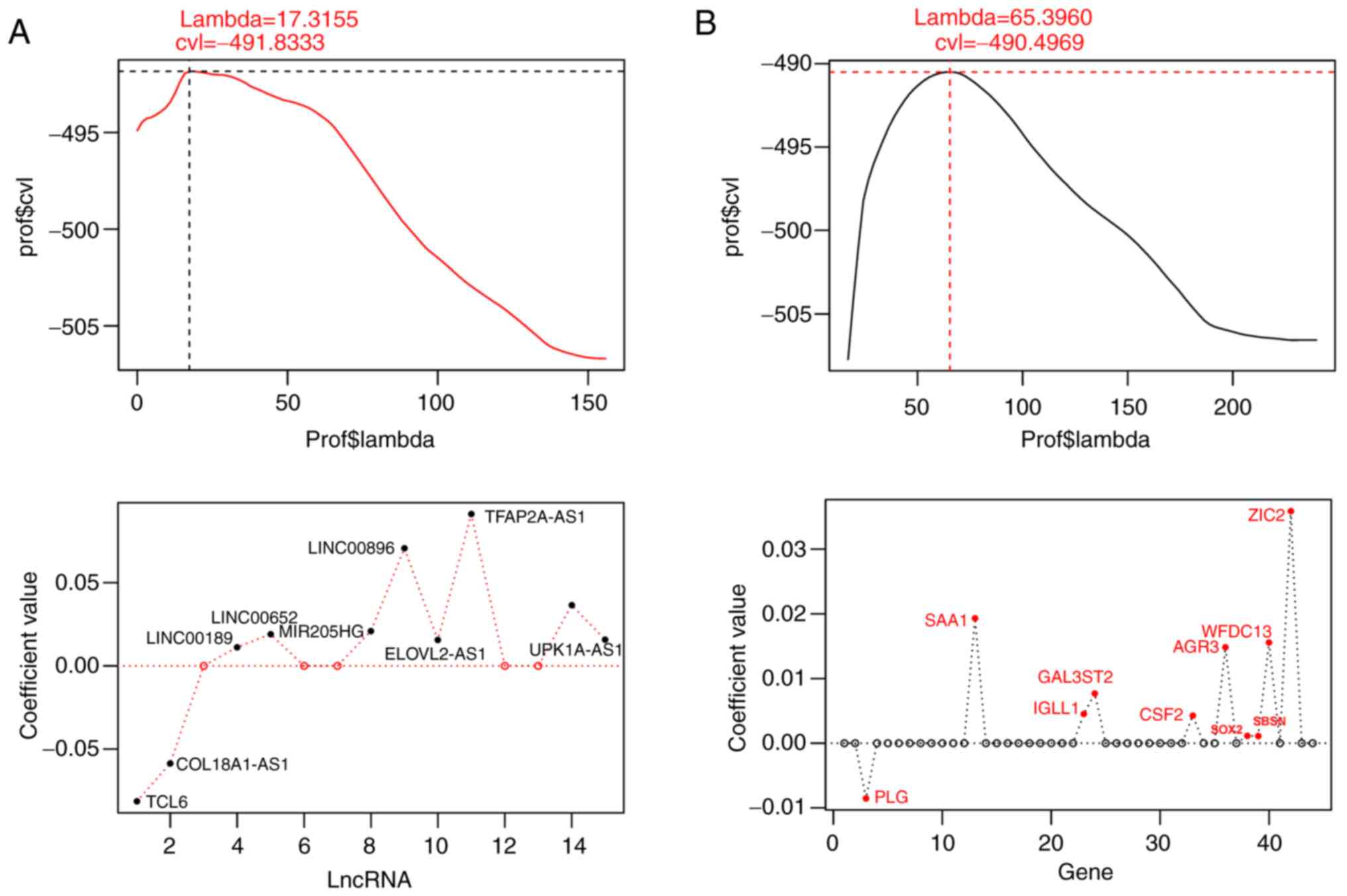
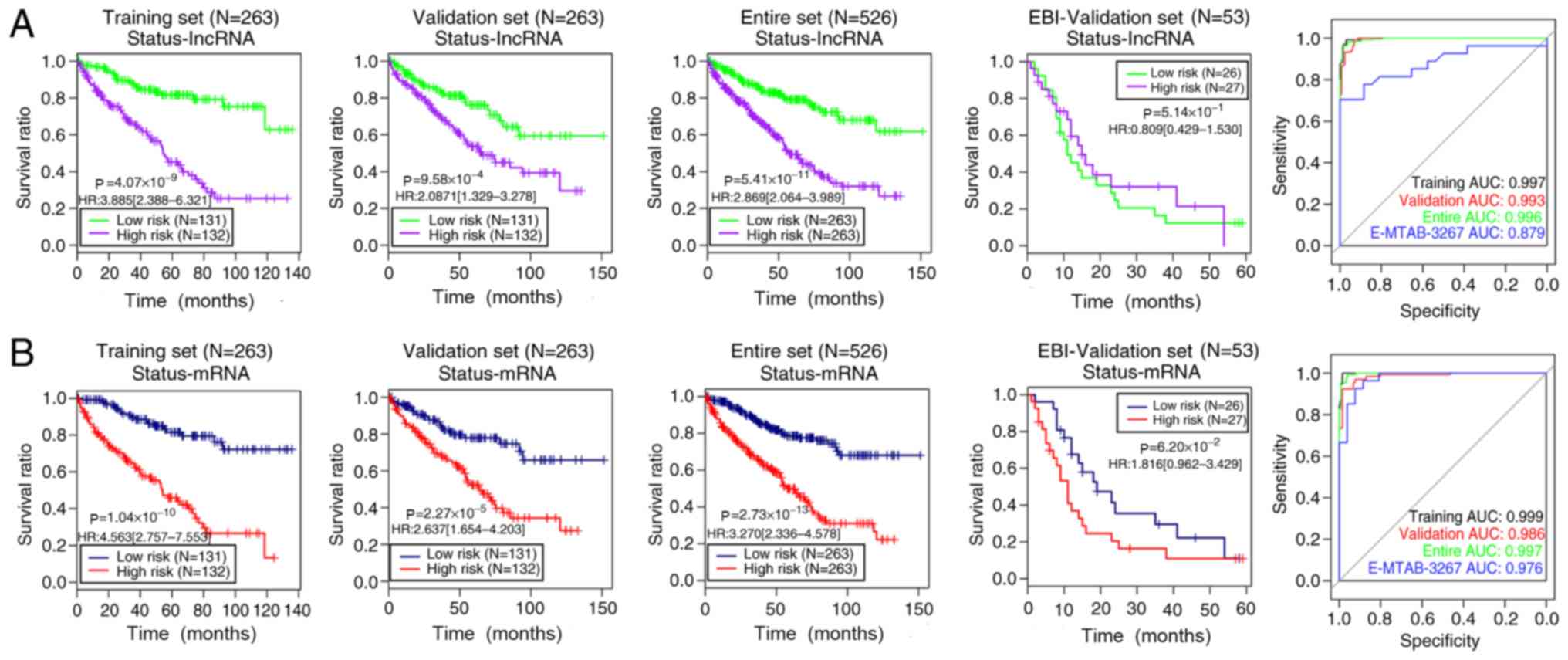
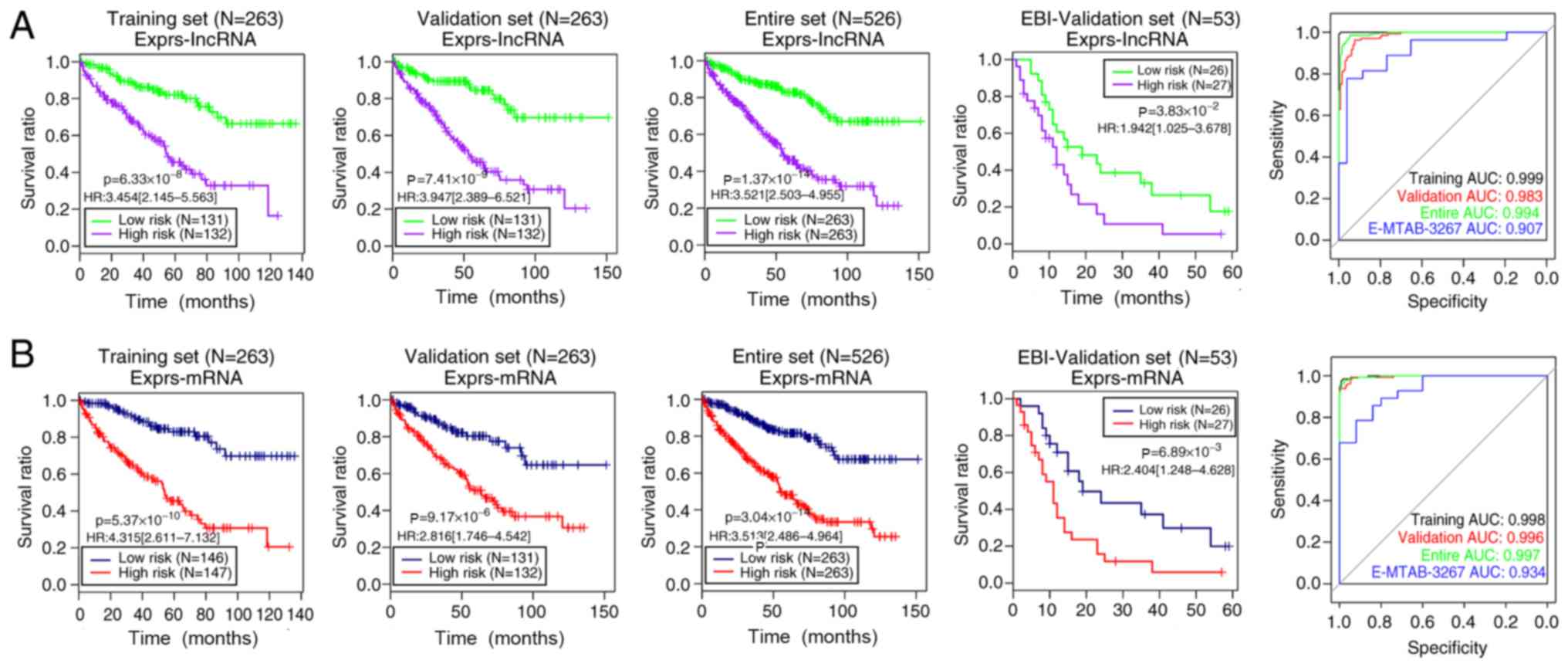
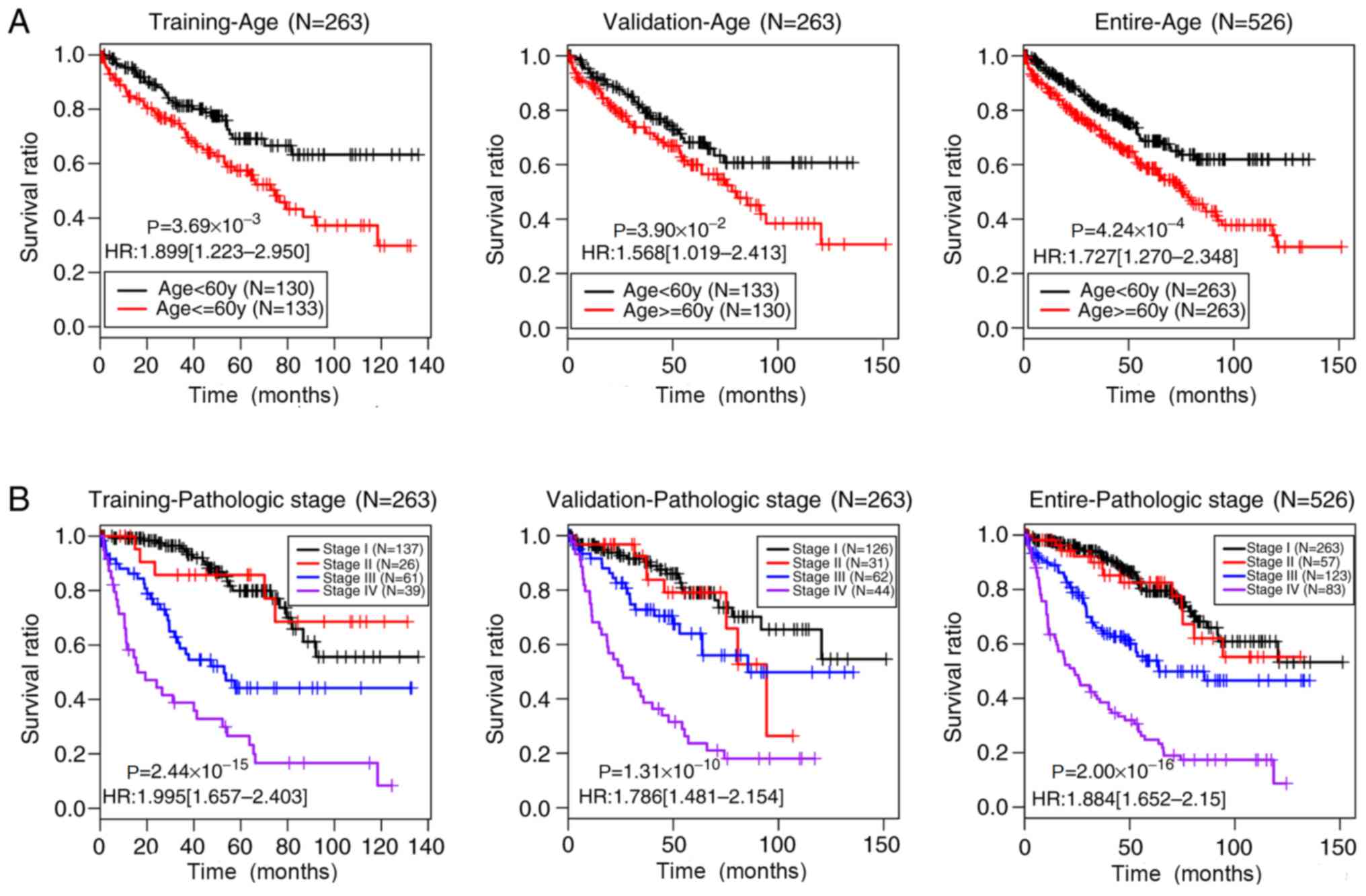
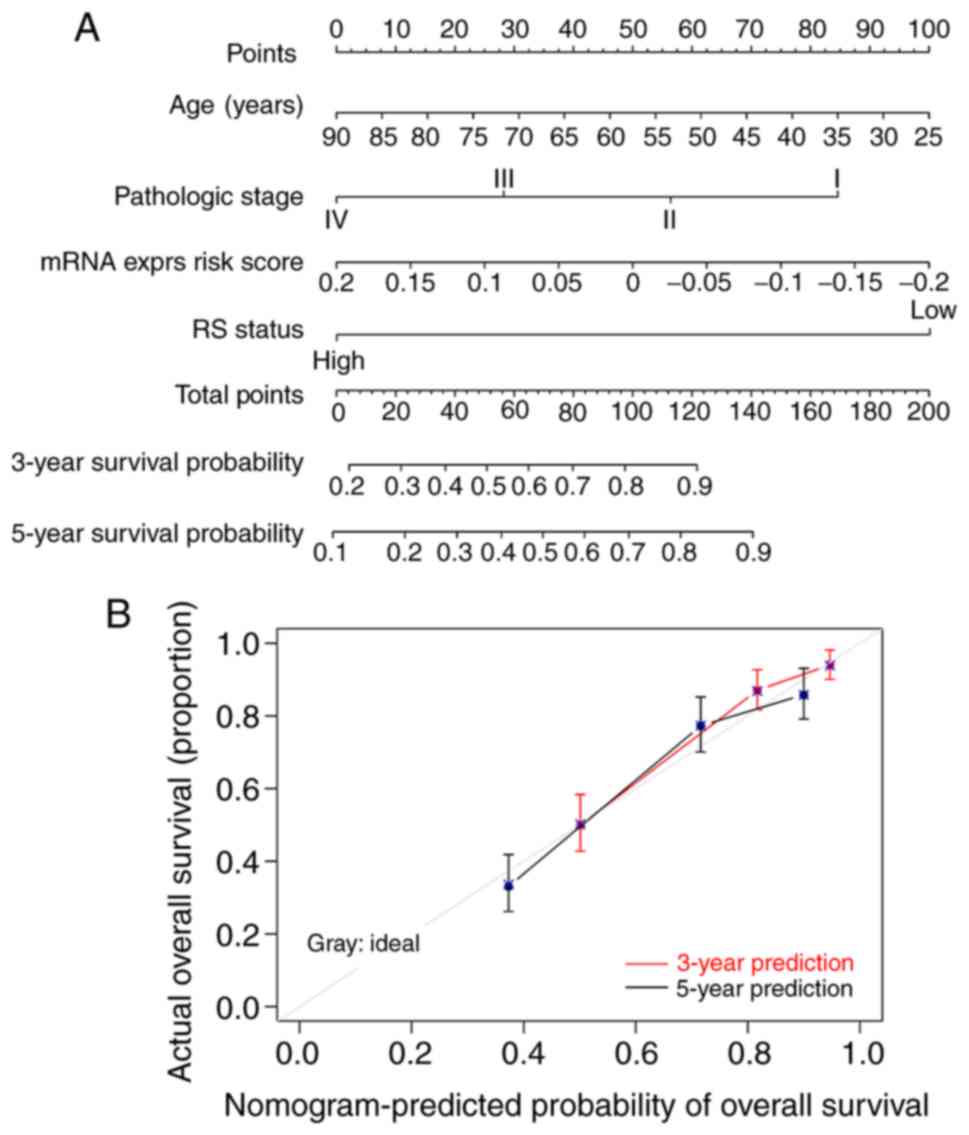
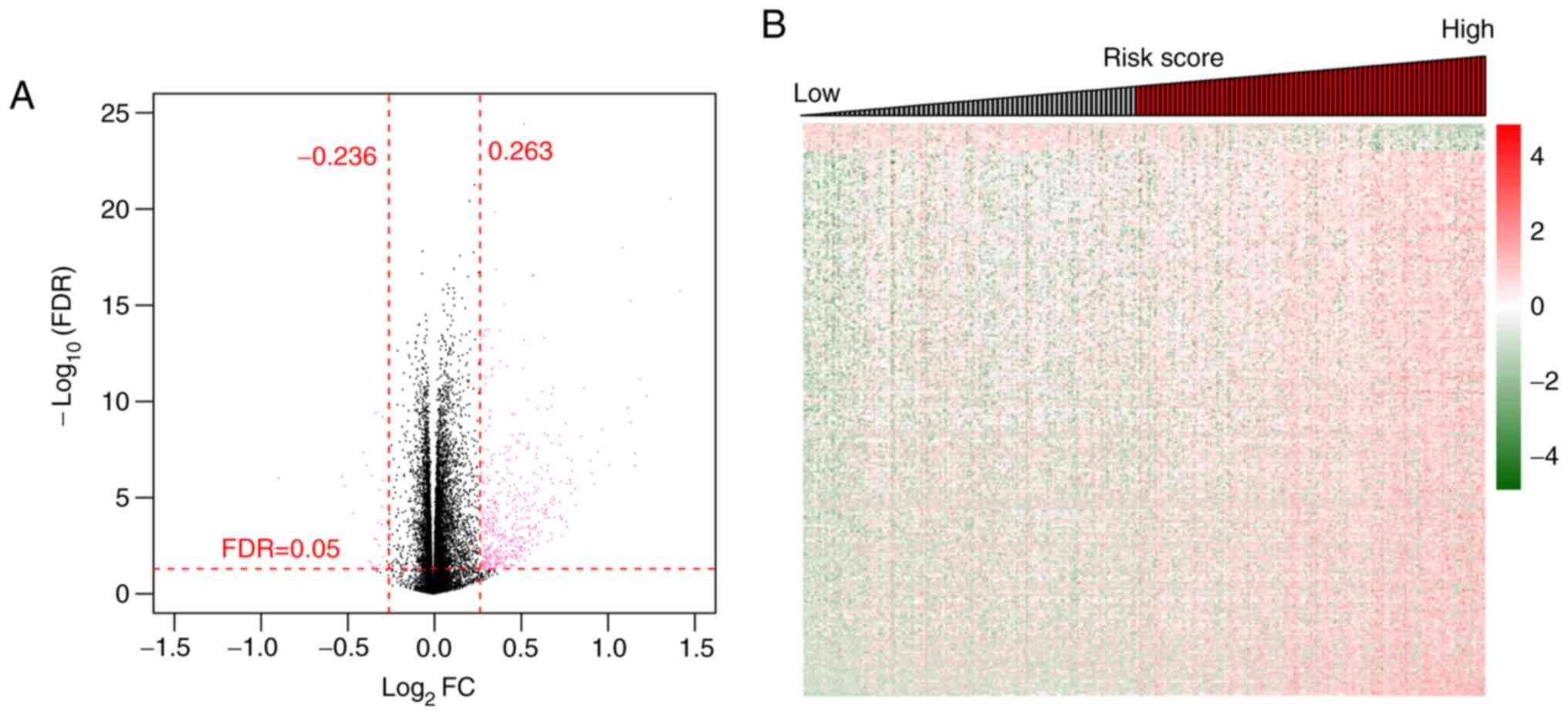
|
1
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of clear cell renal cell
carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valera VA and Merino MJ: Misdiagnosis of
