Introduction
Cancers of the ovaries, most of which are carcinomas
(OC), are the eighth most common malignancy in women and the most
lethal one. In the year 2018, 295,414 new cases were diagnosed and
184,799 deaths occurred from ovarian cancer worldwide (1). OC can be subdivided into various
histological subtypes, each showing distinct genomic and epigenomic
characteristics (2). High-grade
serous carcinoma (HGSC) is the most frequent and aggressive
histotype, comprising 70% of newly diagnosed cases. Less frequent
are endometrioid carcinoma (EC, 15%), clear cell carcinoma (CCC,
12%), low-grade serous carcinoma (LGSC, <10%), and mucinous
carcinoma (MC, 3%) (3).
Carcinosarcomas (CS) of the female genital tract are biphasic
tumors containing some areas showing carcinomatous growth, mostly
HGSC, and others displaying sarcomatous differentiation. CS are
rare but aggressive tumors that often prove fatal within 1–2 years
of diagnosis (4).
The majority of malignant ovarian effusions stem
from carcinomas or CS (5,6). They are an almost universal clinical
finding in advanced-stage OC, i.e., stage III–IV according to the
International Federation of Gynaecology and Obstetrics (FIGO),
reflecting widespread intra-abdominal disease with a large number
of metastatic tumor cells. OC cells in effusions probably represent
a chemoresistant population rendering the disease untreatable and
fatal (7,8).
Different cytologic biomarkers are used as adjuncts
to morphologic examination to diagnose cancer cells in effusions
(5). Studies focusing on molecules
that promote the process of invasion and metastasis, as well as
influence intracellular signalling pathways and/or act as
transcription factors, have provided a better understanding of the
biological events behind formation of malignant effusions (5,8);
however, this knowledge is still far from complete. Although a
growing number of investigations have defined optimal panels for
routine cytologic diagnosis of carcinoma cells in effusions, only
few studies focused on the molecular alterations and genetic
mechanisms behind effusions (5,9,10). And yet, the identification of genetic
mutations and genomic imbalances in tumor cells has become
increasingly important in the management of different cancer types
and also allows us to assess the cells' proneness to develop
metastases (11,12).
We investigated the mutation status of the tumor
suppressor gene TP53, the
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha (PIK3CA), the protooncogenes of the Ras family-ki-ras2
kirsten rat sarcoma viral oncogene homolog (KRAS), Harvey
rat sarcoma viral oncogene homolog (HRAS), the neuroblastoma
RAS viral (V-Ras) oncogene homolog (NRAS)-and the v-raf
murine sarcoma viral oncogene homolog (BRAF) in a series of
103 ovarian effusions. Furthermore, we performed array comparative
genomic hybridization (aCGH) to characterize the genomic imbalances
incurred by the cells of 20 effusions from HGSC, of which ten
tumors showed TP53 mutations whereas the remaining ten had
wild-type TP53.
Materials and methods
Tumor material
The material consisted of 103 effusions from ovarian
cancers, including 84 HGSC, 10 LGSC, two CCC, one EC, and six CS.
All patients were treated at The Norwegian Radium Hospital between
2000 and 2015. The diagnoses were reached using a combination of
cytological, morphological, and immunohistochemistry (IHC)
investigations according to World Health Organization (WHO) 2014
guidelines (3). The study was
approved by the Regional Committee for Medical and Health Research
Ethics (REK, project number S-04300; http://helseforskning.etikkom.no), the
government-appointed committee responsible for overseeing medical
ethics in the South-East region of Norway. Informed consent,
including consent for publication, was obtained according to
national and institutional guidelines. An overview of the cohort
used and the clinical and pathological data are given in Table I.
 | Table I.Clinicopathologic parameters of the
103 ovarian effusions investigated. |
Table I.
Clinicopathologic parameters of the
103 ovarian effusions investigated.
| Parameter | Distribution
(n) |
|---|
| Histology |
|
|
HGSC | 84 |
| CS | 6 |
|
LGSC | 10 |
|
CCC | 2 |
| EC | 1 |
| Age |
|
|
≤60 | 42 |
|
>60 | 61 |
| FIGO stage |
|
| I | 1 |
| II | 1 |
|
III | 68 |
| IV | 33 |
| Residual
disease |
|
| 0
cm | 23 |
| ≤1
cm | 32 |
| >1
cm | 25 |
|
N/A | 23 |
| Chemoresponse after
primary treatmenta |
|
| CR | 53 |
| PR | 32 |
| SD | 7 |
| PD | 1 |
|
N/A | 10 |
Molecular analyses
DNA was extracted using the Maxwell 16 extractor
(Promega) and Maxwell 16 Cell DNA Purification kit (Promega)
according to the manufacturer's recommendations. The concentration
was measured using QIAxel (Qiagen).
