Introduction
Thyroid cancer (TC) is the most common endocrine
malignancy worldwide; the incidence of TC has increased rapidly
over the past decade, with an estimated 45,000 new cases diagnosed
per year in the United States (1,2).
Papillary thyroid carcinoma (PTC) is the most frequent form of TC,
accounting for 80–90% of all thyroid malignancies (3). PTC variants include conventional,
follicular, oncocytic, solid, tall cell, columnar cell, diffuse
sclerosing and cribriform forms (4,5). Among
these variants, conventional PTC is the main histological variant
(6). PTC is a well-differentiated
papillary carcinoma with a relatively low mortality rate compared
with other types of TC, and the 5-year overall survival rate is
>95% (7). However, the recurrence
or persistence rate of PTC is relatively high (8,9).
Potential tumor biomarkers for TC diagnosis and prognosis may serve
a significant role in the development of new therapies for TC.
Identification of low-risk subsets of patients with PTC may lead to
the development of treatments to reduce morbidity. Additionally,
identification of patients with PTC that are at a high risk of a
poor prognosis may allow for more accurate pre/postoperative
assessments and improve the surgical planning based on to the
results of the prognostic biomarker tests. Therefore, novel
biomarkers or prognostic models are urgently needed to devise new
means for the prediction of survival of patients with PTC.
With the advances of genome sequencing technologies
in recent years, accumulating evidence have demonstrated that
molecular biomarkers, such as protein-coding genes and non-coding
RNAs, are informative for cancer detection and classification.
mRNAs have exhibited a great potential in both participating in the
physiological and pathological processes as well as predicting
prognosis of patients with various types of tumor, such as renal
cell carcinoma and hepatic carcinoma (10,11).
Thus, the dysregulated expression or mutation of mRNAs may be a
promising predictor of poor prognosis in PTC. Previous studies have
documented that numerous key mRNAs, such as checkpoint kinase 2
(CHEK2) and distal-less homeobox 6 (DLX6), are closely associated
with the aggressive pathogenesis of PTC (12–14). It
is likely that multiple mRNAs may allow for the development of a
signature model and provide more statistically predictive results
of PTCs patients. Thus, a prospective clinical trial using a large
cohort is required to investigate specific prognostic classifiers
in patients with PTC.
The Cancer Genome Atlas (TCGA) database has
publicized various mRNA sequence data, which may provide novel
information to improve the understanding of the molecular
mechanisms of action in the tumorigenesis and progression of PTC.
The current study aimed to apply advanced RNA sequencing analysis
to identify differentially expressed mRNAs (DEMs) between PTC
samples and normal samples. In additional, the association between
prognostic information and expression of these DEMs were evaluated.
Functional enrichment analyses were also performed to investigate
the mechanisms of action. Furthermore, the application value and
reliability of the 5-mRNA signature model to predict PTC prognosis
were investigated.
Materials and methods
Data collection and
pre-processing
mRNA expression information and the corresponding
clinical data of 551 samples, including 58 normal and 493 PTC
samples, were obtained from TCGA database using the keyword
‘thyroid cancer’ (https://tcga-data.nci.nih.gov) (15) prior to July 15th, 2019. As the
clinical data and mRNA expression information were downloaded from
TCGA database, the current study did not require ethical approval.
The mRNAs that were differentially expressed between normal and PTC
samples were assessed using R Studio software (RStudio version
1.1.463; http://www.r-project.org) (16) with the ‘limma’ package (http://www.bioconductor.org/packages).
mRNAs that satisfied the criteria |log2 fold change
(FC)|>2 and P<0.05 were considered for subsequent analysis
(17).
Survival analysis
The Kaplan-Meier analysis and log-rank test were
applied to identify the prognostic DEMs. The ‘survival’ package in
the R software (http://www.bioconductor.org/packages) was used to
construct the survival curves. The survival endpoint was defined as
the overall survival time (OS). P<0.05 was considered to
indicate a statistically significant difference. In addition, the
median expression value was used as the classification cut-off
value for high and low expression.
Risk stratification and receiver
operating characteristic (ROC) curves
To further investigate the crucial DEMs closely
associated to OS, univariate Cox models were performed.
Subsequently, multivariate COX regression analysis of selected
candidate DEMs was performed. DEMs with P<0.05 were defined as
candidate genes for further multivariate Cox regression analysis.
Owing to the data processing above, 5 key DEMs were eventually
subsumed into the multivariate Cox regression model to calculate
their respective coefficients. Then, the score model composed of
DEMs' respective coefficients and expression value was constructed
as follows:
Survival Risk Score=∑i=1k(C1×Vi)
Where k is the number of prognostic RNAs,
Ci represents the coefficient of the RNA in the
multivariate Cox regression analysis, and Vi is the
expression value of the RNA. The RNAs with Ci >0 were
defined as high-risk markers, whereas those with Ci
<0 were defined as protective RNAs (18). The median risk score was used as the
classification cut-off value between high- and low-risk PTC
subgroups in 5-mRNA signature model as previously described
(19). Between the high- and
low-risk PTC groups, the difference in OS was compared and analyzed
using Kaplan-Meier analysis with the log-rank test. The specificity
and sensitivity of the 5-mRNA prognostic signature model in the
present study were assessed using an ROC curve and area under the
ROC curve (AUC). The ‘timeROC’ package (https://cran.r-project.org/web/packages/timeROC/)
was used to conduct all the analyses.
Prediction of target genes and
functional enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) and
Gene ontology (GO) were used to perform functional enrichment
analyses of DEMs and to elucidate the mechanisms of action in PTC
tumorigenesis using The Database for Annotation, Visualization and
Integrated Discovery (https://david.ncifcrf.gov/) (20).