clear cell renal cell carcinoma. Nat Rev Urol. 8:321–333. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aref S, Al Khodary T, Zeed TA, El Sadiek
A, El Menshawy N and Al Ashery R: The prognostic relevance of BAALC
and ERG expression levels in cytogenetically normal pediatric acute
myeloid leukemia. Indian J Hematol Blood Transfus. 31:21–28. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L, Feng T, Qu J, Feng N, Wang Y, Ma RN,
Li X, Zheng ZJ, Yu H and Qian B: LncRNA expression signature in
prediction of the prognosis of lung adenocarcinoma. Genet Test Mol
Biomarkers. 22:20–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miller A, McLeod L, Alhayyani S, Szczepny
A, Watkins DN, Chen W, Enriori P, Ferlin W, Ruwanpura S and Jenkins
BJ: Blockade of the IL-6 trans-signalling/STAT3 axis suppresses
cachexia in Kras-induced lung adenocarcinoma. Oncogene.
36:3059–3066. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tripathi MK, Doxtater K, Keramatnia F,
Zacheaus C, Yallapu MM, Jaggi M and Chauhan SC: Role of lncRNAs in
ovarian cancer: Defining new biomarkers for therapeutic purposes.
Drug Discov Today. 23:1635–1643. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao J, Chen Y, Wang Y, Liu S, Yuan X, Pan
F and Geng P: Decreased expression of a novel lncRNA CADM1-AS1 is
associated with poor prognosis in patients with clear cell renal
cell carcinomas. Int J Clin Exp Pathol. 7:2758–2767.
2014.PubMed/NCBI
|
|
9
|
Hakimi AA, Reznik E, Lee C-H, Creighton
CJ, Brannon AR, Luna A, Aksoy BA, Liu EM, Shen R, Lee W, et al: An
integrated metabolic atlas of clear cell renal cell carcinoma.