Mutational analysis of TP53, PIK3CA, KRAS,
HRAS, and NRAS was performed according to previously
described protocols, using M13-linked PCR primers designed to flank
and amplify targeted sequences (13,14). The
primer combinations BRAF-F1 (5′TGCTTGCTCTGATAGGAAAATGAGATCT3′) and
BRAF-R1 (5′ATCTCAGGGCCAAAAATTTAATCAGTG3′) were used to detect the
mutation status of BRAF. The thermal cycling for BRAF
included an initial step at 95°C for 10 min followed by 35 cycles
at 96°C for 3 sec, 58°C for 15 sec, 30 sec at 68°C, and a final
step at 72°C for 2 min. Direct sequencing was performed using a
3500 Genetic Analyzer (Applied Biosystems).
The genes were selected based on the information
reported in the COSMIC database (Catalogue of Somatic Mutations in
Cancer, at http://cancer.sanger.ac.uk/cosmic) (15). According to COSMIC, there is no
information on mutations in effusions; however, it contains data on
the most frequently mutated genes in ovarian carcinoma. Since
KRAS was in the top list, we decided to investigate also the
other member genes of the RAS and RAF families, i.e., HRAS,
NRAS and BRAF.
The BLAST (http://blast.ncbi.nlm.nih.gov/blast.cgi) and BLAT
(http://genome.ucsc.edu/cgi-bin/hgblat) programs were
used for computer analysis of sequence data. The reference
sequences used for TP53 was NM_000546.5.
The difference between mutation and polymorphism was
evaluated by the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org/variant/11-534242-A-G).
Whole genome investigation by means of aCGH was
performed using the CytoSure Consortium Cancer + SNP arrays (Oxford
Gene Technology) according the manufacturers' recommendation. Data
were analysed using Agilent Feature Extraction Software (version
10.7.3.1) and CytoSure Interpret Software (version 4.9.40, Oxford
Gene Technology). The genomic imbalances were identified using the
Circular Binary Segmentation (CBS) algorithm and adding a
custom-made aberration filter defining a copy number aberration
(CNA) as a region with minimum five probes gained/lost (16). Annotations are based on human
reference sequence GRCh37/hg19.
Twenty samples were selected for aCGH investigation,
