Introduction
Increasing evidence supports that RNA methylation is
a widespread phenomenon and a critical regulator of transcript
expression (1,2). The most prevalent RNA methylation,
N6-methyladenosine (m6A), is a reversible RNA
post-transcriptional modification and occurs in approximately 25%
of transcripts at the genome-wide level (1). RNA m6A modification regulates RNA
splicing, translocation, stability, and translation into protein
(3–6). Dynamic regulation of the m6A
epitranscriptome is involved in diverse cellular functions,
including heat shock, DNA damage, cancer, stem cell
differentiation, circadian rhythm, spermatogenesis and oogenesis,
response to interferon-γ, and viral infections (2,3,7).
m6A dynamics and functions are executed by three
groups of proteins: Methyltransferases or ‘writers’, demethylases
or ‘erasers’, and m6A-binding proteins or ‘readers’ (2,3,7). In most cell types, m6A
modification is catalyzed by the methyltransferase complex
consisting of the methyltransferase like 3 (METTL3) and METTL14 and
their cofactors (3,7). The erasers include the fat mass and
obesity-associated protein (FTO) and alpha-ketoglutarate-dependent
dioxygenase alkB homolog 3 (ALKBH3) and ALKBH5 (2,3,7). Increasing evidence supports that m6A
modification may play an important role in tumorigenesis (8,9). For
example, METTL3 has been found to be upregulated in human
glioblastoma tissues, and high levels of METTL3 expression in tumor
tissues predict poor patient survival (10). Suppression of METTL3 or METTL14
decreases hepatocellular carcinoma cell proliferation, migration
and colony formation in vitro, as well as hepatocellular
carcinoma tumorigenicity and lung metastasis in vivo
(11). FTO is over-expressed in
human cervical squamous cell carcinoma tissues, and high levels of
FTO expression correlate with poor patient prognosis (12). ALKBH5 is highly expressed in
glioblastoma stem-like cells and demethylates FOXM1 nascent
transcripts, leading to FOXM1 over-expression, stem-like cell
proliferation and tumorigenesis (13).
Gastric cancer is one of the most prevalent and
deadly malignancies that threatens global health (14). Previous studies demonstrated that the
expression level of METTL3 is elevated in many gastric cancer cell
lines and tumor tissues (15,16). The
elevated level of METTL3 expression is clinically correlated with
the procession of gastric cancer (15,16).
Results from previous studies indicate that gastric cancer cell
proliferation is associated with aberrant expression of various
effector molecules, such as leucine rich repeat containing G
protein coupled receptor 5 (LGR5), RAD17 checkpoint clamp loader
component (RAD17), facilitated trehalose transporter Tret1-2
homolog (TRET1-2), ATPase Na+/K+ transporting subunit beta 1
(ATP1B1), matrix metallopeptidase 3 (MMP3), HEPANAS_3, interferon
induced transmembrane protein 3 (IFITM3), and S100 calcium binding
protein A4 (S100A4) (17–19). In addition, increasing evidence
supports that the activity of the suppressor of cytokine signaling
(SOCS) family proteins correlates with the progression and poor
prognosis in various cancers, including gastric cancer (20). SOCS2 is well defined as a negative
feedback regulator in multiple proliferation-related pathways and
may act as a tumor suppressor in multiple malignancies (21–26).
However, the role of METTL3 in gastric cancer progression and
whether METTL3 can modulate SOCS expression to regulate gastric
cancer cell proliferation are still not fully understood. In this
study, we report that upregulation of METTL3 in gastric cancer may
maintain gastric cancer tumorigenicity through suppressing SOCS2 to
promote cell proliferation.
Materials and methods
Gastric cancer cell line culture
The AGS cells (the gastric cancer cell line) were
purchased from American Type Culture Collection (ATCC). Cells were
cultured in the medium with L-15 medium supplemented with 10% fetal
bovine serum and 100 U penicillin/streptomycin as recommended.
CRISPR-Cas9 knockout METTL3
We took the CRISPR/Cas9 approach to knock out the
METTL3 gene (NCBI Gene ID 56339) in AGS cells and generate
stable cell lines. AGS cells were transfected with the METTL3
CRISPR/Cas9 and Homology-Directed Repair (HDR) plasmids (Santa Cruz
Biotech, Inc.). Stably transfected cells were selected, collected
and further confirmed and validated by real-time PCR and Western
blot analysis, as in our previous studies (27).
Cell transfection
The specific siRNA to SOCS2 (si-SOCS2) was obtained
from Santa Cruz Biotech, Inc. (sc-40998). A non-specific siRNA of a
scrambled sequence from Santa Cruz was used as the control. siRNA
was mixed with Lipofectamine RNAiMax (Thermo Fisher Scientific) and
transfected into cells, as we previously reported (28). The pCMV6 plasmid with full-length
SOCS2 sequence (pCMV6-SOCS2) was purchased from Origene (RC203163).
The pCMVv6 empty vector from Origene was used as the control. AGS
cells were transfected with the plasmid via Lipofectamine 2000
(Thermo Fisher Scientific) (28).
Knockdown or overexpression of SOCS2 in AGS cells was validated by
real-time PCR at 48 h after transfection.
Cell proliferation and apoptotic death
assays
AGS cells were initially seeded in 96-well plate at
a density of 1×103 cells/well. At various culture hours
of incubation after seeding, the
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
(MTS) reagent was added cells (Promega) for MTS assay. After 1 h of
incubation, the OD490 value was measured with a SpectraMax
(Molecular Devices Corporation). AGS proliferation was also
measured by direct count of cell numbers. Cells were plated at
1×104 per well in 12-well plate. At various culture
hours of incubation after seeding, cells were trypsinized and total
numbers were counted in a double-blind manner. The Annexin V-FITC
apoptosis staining/detection kit (Abcam) was used to detect
apoptotic cell death.
Real-time PCR
RNA expression levels were quantified by using
real-time PCR. Total RNA was extracted with the Trizol reagent
(Invitrogen). A total of 500 ng RNA was used for reverse
transcription (Invitrogen). PCR reactions were performed using the
SYBR Green reagent (Bio-Rad) on a Bio-Rad CFX96 platform. The
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as
internal control. Each sample was run in triplicate. The relative
abundance of each RNA was calculated using the 2−ΔΔCt
method and normalized to GAPDH as we previously reported (27,28). The
sequences of all the primers were listed in Table I.
 | Table I.The sequences of real-time PCR
primers. |
Table I.