Cancer Cell. 29:104–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang H, Ling W, Qiu T and Luo Y:
Ultrasonographic features of testicular metastasis from renal clear
cell carcinoma that mimics a seminoma: A case report. Medicine
(Baltimore). 97:e127282018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Chen Y, Gu L, Li X, Gao Y, Lyu X,
Chen L, Luo G, Wang L, Xie Y, et al: LncRNAs act as prognostic and
diagnostic biomarkers in renal cell carcinoma: A systematic review
and meta-analysis. Oncotarget. 7:74325–74336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng M, Blondeau JJ, Schmidt D, Perner S,
Müller SC and Ellinger J: Identification of novel differentially
expressed lncRNA and mRNA transcripts in clear cell renal cell
carcinoma by expression profiling. Genom Data. 5:173–175. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ning L, Li Z, Wei D, Chen H and Yang C:
LncRNA, NEAT1 is a prognosis biomarker and regulates cancer
progression via epithelial-mesenchymal transition in clear cell
renal cell carcinoma. Cancer Biomark. 19:75–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen J-M, Cooper DN, Chuzhanova N, Férec C
and Patrinos GP: Gene conversion: Mechanisms, evolution and human
disease. Nat Rev Genet. 8:762–775. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iida M, Ikeda F, Hata J, Hirakawa Y, Ohara
T, Mukai N, Yoshida D, Yonemoto K, Esaki M, Kitazono T, et al:
Development and validation of a risk assessment tool for gastric
cancer in a general Japanese population. Gastric Cancer.
21:383–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hung YC, Lin CL, Liu CJ, Hung H, Lin SM,
Lee SD, Chen PJ, Chuang SC and Yu MW: Development of risk scoring
system for stratifying population for hepatocellular carcinoma
screening. Hepatology. 61:1934–1944. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hussein AA, Ghani KR, Peabody J, Sarle R,
Abaza R, Eun D, Hu J, Fumo M, Lane B, Montgomery JS, et al Michigan
Urological Surgery Improvement Collaborative and Applied Technology
Laboratory for Advanced Surgery Program, : Development and
validation of an objective scoring tool for robot-assisted radical
prostatectomy: Prostatectomy assessment and competency evaluation.
J Urol. 197:1237–1244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Klatte T, Seligson DB, LaRochelle J, Shuch
B, Said JW, Riggs SB, Zomorodian N, Kabbinavar FF, Pantuck AJ and
Belldegrun AS: Molecular signatures of localized clear cell renal
cell carcinoma to predict disease-free survival after nephrectomy.
Cancer Epidemiol Biomarkers Prev. 18:894–900. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heinzelmann J, Henning B, Sanjmyatav J,
Posorski N, Steiner T, Wunderlich H, Gajda MR and Junker K:
Specific miRNA signatures are associated with metastasis and poor
prognosis in clear cell renal cell carcinoma. World J Urol.
29:367–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takahashi M, Rhodes DR, Furge KA, Kanayama
H, Kagawa S, Haab BB and Teh BT: Gene expression profiling of clear
cell renal cell carcinoma: Gene identification and prognostic
classification. Proc Natl Acad Sci USA. 98:9754–9759. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Beuselinck B, Job S, Becht E, Karadimou A,
Verkarre V, Couchy G, Giraldo N, Rioux-Leclercq N, Molinié V,
Sibony M, et al: Molecular subtypes of clear cell renal cell
carcinoma are associated with sunitinib response in the metastatic
setting. Clin Cancer Res. 21:1329–1339. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parkinson H, Kapushesky M, Kolesnikov N,
Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam
I, Farne A, et al: ArrayExpress update--from an archive of
functional genomics experiments to the atlas of gene expression.
Nucleic Acids Res. 37:(Database). D868–D872. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wright MW: A short guide to long
non-coding RNA gene nomenclature. Hum Genomics. 8:72014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
R Development Core Team R, . A language
and environment for statistical computing. R Foundation for
Statistical Computing. (Vienna). 2011.
|
|
27
|
Ito K and Murphy D: Application of ggplot2
to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol.
2:e792013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H, et al: RNA-seq analyses of multiple
meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goeman JJ: L1 penalized estimation in the
Cox proportional hazards model. Biom J. 52:70–84. 2010.PubMed/NCBI
|
|
32
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gettman MT, Blute ML, Spotts B, Bryant SC
and Zincke H: Pathologic staging of renal cell carcinoma:
Significance of tumor classification with the 1997 TNM staging
system. Cancer. 91:354–361. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Delahunt B, Eble JN, Egevad L and
Samaratunga H: Grading of renal cell carcinoma. Histopathology.
74:4–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Anderson WI, Schlafer DH and Vesely KR:
Thyroid follicular carcinoma with pulmonary metastases in a beaver
(Castor canadensis). J Wildl Dis. 25:599–600. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eng KH, Schiller E and Morrell K: On
representing the prognostic value of continuous gene expression
biomarkers with the restricted mean survival curve. Oncotarget.
6:36308–36318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Iasonos A, Schrag D, Raj GV and Panageas
KS: How to build and interpret a nomogram for cancer prognosis. J
Clin Oncol. 26:1364–1370. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang G, Zhao G, Xia J, Wei Y, Chen F,
Chen J and Shi J: FGF2 and FAM201A affect the development of
osteonecrosis of the femoral head after femoral neck fracture.