ten bearing TP53 mutation in their genome and ten wild-type.
The average copy number alteration (ANCA) index was calculated as
the total number of aberrations divided by the samples number
between the two groups (17). The
statistical analysis was performed using the Mann-Whitney U
test.
Results
All effusions analyzed for TP53, PIK3CA, KRAS,
HRAS, NRAS, and BRAF mutation status gave informative
results. TP53 was found mutated in 41 out of 84 HGSC (49%),
in two out of 10 LGSC (20%), in the only case of EC examined, and
in one out of six CS. A detailed overview of the TP53
findings is shown in Table II. Two
novel mutation sites were identified for TP53:
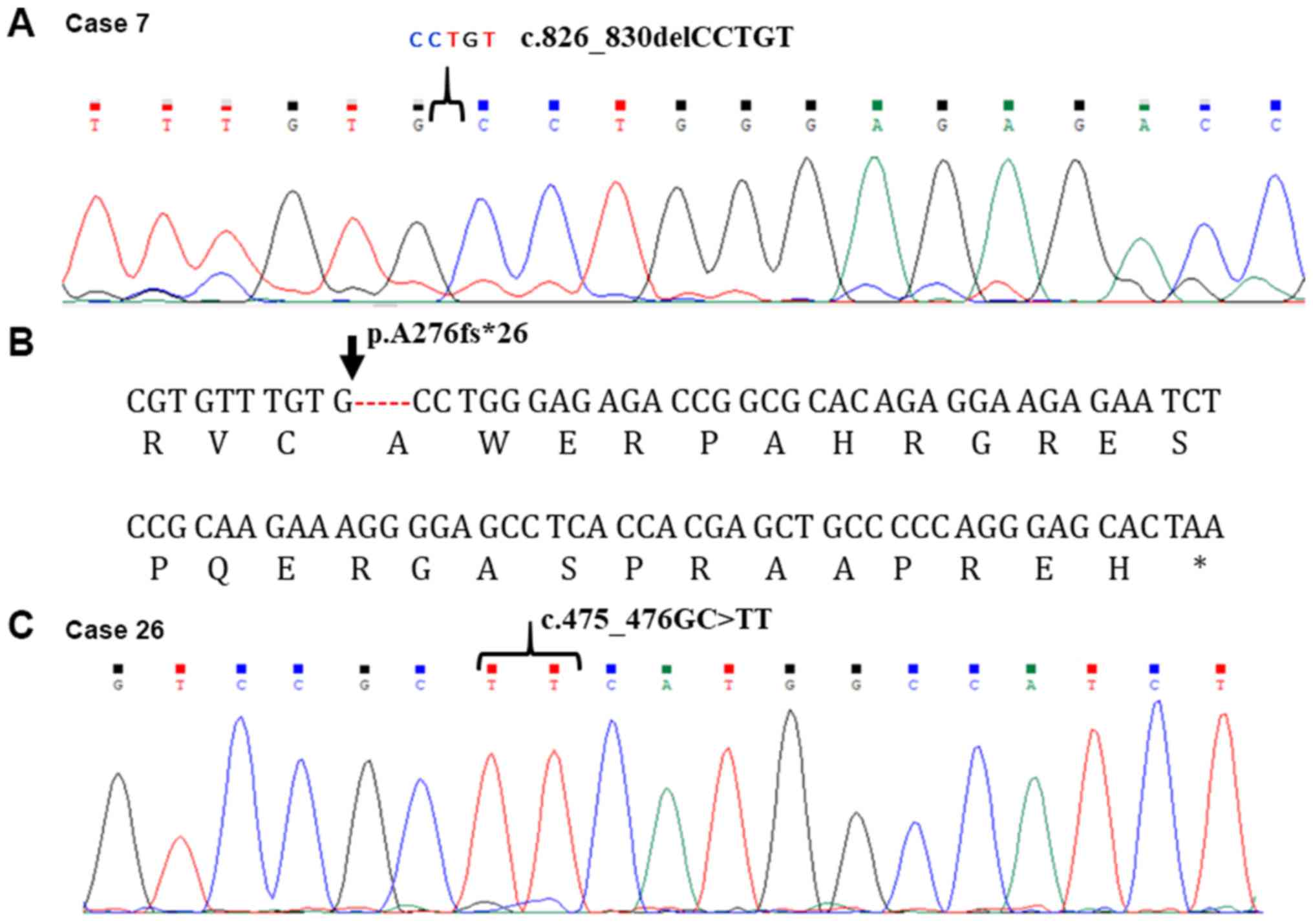
c.826_830delCCTGT in case 7 and c.475-476GC>TT in case 26
(Fig. 1). PIK3CA mutations
were found in four HGSC of 103, in which a c.1634A>C (cases 2,
56, and 58) and a c.3155C>T mutation (case 79) were seen. We
identified the c.34G>T and c.183A>C KRAS mutations in
two of 103 specimens (cases 10, a HGSC, and 85, an LGSC,
respectively). The HRAS mutation c.173C>T was also
detected in two tumors (2%; cases 16 and 23), both of them HGSC.
Finally, we identified an HRAS polymorphism, c.81T>C, in
38 effusions (37.5%) of all histotypes. None of the tumors showed a
mutated sequence for NRAS or BRAF.
 | Table II.Mutation status of TP53. |
Table II.
Mutation status of TP53.
| Case | Histology | TP53 |
|---|
| 1 | HGSC | c.437G>A;
p.W146*; COSM43609 |
| 2 | HGSC | c.584T>C;
p.I195T; COSM11089 |
| 3 | HGSC | c.273G>A;
p.W91*COSM44492 |
| 4 | HGSC |
|
| 5 | HGSC | c.916C>T;
p.R306*; COSM10663 |
| 6 | HGSC |
|
| 7a | HGSC |
c.826_830delCCTGT |
| 8 | HGSC | c.818G>A;
p.R273H; COSM10660 |
| 9 | HGSC | c.797G>A;
p.G266E; COSM10867 |
| 10 | HGSC |
|
| 11 | HGSC | c.488A>G;
p.Y163C; COSM10808 |
| 12 | HGSC | c.524G>A;
p.R175H; COSM10648 |
| 13 | HGSC | c.844C>T;
p.R282W; COSM10704 |
| 14 | HGSC | c.574C>T;
p.Q192*; COSM10733 |
| 15 | HGSC | c.527G>T;
p.C176F; COSM10645 |
| 16 | HGSC | c.469G>T;
p.V157F; COSM10670 |
| 17 | HGSC | c.527G>A;
p.C176Y; COSM10687 |
| 18 | HGSC |
|
| 19 | HGSC | c.754del; p.