Results
DEMs in PTCs
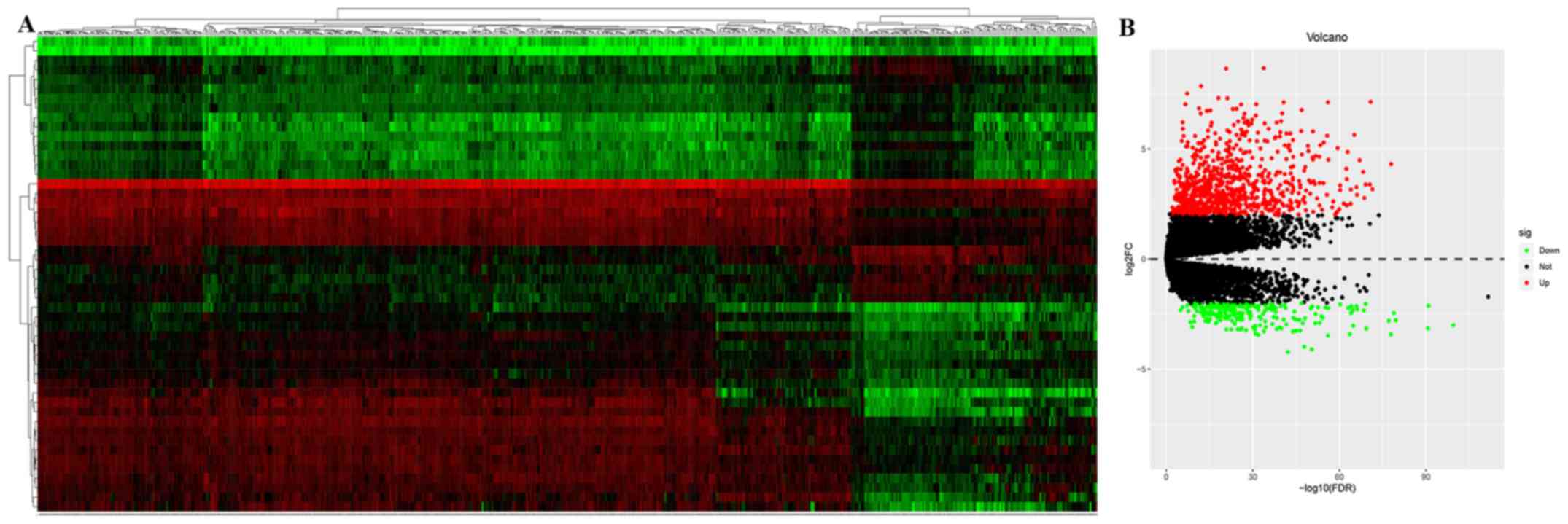
The mRNA expression profiles between PTC and normal
samples from TCGA database were downloaded and analyzed. A total of
853 upregulated and 232 downregulated DEMs that met the criteria
|log2FC|>2 and P<0.05 were identified in PTC
compared with normal samples. The volcano plot and heatmap of the
DEMs in PTC are presented in Fig. 1.
The top 10 upregulated and downregulated DEMs were selected for
further analysis based on the false discovery rate (Table I).
 | Table I.Top 10 upregulated and downregulated
differentially expressed mRNAs. |
Table I.
Top 10 upregulated and downregulated
differentially expressed mRNAs.
| A, Upregulated
mRNAs |
|---|
|
|---|
| Gene symbol | Log2FC | FDR | P-value |
|---|
| ARHGAP36 | 8.670156159 |
1.72×10−34 |
5.05×10−36 |
| SFTPA1 | 8.648973385 |
1.87×10−21 |
1.67×10−22 |
| WIF1 | 7.858016530 |
8.73×10−13 |
1.70×10−13 |
| SERPINB11 | 7.516327014 |
6.02×10−08 |
1.90×10−08 |
| SLC18A3 | 7.315944162 |
8.45×10−19 |
9.47×10−20 |
| SFTPA2 | 7.309149505 |
6.95×10−22 |
6.00×10−23 |
| GABRB2 | 7.144823578 |
1.53×10−71 |
1.04×10−74 |
| TMPRSS6 | 7.123312367 |
2.06×10−41 |
3.30×10−43 |
| SLC22A31 | 7.123263970 |
9.85×10−57 |
3.97×10−59 |
| DPRX | 7.030322665 |
2.35×10−07 |
7.92×10−08 |
|
| B, Downregulated
mRNAs |
|
| Gene
symbol | Log2FC | FDR |
P-values |
|
| OR2V1 | −4.212774737 |
6.32×10−43 |
8.82×10−45 |
| KCNA1 | −4.091346010 |
3.91×10−51 |
2.59×10−53 |
| SLC6A15 | −3.985283867 |
1.61×10−48 |
1.33×10−50 |
| GRIA1 | −3.470505834 |
9.76×10−57 |
3.87×10−59 |
| VSTM2A | −3.439852831 |
1.18×10−32 |
3.94×10−34 |
| UGT2B11 | −3.421202686 |
1.48×10−78 |
6.70×10−82 |
| RELN | −3.414498465 |
2.42×10−64 |
5.35×10−67 |
| MYOC | −3.390338222 |
1.13×10−31 |
4.03×10−33 |
| DPT | −3.282655507 |
1.28×10−44 |
1.51×10−46 |
| SEC14L3 | −3.276717193 |
1.48×10−34 |
4.33×10−36 |
Survival analysis
Kaplan-Meier univariate survival analyses were
performed in the present study to investigate gene expression
profiles on OS. Among the 1,085 DEMs examined in the present study,
361 mRNAs, including adhesion G protein-coupled receptor
D1(ADGRD1), ankyrin 2 (ANK2), anoctamin 5 (ANO5), activity
regulated cytoskeleton associated protein (ARC), acid sensing ion
channel subunit 1 (ASIC1), checkpoint kinase 2 (CHEK2), cytochrome
c oxidase subunit 7A1 (COX7A1), casein beta (CSN2), DDB1 and CUL4
associated factor 8 like 1 (DCAF8L1), distal-less homeobox 6
(DLX6), dipeptidyl peptidase like 6 (DPP6), desmocollin 3 (DSC3),
elastase, neutrophil expressed (ELANE), erythroferrone (ERFE),
coagulation factor X (F10), ficolin 3 (FCN3), FEZ family zinc
finger 1 (FEZF1), surfactant protein A1 (SFTPA1), surfactant
protein A2 (SFTPA2) and solute carrier family 22 member 31
(SLC22A31), were associated OS in patients with PTC. The top 20
mRNAs based on false discovery rate (FDR) value are presented in
Fig. S1.