The sequences of real-time PCR
primers.
| Primer | Sequence
(5′-3′) |
|---|
| METTL3-F |
CTTTGCCAGTTCGTTAGTCTC |
| METTL3-R |
CTGACCTTCTTGCTCTGTTGT T |
| SOCS1-F |
TCTGTAGGATGGTAGCACACA |
| SOCS1-R |
GGAAGAGGAGGAAGGTTCTG |
| SOCS3-F |
GACGATAGCAACCACAAGTG |
| SOCS3-R |
AGATTCCCTGGCAGTTCTC |
| SOCS2-si-F |
AGGGAATGGCAGAGACACT |
| SOCS2-si-R |
TGGCAGAGAGAGAAGGGAT |
| SOCS2-OE-F |
CGAATACCAAGACGGAAA |
| SOCS2-OE-R |
CAGATGAACCACACTGTCA |
| MMP7-F |
ATGAGGATGAACGCTGGA |
| MMP7-R |
TGTCCCATACCCAAAGAATG |
| S100A4-F |
AGAGGGTGACAAGTTCAAGC |
| S100A4-R |
GTCCAAGTTGCTCATCAGC |
| TERT-F |
GCTCCCATTTCATCAGCA |
| TERT-R |
CTGCGTTCTTGGCTTTCA |
| Rad17-F |
TGCCATACCTTGCTCTACTAAC |
| Rad17-R |
TTCAATCTTCCAAAGTGTCG |
| Heparanase-F |
TGCTATCCGACACCTTTGC |
| Heparanase-R |
CTTGCCTCATCACCACTTCTAT |
| MMP7-F |
CCAACCTATGGAAATGGAGA |
| MMP7-R |
GAATGGATGTTCTGCCTGA |
| ATP1B1-F |
TCAAACCTAAGCCTCCCA |
| ATP1B1-R |
CTCCACATTTCCAACTTTATCC |
| IFITM3-F |
TCGCCTACTCCGTGAAGTCT |
| IFITM3-R |
GGATGACGATGAGCAGAATG |
| LGR5-F |
CTACATGGTCGCTCTCATCTTG |
| LGR5-R |
ATATTCTCCAGGTCTCCCTTGTC |
| GAPDH-F |
TGCACCACCAACTGCTTAGC |
| GAPDH-R |
GGCATGGACTGTGGTCATGAG |
Western blot
AGS cell cultures were harvested and lysed with the
M-PER mammalian protein extraction reagent (Thermo Fisher
Scientific). Samples were run in 10% acrylamide gel (30 µg total
protein/well) and transferred to the PVDF membranes (Millipore).
Membranes were then subjected to the following primary antibody at
4°C overnight: Anti-METTL3 (1:1,000, 15073-1-AP; Proteintech),
anti-SOCS1 (Proteintech), anti-SOCS2 (1:1,000, 2779, CST, MA, USA),
anti-SOCS3 (Proteintech), anti-tyrosine phosphorylation (Santa Cruz
Biotech Inc.), anti-beta actin (Santa Cruz Biotech Inc.) or
anti-GAPDH (Santa Cruz Biotech Inc.). Details for Western blot are
as described in our previous studies (28,29).
m6A quantification
Total RNA extracted from cells was purified as
described above for real-time PCR analysis. The rRNA and noncoding
RNAs were removed using the Dynabeads mRNA Purification kit (Thermo
Fisher Scientific). A total of 200 ng mRNA of each sample was used
for m6A quantification using the EpiQuik m6A Methylation
Quantification kit (Colorimetric; Epigentek), according to the
manufacturer's protocol.
RNA stability
AGS cells were seeded in 12-well plates at a density
of 2×104/well. After overnight adhesion, cells were
treated with actinomycin (A9415; Sigma Aldrich) at a final
concentration of 10 µg/ml. Cells were collected and total RNA
isolated at different time points after actinomycin treatment (0,
30 min or 2 h). Expression levels of specific genes of interest
were quantified by using real-time PCR.
Statistical analysis
All experiments were performed as three independent
replications. Experiment data were presented as the mean ± SEM.
Student's t-test (for differences between two groups) or
one-way ANOVA (for multiple group comparisons) was used to analyze
the data using SPSS software (v20.0; SPSS, Inc.). Tukey post hoc
tests were conducted following one-way ANOVA to determine
significant differences between specific groups in datasets
containing 3 or more groups. P<0.05 was considered to be
statistically significant.
Results
The establishment of stable
METTL3-knockout (METTL3-KO) gastric cancer cell lines
We took the CRISPR-Cas9 approach to generate stable
knockout gastric cancer cells using the well-documented human AGS
cell line. Two generated cell lines of METTL3-KO, METTL3-KO-C20 and
METTL3-KO-C43, were generated and knockout of METTL3 in the cells
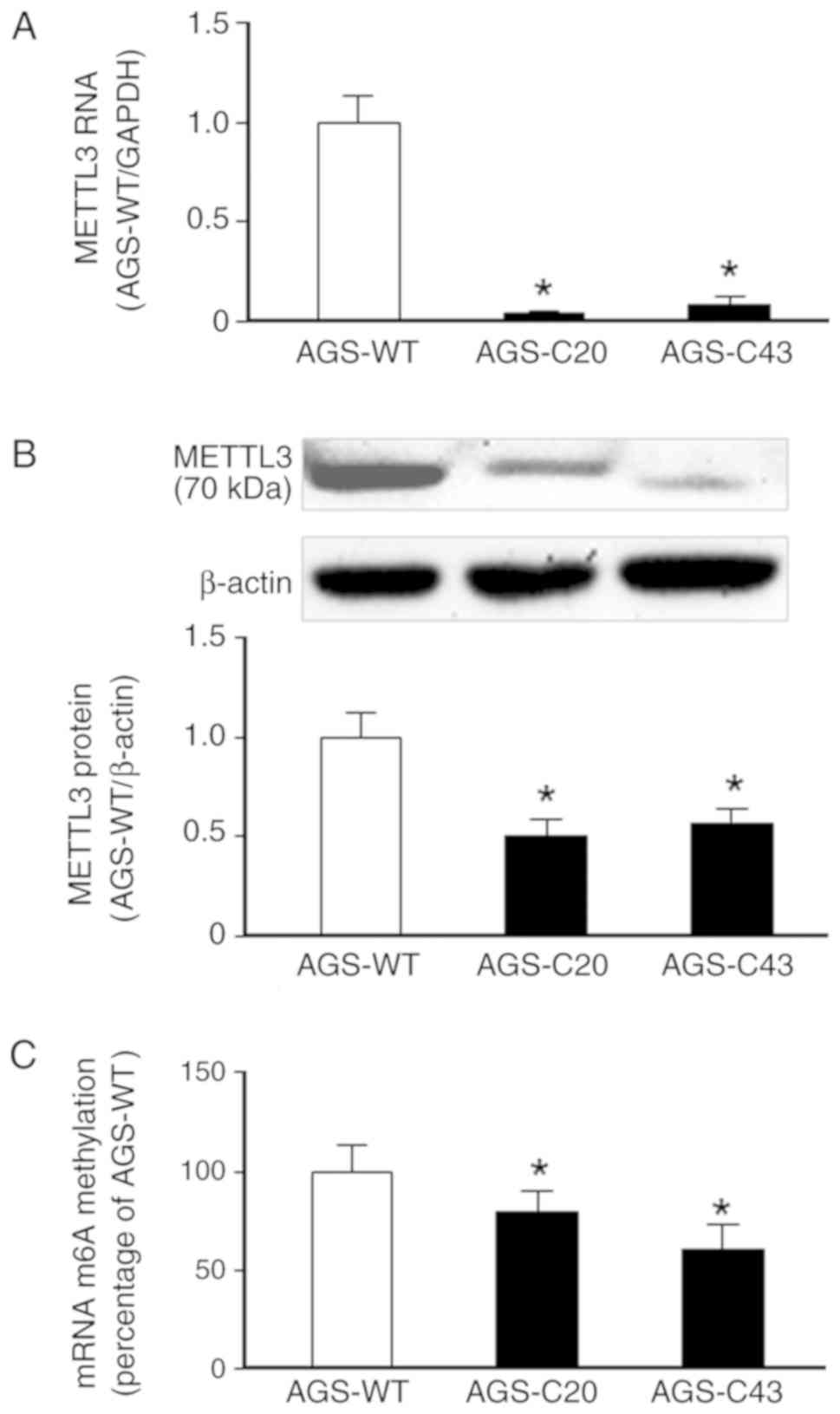
was confirmed by using real-time PCR and Western blot (Fig. 1A and B). No real-time PCR product was
detected in the KO cells using the primer sets covering the
targeted region of METTL3 (Fig. 1A).