L252fs*93; COSM45215 |
| 20 | HGSC | c.403del;
p.C135fs*35; COSM44670 |
| 21 | HGSC |
|
| 22 | HGSC | c.394A>T;
p.K132*; COSM44641 |
| 23 | HGSC | c.832C>G;
p.P278A; COSM10814 |
| 24 | HGSC | c.814G>A;
p.V272M; COSM10891 |
| 25 | HGSC | c.394A>G;
p.K132E; COSM10813 |
| 26a | HGSC |
c.475_476GC>TT |
| 27 | HGSC |
|
| 28 | HGSC | c.797G>A;
p.G266E; COSM10867 |
| 29 | HGSC | c.108G>A;
p.P36P; COSM6474191 |
|
|
| c.737T>A;
p.M246K; COSM44103 |
| 30 | HGSC | c.742C>T;
p.R248W; COSM10656 |
| 31 | HGSC |
|
| 32 | HGSC | c.488A>G;
p.Y163C; COSM10808 |
| 33 | HGSC | c.836G>A;
p.G279E; COSM43714 |
| 34 | HGSC |
|
| 35 | HGSC | c.818G>A;
p.R273H; COSM10660 |
| 36 | HGSC |
|
| 37 | HGSC |
|
| 38 | HGSC |
|
| 39 | HGSC | c.524G>A;
p.R175H; COSM10648 |
| 40 | HGSC |
|
| 41 | HGSC | c.711G>A;
p.M237I; COSM10834 |
| 42 | HGSC |
|
| 43 | HGSC | c.166G>T;
p.E56*; COSM12168 |
| 44 | HGSC | c.524G>A;
p.R175H; COSM10648 |
| 45 | HGSC |
|
| 46 | HGSC |
|
| 47 | HGSC |
|
| 48 | HGSC |
|
| 49 | HGSC |
|
| 50 | HGSC |
|
| 51 | HGSC |
|
| 52 | HGSC | c.434T>C;
p.L145P; COSM43899 |
| 53 | HGSC |
|
| 54 | HGSC |
|
| 55 | HGSC | c.475G>C;
Pa159P; COSM43836 |
| 56 | HGSC |
|
| 57 | HGSC |
|
| 58 | HGSC |
|
| 59 | HGSC |
|
| 60 | HGSC | c.844C>T;
p.R282W; COSM10704 |
| 61 | HGSC | c.646G>A;
p.V216M; COSM10667 |
| 62 | HGSC | c.832 C>T;
p.P278S; COSM10939 |
| 63 | HGSC |
|
| 64 | HGSC |
|
| 65 | HGSC |
|
| 66 | HGSC |
|
| 67 | HGSC |
|
| 68 | HGSC |
|
| 69 | HGSC |
|
| 70 | HGSC |
|
| 71 | HGSC | c.527G>T;
p.C176F; COSM10645 |
| 72 | HGSC |
|
| 73 | HGSC |
|
| 74 | HGSC |
|
| 75 | HGSC | c.578A>G;
p.H193R; COSM10742 |
| 76 | HGSC |
|
| 77 | HGSC |
|
| 78 | HGSC |
|
| 79 | HGSC |
|
| 80 | HGSC |
|
| 81 | HGSC | c.796G>A;
p.G266R; COSM10794 |
| 82 | HGSC | c.844C>T;
p.R282W; COSM10704 |
| 83 | HGSC |
|
| 84 | HGSC |
|
| 85 | LGSC | c.750del; p.
I251fs*94; COSM44064 |
| 86 | LGSC |
|
| 87 | LGSC | c.714T>A;
p.C238*; COSM45677 |
| 88 | LGSC |
|
| 89 | LGSC |
|
| 90 | LGSC |
|
| 91 | LGSC |
|
| 92 | LGSC |
