Introduction
According to GLOBOCAN 2018 data, colorectal cancer
(CRC) is the fourth most common cancer and the third leading cause
of cancer-associated death (1). It
has been reported that 20–25% of patients have metastases at the
time of diagnosis (2) and remains
the primary reason of poor prognosis and cause of CRC-associated
death (3). The most common sites of
distant metastases from CRC are the liver and lung (4,5), which
affects the prognosis and survival of patients with CRC (6). In recent years, it has been
demonstrated that CRC treatment should be tailored to the
individual patient due to the wide variety of risk factors, such as
sex, age, tumor-node-metastases (TNM) stage and tumor location,
genetic factors and surgical complexity (7). Therefore, it is important to identify
clinicopathological features and genetic mutations in patients with
CRC.
RAS gene mutations serve a role in the
carcinogenesis of CRC (8).
KRAS and NRAS are different mutant forms. KRAS
mutations are observed in 43% of patients with metastatic CRC
(mCRC) and have a less favorable prognosis in patients with mCRC.
Amado et al (9) demonstrated
the treatment effect of anti-epidermal growth factor receptor
(EGFR) monoclonal antibody (panitumumab) on progression-free
survival (PFS) in the wild-type (WT) KRAS group was
significantly greater compared with the mutant group. WT
KRAS patients had longer overall survival (OS) (9). NRAS mutations affect patients
with mCRC prognosis and predict lack of response to anti-epidermal
growth factor receptors (10). The
prognostic role of RAS mutations has been investigated
previously and several studies have focused on stage II and III CRC
(11–13); the effect of RAS mutations on
the efficacy of mCRC treatment remains uncertain. Few studies have
assessed the association between RAS gene mutation status,
characteristics and survival outcome of patients with the
synchronous liver-metastasis only (LiM-only) and lung-metastasis
only (LuM-only) mCRC.
EGFR is a transmembrane protein. Overexpression of
EGFR has been described to have an association with disease
progression, poor prognosis, metastatic spread and drug resistance
in colorectal cancers (14–16). The efficacy of anti-EGFR monoclonal
antibody (mAb) has been evaluated as monotherapy or combined with
different types of chemotherapy in patients with mCRC (14). There were three methods to detect the
EGFR status: Protein expression by immunohistochemistry (IHC), gene
copy number by fluorescence in situ hybridization (FISH) and
mutation analysis using the Scorpion amplification refractory
mutation system (ARMS) (17).
However, these 3 methods were closely related to each other
(17). In the present study, the
expression of EGFR was analyzed by immunohistochemical
staining.
The aim of the present retrospective study was to
investigate the clinicopathological and genetic characteristics and
survival outcomes in patients with synchronous LiM- and LuM-only
mCRC.
Materials and methods
Patient selection
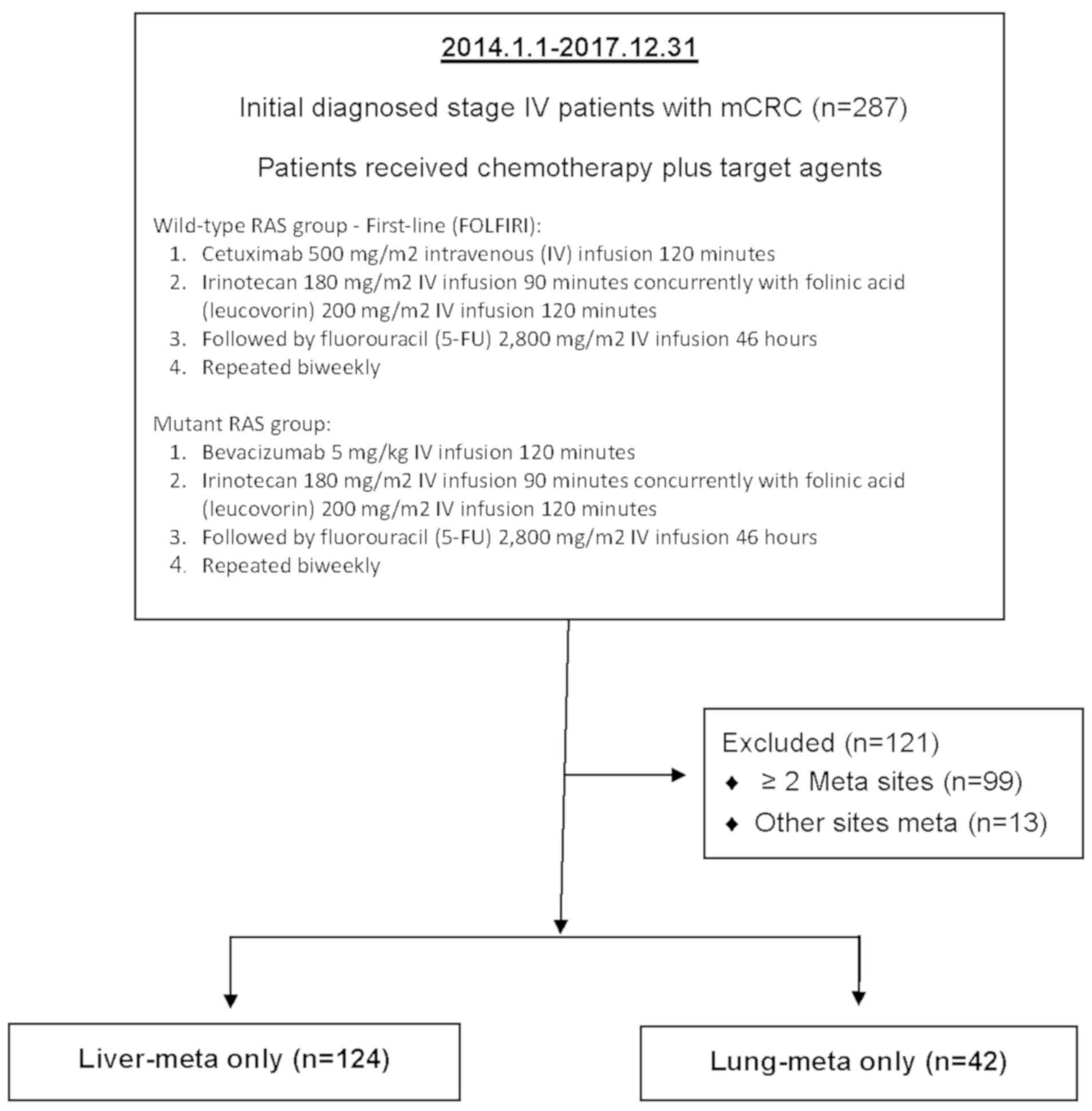
According to the 7th edition of AJCC (American Joint
Committee on Cancer staging system in 2010) (18), the retrospective cohort included 287
consecutive patients registered with a pathological proof stage IV
mCRC at the Cancer Center of Kaohsiung Medical University Hospital
(Kaohsiung, Taiwan) within a 4-year period (from January 2014 to
December 2017). The inclusion criteria of this study were patients
with mCRC with synchronous liver-only or lung-only metastasis and
aimed to explore the effect of KRAS and NRAS
mutations on the prognosis of patients with synchronous mCRC
presenting with liver-only (LiM-only) and lung-only (LuM-only)
metastases. Patients with >2 sites metastases (n=99), other
sites of metastases other than the liver and lung, such as bone,
spleen and brain (n=13), and peritoneal metastases only (n=9) were
excluded. Ultimately, a total of 166 eligible patients were
analyzed, including 124 synchronous LiM-only and 42 synchronous
LuM-only patients with CRC (Fig. 1).