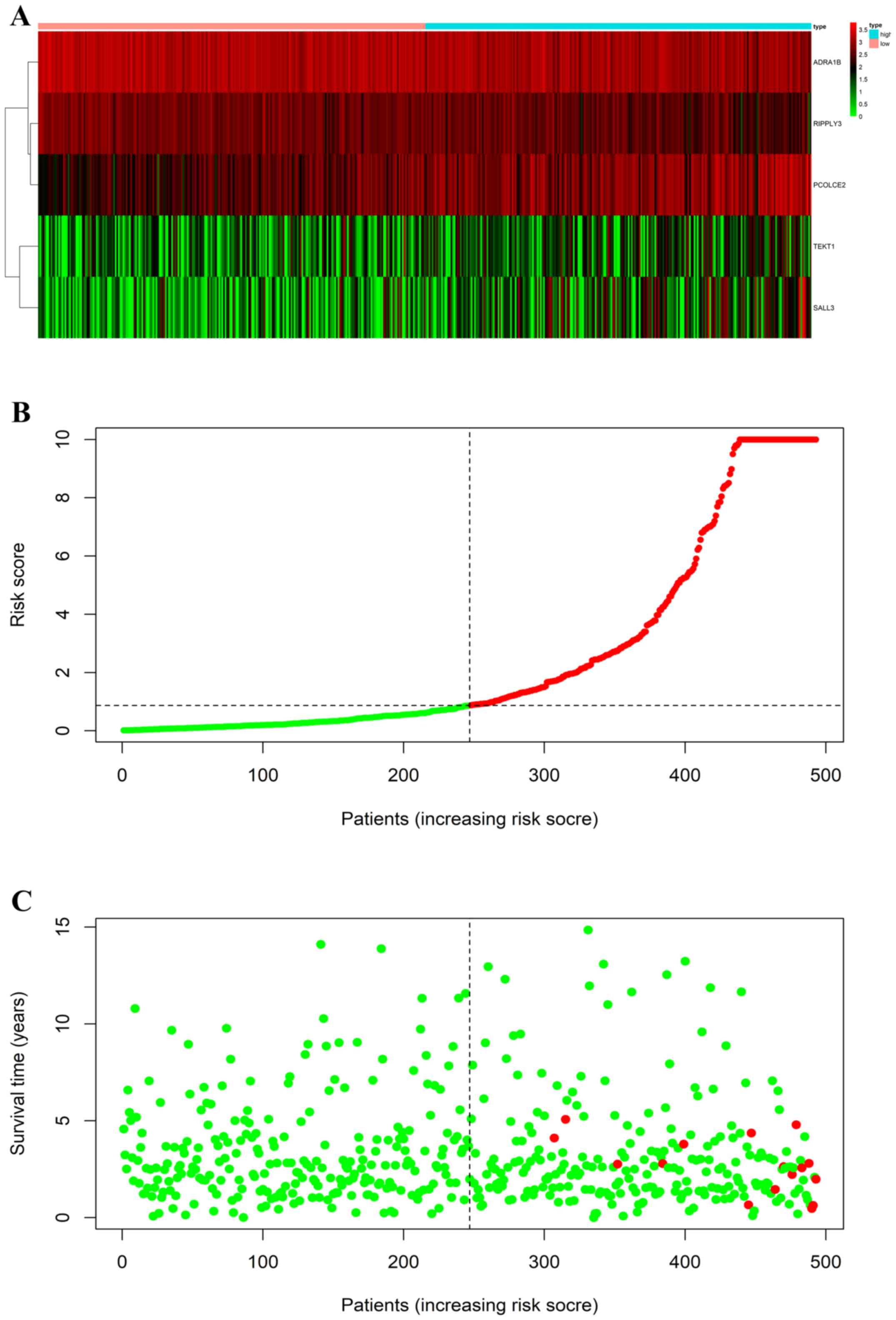
Establishment of a five-mRNA signature
model
Uni- and multivariate analyses were performed using
Cox regression analysis to assess the association between OS and
the expression levels of the identified DEMs in patients with PTC
(Table SI; Table II). A predictive signature model was
established based on five DEMs, namely ADRA1B, RIPPLY3, PCOLCE,
TEKT1 and SALL3, that were selected from the multivariate Cox
regression analysis. The signature model in the current study was
defined as follows: Survival risk score=(−0.771870966 ×
VADRA1B) + (−0.782948964 × VRIPPLY3) +
(0.708200312 × VPCOLCE2) + (0.241743456 ×
VTEKT1) + (0.378467099 × VSALL3), and the V
was the expression value (Table
II).
 | Table II.Multivariate cox regression analysis
between OS and the expression levels of DEMs. |
Table II.
Multivariate cox regression analysis
between OS and the expression levels of DEMs.