Western blot showed a decrease in signaling of METTL3 band in the
METTL3-KO-C20 cells and a truncated form of METTL3 protein for the
METTL3-KO-C43 cells (Fig. 1B). A
significant decrease in the global m6A levels was detected in
METTL3-KO-C20 and METTL3-KO-C43 cells, compared with the wild-type
AGS cells (Fig. 1C).
METTL3-KO cells elicit an inhibitory
effect on cell proliferation
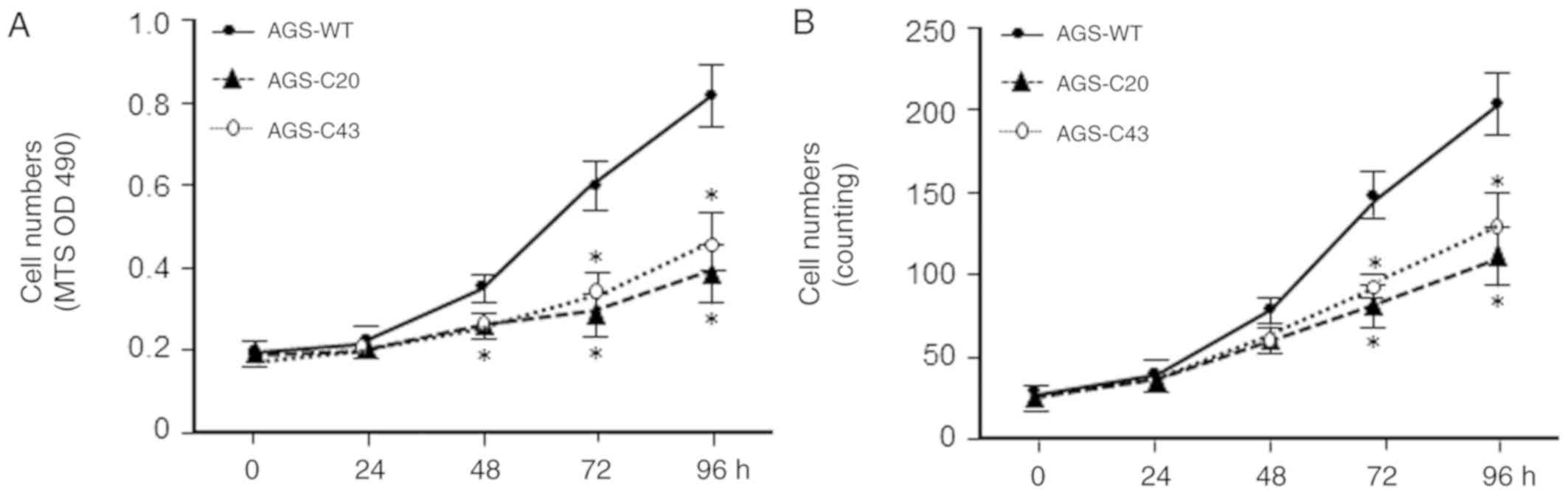
We then examined the effects of METTL3 KO on AGS
cell proliferation. The METTL3-KO-C20 and -C43 cells showed a
decrease in cell growth as revealed by cell number counting and by
MTS assay, respectively (Fig. 2A and
B). Decrease in cell number of METTL3-KO AGS cells is not due
to increased cell death as no significant difference in apoptotic
cell death was observed in the wild-type and METTL3-KO cells (data
not shown). These data suggest that METTL3 may play an important
role in regulating AGS cell proliferation.
Altered expression pattern of effector
molecules associated in cell proliferation in METTL3 KO AGS
cells
To explore the underlying mechanisms of
METTL3-mediated cell proliferation suppression in AGS cells, we
asked if METTL3 can affect the expression levels of these effector
molecules that are associated with gastric cancer cell
proliferation demonstrated from previous studies (17–19). We
screened the expression levels of these effector molecules in our
METTL3-KO AGS cells, compared with that in the wild-type cells.
Interestingly, many of them showed an increased expression level in
METTL3-KO-C20 cells (Fig. 3A) and
METTL3-KO-C43 cells (data not shown). Whereas a decrease in
HEPANAS_3 was detected in the METTL3-KO AGS cells, no detectable
changes in IFITM3 and S100A4 expression levels was observed between
METTL3-KO cells and wild-type AGS cells (Fig. 3A). The above data suggest that
METTL3-mediated RNA methylation may impact the expression of genes
that can subsequently influence AGS cell proliferation.