There were 95 males and 71 females, with a median age of 63.3 years
(range, 26–90 years). The clinical outcomes and survival status of
the patients were regularly followed up every 3 months during a
clinic visit either until February 2019 or until their death, with
a median follow-up time of 19.2 months (1.0–57.1 months). The PFS
and OS of the patients, according to RAS mutation status
were investigated. For genetic analysis of KRAS and
NRAS, all samples were collected immediately after surgical
resection, frozen instantly in liquid nitrogen and then stored in
−80°C freezer until analyzed.
Clinicopathological features
The clinical and pathological records of each
patient were obtained from medical records. The clinical
information included demographic data (age and sex) and clinical
parameters, such as the location of primary tumor, preoperative
serum carcinoembryonic antigen (CEA) level, cancer cell
differentiation according to pathological report (well
differentiated, moderately differentiated and poorly
differentiated), and duration of liver or lung metastases (Table I).
 | Table I.Characteristics of 166 patients with
metastatic colorectal cancer at diagnosis and gene mutation
profiles. |
Table I.
Characteristics of 166 patients with
metastatic colorectal cancer at diagnosis and gene mutation
profiles.
| Characteristic | Liver-meta only
(n=124) | Lung-meta only
(n=42) | P-value |
|---|
| Median age, years
(range) | 62.2 (26–90) | 66.6 (41–86) | 0.050 |
| Sex,
male:female | 76:48 | 19:23 | 0.069 |
| Location of primary
tumor, n |
|
| 0.388 |
| Right
colon | 30 | 13 |
|
| Left
colon | 94 | 29 |
|
| CEA level, ng/ml,
n |
|
| 0.589 |
|
<5 | 41 | 12 |
|
| ≥5 | 83 | 30 |
|
| Differentiated,
n |
|
| 0.713 |
| WD | 9 | 1 |
|
| MD | 89 | 34 |
|
| PD | 9 | 3 |
|
| Not
Determined | 17 | 4 |
|
| KRAS
mutation, % (n/total) | 41.0 (43/105) | 35.1 (13/37) | 0.534 |
| NRAS
mutation, % (n/total) | 5.0 (5/101) | 8.1 (3/37) | 0.482 |
| EGFR
overexpression, % (n/total) | 86.6 (58/67) | 80.0 (12/15) | 0.515 |
DNA extraction and KRAS/NRAS direct
sequencing
Not all patients were routinely checked for their
genetic profile, as some patients did not undergo surgery or
biopsies. 105/124 (84.7%) LiM-only and 37/42 (88.1%) LuM-only
patients received KRAS genetic testing, 101/124 (81.5%)
LiM-only and 37/42 (88.1%) LuM-only patients received NRAS
genetic testing. Genomic DNA was isolated from frozen primary CRC
tissues using proteinase-K (Stratagene; Agilent Technologies, Inc.)
digestion and phenol/chloroform extraction, as described previously
(19). The designed sequences of the
oligonucleotide primers for KRAS and NRAS exons 2–4
and the operational procedure for direct sequencing were based on
those reported in our previous studies (20,21). For
KRAS and NRAS genotyping, the frozen primary CRC
tissues were fixed with 10% formalin for 24–48 h at room
temperature, then deparaffinized (the procedure included 3 washes
in xylene for 3 min followed by 3 washes in 99.8% ethanol for 3
min) and sliced into 5-µm tissue sections. Following
deparaffinization and rehydration, the DNA was isolated using a
Gentra Puregene Tissue kit (Qiagen NV, Thermo Fisher Scientific
Inc.) according to the manufacturer's instructions. The primers
used were designed using the Primer 3 v.0.4.0 free software program
(http://primer3 ut.ee) and the sequences are as
follows: KRAS forward, 5′-TCATTATTTTTATTATAAGGCCTGCTGAA-3′
and reverse, 5′-CAAAGACTGGTCCTGCACCAGTA-3′; NRAS forward,
5′-GATGTGGCTCGCCAATTAAC-3′ and reverse,
5′-GAATATGGGTAAAGATGATCCGA-3′. Polymerase chain reaction (PCR)
amplification of 0.5 µg DNA with 2.5U of Pro Taq Plus DNA
Polymerase (Protech Technology Enterprise Co., Ltd) in the presence
of 200 µM dNTPs, 0.2 µM primers, and 1X reaction buffer was carried
out in an Applied Biosystems 2720 Thermal Cycler (Applied
Biosystems; Thermo Fisher Scientific Inc.). The PCR reaction volume
was 40 µl and the PCR conditions for KRAS and NRAS
were as follows: 94.0°C for 10 min, 35 cycles of denaturation for
30 sec at 94.0°C, annealing for 60 sec at 56.0°C, primer extension
for 30 sec at 72.0°C and a final extension for 7 min at 72.0°C. A
fragment analysis of the PCR products was conducted to verify the
KRAS and NRAS genotypes using automated capillary
electrophoresis using an ABI PRISM 310 Genetic Analyzer system and
Taqman Genotyper v.1.6 software (Applied Biosystems; Thermo Fisher
Scientific, Inc.). According to RAS status, patients were
categorized into the WT or mutant RAS group. There were 59
WT and 42 mutant RAS in the LiM-only group, and 22 WT and 15
mutant RAS in the LuM-only group.
Treatment protocol
For the WT RAS group, the majority of
patients received biweekly cetuximab, at a dose of 500
mg/m2 in a 2-h infusion, followed by folinic acid
(leucovorin), fluorouracil (5-FU) and irinotecan (FOLFIRI) on day 1
of a 14-day cycle as the first-line treatment. The FOLFIRI regimen
consisted of 180 mg/m2 irinotecan as a 90-minute
infusion, and 200 mg/m2 leucovorin (LV), and 2,800
mg/m2 5-FU as a 46-h infusion injection on days 1 and 2.
In contrast, for the mutant RAS group, the treatment regimen
comprised of bevacizumab [5 mg/kg as a 120-min intravenous (IV)
infusion] on day 1, followed by irinotecan (180 mg/m2 as
a 90-min IV infusion), LV (200 mg/m2 as an IV infusion
over 2 h) and 5-FU (2,800 mg/m2 as an IV infusion over a
46-h period), which was repeated biweekly.
Immunohistochemical (IHC) analysis of
epidermal growth factor receptor (EGFR) expression
The IHC analysis of EGFR expression was based on
that from our previous studies (22,23). The
tissues were fixed with 10% formalin for 24 h at room temperature.
Following fixation, the tissues were paraffin embedded as follows
(total duration, 16 h): 70% ethanol, two changes for 1 h each; 80%
ethanol, one change for 1 h; 95% ethanol, one change for 1 h; 100%
ethanol, three changes for 1.5 h each; xylene or xylene substitute,
three changes for 1.5 h each; paraffin wax, two changes for 2 h
each at 60°C. Formalin-fixed and paraffin-embedded tissue blocks
were cut into 3-µm sections to retrieve antigens. The sections were
deparaffinized by performing 2–3 changes of xylene, 10 min each at
room temperature. The tissues were rehydrated in a descending
alcohol series which comprised of 2 changes of 100% ethanol for 3
min each followed by 95 and 80% ethanol for 1 min each. The
sections were then rinsed with distilled water. Endogenous
peroxidase activity was blocked with methanol containing 0.1%
H2O2 for 30 min. After washing with a
Tris-buffer solution, the sections were incubated for 30 min at
room temperature with EGFR primary antibody (1:200; cat. no.