|
| 93 | LGSC |
|
| 94 | LGSC |
|
| 95 | CCC |
|
| 96 | CCC |
|
| 97 | EC | c.1024C>T;
p.R342*; COSM11073 |
| 98 | CS | c.796G>A;
p.G266R; COSM10794 |
| 99 | CS |
|
| 100 | CS |
|
| 101 | CS |
|
| 102 | CS |
|
| 103 | CS |
|
aCGH analysis for genomic imbalances was performed
on 20 effusions from patients with HGSC, comparing 10 tumors
bearing TP53 mutations (cases 1, 3, 5, 7, 8, 13, 14, 15, 19,
and 32) and 10 which had a wild-type TP53 sequence (cases
18, 27, 31, 36, 37, 38, 42, 45, 47, and 48). Overall, the aCGH
analysis revealed highly imbalanced genomes in all tumors analysed
with many gains and/or losses (Table
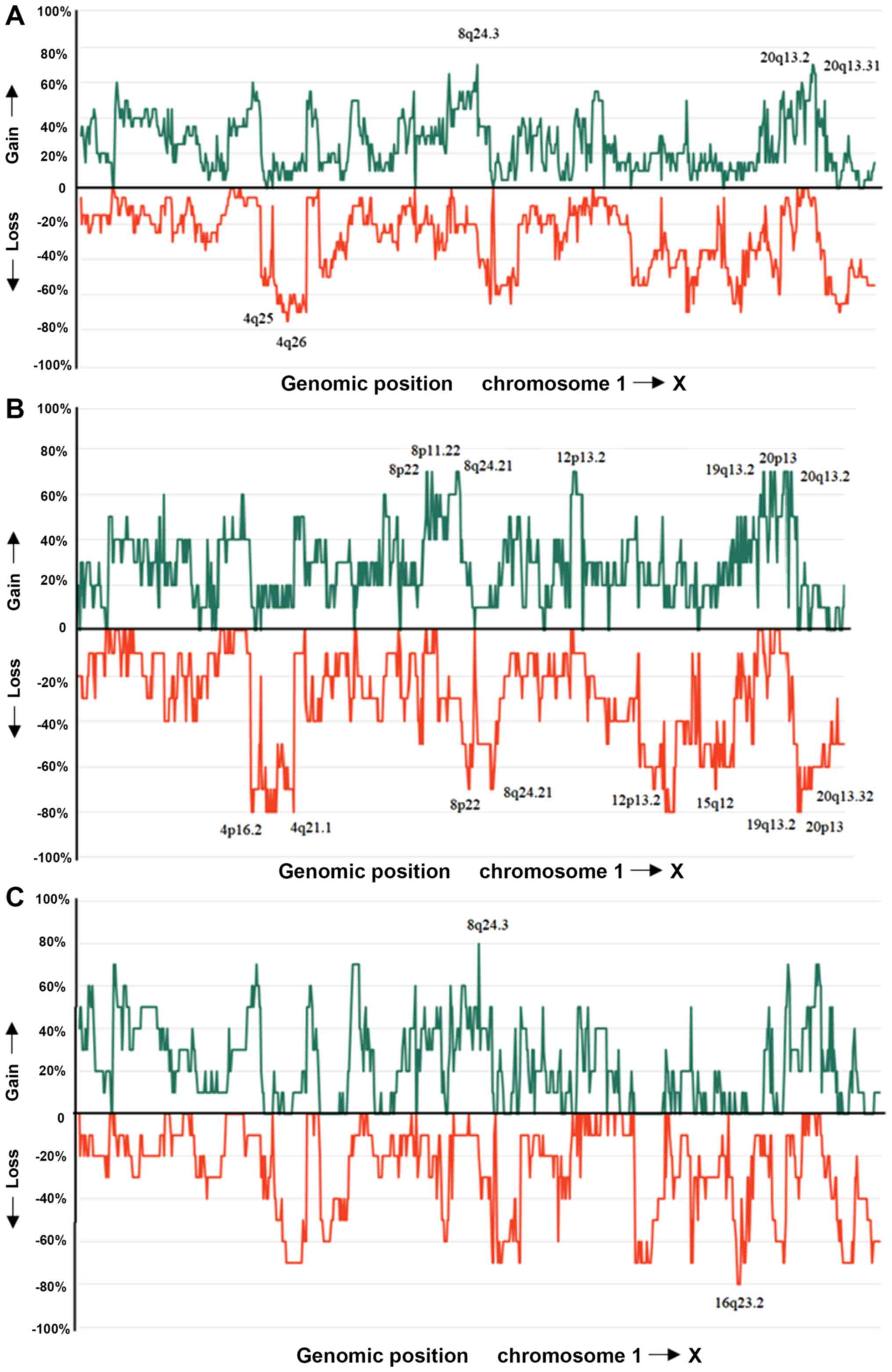
SI). The most frequent gains were scored at 8q24.3, 20q13.2,
and 20q13.31 (70%) whereas the most frequent losses were scored at
4q25 and 4q26 (75%) (Fig. 2).
Amplifications mostly involved chromosomal band 19q11 followed by
the segment 3q22q29. The two subgroups of effusions, i.e., with and
without TP53 mutation, were both very complex and similar
with regard to imbalances. The ANCA index calculated for tumors
(18) with TP53 mutation was
83.2 but 66.3 for tumors with wild-type TP53 (P=0.14).
Discussion
Molecular profiles of different tumor types have
helped manage cancer patients with regard to diagnosis, prognosis,
and lately also choice of treatment (19). A similar molecular characterization
of effusions from ovarian cancer might highlight the mechanisms
behind development of metastasis and possibly, further down the
road, help decide among different personalized therapies (5). Since the number of studies focusing on
molecular analysis of ovarian cancers at such advanced stage that
effusions have already developed, is low, and since chemoresistance
is one of the main characteristics of these malignancies, we aimed
to add to the existing knowledge by performing mutation analyses of
selected genes as well as determining copy number profiles of two
groups of patients, those whose tumors did or did not have
TP53 mutations.