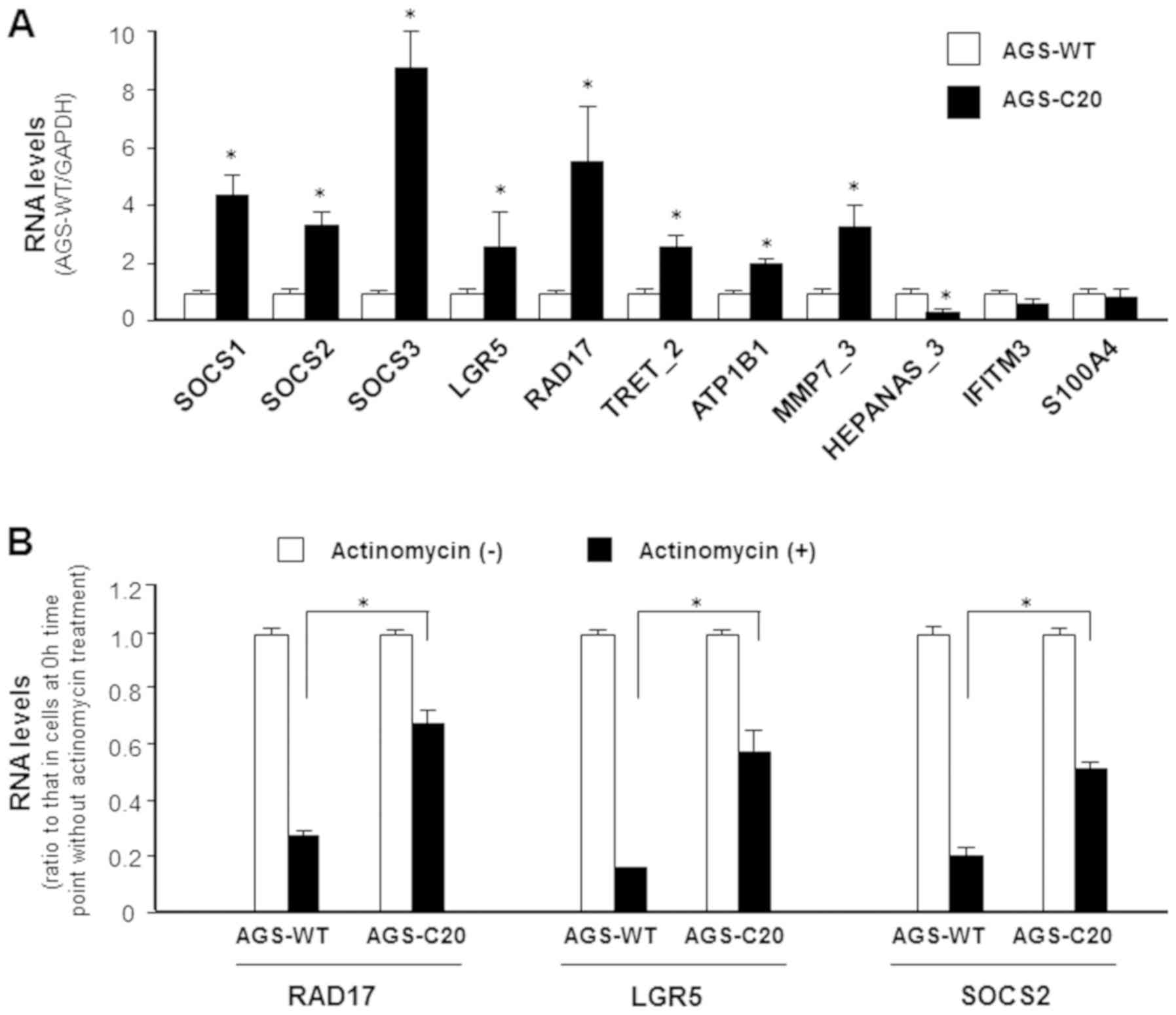 | Figure 3.Expression of effector molecules
associated with AGS cell proliferation in METTL3-KO AGS cells. (A)
Expression of selected genes associated with AGS cell proliferation
were measured using reverse transcription-quantitative PCR. Several
of these genes, including SOCS1, SOCS2, SOCS3 and LGR5, exhibited
decreased expressions in METTL3-KO cells when compared with
METTL3-WT cells. (B) Inhibition of RAD17, LGR5 and SOCS2 RNA
stability in METTL3-KO AGS cells. AGS-WT and AGS METTL3-KO cells
were cultured in the presence of actinomycin for 2 h. Levels of
RAD17, LGR5 and SOCS2 were measured in cells following actinomycin
treatment and compared with cells tested prior to actinomycin
administration. Reduced RNA levels represents degradation rate. A
lower degradation rate was observed for RAD17, LGR5 and SOCS2 in
METTL3-KO AGS cells compared with AGS-WT. *P<0.01 vs. AGS-WT.
METTL3-KO, methyltransferase like 3 knocked down; WT, wild type;
SOCS, suppressor of cytokine signaling; LGR5, leucine-rich
repeat-containing G-protein coupled receptor 5; RAD17, RAD17
checkpoint clamp loader component. |
We then aimed to explore the underlying molecular
mechanisms by which METTL3 may control expression of these genes.
Previous studies show that RNA m6A methylation exerts its function
via modulation of RNA stability (4).
To investigate this potential, we carried out the RNA stability
assay. After blocking new RNA synthesis with actinomycin, a slower
decay rate for RAD17, LGR5 and SOCS2 RNAs was observed in METTL3-KO
AGS cells (Fig. 3B), suggesting
modulation of their stability through m6A modification.
Aberrant expression of SOCS2 may
contribute to METTL3-mediated cell proliferation in AGS cells
Given the fact that several SOCS family members are
among these effector molecules that are upregulated in the
METTL3-KO AGS cells (Fig. 3), we
hereafter focused on the SOCS family members to explore their
potential role in METTL3-associated AGS cell proliferation. To
further confirm the expression of SOCSs associated with METTL3 in
AGS cells, we expanded our measurement of SOCS1, SOCS2, and SOCS3
at both RNA level and protein levels in METTL3-KO cells. The
expression of SOCS1, SOCS2, and SOCS3 at the RNA level was
significantly higher in the METTL3-KO cells, compared with that in
the wild-type AGS cells (Fig. 4A-C).
Nevertheless, at the protein level, only SOCS2, but not SOCS1 and
SOCS3, showed an increased content in the METTL3-KO cells, compared
with that in the wild-type AGS cells (Fig. 4D-F).