NCL-L-EGFR-384; Leica Microsystems Trading Co., Ltd.) and then DAKO
REAL EnVision Detection System-HRP (1:100 dilution; Agilent
Technologies Inc.) was added for 30 min at room temperature
according to the manufacturer's instructions. The slides were
washed with Tris-buffered saline between incubations. Finally, the
sections were incubated in 0.5% 3′,3′-diaminobenzidine for 15–20
min at room temperature before being counterstained with Mayer's
hematoxylin. Negative controls consisted of incubating the slides
with negative control rabbit immunoglobulin (Agilent Technologies
Inc.) in the absence of the primary antibody. The immunoreactivity
of EGFR was evaluated by two independent researchers blinded to the
outcomes of the patients. The expression patterns of EGFR were
determined in a semiquantitative manner using light microscopy
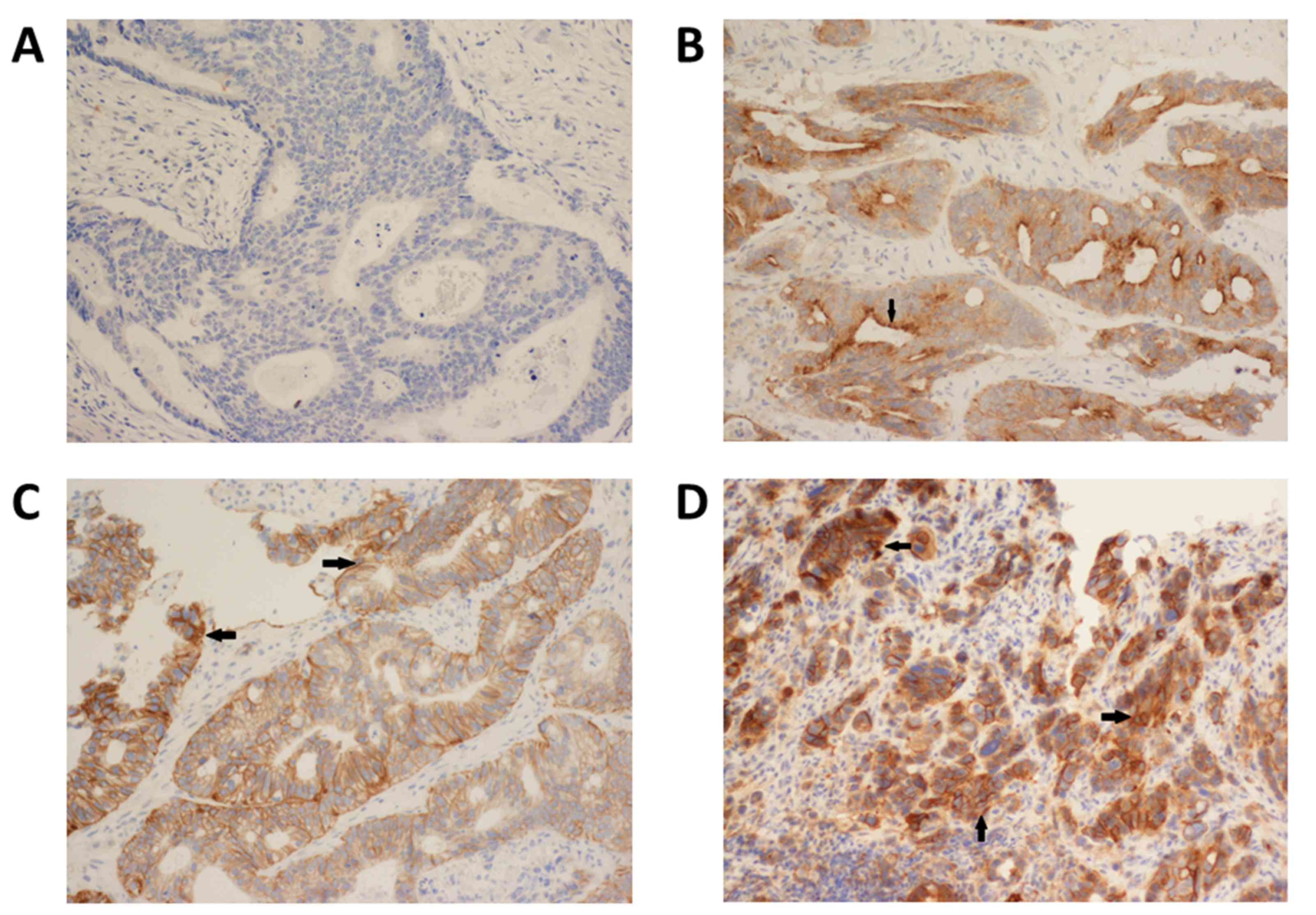
(magnification, ×100). Immunoreactivity for EGFR (membrane
staining) was categorized according to the presence of tumor cell
staining and staining intensity. The intensity of EGFR
immunoreactivity was scored using a three-tier system as follows:
1+ (weak intensity, faint brown membranous staining); 2+ (moderate
intensity, brown membranous staining of intermediate darkness
producing a complete or incomplete circular outline of the
neoplastic cell) and 3+ (strong intensity, dark brown or black
membranous staining producing a thick outline, complete or
incomplete of the neoplastic cell) (24). Negative EGFR expression was defined
as the absence of membrane staining above the background in all
tumor cells, whereas positive EGFR expression was defined the
complete or incomplete IHC membrane staining of tumor cells,
including intensities of 1+, 2+, or 3+ (Fig. 2).
Statistical analysis
All data were statistically analyzed using SPSS
version 22.0 (IBM Corp.). Values are presented as the mean ±
standard deviation for continuous variables and were compared using
an unpaired Student's t-test. The χ2 test was used to
determine the difference of clinicopathological characteristics,
KRAS/NRAS mutation and EGFR overexpression status between
the LiM-only and LuM-only groups. All probability values were
two-tailed. The clinicopathological characteristics of the WT and
mutant RAS/KRAS/NRAS groups were compared using the
Pearson's χ2 test and the survival rates were calculated
using the Kaplan-Meier method and log-rank tests were used to
analyze survival distribution. P<0.05 was considered to indicate
a statistically significant difference. The mutations identified in
the present study were compared with that in the TCGA dataset
(https://cancer.gov/tcga; accessed on 2019/12/31)
(25,26). The selection criterias used for this
dataset include colon cancer, colorectal cancer, KRAS, NRAS
and BRAF.
Results
Clinical characteristics
The clinicopathological characteristics of the 166
patients, including 124 LiM-only and 42 LuM-only, are summarized in
Table I. The incidence of LiM-only
was nearly 3× that of LuM-only in stage IV CRC. Patients with
LuM-only were significantly older compared with that in patients
with LiM-only (P=0.050). The status of LiM- or LuM-only was not
significantly associated with the location of the primary tumor,
serum CEA level and differentiation of the primary tumor. Moreover,
in 94 (75.8%) patients with LiM-only and 29 (69.0%) patients with
LuM-only, the primary tumor originated from the left colon. A total
of 84.7, 81.5 and 54.0% of patients with LiM-only and 88.1, 88.1
and 35.7% of patients with LuM-only were analyzed for KRAS,
NRAS mutations and EGFR overexpression status, respectively.
KRAS and NRAS mutations and EGFR overexpression rates
were 41.0, 5.0 and 86.6% in patients with LiM-only, respectively,
and 35.1, 38.1 and 80.0% in patients with LuM-only,
respectively.