The tumor suppressor gene TP53 has been found
mutated in many different malignancies (20), including those arising in the
ovaries, at a frequency of 66% in the most aggressive serous
carcinomas (21). The rate of
TP53 mutation detected in our series was 46% for effusions
from HGSC and LGSC. The seeming discrepancy between the frequencies
recorded in the present series and in the literature could be due
to methodological limitations, see below. In HGSC, we identified
two novel sites for TP53 mutation: A deletion of the CCTGT
sequence was found in position c.826_830 of case 7 (stage III
tumor), whereas a substitution GC>TT in position 475_476GC was
identified in case 26 (stage IV tumor). The c.826_830del CCTGT is
an out-of-frame change resulting in a frameshift of 26 amino acids
(aa) (p. A276fs*26) (Fig. 1) after
which a stop codon occurs. The predicted protein would consist of
156 aa. The substitution c.475_476GC>TT results in a change from
alanine (A) to phenylalanine (F) (p.A159F). The mutation is at
present of unknown pathogenicity in ovarian cancer. However, other
mutations on c.475 have been reported as pathogenic in the COSMIC
database, e.g., in tumors of the lung and liver (https://cancer.sanger.ac.uk/cosmic). The impact
of the new mutation sites in relation to different clinical
parameters awaits further studies, ideally of larger series of
patients. The two patients here examined had received upfront
surgery and standard chemotherapy; case 7 showed a residual disease
of 6 cm whereas case 26 had no residual disease at primary
operation. Furthermore, both cases showed relatively long survival:
Case 7 had 13 months progression-free survival (PFS) and overall
survival (OS) of 81 months, whereas case 26 had PFS of 27 months
and OS of 45 months.
PIK3CA belongs to the family of genes
encoding phosphatidylinositol 3-kinases (PI3Ks). It is activated
through the PI3K/AKT signalling pathway in 70% of ovarian cancers,
promoting cellular growth, proliferation, and cell survival
(22). Somatic mutations of this
gene have been detected in different cancer types (23). In ovarian cancer, it occurs in 30% of
all tumors, but reaches 45% in EC and CCC (24). We found PIK3CA mutated in 4%
of the HGSC effusions examined, which is in line with what is
reported in the COSMIC database. Unfortunately, the number of EC
and CCC samples was too low to allow statistical conclusions. A
number of clinical studies have focused on the PI3K/AKT/mTOR
signaling pathway as a therapeutic target for patients with ovarian
cancer (25,26); the identification of patients
carrying PIK3CA mutation may therefore be important for the
choice of therapy. Important to note in this regard is the fact
that also other genes of the PI3K/AKT/mTOR signaling pathway should
be investigated for their mutation status as they, too, may be
involved pathogenetically (26).
KRAS and HRAS are principal members of
the RAS family and have frequently been implicated in the
development of different types of tumors (27). In ovarian carcinomas, the incidence
of KRAS point mutations was found to be 13% (21). Previous studies have demonstrated an
association between KRAS mutations and well-differentiated,
clinically less advanced cancers (28,29).
KRAS mutation was in ovarian serous carcinoma found more
frequently in LGSC than in HGSC (30–32).
HRAS mutations are rare in ovarian tumors
(33,34). We found an HRAS mutation in
only two HGSC: However, our study showed presence of the 81T>C
polymorphism in the coding region of HRAS in 38 out of 103
tumors (37%) of all histotypes. The Genome Aggregation Database,
gnomAD, reports that SNP 81T>C is a polymorphism seen in 30% of
the normal population. Both tumors with HRAS mutation also
showed TP53 mutation. In each case, one can hypothesize a
scenario in which the mutations represent a primary and a secondary
event either in the same cell or in different cells/clones.
Information on effusions from CS arising in the
female genital tract is limited to data generated by
immunohistochemical techniques (35). This is the first time that mutation
analyses have been performed on such metastatic cells. It seems,
however, that the genes investigated in the present study are not
relevant in cells from effusions since we found only one CS with
TP53 mutation.
The mutation rates for the analysed genes in the
present study differ slightly from those reported in the
literature, something that may be attributable to the molecular
methods applied. We used PCR followed by Sanger sequencing. It is
known that Sanger sequencing cannot detect mutation if the level of
abnormal cells is below 15% (36),
whereas next generation sequencing (NGS) or exome sequencing, used
in most published studies (37), is
more sensitive, i.e., has a higher resolution level. NGS, on the
other hand, cannot discriminate between a ‘real’ mutation and a
polymorphism. Taking into account these two factors, one would
indeed expect higher mutation rates to be detected by NGS compared
to Sanger sequencing, as was observed.
aCGH data showed highly imbalanced genomes both in
tumors with mutated and wild-type TP53. The genomic regions
involved are in agreement with the results of previous studies
where primary OC were investigated (38). The ANCA index detected in the
TP53 mutated subgroup was 83.2 whereas it was 66.3 in the
subgroup with wild-type TP53. The difference between the two
groups was not found statistically significant using the
Mann-Whitney U test.