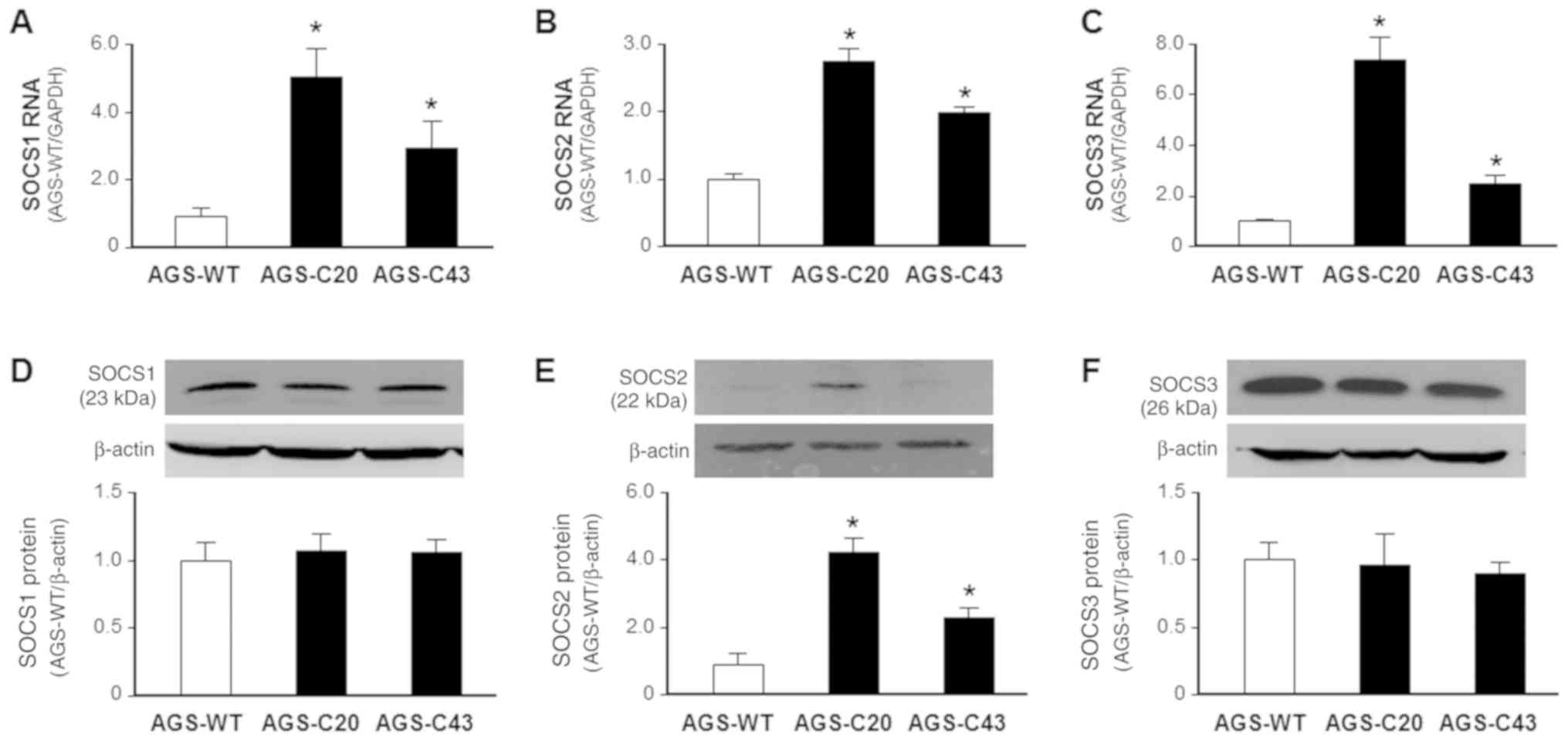 | Figure 4.Expression of SOCS1, SOCS2 and SOCS3
at the RNA and protein levels in METTL3-WT and METTL3-KO AGS cells.
Elevated RNA expressions of (A) SOCS1, (B) SOCS2 and (C) SOCS3 were
detected in METTL3-KO AGS cells, as determined via reverse
transcription-quantitative PCR and compared with METTL3-WT AGS
cells. METTL3-KO resulted in increased levels of (E) SOCS2, but not
(D) SOCS1 and (F) SOCS3 (F) protein in AGS cells, as determined via
western blotting and compared with METTL3-WT AGS cells. β-actin was
also immunoblotted to ensure equal loading of proteins to each
lane. Representative blotting images are presented. Densitometric
levels of positive bands for SOCS1, SOCS2 and SOCS3 were quantified
and are expressed as the ratio to β-actin. *P<0.01 vs. AGS-WT.
SOCS, suppressor of cytokine signaling; METTL3, methyltransferase
like 3; WT, wild type; KO, knocked down. |
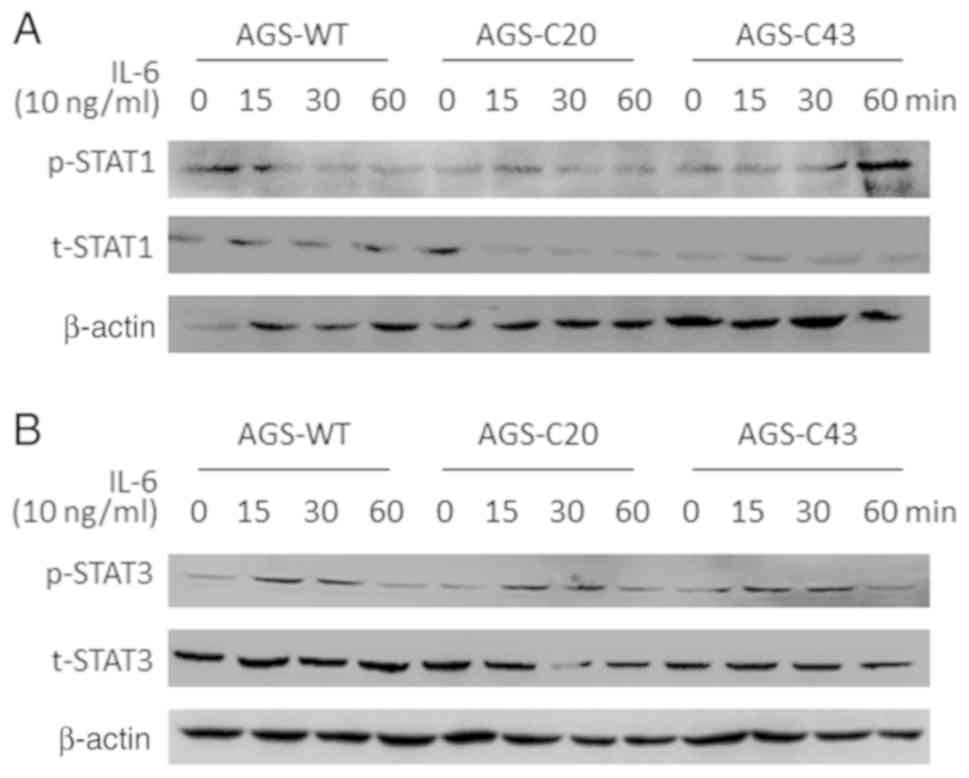
To further exclude the potential involvement of
SOCS1 and SOCS3 in METTL3-mediated cell proliferation, we measured
the activation of SOCS1 and SOCS3 in METTL3-KO AGS cells. One of
its key downstream transcription factors for the SOCS signaling is
the STAT family proteins (30). The
activation of the STAT signaling pathway involves tyrosine
phosphorylation of STAT proteins (31). We then measured by Western blot the
expression of STAT1 and STAT3 at the protein level in METTL3-KO AGS
cells in response to IL6 stimulation. No detectable changes in the
protein content of STAT1 (Fig. 5A)
and STAT3 (Fig. 5B) was observed in
METTL3-KO AGS cells following IL-6 stimulation, compared with the
wild-type cells. Moreover, we also blotted for the tyrosine
phosphorylation ratio relative to the total protein levels in
METTL3-KO AGS cells. We failed to detect any difference in the
tyrosine phosphorylation status of STAT1 and STAT3 in IL-6-treated
MELLT3-KO cells, compared with that in the wild-type AGS cells
(Fig. 5A and B).