| Gene symbol | Coef | P-value |
|---|
| ADRA1B | −0.771870966 | 2.18×10-3 |
| RIPPLY3 | −0.782948964 | 9.03×10-4 |
| TEKT1 | 0.241743456 | 9.43×10-2 |
| PCOLCE2 | 0.708200312 | 9.17×10-5 |
| SALL3 | 0.378467099 | 1.67×10-4 |
Risk stratification and ROC curve
analysis
The risk scores of 493 patients with PTC from TCGA
database were calculated using the 5-mRNA prediction model. All
patients with PTC were assigned to the low-risk (risk score >1;
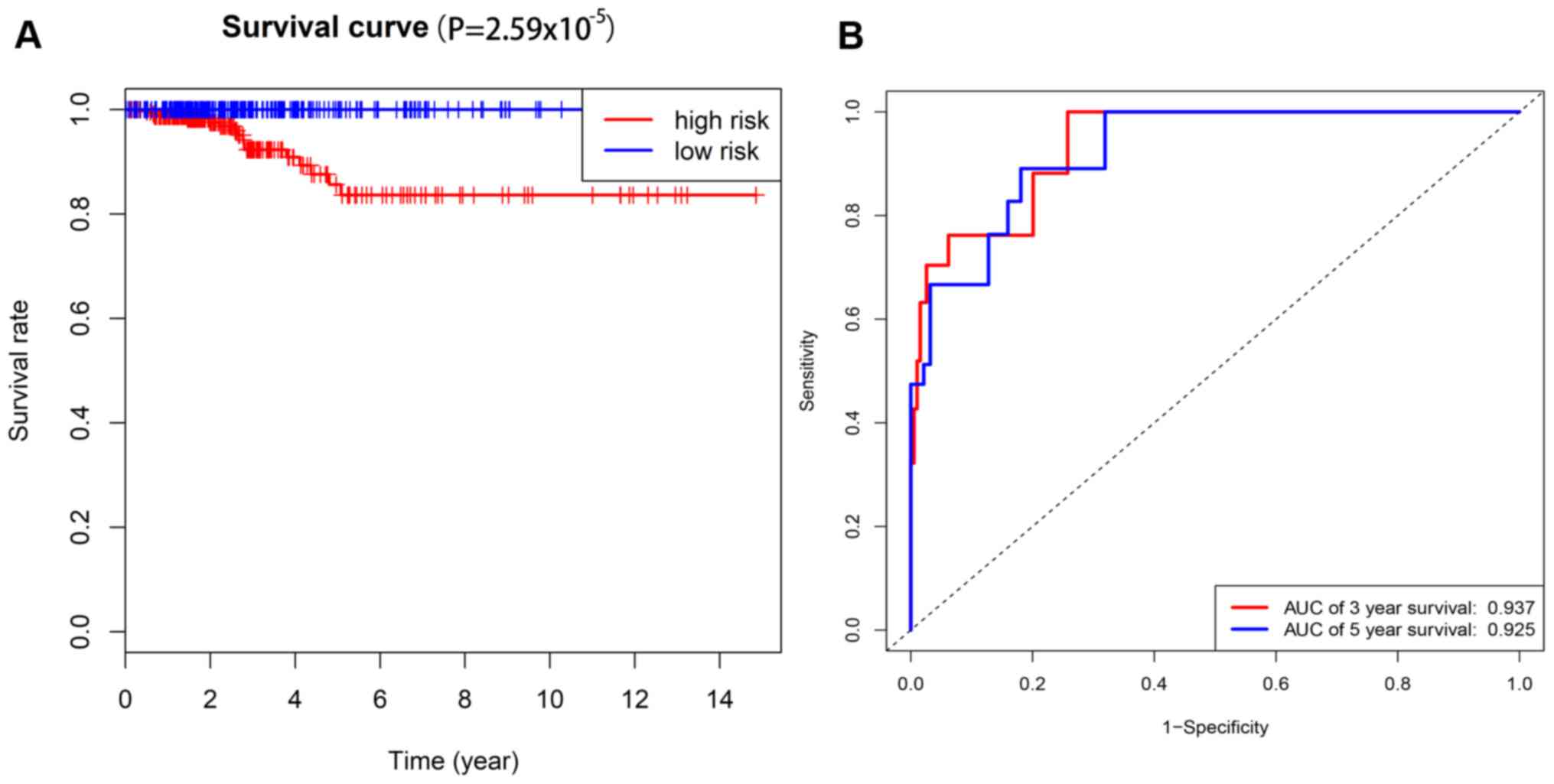
n=264) or the high-risk group (risk score ≤1; n=229; Fig. 2). The prognostic power of the 5-mRNA
signature model was evaluated using the AUC value of the ROC curve
(Fig. 3). The 3- and 5-year AUC
values in the present study were 0.937 and 0.925, respectively,
which demonstrated a high sensitivity and specificity of the 5-mRNA
signature model for predicting the risk levels in patients with
PTC. This signature model may be used to identify patients with
high risk scores who may benefit from more radical, effective and
individualized therapy.
Functional enrichment analysis
The GO analysis results demonstrated that prognostic
mRNAs were mainly associated with ‘calcium ion binding’, ‘enzyme
inhibitor activity’, ‘carbohydrate binding’, ‘transcriptional
activator activity’, ‘RNA polymerase II core promoter proximal
region sequence-specific binding’ and ‘glutathione transferase
activity’ (Table III). Using KEGG
analysis, five statistically significant pathways were identified,
namely ‘pertussis’, ‘ascorbate and aldarate metabolism’, ‘systemic
lupus erythematosus’, ‘drug metabolism-cytochrome P450’ and
‘complement and coagulation cascades’ (Table III). These results suggested that
the prognostic mRNA are key players in in many cellular and
biochemical processes.
 | Table III.Top 10 enriched GO terms, KEGG
pathways and target mRNAs. |
Table III.
Top 10 enriched GO terms, KEGG
pathways and target mRNAs.