In the present study, the mutation spectra of
RAS isoforms: KRAS and NRAS that mutationally
activated at codons 12, 13 or 61 were studied. 69.6% (39/56) of
KRAS mutations occur at codon 12 whereas 5.4% (3/56)
mutations are observed at codon 61. In contrast, 57.1% (4/7) of
NRAS tumors harbor mutations at codon 61 vs. 28.6% (2/7) at
codon 12 (Table II). KRAS
exon 2 mutations were detected in 39.4% (56/142) of patients, with
both LiM and LuM-only CRC, which included 24 (42.9%) patients with
G12D, 8 (14.3%) patients with G12V, 7 (12.5%) patients with A146T,
6 (10.7%) with G13D, 3 (5.4%) patients with G12C, 3 (5.4%) patients
with codon 61, 2 (3.6%) patients with G12A, 1 (1.8%) patients with
G13R, 1 (1.8%) patient with G12S and 1 (1.8%) patient with G12D,
G13D mutations. NRAS mutation status was analyzed in 83.1%
(138/166) of patients and only 5.8% (8/138) of patients harbored a
mutation, including 3 (37.5%) patients with codon 61, 2 (25.0%)
with codon 12, 1 (12.5%) patient with codon 146, 1 (12.5%) patient
with Q61K and 1 (12.5%) patient with T58I and Q61L mutations
(Table II). The TCGA data set
showed that there were 176 KRAS mutations and 41 NRAS
mutations among 10,202 papules. KRAS mutations included 49
(27.8%) with G12D, 33 (18.8%) with G12V, 31 (17.6%) with G13D, 13
(7.4%) with A146T, 10 (5.7%) with G12C, 8 (4.6%) with G12A, 7
(4.0%) with G12S and 3 (1.7%) with Q61K mutations. NRAS
mutation included 4 (9.8%) with Q61K, 4 (9.8%) with G12D, 2 (4.9%)
with G13R, and 1 (2.4%) with R164C, G12A, G12C, Q61R, G12V, NRAS
L6L, E132K, E76K and Q61L mutations (Table II). Notably, the distributions of
the different mutations were similar between the TCGA dataset and
our study. 67/124 (54.0%) of LiM-only and 15/42 (35.7%) LuM-only
patients underwent EGFR expression tests. The results demonstrated
that 86.6% of LiM-only and 80.0% of LuM-only patients had EGFR
overexpression. There was no significant difference between the
LiM-only group and LuM-only group (P=0.515) (Table I).
 | Table II.Sites and occurrence of mutations in
cases in the present study compared with The Cancer Genome Atlas
dataset. |
Table II.
Sites and occurrence of mutations in
cases in the present study compared with The Cancer Genome Atlas
dataset.
| A, LiM-only,
n=124 |
|---|
|
|---|
| KRAS mut
41.0% (43/105) | NRAS mut
5.0% (5/101) | BRAF mut
7.1% (6/85) |
|---|
|
|---|
| Mut site (n,
%) | Mut site (n,
%) | Mut site (n,
%) |
|---|
| G12D (20,
46.5) | Codon 61 (1,
1) | Codon V600E (5,
5.9) |
| G12V (7, 16.3) | Q61K (1, 1) | Codon V601E (1,
1.2) |
| G13D (5, 11.6) | Codon 12 (1,
1) | – |
| A146T (4, 9.3) | Codon 146
(1,1) | – |
| G12C (2, 4.7) | T58I and Q61L (1,
1) | – |
| G12A (2, 4.7) | – | – |
| G12D. G13D (1,
2.3) | – | – |
| G12S (1, 2.3) | – | – |
| G13R (1, 2.3) | – | – |
|
| B, LuM-only
(n=42) |
| KRAS mut
35.1% (13/37) | NRAS mut
8.1% (3/37) | BRAF mut
3.2% (1/31) |
|
| Mut site (n,
%) | Mut site (n,
%) | Mut site (n,
%) |
|
| G12D (4, 10.8) | Codon 61 (2,
5.4) | Codon V600E (1,
3.2) |
| A146T (3, 8.1) | Codon 12 (1,
2.7) | – |
| Codon 61 (3,
8.1) | – | – |
| G12C (1, 2.7) | – | – |
| G12V (1, 2.7) | – | – |
| G13D (1, 2.7) | – | – |
|
| C, The Cancer
Genome Atlas dataset (N=10,202) |
| KRAS mut
(n=176) | NRAS mut
(n=41) | BRAF
mut |
|
| Mut site (n,
%) | Mut site (n,
%) |
|
|
| G12D (49,
27.8) | Q61K (4, 9.8) | – |
| G12V (33,
18.8) | G12D (4, 9.8) | – |
| G13D (31,
17.6) | G13R (2, 4.9) | – |
| A146T (13,
7.4) | R164C (1, 2.4) | – |
| G12C (10, 5.7) | G12A (1, 2.4) | – |
| G12A (8, 4.6) | G12C (1, 2.4) | – |
| G12S (7, 4.0) | Q61R (1, 2.4) | – |
| Q61K (3, 1.7) | G12V (1, 2.4) | – |
| – | NRAS L6L (1,
2.4) | – |
| – | E132K (1, 2.4) | – |
| – | E76K (1, 2.4) | – |
| – | Q61L (1, 2.4) | – |
Survival analysis
Survival data were collected for all patients. The
median follow-up time was 19.2±5.3 months (1.0–57.1 months). The
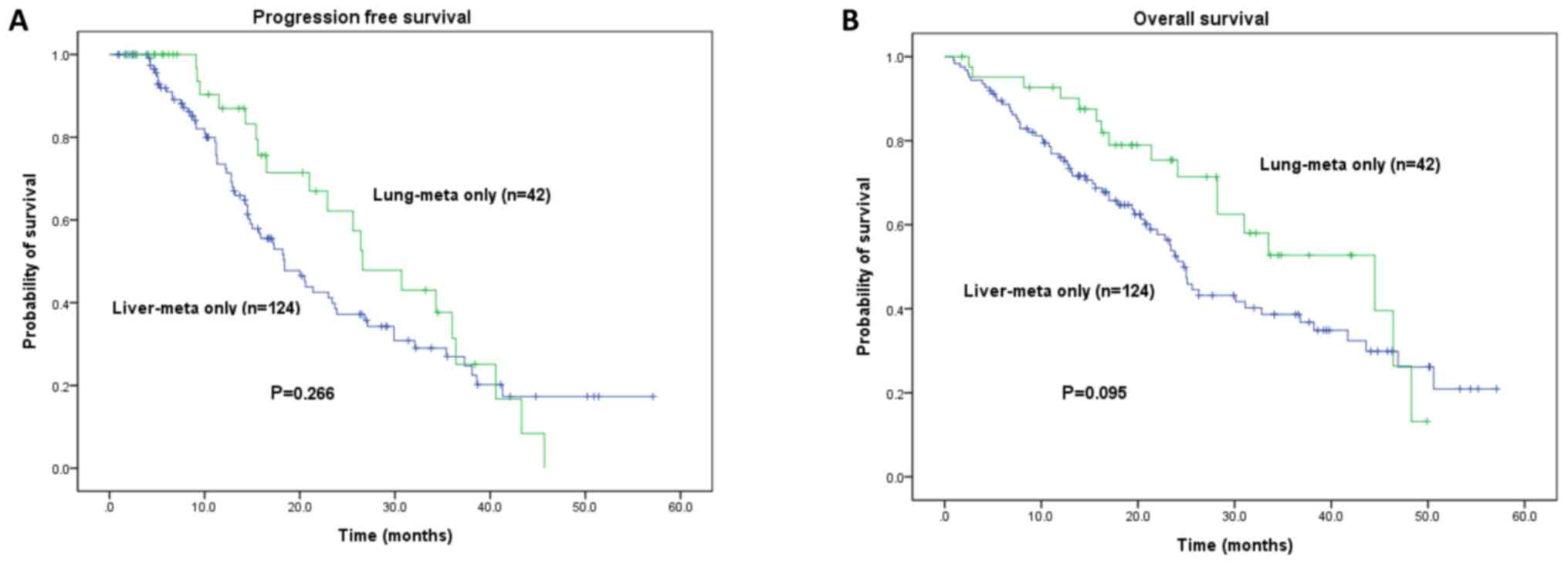
median PFS and OS times for LiM-only vs. LuM-only were 18.4±2.1 vs.
26.6±3.7 months (P=0.266) and 24.7 ±1.1 vs. 44.5±10.0 months
(P=0.095), respectively (Fig. 3).