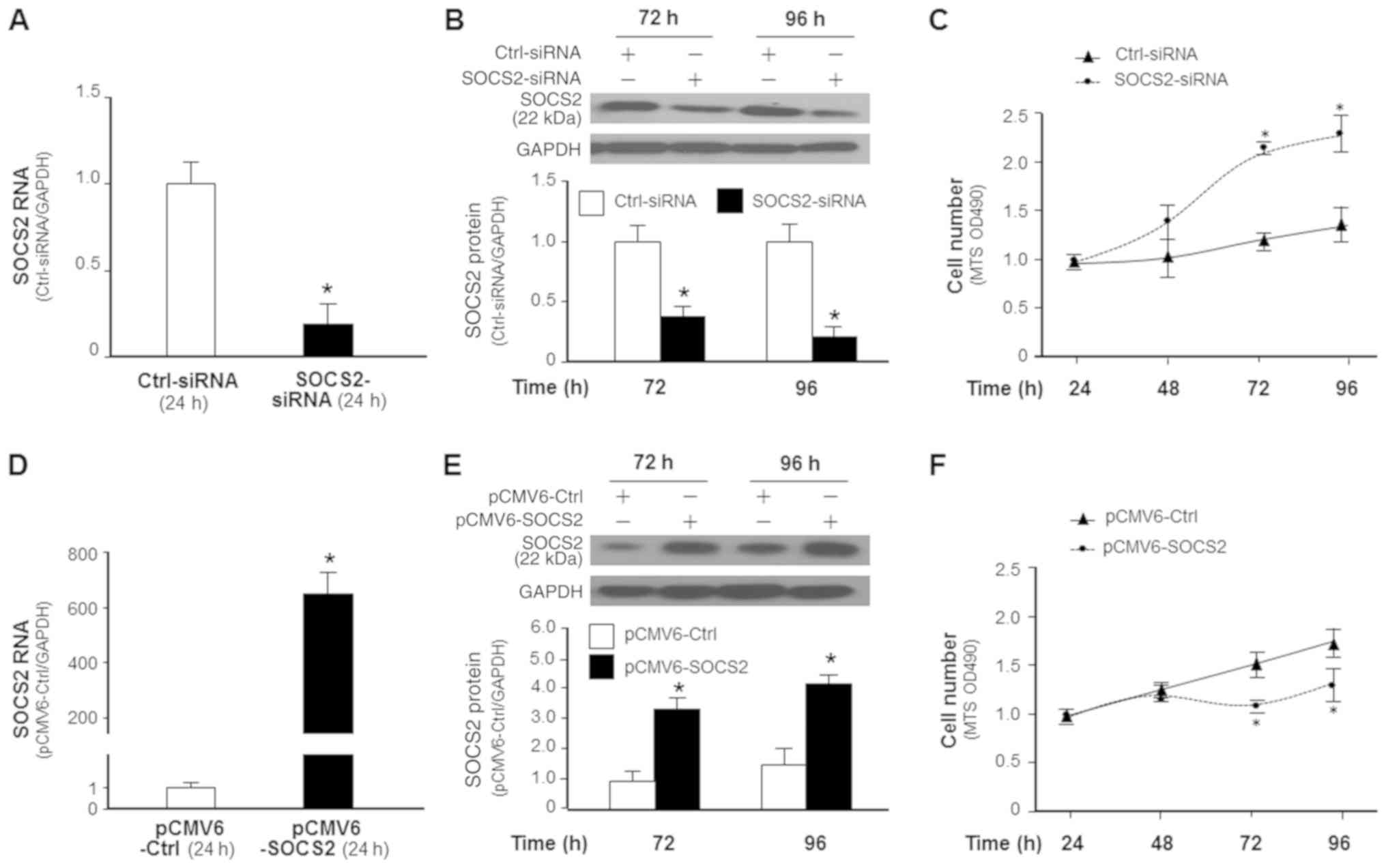
Given the significant increase in SOCS2 protein
level in METTL3-KO in AGS cells, we further tested the potential
role of SOCS2 in METTL3-mediated cell proliferation by manipulating
its expression. We took the RNAi approach to knockdown SOCS2 and
used the pCMV6-SOCS2 plasmid to transfect cells to upregulate SOCS2
expression in AGS cells (Fig. 6A).
Accordingly, a significant increase or decrease in cell
proliferation was detected in the SOCS2 siRNA-treated AGS cells and
cells overexpressing SOCS2, respectively (Fig. 6B), further supporting the role of
SOCS2 in regulating AGS cell proliferation.
Discussion
m6A modification is the most prevalent internal
modification decorating RNA molecules and may play a critical
regulatory role in a series of biological processes, such as RNA
decay, splicing, translation and transportation (3–6).
Emerging evidence indicates that the cellular status of m6A
modification may impact the pathologic processes of various cancers
(8,9,32).
METTL3, a major writer of m6A modification, is often upregulated in
numerous malignancies and can epigenetically silence specific gene
expression, eliciting a potential oncogenic function (11,33).
Previous studies demonstrated that expression levels of METTL3 can
critically change the cell cycle and apoptosis processes in hepatic
cancer and acute myeloid leukemia (11,34).
METTL3 is usually upregulated in gastric cancer, which should
maintain a higher m6A RNA modification level in tumor cells. In
this report, we demonstrate that knockdown of METTL3 in gastric
cancer cells results in suppression of cell proliferation through
induction of SOCS2, suggesting a potential role for elevated METTL3
expression in gastric cancer progression.
Given the elevated expression level of METTL3 in
gastric cancer tissues (15,16), we took the CRISPR/Cas9 approach to
generate stable cell lines of gastric cancer cells deficient in
METTL3 function. The CRISPR/Cas9 approach may provide several
advantages over the siRNA approach, as it may be more specific and
can generate stable KO cells (35).
Of these stable METTL3-KO cells that generated from this approach,
we selected two clones, METTL3-KO-C20 and METTL3-KO-C43, which
showed an almost complete deletion of METTL3 RNA expression as
evident by the PCR analysis using the primer set covering the
CRISP/Cas9 target region of METTL3. Interestingly, Western blot
indicated a significant decrease in signaling of METTL3 band in the
METTL3-KO-C20 cells and a truncated form of METTL3 protein for the
METTL3-KO-C43 cells. Nevertheless, both cell lines demonstrated a
significant suppression of global m6A methylation, compared with
the wild-type cells. Functionally, knockdown of METTL3
significantly reduced AGS cell proliferation.