| A, GO terms |
|---|
|
|---|
| Term | ID | FDR | Target mRNAs |
|---|
| Calcium ion
binding | 0005509 | 4.705418 |
PCDHGA12/F10/ADGRE3/MYL3/ENPP2/REG4/CRTAC1/DMP1/MMP19/MMP27/CABP7/PCDH12/PCDH8/MMP28/PADI4/DNASE1L3/PCDH19/DSC3/RGN/CALML6/PCDHA12/SGCA/CSN2/AOC3 |
| Enzyme inhibitor
activity | 0004857 | 19.02155 |
SLN/SCG5/UGT1A1/CSN2 |
| Carbohydrate
binding | 0030246 | 26.01117 |
EMCN/CHIA/OLR1/FCN3/SFTPA2/CHODL/SFTPA1/CLEC4D/CLEC4M |
| Transcriptional
activator activity, RNA polymerase II core promoter proximal region
sequence-specific binding | 0001077 | 27.08023 |
TCF21/OTX1/DLX5/ONECUT3/SOX2/TP63/NOBOX/PITX3/GRHL1/FOXD1 |
| Glutathione
transferase activity | 0004364 | 27.42761 |
GSTA1/CLIC3/ALOX5AP/CLIC5 |
| RNA polymerase II
core promoter proximal region sequence-specific DNA binding | 0000978 | 48.17595 |
FEZF1/MUC1/FEZF2/OTX1/DLX5/NKX3-2/KLF17/NOBOX/PITX3/GRHL1/DDN/FOXD1 |
| Interleukin-8
binding | 0019959 | 52.49444 | A2M/CXCR1 |
| Heparin
binding | 0008201 | 58.36638 |
CCL2/REG4/ELANE/APOH/SERPIND1/GREM2/SOD3 |
| Structural molecule
activity | 0005198 | 62.23154 |
KRT25/DES/KRT74/LAMC3/KRTAP3-2/SYNC/CLDN5/LCE2B/LELP1 |
| Chloride channel
activity | 0005254 | 62.51447 |
CLCA2/GABRR1/CLIC3/CLIC5 |
|
| B, KEGG
pathways |
|
| Pathway | ID | FDR | Target
mRNAs |
|
| Ascorbate and
aldarate metabolism | hsa00053 | 13.65207 |
ALDH2/RGN/UGT2B4/UGT1A1 |
| Systemic lupus
erythematosus | hsa05322 | 32.92029 |
HIST1H2BO/ELANE/HIST1H2BH/C2/HIST1H2AJ/HIST1H3G/HIST1H4J |
| Drug
metabolism-cytochrome P450 | hsa00982 | 33.51738 |
GSTA1/MAOB/UGT2B4/ADH7/UGT1A1 |
| Complement and
coagulation cascades | hsa04610 | 34.81476 |
A2M/F10/C2/SERPIND1/PLAU |
| Metabolism of
xenobiotics by cytochrome P450 | hsa00980 | 41.43999 |
GSTA1/SULT2A1/UGT2B4/ADH7/UGT1A1 |
| Chemical
carcinogenesis | hsa05204 | 49.49964 |
GSTA1/SULT2A1/UGT2B4/ADH7/UGT1A1 |
| Salmonella
infection | hsa05132 | 53.48304 |
PFN2/IL6/WASF1/CXCL2/NOS2 |
| Morphine
addiction | hsa05032 | 63.62918 |
ADCY4/GABRR1/PDE1B/PDE2A/GABRP |
| Alcoholism | hsa05034 | 70.92004 |
HIST1H2BO/HIST1H2BH/MAOB/CALML6/HIST1H2AJ/HIST1H3G/HIST1H4J |
Discussion
PTC is a well-differentiated papillary carcinoma
with a relatively low mortality rate among types of TC (14). However, the persistence and
recurrence rates in patients with PTC are high, ≥30% in certain
demographics (8,9). Investigation into prognostic biomarkers
for predicting the risk for patients with PTC may help develop
clear and effective treatment strategies. mRNAs have been
considered as prognostic biomarkers for the diagnosis and prognosis
for patients with PTC in recent years (21). For instance, zinc finger E-box
binding homeobox 2 has been reported to be highly expressed in
ovarian tumor tissues, which is significantly associated with a
poor prognosis (22). The expression
of Forkhead box Q1 is also negatively associated with patient
survival (23). It has been reported
that the mRNA PFKFB4 is associated with tumorigenesis in TC, with
PFKFB4 overexpression promoting migration and invasion of TC cells
(24). In addition, Huang et
al (25) have demonstrated that
the overexpression of JAZF zinc finger 1 mRNA significantly
inhibited proliferation, caused cell cycle arrest at the
G0/G1 phase and promoted apoptosis in PTC
cells. A previous study has suggested that the upregulation of the
cytokine receptor factor-like 1 gene is associated with aggressive
clinicopathological features and poor disease-free survival rates
in patients with PTC (26).
Additionally, Vecchio et al (27) reported that the downregulation of
α-L-fucosidase 1 is associated with the increased aggressiveness of
TC. A previous study has also identified an association between
high expression levels of endo-5′-nucleotidase and the development
of metastatic lymph nodes as well as tumor microinvasion in PTC
(28). However, various mRNA-chip
platforms or small number of patients were the major limitations of
the aforementioned studies. PTC is a multifactorial disease, in
which numerous dysregulated mRNAs participate in the tumorigenesis
and progression of the tumor (29).
Therefore, a signature model including multiple mRNAs may provide a
more accurate and detailed prediction system for the diagnosis and
prognosis of PTC compared with the use of a single mRNA
biomarker.
A total of 853 upregulated and 232 downregulated
DEMs in PTC compared with normal thyroid tissues were identified in
the present study. Among them, 361 DEMs were associated with OS in
patients with PTC. A number of DEMs identified in the present study
have been previously investigated in TC. For example, CHEK2
mutations predispose patients to TC, familial breast cancer and
double primary cancers of the breast and thyroid (30). Variants in the ATM-CHEK2-BRCA1 axis
determine genetic predispositions and clinical presentations of PTC
(31). The ANK2 gene has been
demonstrated to exhibit a decrease in expression levels in novel
tumors that were identified in pediatric post-Chernobyl tumor cases
(32). A previous study has
demonstrated that ANO5 was downregulated in PTC and follicular
thyroid cancer when compared to adjacent non-cancerous tissues
(33). ANO5 knockdown was also
demonstrated to promote thyroid cancer cell invasion and migration
in vitro; overexpression of ANO5 inhibited these phenotypes
(33). Tenbaum et al
(34) detected two Alien-specific
mRNAs and two Alien-specific proteins in vivo and in cell
lines; one of the two forms represented the CSN2 subunit of the
COP9 signalosome.