The origin of ovarian carcinomas has lately been
debated but, according to the latest WHO classification, the
majority of HGSC are thought to originate in the tubes whereupon
metastatic spreading occurs to the ovaries (39,40). In
the light of this concept, it is not surprising that ovarian
carcinomas show the same imbalances as do ovarian cancer cells
found in effusions, since both represent late evolutionary stages
in carcinoma development.
Supplementary Material
Supporting Data
Acknowledgements
The authors wish to thank Miss Margrethe Stoltenberg
and Dr Rønnaug A. U. Strandabø, both from the Section for Cancer
Cytogenetics, Institute for Cancer Genetics and Informatics, Oslo
University Hospital, for technical assistance.
Funding
This work was supported by grants from the
South-East Norway Regional Health Authority (Helse Sør-Øst) and
Radiumhospitalets Legater.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MB performed molecular experiments and wrote the
manuscript. IP participated in performing molecular experiments and
interpretation of data. IK participated in performing data
analysis. BD provided clinical data and specimens. SH assisted with
writing of the article and experimental design. FM designed the
study and supervised the writing of the manuscript. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The ethical approval was granted by the Regional
Committee for Medical and Health Research Ethics (REK; http://helseforskning.etikkom.no); for further
information, please see this website: http://www.eurecnet.org/information/norway.html.
Patient consent for publication
Consent for publication of data was provided by all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Prat J, D'Angelo E and Espinosa I: Ovarian
carcinomas: At least five different diseases with distinct
histological features and molecular genetics. Hum Pathol. 80:11–27.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kurman RJ, Carcangiu ML, Herrington CS and
Young RH: WHO classification of tumors of female reproductive
organs. IARC. 2014.
|
|
4
|
D'Angelo E and Prat J: Pathology of mixed
Mullerian tumours. Best Pract Res Clin Obstet Gynaecol. 25:705–718.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davidson B: Ovarian and primary peritoneal
carcinoma. Serous Effusions - Etiology, Diagnosis, Prognosis and
Therapy. Davidson B, Firat P and Michael CW: Springer; London, UK:
pp. 47–68, 167-204. 2011
|
|
6
|
Piche A: Malignant peritoneal effusion
acting as a tumor environment in ovarian cancer progression: Impact
and significance. World J Clin Oncol. 9:167–171. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davidson B: Recently identified drug
resistance biomarkers in ovarian cancer. Expert Rev Mol Diagn.
16:569–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davidson B: Biomarkers of drug resistance
in ovarian cancer-an update. Expert Rev Mol Diagn. 19:469–476.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brunetti M, Holth A, Panagopoulos I, Staff
AC, Micci F and Davidson B: Expression and clinical role of the
dipeptidyl peptidases DPP8 and DPP9 in ovarian carcinoma. Virchows
Archiv. 474:177–185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davidson B, Stavnes HT, Holth A, Chen X,
Yang Y, Shih IeM and Wang TL: Gene expression signatures
differentiate ovarian/peritoneal serous carcinoma from breast
carcinoma in effusions. J Cell Mol Med. 15:535–544. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shah RH, Scott SN, Brannon AR, Levine DA,
Lin O and Berger MF: Comprehensive mutation profiling by
next-generation sequencing of effusion fluids from patients with
high-grade serous ovarian carcinoma. Cancer Cytopathol.
123:289–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nagel H, Schulten HJ, Gunawan B, Brinck U
and Fuzesi L: The potential value of comparative genomic
hybridization analysis in effusion-and fine needle aspiration
cytology. Mod Pathol. 15:818–825. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Malcikova J, Tausch E, Rossi D, Sutton LA,
Soussi T, Zenz T, Kater AP, Niemann CU, Gonzalez D, Davi F, et al:
ERIC recommendations for TP53 mutation analysis in chronic
lymphocytic leukemia-update on methodological approaches and
results interpretation. Leukemia. 32:1070–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brunetti M, Agostini A, Staurseth J,
Davidson B, Heim S and Micci F: Molecular characterization of
carcinosarcomas arising in the uterus and ovaries. Oncotarget.
10:3614–3624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tate JG, Bamford S, Jubb HC, Sondka Z,
Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E,
et al: COSMIC: The catalogue of somatic mutations in cancer.
Nucleic Acids Res. 47:D941–D947. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Olshen AB, Venkatraman ES, Lucito R and
Wigler M: Circular binary segmentation for the analysis of
array-based DNA copy number data. Biostatistics. 5:557–572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ried T, Heselmeyer-Haddad K, Blegen H,
Schröck E and Auer G: Genomic changes defining the genesis,
progression, and malignancy potential in solid human tumors: A
phenotype/genotype correlation. Genes Chromosomes Cancer.