The process of AGS cell proliferation involves a
complex regulatory network of multiple signaling pathways,
involving key effector molecules, such as SOCS1, SOCS2, SOCS3,
LGR5, RAD17, TRET_2, ATP1B1, MMP7_3, HEPANAS_3, IFITM3, and S100A4
(17–19). Interestingly, knockdown of METTL3 in
AGS cells showed an increased expression level for many of these
effector molecules, implicating that METTL3-mediated RNA
methylation may influence the expression of key components of
multiple signaling pathways to modulate AGS cell proliferation.
Regulation of RNA stability, particularly for SOCS2, may be
involved in METTL3-mediated gene expression associated with AGS
cell proliferation.
SOCS2, a member of the SOCS family that regulates
multiple cytokine-induced intracellular signal pathways, is
involved in the regulation of numerous biological processes,
including immune responses (36,37).
SOCS2 can be activated through tyrosine phosphorylation and may act
as a downstream factor of the JAK/STAT pathway, providing a
negative feedback regulation to this signaling pathway (38). Indeed, the JAK/STAT signal pathway is
often dysregulated in gastric cancer (39). The JAK/STAT signaling pathway
participates in multiple cancerous cellular processes: apoptosis,
proliferation, metastasis and inflammation (40–43).
Aberrant expression of SOCS2 is of important pathologic
significance in development of many diseases (38,44).
Overexpression of SOCS2 showed a proliferation-suppression effect
in gastric cancer cells (45). In
this study using AGS cells in culture, we failed to detect any
difference in the tyrosine phosphorylation status of STAT1 and
STAT3 in the MELLT3-KO AGS cells, compared with that in the
wild-type cells. Instead, we observed the negative association of
SOCS2 and cell proliferation in gastric cancer cells. Knockdown of
METTL3 resulted in an increased expression of SOCS2, accompanied
with a decreased cell proliferation in AGS cells. Experimentally
induced suppression or forced expression of SOCS2 caused reciprocal
alterations in AGS cell proliferation, indicating that SOCS2
downregulation, at least partially, contributes to METTL3-mediated
cell proliferation in cultured AGS cells. Several recent reports
indicate that METTL3 may contribute to the proliferation and
migration of gastric cancer (16,46–48).
Results from our study further support that an elevated expression
of METTL3 may increase AGS cell proliferation, promoting cancer
progression. Therefore, targeting METTL3-mediated RNA methylation
and SOCS signaling may be of therapeutic potential for gastric
cancer. Future experiments should investigate the molecular
mechanisms of METTL3-mediated gastric cancer progression, including
in vivo experiments on the role of SOCS2 in METTL3-mediated
cancer cell proliferation.
Acknowledgements
The authors would like to thank Ms. Barbara L.
Bittner (Creighton University) for her assistance in English
language editing with the manuscript.
Funding
The present study was supported by the Nebraska
Cancer and Smoking Disease Research Program (grant no. LB595; to
XMC).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LJ, TC, LX, JHX, AYG and XMC conceived the project.
LJ, TC, LX, JHX, BD, GW, KZ, EL and AYG conducted the experiments.
LJ, JHX, TC, LX, NWM, AYG and XMC analyzed and interpreted the
data. LJ, JHX, NWM and XMC wrote the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
m6A
|
N6-methyladenosine
|
|
METTL3
|
methyltransferase like 3
|
|
METTL14
|
methyltransferase like 14
|
|
FTO
|
fat mass and obesity-associated
protein
|
|
ALKBH5
|
a-ketoglutarate-dependent dioxygenase
alkB homolog 5
|
|
SOCS
|
suppressor of cytokine signaling
|
|
KO
|
knockout
|
|
LGR5
|
leucine rich repeat containing G
protein-coupled receptor 5
|
|
ACD
|
Actinomycin D
|
References
|
1
|
Yue Y, Liu J and He C: RNA
N6-methyladenosine methylation in post-transcriptional
gene expression regulation. Genes Dev. 29:1343–1355. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2013. View Article : Google Scholar
|
|
3
|
Batista PJ: The RNA modification
N6-methyladenosine and its implications in human
disease. Genomics Proteomics Bioinformatics. 15:154–163. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Cgen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang S, Sun C, Li J, Zhang E, Ma Z, Xu W,
Li H, Qiu M, Xu Y, Xia W, et al: Roles of RNA methylation by means
of N6-methyladenosine (m6A) in human cancers. Cancer Lett.
408:112–120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lan Q, Liu PY, Haase J, Bell JL,
Hüttelmaier S and Liu T: The critical role of RNA m6A methylation
in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m6A modification in glioma stem-like cells
maintenance and radioresistance. Oncogene. 37:522–533. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R,
Wang YY and Zhe H: FTO regulates the chemo-radiotherapy resistance
of cervical squamous cell carcinoma (CSCC) by targeting β-catenin
through mRNA demethylation. Mol Carcinog. 57:590–597. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m(6)A demethylase
ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by
sustaining FOXM1 expression and cell proliferation program. Cancer
Cell. 31:591–606.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lyons K, Le LC, Pham YT, Borron C, Park
JY, Tran CTD, Tran TV, Tran HT, Vu KT, Do CD, et al: Gastric
cancer: Epidemiology, biology, and prevention: A mini review. Eur J
Cancer Prev. 28:397–412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Zheng D, Wang F, Xu Y, Yu H and
Zhang H: Expression of demethylase genes, FTO and ALKBH1, is
associated with prognosis of gastric cancer. Dig Dis Sci.
64:1503–1513. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Yang S, Sui J, Xu SY, Cheng YP,
Shen B, Zhang Y, Zhang XM, Yin LH, Pu YP and Liang GY: Dysregulated
N6-methyladenosine methylation writer METTL3 contributes to the
proliferation and migration of gastric cancer. J Cell Physiol.
235:548–562. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Katoh M: Canonical and non-canonical WNT
signaling in cancer stem cells and their niches: Cellular
heterogeneity, omics reprogramming, targeted therapy and tumor
plasticity (Review). Int J Onco. 51:1357–1369. 2017. View Article : Google Scholar
|
|
18
|
Aggarwal A, Leong SH, Lee C, Kon O and Tan
P: Wavelet transformations of tumor expression profiles reveals a
pervasive genome-wide imprinting of aneuploidy on the cancer
transcriptome. Cancer Res. 65:186–194. 2005.PubMed/NCBI
|
|
19
|
Xu QW, Zhao W, Wang Y, Sartor MA, Han DM,
Deng J, Ponnala R, Yang JY, Zhang QY, Liao GQ, et al: An integrated
genome-wide approach to discover tumor-specific antigens as
potential immunologic and clinical targets in cancer. Cancer Res.
72:6351–6361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li G, Xu J, Wang Z, Yuan Y, Li Y, Cai S
and He Y: Low expression of SOCS-1 and SOCS-3 is a poor prognostic
indicator for gastric cancer patients. J Cancer Res Clin Oncol.