Information regarding the regulatory role of ADGRD1,
ARC, ASIC1, COX7A1, DCAF8L1, DLX6, DPP6, DSC3, ELANE, ERFE, F10,
FCN3, FEZF1, SFTPA1, SFTPA2 and SLC22A31 in PTC is currently
limited. However, these DEMs have been reported to be associated
with the oncogenesis of other types of cancer in previous studies.
For example, the ASIC1 gene is upregulated in pancreatic cancer
tissues and expressed on the membrane of pancreatic cancer cells
(35). Ceder et al (36) conducted a study that reported that
high COX7A1 expression was associated with worse outcome in head
and neck squamous cell carcinoma. The DLX6, HOXB7, ELF3, EN1, ETV1,
IRX4, BARX1, FOXE1, IRX5 and SALL1 genes are potential oncogenic
transcription factors in non-small cell lung cancer cells (37). Overexpression of DSC3 reduced cell
colony formation and proliferative ability to promote apoptosis and
induce G0/G1 cell cycle arrest in colorectal cancer cells (38). In addition, the ERFE gene was
demonstrated to be associated with OS in colon cancer using
univariate Cox regression analysis (39). Of note, the FEZF1 gene is an
independent predictive factor for cervical cancer recurrence and
promotes cervical cancer cell migration and proliferation (40). Of note, SLC22A31 mRNA expression
levels are associated with OS in right-sided colon cancer, as
determined using TCGA data (39).
GO and KEGG analyses of 361 prognostic DEMs was
performed in the present study to identify the molecular mechanisms
of action in PTC. GO and KEGG analyses results suggested that
prognostic DEMs are key players in in numerous cellular and
biochemical processes. Several signaling pathways and biological
processes identified in the present study have been investigated in
previous studies on PTC. The stress-activated MAPK signaling
pathway is a common pro-oncogenic signaling pathway and tumor
suppressor during the process of tumorigenesis in different
malignancies including PTC, colorectal cancer, liver cancer and
ovarian cancer (41,42). Of note, Li et al (43) reported that greater heparin binding
was achieved by differentially expressed genes in PTC tissues
compared with that in normal thyroid tissues. The results of the
present study demonstrated that the signaling pathways associated
with the complement and coagulation cascades as well as thyroid
cancer may also serve important roles in the development of PTC. A
previous study revealed that genes involved in a competing
endogenous RNA network were significantly enriched during the
metabolism of xenobiotics by cytochrome P450 in PTC (44). The aforementioned evidence was
partially consistent with the results of the present GO and KEGG
analyses.
Differential gene expression analysis results in PTC
samples demonstrated that the intrathyroidal iodine metabolism
machinery, methylation-induced gene silencing of tumor suppressor
genes, upregulation of Glut-1 mRNA and pro-angiogenetic proteins
such as VEGF were all associated with the impairment of the
expression of BRAF genes (45). In
the present study, it was demonstrated that the 5-mRNAs signature
model constructed using TCGA data may be valuable prognostic tool
to distinguish patients with PTC into low-risk and high-risk
subgroups. Different therapeutic strategies may be selected
according to the different risk levels based on the 5-mRNAs
signature model, which may help improve the survival of patients
with PTC. Patients with PTC in the high-risk group may demonstrate
poor clinical outcomes, including aggressive tumor behavior,
disease recurrence and disease-specific mortality; therefore,
treatment strategies such as prophylactic central neck dissection,
radioactive iodine therapy and close long-term follow-up may be
applied. Wu et al (46)
identified DEGs related to the progression-free interval (PFI) and
applied lasso Cox regression to establish a prognostic gene
signature which could predict the PFI of PTC. Similar multi-mRNA
prognostic models have also been constructed in other tumor types.
A previous study has demonstrated that a 3-mRNA signature (CLEC3B,
C6 and CLCN1) model can successfully predict the survival of
patients with oral squamous cell carcinoma (47). A 24-mRNA signature model was also
constructed for early-relapse prediction of patients with
hepatocellular carcinoma, which may help facilitate disease
management for individual patients (48). A metastasis-related 4-mRNA gene
signature model has been demonstrated to effectively classify
patients with breast cancer into high- and low-risk groups
(49). In addition, Cui et al
(38) developed a 6-mRNA signature
model that may potentially improve the risk stratification of
patients with head and neck squamous cell carcinoma (50). Liang et al (39) reported an effective 6-gene prognostic
signature model to act as a specific, robust and clinically
practical risk stratification model for the survival outcome of
patients with cytogenetically normal-acute myeloid leukemia
(51). Additionally, a prognostic
9-mRNA gene signature model was constructed based on survival data
from patients with glioma according to Chinese Glioma Genome Atlas
database (52).
The present study had certain limitations. First,
the mRNA expression data used for the signature model were
downloaded from a single database (TCGA) rather than from a number
of databases. Additional validation with independent external mRNA
expression data is needed to investigate the performance of the
5-mRNAs signature model. Second, the 5-mRNA signature model was
constructed using bioinformatics data. Further experimental and
clinical studies are required to validate the functions and the
predictive value of the 5-mRNA signature model.
In conclusion, in the present study, a 5-mRNA model
was constructed to predict the prognosis for patients with PTC.
mRNAs with significantly altered expression patterns in PTC may act
as prognostic biomarkers. The signature model developed in the
present study may improve the prediction of PTC survival and
progression.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor TianXiong
Shi (Department of General Surgery, Zhongshan City People's
Hospital Affiliated to Sun Yat-sen University) for technical
support.
Funding
This research was supported by Guangzhou Medicine
and Healthcare Technology projects (grant nos. 20141A011011,
20151A011007 and 20161A011008).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LKZ and BX conceived the study. LKZ, XXG and XYD
designed the study, drafted and revised the manuscript. FS acquired
the data. JHM, JHF, WSC, QYL and BXZ analyzed the data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Howlader N, Noone AM, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al:
SEER Cancer Statistics Review, 1975–2016. Bethesda, MD: National
Cancer Institute; based on
November 2018 SEER data submission, posted to the SEER web site.