25:195–204. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Micci F, Teixeira MR, Haugom L, Kristensen
G, Abeler VM and Heim S: Genomic aberrations in carcinomas of the
uterine corpus. Genes Chromosomes Cancer. 40:229–246. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jackson SE and Chester JD: Personalised
cancer medicine. Int J Cancer. 137:262–266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang W, Edwards A, Flemington EK and
Zhang K: Significant prognostic features and patterns of somatic
TP53 mutations in human cancers. Cancer Inform.
16:11769351176912672017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
simplewww.sanger.ac.uk/genetics/CGP/cosmic
|
|
22
|
Li H, Zeng J and Shen K: PI3K/AKT/mTOR
signaling pathway as a therapeutic target for ovarian cancer. Arch
Gynecol Obstet. 290:1067–1078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Samuels Y and Waldman T: Oncogenic
mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol.
347:21–41. 2010.PubMed/NCBI
|
|
24
|
Campbell IG, Russell SE, Choong DY,
Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB and
Phillips WA: Mutation of the PIK3CA gene in ovarian and breast
cancer. Cancer Res. 64:7678–7681. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mabuchi S, Kuroda H, Takahashi R and
Sasano T: The PI3K/AKT/mTOR pathway as a therapeutic target in
ovarian cancer. Gynecol Oncol. 137:173–179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gasparri ML, Bardhi E, Ruscito I, Papadia
A, Farooqi AA, Marchetti C, Bogani G, Ceccacci I, Mueller MD and
Benedetti Panici P: PI3K/AKT/mTOR pathway in ovarian cancer
treatment: Are we on the right track? Geburtshilfe Frauenheilkd.
77:1095–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fernández-Medarde A and Santos E: Ras in
cancer and developmental diseases. Genes Cancer. 2:344–358. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nodin B, Zendehrokh N, Sundstrom M and
Jirstrom K: Clinicopathological correlates and prognostic
significance of KRAS mutation status in a pooled prospective cohort
of epithelial ovarian cancer. Diagn Pathol. 8:1062013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dobrzycka B, Terlikowski SJ, Kowalczuk O,
Niklińska W, Chyczewski L and Kulikowski M: Mutations in the KRAS
gene in ovarian tumors. Folia Histochem Cytobiol. 47:221–224. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Della Pepa C, Tonini G, Santini D, Losito
S, Pisano C, Di Napoli M, Cecere SC, Gargiulo P and Pignata S: Low
grade serous ovarian carcinoma: From the molecular characterization
to the best therapeutic strategy. Cancer Treat Rev. 41:136–143.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Singer G, Shih Ie M, Truskinovsky A,
Umudum H and Kurman RJ: Mutational analysis of K-ras segregates
ovarian serous carcinomas into two types: Invasive MPSC (low-grade
tumor) and conventional serous carcinoma (high-grade tumor). Int J
Gynecol Pathol. 22:37–41. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singer G, Oldt R III, Cohen Y, Wang BG,
Sidransky D, Kurman RJ and Shih IeM: Mutations in BRAF and KRAS
characterize the development of low-grade ovarian serous carcinoma.
J Natl Cancer Inst. 95:484–486. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hunter SM, Anglesio MS, Ryland GL, Sharma
R, Chiew YE, Rowley SM, Doyle MA, Li J, Gilks CB, Moss P, et al:
Molecular profiling of low grade serous ovarian tumours identifies
novel candidate driver genes. Oncotarget. 6:37663–37677. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hollis RL and Gourley C: Genetic and
molecular changes in ovarian cancer. Cancer Biol Med. 13:236–247.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ikeda K, Tate G, Suzuki T and Mitsuya T:
Effusion cytodiagnosis of carcinosarcoma derived from the female
genital tract: Immunohistochemical features of MMP-7 and Ki-67 and
immunofluorescence double staining analyses of eight cases. Gynecol
Oncol. 97:323–329. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rohlin A, Wernersson J, Engwall Y, Wiklund
L, Bjork J and Nordling M: Parallel sequencing used in detection of
mosaic mutations: Comparison with four diagnostic DNA screening
techniques. Hum Mutat. 30:1012–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Salk JJ, Schmitt MW and Loeb LA: Enhancing
the accuracy of next-generation sequencing for detecting rare and
subclonal mutations. Nat Rev Genet. 19:269–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Micci F, Haugom L, Abeler VM, Davidson B,
Trope CG and Heim S: Genomic profile of ovarian carcinomas. BMC
cancer. 14:3152014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim J, Park EY, Kim O, Schilder JM, Coffey
DM, Cho CH and Bast RC Jr: Cell origins of high-grade serous
ovarian cancer. Cancers (Basel). 10:4332018. View Article : Google Scholar
|
|
40
|
Lengyel E: Ovarian cancer development and
metastasis. Am J Pathol. 177:1053–1064. 2010. View Article : Google Scholar : PubMed/NCBI
|