Gene. 652:39–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Klatte T and Rossi SH: Prognostic factors
and prognostic models for renal cell carcinoma: a literature
review. World J Urol. 36:1943–1952. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Adam PJ, Boyd R, Tyson KL, Fletcher GC,
Stamps A, Hudson L, Poyser HR, Redpath N, Griffiths M, Steers G, et
al: Comprehensive proteomic analysis of breast cancer cell
membranes reveals unique proteins with potential roles in clinical
cancer. J Biol Chem. 278:6482–6489. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
King ER, Tung CS, Tsang YTM, Zu Z, Lok GT,
Deavers MT, Malpica A, Wolf JK, Lu KH, Birrer MJ, et al: The
anterior gradient homolog 3 (AGR3) gene is associated with
differentiation and survival in ovarian cancer. Am J Surg Pathol.
35:904–912. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Obacz J, Takacova M, Brychtova V, Dobes P,
Pastorekova S, Vojtesek B and Hrstka R: The role of AGR2 and AGR3
in cancer: Similar but not identical. Eur J Cell Biol. 94:139–147.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qiu C, Wang Y, Wang X, Zhang Q, Li Y, Xu
Y, Jin C, Bu H, Zheng W, Yang X, et al: Combination of TP53 and
AGR3 to distinguish ovarian high-grade serous carcinoma from
low-grade serous carcinoma. Int J Oncol. 52:2041–2050.
2018.PubMed/NCBI
|
|
44
|
He J-Q, Shumansky K, Connett JE,
Anthonisen NR, Paré PD and Sandford AJ: Association of genetic
variations in the CSF2 and CSF3 genes with lung function in
smoking-induced COPD. Eur Respir J. 32:25–34. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee Y-Y, Wu W-J, Huang C-N, Li CC, Li WM,
Yeh BW, Liang PI, Wu TF and Li CF: CSF2 overexpression is
associated with STAT5 phosphorylation and poor prognosis in
patients with urothelial carcinoma. J Cancer. 7:711–721. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi B-Z, Hu P, Geng F, He P-J and Wu X-Z:
Gal3ST-2 involved in tumor metastasis process by regulation of
adhesion ability to selectins and expression of integrins. Biochem
Biophys Res Commun. 332:934–940. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guerra LN, Suárez C, Soto D, Schiappacasse
A, Sapochnik D, Sacca P, Piwien-Pilipuk G, Peral B and Calvo JC:
GAL3ST2 from mammary gland epithelial cells affects differentiation
of 3T3-L1 preadipocytes. Clin Transl Oncol. 17:511–520. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qin F, Song Z, Chang M, Song Y, Frierson H
and Li H: Recurrent cis-SAGe chimeric RNA, D2HGDH-GAL3ST2, in
prostate cancer. Cancer Lett. 380:39–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Turnell W, Sarra R, Glover ID, Baum JO,
Caspi D, Baltz ML and Pepys MB: Secondary structure prediction of
human SAA1. Presumptive identification of calcium and lipid binding
sites. Mol Biol Med. 3:387–407. 1986.PubMed/NCBI
|
|
50
|
Sung H-J, Ahn J-M, Yoon Y-H, Rhim TY, Park
CS, Park JY, Lee SY, Kim JW and Cho JY: Identification and
validation of SAA as a potential lung cancer biomarker and its
involvement in metastatic pathogenesis of lung cancer. J Proteome
Res. 10:1383–1395. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mattarollo SR and Smyth MJ: A novel axis
of innate immunity in cancer. Nat Immunol. 11:981–982. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Milan E, Lazzari C, Anand S, Floriani I,
Torri V, Sorlini C, Gregorc V and Bachi A: SAA1 is over-expressed
in plasma of non small cell lung cancer patients with poor outcome
after treatment with epidermal growth factor receptor
tyrosine-kinase inhibitors. J Proteomics. 76:91–101. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alam MT, Nagao-Kitamoto H, Ohga N, Akiyama
K, Maishi N, Kawamoto T, Shinohara N, Taketomi A, Shindoh M, Hida
Y, et al: Suprabasin as a novel tumor endothelial cell marker.