Patients with LuM-only had a favorable PFS and OS compared with
that in patients with LiM-only, however this difference was not
significant (P=0.266 and P=0.095).
The results of the subgroup analysis of PFS and OS
times conducted according to RAS, KRAS and NRAS
mutation status in the LiM-only and LuM-only groups are presented
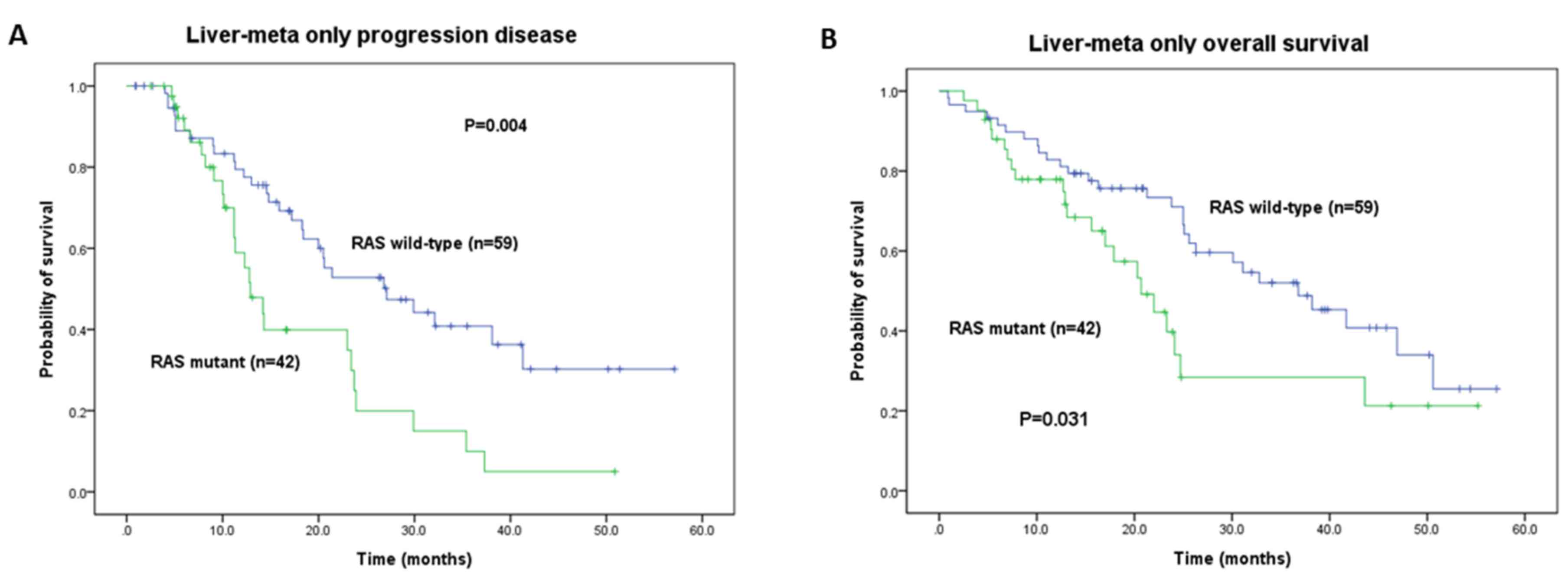
in Tables III and IV. In the LiM-only group, the median PFS
of the RAS WT group was significantly more favorable
compared with that of the mutant group (27.1 vs. 12.9 months;
P=0.004; Table III). The OS was
also more favorable in the RAS WT group compared with the
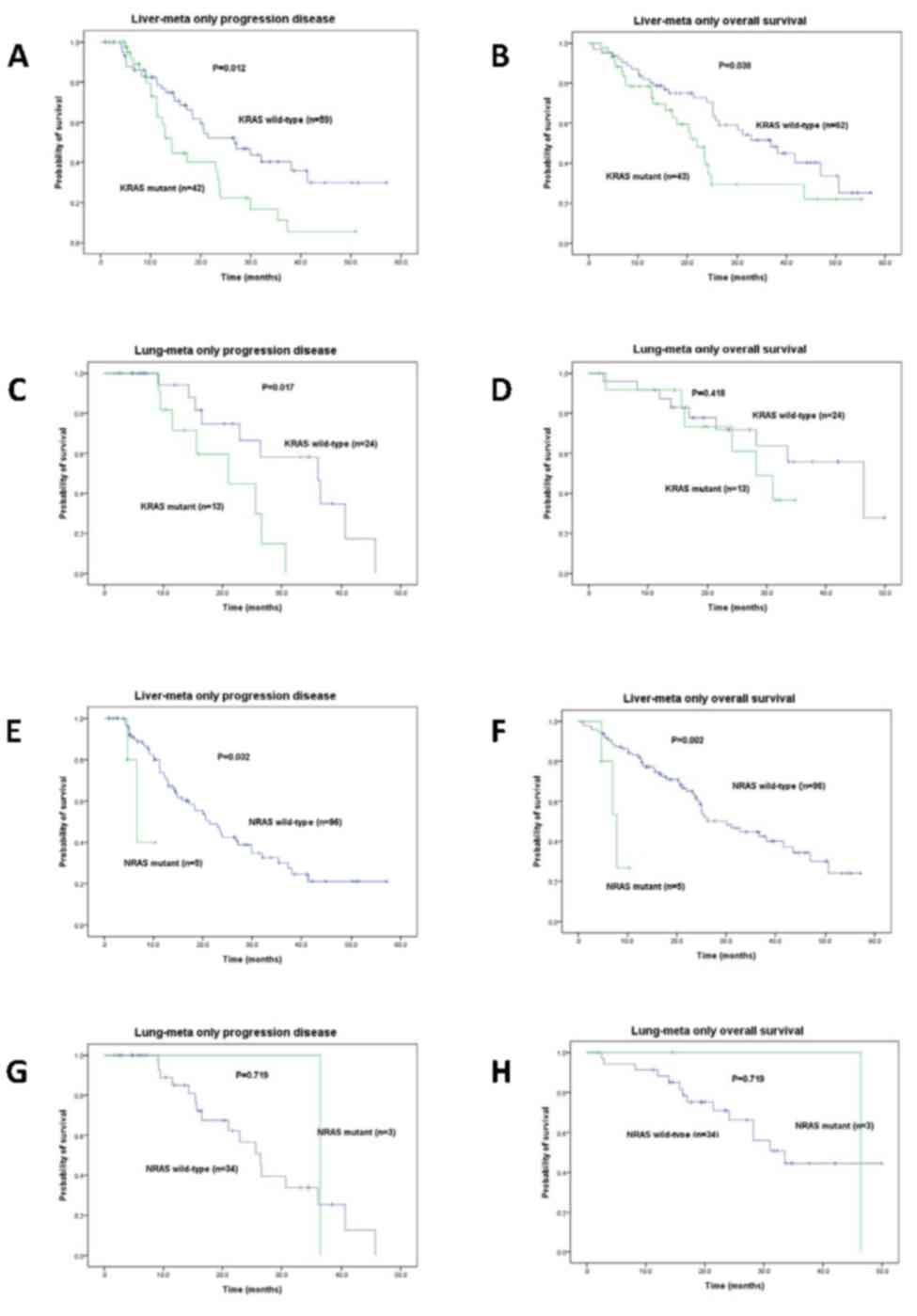
mutant group (36.8 vs. 20.7 months, P=0.031; Table IV). In the LuM-only group, the
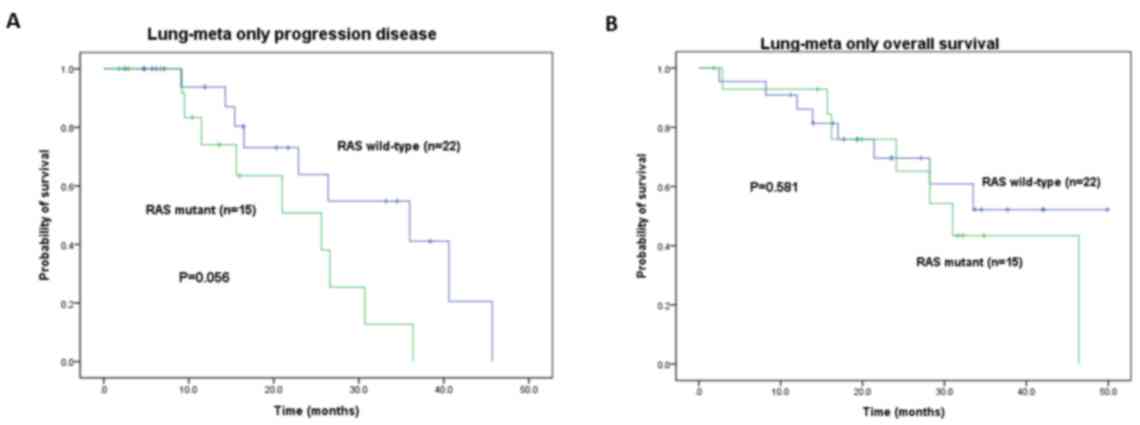
median PFS of the RAS WT group was not significantly more
favorable compared with that of the mutant group (36.0 vs. 25.6
months, P=0.056; Table III) but
was significantly more favorable in the KRAS WT group
compared with patients with KRAS mutation (36.0 vs. 21.0
months, P=0.017; Table III).