April. 2019
|
|
3
|
Alzahrani AS and Xing M: Impact of Iymph
node metastases identified on central neck dissection (CND) on the
recurrence of papillary thyroid cancer: Potential role of
BRAFV600E mutation in defining CND. Endocr Relat Cancer.
20:13–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carcangiu ML, Zampi G, Pupi A, Castagnoli
A and Rosai J: Papillary carcinoma of the thyroid. A
clinicopathologic study of 241 cases treated at the University of
Florence, Italy. Cancer. 55:805–828. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Albores-Saavedra J and Wu J: The many
faces and mimics of papillary thyroid carcinoma. Endocr Pathol.
17:1–18. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen X, Liu R and Xing M: A six-genotype
genetic prognostic model for papillary thyroid cancer. Endocr Relat
Cancer. 24:41–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Omry-Orbach G: Risk stratification in
differentiated thyroid cancer: An ongoing process. Rambam
Maimonides Med J. 7:e00032016. View Article : Google Scholar
|
|
9
|
Luster M, Clarke SE, Dietlein M, Lassmann
M, Lind P, Oyen WJ, Tennvall J and Bombardieri E; European
Association of Nuclear Medicine (EANM), : Guidelines for
radioiodine therapy of differentiated thyroid cancer. Eur J Nucl
Med Mol Imaging. 35:1941–1959. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng G, Ma HM, Huang HB, Li YW, Zhang P,
Huang JJ, Cheng L and Li GR: Overexpression of COL5A1 promotes
tumor progression and metastasis and correlates with poor survival
of patients with clear cell renal cell carcinoma. Cancer Manag Res.
11:1263–1274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tschirdewahn S, Panic A, Püllen L, Harke
NN, Hadaschik B, Riesz P, Horváth A, Szalontai J, Nyirády P, Baba
HA, et al: Circulating and tissue IMP3 levels are correlated with
poor survival in renal cell carcinoma. Int J Cancer. 145:531–539.
2019.PubMed/NCBI
|
|
12
|
Gąsior-Perczak D, Kowalik A, Walczyk A,
Siołek M, Gruszczyński K, Pałyga I, Mikina E, Trybek T, Kopczyński
J, Mężyk R, et al: Coexisting germline CHEK2 and somatic
BRAFV600E mutations in papillary thyroid cancer and
their association with clinicopathological features and disease
course. Cancers (Basel). 11:17442019. View Article : Google Scholar
|
|
13
|
Cancer Genome Atlas Research Network, .
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Y, Yu T, Chen L, Xie D, Wang F, Fu L,
Cheng C, Li Y, Zhu X and Miao G: A germline CHEK2 mutation in a
family with papillary thyroid cancer. Thyroid. 30:924–930. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
16
|
R Core Team: R: A language and environment
for statistical computing. Computing. 1:12–21. 2015.
|
|
17
|
Gong Y, Zou B, Chen J, Ding L, Li P, Chen
J, Chen J, Zhang B and Li J: Potential Five-MicroRNA signature
model for the prediction of prognosis in patients with Wilms tumor.
Med Sci Monit. 25:5435–5444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan Q and Liu B: Identification of a
RNA-Seq based 8-long non-coding RNA signature predicting survival
in esophageal cancer. Med Sci Monit. 22:5163–5172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tian S, Meng G and Zhang W: A six-mRNA
prognostic model to predict survival in head and neck squamous cell
carcinoma. Cancer Manag Res. 11:131–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang J, Kong D, Cui Q, Wang K, Zhang D,
Yuan Q, Liao X, Gong Y and Wu G: Bioinformatic analysis and
identification of potential prognostic microRNAs and mRNAs in
thyroid cancer. PeerJ. 6:e46742018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Prislei S, Martinelli E, Zannoni GF,
Petrillo M, Filippetti F, Mariani M, Mozzetti S, Raspaglio G,
Scambia G and Ferlini C: Role and prognostic significance of the
epithelial-mesenchymal transition factor ZEB2 in ovarian cancer.
Oncotarget. 6:18966–18979. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhan HX, Xu JW, Wang L, Wu D, Zhang GY and
Hu SY: FoxQ1 is a novel molecular target for pancreatic cancer and
is associated with poor prognosis. Curr Mol Med. 15:469–477. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu H, Chen S, You Z, Xie C, Huang S and Hu
X: PFKFB4 negatively regulated the expression of histone
acetyltransferase GCN5 to mediate the tumorigenesis of thyroid
cancer. Dev Growth Differ. 62:129–138. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang L, Cai Y, Luo Y, Xiong D, Hou Z, Lv
J, Zeng F, Yang Y and Cheng X: JAZF1 suppresses papillary thyroid
carcinoma cell proliferation and facilitates apoptosis via
regulating TAK1/NF-κB pathways. Onco Targets Ther. 12:10501–10514.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu ST, Zhong Q, Chen RH, Han P, Li SB,
Zhang H, Yuan L, Xia TL, Zeng MS and Huang XM: CRLF1 promotes
malignant phenotypes of papillary thyroid carcinoma by activating
the MAPK/ERK and PI3K/AKT pathways. Cell Death Dis. 9:3712018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vecchio G, Parascandolo A, Allocca C,
Ugolini C, Basolo F, Moracci M, Strazzulli A, Cobucci-Ponzano B,
Laukkanen MO, Castellone MD and Tsuchida N: Human a-L-fucosidase-1
attenuates the invasive properties of thyroid cancer. Oncotarget.
8:27075–27092. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bertoni APS, Bracco PA, de Campos RP, Lutz
BS, Assis-Brasil BM, Meyer ELS, Saffi J, Braganhol E, Furlanetto TW
and Wink MR: Activity of ecto-5′-nucleotidase (NT5E/CD73) is
increased in papillary thyroid carcinoma and its expression is
associated with metastatic lymph nodes. Mol Cell Endocrinol.