Cancer Sci. 105:1533–1540. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Park GT, Lim SE, Jang S-I and Morasso MI:
Suprabasin, a novel epidermal differentiation marker and potential
cornified envelope precursor. J Biol Chem. 277:45195–45202. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Formolo CA, Williams R, Gordish-Dressman
H, MacDonald TJ, Lee NH and Hathout Y: Secretome signature of
invasive glioblastoma multiforme. J Proteome Res. 10:3149–3159.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Glazer CA, Smith IM, Ochs MF, Begum S,
Westra W, Chang SS, Sun W, Bhan S, Khan Z, Ahrendt S, et al:
Integrative discovery of epigenetically derepressed cancer testis
antigens in NSCLC. PLoS One. 4:e81892009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shao C, Tan M, Bishop JA, Liu J, Bai W,
Gaykalova DA, Ogawa T, Vikani AR, Agrawal Y, Li RJ, et al:
Suprabasin is hypomethylated and associated with metastasis in
salivary adenoid cystic carcinoma. PLoS One. 7:e485822012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhu J, Wu G, Li Q, Gong H, Song J, Cao L,
Wu S, Song L and Jiang L: Overexpression of suprabasin is
associated with proliferation and tumorigenicity of esophageal
squamous cell carcinoma. Sci Rep. 6:215492016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Boumahdi S, Driessens G, Lapouge G, Rorive
S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E,
et al: SOX2 controls tumour initiation and cancer stem-cell
functions in squamous-cell carcinoma. Nature. 511:246–250. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bass AJ, Watanabe H, Mermel CH, Yu S,
Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et
al: SOX2 is an amplified lineage-survival oncogene in lung and
esophageal squamous cell carcinomas. Nat Genet. 41:1238–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Leis O, Eguiara A, Lopez-Arribillaga E,
Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R
and Martin AG: Sox2 expression in breast tumours and activation in
breast cancer stem cells. Oncogene. 31:1354–1365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Elms P, Siggers P, Napper D, Greenfield A
and Arkell R: Zic2 is required for neural crest formation and
hindbrain patterning during mouse development. Dev Biol.
264:391–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chan DW, Liu VW, Leung LY, Yao KM, Chan
KK, Cheung AN and Ngan HY: Zic2 synergistically enhances Hedgehog
signalling through nuclear retention of Gli1 in cervical cancer
cells. J Pathol. 225:525–534. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Marchini S, Poynor E, Barakat RR, Clivio
L, Cinquini M, Fruscio R, Porcu L, Bussani C, D'Incalci M, Erba E,
et al: The zinc finger gene ZIC2 has features of an oncogene and
its overexpression correlates strongly with the clinical course of
epithelial ovarian cancer. Clin Cancer Res. 18:4313–4324. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhu P, Wang Y, He L, Huang G, Du Y, Zhang
G, Yan X, Xia P, Ye B, Wang S, et al: ZIC2-dependent OCT4
activation drives self-renewal of human liver cancer stem cells. J
Clin Invest. 125:3795–3808. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Smith VJ: Phylogeny of whey acidic protein
(WAP) four-disulfide core proteins and their role in lower
vertebrates and invertebrates. Biochem Soc Trans. 39:1403–1408.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chen Y, Mu X, Wang S, Zhao L, Wu Y, Li J
and Li M: WAP four-disulfide core domain protein 2 mediates the
proliferation of human ovarian cancer cells through the regulation
of growth- and apoptosis-associated genes. Oncol Rep. 29:288–296.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang J, Zhang C, He W and Gou X:
Construction and comprehensive analysis of dysregulated long
non-coding RNA-associated competing endogenous RNA network in clear
cell renal cell carcinoma. J Cell Biochem. 120:2576–2593. 2018.
View Article : Google Scholar
|
|
69
|
Liu T, Sui J, Zhang Y, Zhang XM, Wu WJ,
Yang S, Xu SY, Hong WW, Peng H, Yin LH, et al: Comprehensive
analysis of a novel lncRNA profile reveals potential prognostic
biomarkers in clear cell renal cell carcinoma. Oncol Rep.
40:1503–1514. 2018.PubMed/NCBI
|
|
70
|
Su H, Sun T, Wang H, Shi G, Zhang H, Sun F
and Ye D: Decreased TCL6 expression is associated with poor
prognosis in patients with clear cell renal cell carcinoma.
Oncotarget. 8:5789–5799. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jiang W, Guo Q, Wang C and Zhu Y: A
nomogram based on 9-lncRNAs signature for improving prognostic
prediction of clear cell renal cell carcinoma. Cancer Cell Int.
19:208. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Y, Yang F and Zhuang Y:
Identification of a progression-associated long non-coding RNA
signature for predicting the prognosis of lung squamous cell
carcinoma. Exp Ther Med. 15:1185–1192. 2018.PubMed/NCBI
|
|
73
|
Zhang Y, Zhang X, Zhu H, Liu Y, Cao J, Li
D, Ding B, Yan W, Jin H and Wang S: Identification of Potential
prognostic long non-coding RNA biomarkers for predicting recurrence
in patients with cervical cancer. Cancer Manag Res. 12:719–730.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yan J, Huang X, Zhang X, Chen Z, Ye C,
Xiang W and Huang Z: LncRNA LINC00470 promotes the degradation of
PTEN mRNA to facilitate malignant behavior in gastric cancer cells.
Biochem Biophys Res Commun. 521:887–893. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao
WZ, Peng H, Hong WW, Yin LH, Pu YP, et al: Integrated analysis of
long non-coding RNA-associated ceRNA network reveals potential
lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol.
49:2023–2036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Luo Q, Cui M, Deng Q and Liu J:
Comprehensive analysis of differentially expressed profiles and
reconstruction of a competing endogenous RNA network in papillary
renal cell carcinoma. Mol Med Rep. 19:4685–4696. 2019.PubMed/NCBI
|