 | Table III.Gene mutation status and
progression-free survival analysis of patients with metastatic
colorectal cancer. |
Table III.
Gene mutation status and
progression-free survival analysis of patients with metastatic
colorectal cancer.
|
|
Liver-meta only | Lung-meta only |
|---|
|
|
|
|
|---|
| Gene | n | Median PFS
(months) | P-value | n | Median PFS
(months) | P-value |
|---|
| RAS |
|
| 0.004 |
|
| 0.056 |
|
Wild-type | 59 | 27.1 |
| 22 | 36.0 |
|
|
Mutant | 42 | 12.9 |
| 15 | 25.6 |
|
| KRAS |
|
| 0.012 |
|
| 0.017 |
|
Wild-type | 62 | 26.8 |
| 24 | 36.0 |
|
|
Mutant | 43 | 14.2 |
| 13 | 21.0 |
|
| NRAS |
|
| 0.032 |
|
| 0.719 |
|
Wild-type | 96 | 21.4 |
| 34 | 26.4 |
|
|
Mutant | 5 |
6.6 |
| 3 | 36.4 |
|
| EGFR
overexpression |
|
| 0.628 |
|
| 0.942 |
|
Negative | 9 | 32.1 |
| 3 | 26.6 |
|
|
Positive | 58 | 23.4 |
| 12 | 36.0 |
|
 | Table IV.Gene mutation status and overall
survival analysis of patients with metastatic colorectal
cancer. |
Table IV.
Gene mutation status and overall
survival analysis of patients with metastatic colorectal
cancer.
|
|
Liver-meta only | Lung-meta only |
|---|
|
|
|
|
|---|
| Gene | n | Median OS
(mon) | P-value | n | Median OS
(mon) | P-value |
|---|
| RAS |
|
| 0.031 |
|
| 0.581 |
| Wild
type | 59 | 36.8 |
| 22 | 33.5 |
|
|
Mutant | 42 | 20.7 |
| 15 | 31.0 |
|
| KRAS |
|
| 0.038 |
|
| 0.418 |
| Wild
type | 62 | 36.8 |
| 24 | 46.4 |
|
|
Mutant | 43 | 22.0 |
| 13 | 28.2 |
|
| NRAS |
|
| 0.002 |
|
| 0.719 |
| Wild
type | 96 | 30.1 |
| 34 | 33.5 |
|
|
Mutant | 5 | 7.8 |
| 3 | 46.4 |
|
| EGFR
overexpression |
|
| 0.876 |
|
| 0.449 |
|
Negative | 9 | 36.8 |
| 3 | 43.1 |
|
|
Positive | 58 | 31.1 |
| 12 | 36.6 |
|
The PFS and OS times in the RAS WT group were
significantly improved compared with the RAS mutant cohorts
(27.1 vs. 12.9 months, P=0.004 and 36.8 vs. 20.7 months, P=0.031,
respectively) in the LiM-only group (Fig. 4A and B). Conversely, the RAS
WT and mutant groups exhibited no significant PFS and OS difference
in the LuM-only group (36.0 vs. 25.6 months, P=0.056 and 33.5 vs.
31.0 months, P=0.581, respectively; Fig.
5A and B). The PFS and OS times between the WT and mutant
status for KRAS were 26.8 vs. 14.2 months (P=0.012) and 36.8
vs. 22.0 months (P=0.038), respectively in the LiM-only group
(Tables III and IV; Fig. 6A and
B). The KRAS WT and mutant groups showed significant PFS
times (36.0 vs. 21.0 months, P=0.017) but not OS times (46.4 vs.
28.2 months, P=0.418) in the LuM-only group (Tables III and IV; Fig. 6C and
D). Similarly, the PFS and OS times between the WT and mutant
status for NRAS were 21.4 vs. 6.6 months (P=0.032) and 30.1
vs. 7.8 months (P=0.002), respectively in the LiM-only group
(Tables III, IV, Fig. 6E and
F). The NRAS WT and mutant groups showed no significant
PFS times (26.4 vs. 36.4 months, P=0.719) and OS times (33.5 vs.
46.4 months, P=0.719) in the LuM-only group (Tables III and IV; Fig. 6G and
H).
Discussion
In the present study, data were collected over a
4-year period with a median follow-up period of 19.2 months
(1.0–57.1 months) from 166 patients initially diagnosed with mCRC
with LiM- or LuM-only. The results revealed that the incidence of
LiM-only was ~3× that of LuM-only. Overall, the median OS was
19.2±13.7 months (1.0–57.1 months) in patients with LiM- and
LuM-only mCRC. The association between RAS mutation status
and clinicopathological features was also evaluated between the
LiM- and LuM-only groups. The results demonstrated that RAS,
KRAS and NRAS mutations were associated with both the
PFS and OS time of patients with LiM-only. In contrast, RAS
mutation did not affect the PFS and OS time of patients with
LuM-only except for mutant KRAS in PFS. In addition, the
association between EGFR overexpression and survival was examined;
however, there was no significant difference between LiM- and
LuM-only mCRC groups. To the best of our knowledge, the present
study was the first to analyze RAS mutation status and
survival in synchronous LiM-only and LuM-only patients with
CRC.
RAS mutations lead to the constitutive
activation of EGFR signaling through the oncogenic Ras/Raf/Mek/Erk
pathway (27). All types of mutant
KRAS are present in 35–45% of patients with CRC, and codons
12 and 13 are the two most common hotspots, accounting for ~95% of
all mutation types (~80% occurring in codon 12 and 15% in codon 13)
(28). The present study population
was relatively homogenous, all of Han ethnicity and from the same
geographic location.