479:54–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jendrzejewski JP, Liyanarachchi S, Nagy R,
Senter L, Wakely PE, Thomas A, Nabhan F, He H, Li W, Sworczak K, et
al: Papillary thyroid carcinoma: Association between germline DNA
variant markers and clinical parameters. Thyroid. 26:1276–1284.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Siołek M, Cybulski C, Gąsior-Perczak D,
Kowalik A, Kozak-Klonowska B, Kowalska A, Chłopek M, Kluźniak W,
Wokołorczyk D, Pałyga I, et al: CHEK2 mutations and the risk of
papillary thyroid cancer. Int J Cancer. 137:548–552. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wójcicka A, Czetwertyńska M, Świerniak M,
Długosińska J, Maciąg M, Czajka A, Dymecka K, Kubiak A, Kot A,
Płoski R, et al: Variants in the ATM-CHEK2-BRCA1 axis determine
genetic predisposition and clinical presentation of papillary
thyroid carcinoma. Genes Chromosomes Cancer. 53:516–523. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stein L, Rothschild J, Luce J, Cowell JK,
Thomas G, Bogdanova TI, Tronko MD and Hawthorn L: Copy number and
gene expression alterations in radiation-induced papillary thyroid
carcinoma from chernobyl pediatric patients. Thyroid. 20:475–487.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang Z, Cai C, Han D, Gao Y, Li Q, Feng
L, Zhang W, Zheng J, Jin J, Zhang H and Wei Q: Anoctamin5 regulates
cell migration and invasion in thyroid cancer. Int J Oncol.
51:1311–1319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tenbaum SP, Juenemann S, Schlitt T, Bernal
J, Renkawitz R, Muñoz A and Baniahmad A: Alien/CSN2 gene expression
is regulated by thyroid hormone in rat brain. Dev Biol.
254:149–160. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu S, Zhou HY, Deng SC, Deng SJ, He C, Li
X, Chen JY, Jin Y, Hu ZL, Wang F, et al: ASIC1 and ASIC3 contribute
to acidity-induced EMT of pancreatic cancer through activating
Ca2+/RhoA pathway. Cell Death Dis. 8:e28062017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ceder R, Haig Y, Merne M, Hansson A, Zheng
X, Roberg K, Nees M, Iljin K, Bloor BK, Morgan PR, et al:
Differentiation-promoting culture of competent and noncompetent
keratinocytes identifies biomarkers for head and neck cancer. Am J
Pathol. 180:457–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang DL, Qu LW, Ma L, Zhou YC, Wang GZ,
Zhao XC, Zhang C, Zhang YF, Wang M, Zhang MY, et al: Genome-wide
identification of transcription factors that are critical to
non-small cell lung cancer. Cancer Lett. 434:132–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cui T, Yang L, Ma Y, Petersen I and Chen
Y: Desmocollin 3 has a tumor suppressive activity through
inhibition of AKT pathway in colorectal cancer. Exp Cell Res.
378:124–130. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ
and Chen G: Distinguishable prognostic signatures of left- and
right-sided colon cancer: A study based on sequencing data. Cell
Physiol Biochem. 48:475–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lan Y, Xiao X, Luo Y, He Z and Song X:
FEZF1 is an independent predictive factor for recurrence and
promotes cell proliferation and migration in cervical cancer. J
Cancer. 9:3929–3938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Burotto M, Chiou VL, Lee JM and Kohn EC:
The MAPK pathway across different malignancies: A new perspective.
Cancer. 120:3446–3456. 2015. View Article : Google Scholar
|
|
42
|
Nakagawa H, Hirata Y, Takeda K, Hayakawa
Y, Sato T, Kinoshita H, Sakamoto K, Nakata W, Hikiba Y, Omata M, et
al: Apoptosis signal-regulating kinase 1 inhibits
hepatocarcinogenesis by controlling the tumor-suppressing function
of stress-activated mitogen-activated protein kinase. Hepatology.
54:185–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Yin Y and Yu H: Genetic expression
profile-based screening of genes and pathways associated with
papillary thyroid carcinoma. Oncol Lett. 16:5723–5732.
2018.PubMed/NCBI
|
|
44
|
Chen S, Fan X, Gu H, Zhang L and Zhao W:
Competing endogenous RNA regulatory network in papillary thyroid
carcinoma. Mol Med Rep. 18:695–704. 2018.PubMed/NCBI
|
|
45
|
Puxeddu E and Moretti S: Clinical
prognosis in BRAF-mutated PTC. Arq Bras Endocrinol Metabol.
51:736–747. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu M, Yuan H, Li X, Liao Q and Liu Z:
Identification of a five-gene signature and establishment of a
prognostic nomogram to predict progression-free interval of
papillary thyroid carcinoma. Front Endocrinol (Lausanne).
10:7902019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cao R, Wu Q, Li Q, Yao M and Zhou H: A
3-mRNA-based prognostic signature of survival in oral squamous cell
carcinoma. PeerJ. 7:e73602019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cai J, Tong Y, Huang L, Xia L, Guo H, Wu
H, Kong X and Xia Q: Identification and validation of a potent
multi-mRNA signature for the prediction of early relapse in
hepatocellular carcinoma. Carcinogenesis. 40:840–852. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xie X, Wang J, Shi D, Zou Y, Xiong Z, Li
X, Zhou J, Tang H and Xie X: Identification of a 4-mRNA
metastasis-related prognostic signature for patients with breast
cancer. J Cell Mol Med. 23:1439–1447. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo W, Chen X, Zhu L and Wang Q: A
six-mRNA signature model for the prognosis of head and neck
squamous cell carcinoma. Oncotarget. 8:94528–94538. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chuang MK, Chiu YC, Chou WC, Hou HA, Tseng
MH, Kuo YY, Chen Y, Chuang EY and Tien HF: An mRNA expression
signature for prognostication in de novo acute myeloid leukemia
patients with normal karyotype. Oncotarget. 6:39098–39110. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ,
Yan W, Liu YW, Zhang W, Chen L and Jiang T: Prognostic value of a
nine-gene signature in glioma patients based on mRNA expression
profiling. CNS Neurosci Ther. 20:112–118. 2014. View Article : Google Scholar : PubMed/NCBI
|