141:443–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Inagaki-Ohara K, Kondo T, Ito M and
Yoshimura A: SOCS, inflammation, and cancer. JAKSTAT.
2:e240532013.PubMed/NCBI
|
|
22
|
Sutherland KD, Lindeman GJ, Choong DY,
Wittlin S, Brentzell L, Phillips W, Campbell IG and Visvader JE:
Differential hypermethylation of SOCS genes in ovarian and breast
carcinomas. Oncogene. 23:7726–7733. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Slattery ML, Lundgreen A, Hines LM,
Torres-Mejia G, Wolff RK, Stern MC and John EM: Genetic variation
in the JAK/STAT/SOCS signaling pathway influences breast
cancer-specific mortality through interaction with cigarette
smoking and use of aspirin/NSAIDs, the breast cancer health
disparities study. Breast Cancer Res Treat. 147:145–158. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hoefer J, Kern J, Ofer P, Eder IE, Schäfer
G, Dietrich D, Kristiansen G, Geley S, Rainer J, Gunsilius E, et
al: SOCS2 correlates with malignancy and exerts growth-promoting
effects in prostate cancer. Endocr Relat Cancer. 21:175–187. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Das R, Gregory PA, Fernandes RC, Denis I,
Wang Q, Townley SL, Zhao SG, Hanson AR, Pickering MA, Armstrong HK,
et al: MicroRNA-194 promotes prostate cancer metastasis by
inhibiting SOCS2. Cancer Res. 77:1021–1034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vitali C, Bassani C, Chiodoni C, Fellini
E, Guarnotta C, Miotti S, Sangaletti S, Fuligni F, De Cecco L,
Piccaluga PP, et al: SOCS2 controls proliferation and stemness of
hematopoietic cells under stress conditions and its deregulation
marks unfavorable acute leukemias. Cancer Res. 75:2387–2399. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Gong AY, Ma S, Chen X,
Strauss-Soukup JK and Chen XM: Delivery of parasite Cdg7_Flc_0990
RNA transcript into intestinal epithelial cells during
cryptosporidium parvum infection suppresses host cell gene
transcription through epigenetic mechanisms. Cell Microbiol.
19:e127602017. View Article : Google Scholar
|
|
28
|
Ming Z, Gong AY, Wang Y, Zhang XT, Li M,
Mathy NW, Strauss-Soukup JK and Chen XM: Involvement of
cryptosporidium parvum Cdg7_FLc_1000 RNA in the attenuation of
intestinal epithelial cell migration via trans-suppression of host
cell SMPD3. J Infect Dis. 217:122–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Gong AY, Ma S, Chen X, Li Y, Su
CJ, Norall D, Chen J, Strauss-Soukup JK and Chen XM: Delivery of
parasite RNA transcripts into infected epithelial cells during
cryptosporidium infection and its potential impact on host gene
transcription. J Infect Dis. 215:636–643. 2017.PubMed/NCBI
|
|
30
|
Durham GA, Williams JJL, Nasim MT and
Palmer TM: Targeting SOCS proteins to control JAK-STAT signalling
in disease. Trends Pharmacol Sci. 40:298–308. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stark GR and Darnell JE Jr: The JAK-STAT
pathway at twenty. Immunity. 36:503–514. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers: Current
status and perspectives. Cell Res. 28:507–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m(6)A RNA methylation regulates
the self-renewal and tumorigenesis of glioblastoma stem cells. Cell
Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid
differentiation of normal hematopoietic and leukemia cells. Nat
Med. 23:1369–1376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lino CA, Harper JC, Carney JP and Timlin
JA: Delivering CRISPR: A review of the challenges and approaches.
Drug Deliv. 25:1234–1257. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dimitriou ID, Clemenza L, Scotter AJ, Chen
G, Guerra FM and Rottapel R: Putting out the fire: Coordinated
suppression of the innate and adaptive immune systems by SOCS1 and
SOCS3 proteins. Immunol Rev. 224:265–283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Palmer DC and Restifo NP: Suppressors of
cytokine signaling (SOCS) in T cell differentiation, maturation,
and function. Trends Immunol. 30:592–602. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Letellier E and Haan S: SOCS2:
Physiological and pathological functions. Front Biosci (Elite Ed).
8:189–204. 2016.PubMed/NCBI
|
|
39
|
Khanna P, Chua PJ, Wong BSE, Yin C, Thike
AA, Wan WK, Tan PH and Baeg GH: GRAM domain-containing protein 1B
(GRAMD1B), a novel component of the JAK/STAT signaling pathway,
functions in gastric carcinogenesis. Oncotarget. 8:115370–115383.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kong D and Wang Y: Knockdown of lncRNA
HULC inhibits proliferation, migration, invasion, and promotes
apoptosis by sponging miR-122 in osteosarcoma. J Cell Biochem.
119:1050–1061. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lui AJ, Geanes ES, Ogony J, Behbod F,
Marquess J, Valdez K, Jewell W, Tawfik O and Lewis-Wambi J: IFITM1
suppression blocks proliferation and invasion of aromatase
inhibitor-resistant breast cancer in vivo by JAK/STAT-mediated
induction of p21. Cancer Lett. 399:29–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cho KH, Jeong KJ, Shin SC, Kang J, Park CG
and Lee HY: STAT3 mediates TGF-β1-induced TWIST1 expression and
prostate cancer invasion. Cancer Lett. 336:167–173. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Twyman-Saint Victor C, Rech AJ, Maity A,
Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi
PM, et al: Radiation and dual checkpoint blockade activate
non-redundant immune mechanisms in cancer. Nature. 520:373–377.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Trengove MC and Ward AC: SOCS proteins in
development and disease. Am J Clin Exp Immunol. 2:1–29.
2013.PubMed/NCBI
|
|
45
|
Zhou X, Xia Y, Li L and Zhang G: MiR-101
inhibits cell growth and tumorigenesis of helicobacter pylori
related gastric cancer by repression of SOCS2. Cancer Biol Ther.
16:160–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang C, Zhang M, Ge S, Huang W, Lin X,
Gao J, Gong J and Shen L: Reduced m6A modification predicts
malignant phenotypes and augmented Wnt/PI3K-Akt signaling in
gastric cancer. Cancer Med. 8:4766–4781. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin S, Liu J, Jiang W, Wang P, Sun C, Wang
X, Chen Y and Wang H: METTL3 promotes the proliferation and
mobility of gastric cancer cells. Open Med (Wars). 14:25–31. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He H, Wu W, Sun Z and Chai L: MiR-4429
prevented gastric cancer progression through targeting METTL3 to
inhibit m6A-caused stabilization of SEC62. Biochem Biophys Res
Commun. 517:581–587. 2019. View Article : Google Scholar : PubMed/NCBI
|