Several clinical trials have demonstrated that
active KRAS mutations are negative predictors of the
clinical benefit of anti-EGFR therapies in patients with mCRC
(21,29,30). The
prognostic role of KRAS mutations has been previously
investigated; however the prognostic value of RAS mutations
remains uncertain with respect to the treatment of mCRC. Roth et
al (11) prospectively collected
1,404 samples from patients with stage II and III CRC and
demonstrated that KRAS mutations did not have a major
prognostic value regarding the relapse-free survival or OS. There
are several possible explanations for the differences in the
results of these clinical trials, such as the study size and
design, patient population and staging, tumor sampling (primary or
metastatic site), use of archival vs. fresh or frozen material,
laboratory methods, data analyses and distinct chemotherapy
protocol and regimen. In terms of the metastatic sites in mCRC,
numerous studies have reported an association with primary tumor
location and RAS mutation status. For example, left-sided
colon cancer is more likely to metastasize to the liver and lung
(31,32). Kim et al (33) reported that RAS mutation rate
was higher in patients with lung metastases compared with those
with liver and ovary or bladder metastases (P=0.039). Prasanna
et al (4) also revealed that
the incidence of LuM-only was higher with KRAS/RAS
mutations (relative risk, 1.4; P=0.007) when compared with other
site metastasis (including liver, lymph node, brain, bone and
peritoneum). Recently, a retrospective analysis of 899 patients
with mCRC demonstrated that WT KRAS had greater proportion
of liver metastases (78.6 vs. 53.5%; P<0.001) when compared with
mutant KRAS, whereas patients with mutant KRAS had
greater proportion of lung metastases (23.3 vs. 8.7%; P=0.02) when
compared with WT KRAS in the patients with left-sided tumors
(34). In the present study, 75.8%
(94/124) of patients with LiM-only and 69.0% (29/42) with LuM-only
mCRC metastasized from left-sided colon cancer, which was not
significantly associated.
The identification of KRAS mutational status
as a predictive marker of response to anti-EGFR mAb has been one of
the most significant and practice-changing recent advances in
colorectal cancer (35). However, in
a clinical setting, treatment with anti-EGFR mAb is not recommended
for patients with mCRC and KRAS or NRAS mutant forms
of RAS, in which case only anti-VEGF mAb therapy should be
used (36). In addition, EGFR is a
direct downstream target of RAS signaling; however, EGFR
amplification (>2 copies) and protein overexpression in colon
cancer tissues have not been established as reliable biomarkers for
anti-EGFR agents (37,38). Several studies have demonstrated that
KRAS mutations are associated with a higher incidence of CRC
recurrence, metastatic spread and shorter OS time (39–41).
Moreover, various metastatic sites and RAS mutation status
exhibit distinct outcomes. For example, Prasanna et al
(4) indicated that survival of
patients with mCRC was associated with the site of metastases, with
lung-only metastasis displaying a more favorable survival outcome
compared with other single metastatic site diseases. The results
from univariate analysis revealed that the median OS time was
longer when metastases were limited to the lung or liver and
shorter in cases of brain, bone or peritoneal metastases (4).
The genetic analysis of somatic mutation hotspots in
KRAS, NRAS and BRAF is now standard practice for
selecting patients with mCRC eligible for anti-EGFR therapy
(42). Mutations can be assessed
using next-generation sequencing (NGS) or PCR-based assays. The
number of analyzed targets, the speed of the assays and accuracy of
the results are crucial. Several factors may affect the data
produced, such as the quality of the DNA extracted from
paraffin-embedded tissue, tumor heterogeneity, quality control of
laboratories and different DNA polymerase enzymes. Nagakubo et
al (42) compared mutation
detection in KRAS and NRAS genes between the
PCR-reverse sequence-specific oligonucleotide probe method and
bridged nucleic acid-clamp PCR using Sanger sequencing. A total of
three discordant results were obtained and the concordance rate was
94% between the two methods. All mutations identified using
BNA-clamp PCR and Sanger sequencing were also identified using NGS.
This suggested that BNA-clamp PCR using Sanger sequencing detects
somatic mutations in KRAS, NRAS and BRAF with a high
accuracy (42). Gilson et al
(43) used DNA pipetted directly in
the cartridge of the Idylla system, exhibiting a good sensitivity,
specificity, reproducibility and limit of detection, and can be
integrated in a laboratory workflow for samples with little tissue
without compromising the further complete tumor characterization
using NGS. In the present study, the ABI PRISM 310 genetic analyzer
system and Taqman genotyper v.1.6 software (Applied Biosystems;
Thermo Fisher Scientific, Inc.) were used to perform accurate
analysis using automated capillary electrophoresis for
genotyping.
The present study was limited by its relatively
small sample size and retrospective nature. Not all patients were
routinely checked for their genetic profile. Furthermore, this
study did not consider the severity of the patient's comorbidities
and the performance status that may affect survival. Four patients
hesitated to receive treatment, which affects the results. In
addition, the small sample size did not permit evaluation of the
effect of various KRAS mutation subtypes, such as G12D,
G12V, G13D, and A146T.
In the present study, a population of patients with
synchronous LiM- and LuM-only mCRC were evaluated. The
clinicopathological characteristics and RAS mutation status
between these two groups were compared. The results revealed a
difference in the PFS and OS between patients with WT and mutant
forms of KRAS in the LiM-only group but not in the LuM-only
group. RAS mutation is a poor prognostic predictor of less
favorable PFS and OS in synchronous patients with LiM- and LuM-only
mCRC.
Acknowledgements
The authors would like to thank Dr Zhi-Feng Miao,
Division of Colorectal Surgery, Department of Surgery, Kaohsiung
Medical University Hospital (Kaohsiung, Taiwan) for the analysis of
the TCGA dataset.
Funding
This study was supported by grants from The Ministry
of Science and Technology (grant nos. MOST108-2321-B-037-001,
MOST107-2321-B-037-003, MOST107-2314-B-037-116,
MOST107-2314-B-037-022-MY2 and MOST107-2314-B-037-023-MY2), The
Ministry of Health and Welfare (grant nos.
MOHW107-TDU-B-212-123006, MOHW107-TDU-B-212-114026B,
MOHW108-TDU-B-212-133006 and MOHW108-TDU-B-212-124026), the Health
and Welfare Surcharge of Tobacco Products and the Kaohsiung Medical
University Hospital (grant nos. KMUH107-7R28, KMUH107-7R29,
KMUH107-7R30, KMUH107-7M22, KMUH107-7M23, KMUHS10701, KMUHS10801,
KMUHS10804 and KMUHS10807) and The Center for Cancer Research,
Kaohsiung Medical University (grant no. KMU-TC108A04). In addition,
this study was supported by The Grant of Taiwan Precision Medicine
Initiative and Biomarker Discovery in Major Diseases of Taiwan
Project (grant no. AS-BD-108-1).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SC conceived the study and wrote the manuscript. CH
and CM performed the statistical analysis. YC performed the
histological examinations. HT, TC, WS, WH and CK analyzed and
interpreted the clinical data. JW conceived the study and revised
the manuscript for important intellectual content. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
In compliance with the Helsinki Declaration, the
present study was approved by The Institutional Review Board of the
Kaohsiung Medical University Hospital (approval no.
KMUHIRB-20130022). Written informed consent was provided by all
patients prior to enrollment in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferly J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Millikan KW, Staren ED and Doolas A:
Invasive therapy of metastatic colorectal cancer to the liver. Surg
Clin North Am. 77:27–48. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nitzkorski JR, Farma JM, Watson JC,
Siripurapu V, Zhu F, Matteotti RS and Sigurdson ER: Outcome and
natural history of patients with stage IV colorectal cancer
receiving chemotherapy without primary tumor resection. Ann Surg
Oncol. 19:379–383. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prasanna T, Karapetis CS, Roder D, Tie J,
Padbury R, Price T, Wong R, Shapiro J, Nott L, Lee M, et al: The
survival outcome of patients with metastatic colorectal cancer
based on the site of metastases and the impact of molecular markers
and site of primary cancer on metastatic pattern. Acta Oncol.
57:1438–1444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qiu M, Hu J, Yang D, Cosgrove DP and Xu R:
Pattern of distant metastases in colorectal cancer: A SEER based
study. Oncotarget. 6:38658–38666. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zizzo M, Galeone C, Braglia L, Ugoletti L,
Siciliani A, Nachira D, Margaritora S, Pedrazzoli C, Paci M, Lococo
F, et al: Long-Term outcomes after surgical resection for
synchronous or metachronous hepatic and pulmonary colorectal cancer
metastases. Digestion. 101:144–155. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Augestad KM, Merok MA and Ignatovic D:
Tailored treatment of colorectal cancer: Surgical, molecular, and
genetic considerations. Clin Med Insights Oncol.
11:11795549176907662017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Raskov H, Pommergaard HC, Burcharth J and
Rosenberg J: Colorectal carcinogenesis-update and perspectives.
World J Gastroenterol. 20:18151–18164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amado RG, Wolf M, Peeters M, Van Cutsem E,
Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et
al: Wild-type KRAS is required for panitumumab efficacy in patients
with metastatic colorectal cancer. J Clin Oncol. 26:1626–1634.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schirripa M, Cremolini C, Loupakis F,
Morvillo M, Bergamo F, Zoratto F, Salvatore L, Antoniotti C,
Marmorino F, Sensi E, et al: Role of NRAS mutations as prognostic
and predictive markers in metastatic colorectal cancer. Int J
Cancer. 136:83–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roth AD, Tejpar S, Delorenzi M, Yan P,
Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C,
et al: Prognostic role of KRAS and BRAF in stage II and III
resected colon cancer: Results of the translational study on the
PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 28:466–474.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hutchins G, Southward K, Handley K, Magill
L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M,
Kerr D, et al: Value of mismatch repair, KRAS, and BRAF mutations
in predicting recurrence and benefits from chemotherapy in
colorectal cancer. J Clin Oncol. 29:1261–1270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Ma J, Zhang S, Deng G, Wu X, He
J, Pei H, Shen H and Zeng S: A prognostic analysis of 895 cases of
stage III colon cancer in different colon subsites. Int J
Colorectal Dis. 30:1173–1183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fakih M and Wong R: Efficacy of the
monoclonal antibody EGFR inhibitors for the treatment of metastatic
colorectal cancer. Curr Oncol. 17 (Suppl 1):S3–S17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Venook AP: Epidermal growth factor
receptor-targeted treatment for advanced colorectal carcinoma.
Cancer. 103:2435–2446. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bibeau F, Boissiere-Michot F, Sabourin JC,
Gourgou-Bourgade S, Radal M, Penault-Llorca F, Rochaix P, Arnould
L, Bralet MP, Azria D and Ychou M: Assessment of epidermal growth
factor receptor (EGFR) expression in primary colorectal carcinomas
and their related metastases on tissue sections and tissue
microarray. Virchows Arch. 449:281–287. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang Z, Zhang J, Zeng X, Gao J, Wu S and
Liu T: Relationship between EGFR expression, copy number and
mutation in lung adenocarcinomas. BMC Cancer. 10:3762010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sambrook J and Russell DW: The condensed
protocols from molecular cloning: A laboratory manual. Cold Spring
Harbor Laboratory Press, Cold Spring Harbor. (NY). p. v.
8002006.
|
|
20
|
Yen LC, Uen YH, Wu DC, Lu CY, Yu FJ, Wu
IC, Lin SR and Wang JY: Activating KRAS mutations and
overexpression of epidermal growth factor receptor as independent
predictors in metastatic colorectal cancer patients treated with
cetuximab. Ann Surg. 251:254–260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang JY, Hsieh JS, Chen FM, Yeh CS,
Alexandersen K, Huang TJ, Chen DC and Lin SR: High frequency of
activated K-ras codon 15 mutant in colorectal carcinomas from
Taiwanese patients. Int J Cancer. 107:387–393. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang CW, Chen YT, Tsai HL, Yeh YS, Su WC,
Ma CJ, Tsai TN and Wang JY: EGFR expression in patients with stage
III colorectal cancer after adjuvant chemotherapy and on cancer
cell function. Oncotarget. 8:114663–114676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang CW, Tsai HL, Chen YT, Huang CM, Ma
CJ, Lu CY, Kuo CH, Wu DC, Chai CY and Wang JY: The prognostic
values of EGFR expression and KRAS mutation in patients with
synchronous or metachronous metastatic colorectal cancer. BMC
Cancer. 13:5992013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Scartozzi M, Bearzi I, Berardi R,
Mandolesi A, Fabris G and Cascinu S: Epidermal growth factor
receptor (EGFR) status in primary colorectal tumors does not
correlate with EGFR expression in related metastatic sites:
Implications for treatment with EGFR-targeted monoclonal
antibodies. J Clin Oncol. 1:4772–4778. 2004. View Article : Google Scholar
|
|
25
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Sethi NS, Hinoue T, Schneider BG,
Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R,
Islam M, et al: Comparative molecular analysis of gastrointestinal
adenocarcinomas. Cancer Cell. 33:721–735.e8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wee P and Wang Z: Epidermal growth factor
receptor cell proliferation signaling pathways. Cancers (Basel).
9:522017. View Article : Google Scholar
|
|
28
|
Tan C and Du X: KRAS mutation testing in
metastatic colorectal cancer. World J Gastroenterol. 18:5171–5180.
2012.PubMed/NCBI
|
|
29
|
Lo Nigro C, Ricci V, Vivenza D, Granetto
C, Fabozzi T, Miraglio E and Merlano MC: Prognostic and predictive
biomarkers in metastatic colorectal cancer anti-EGFR therapy. World
J Gastroenterol. 22:6944–6954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Stefano A and Carlomagno C: Beyond
KRAS: Predictive factors of the efficacy of anti-EGFR monoclonal
antibodies in the treatment of metastatic colorectal cancer. World
J Gastroenterol. 20:9732–9743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He XK, Wu W, Ding YE, Li Y, Sun LM and Si
J: Different anatomical subsites of colon cancer and mortality: A
population-based study. Gastroenterol Res Pract. 2018:71536852018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Engstrand J, Nilsson H, Stromberg C, Jonas
E and Freedman J: Colorectal cancer liver metastases-a
population-based study on incidence, management and survival. BMC
Cancer. 18:782018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim HS, Heo JS, Lee J, Lee JY, Lee MY, Lim
SH, Lee WY, Kim SH, Park YA, Cho YB, et al: The impact of KRAS
mutations on prognosis in surgically resected colorectal cancer
patients with liver and lung metastases: A retrospective analysis.
BMC Cancer. 16:1202016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yong ZZ, Ching GTH and Ching MTC:
Metastatic profile of colorectal cancer: Interplay between primary
tumor location and KRAS Status. J Surg Res. 246:325–334. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shankaran V, Obel J and Benson AB III:
Predicting response to EGFR inhibitors in metastatic colorectal
cancer: Current practice and future directions. Oncologist.
15:157–167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Helden EJ, Menke-van der Houven van
Oordt CW, Heymans MW, Ket JCF, van den Oord R and Verheul HMW:
Optimal use of anti-EGFR monoclonal antibodies for patients with
advanced colorectal cancer: A meta-analysis. Cancer Metastasis Rev.
36:395–406. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kato S, Okamura R, Mareboina M, Lee S,
Goodman A, Patel SP, Fanta PT, Schwab RB, Vu P, Raymond VM, et al:
Revisiting Epidermal Growth Factor Receptor (EGFR) Amplification as
a Target for Anti-EGFR therapy: Analysis of cell-free circulating
tumor DNA in patients with advanced malignancies. JCO Precis Oncol.
3:102019.
|
|
38
|
Yang ZY, Shen WX, Hu XF, Zheng DY, Wu XY,
Huang YF, Chen JZ, Mao C and Tang JL: EGFR gene copy number as a
predictive biomarker for the treatment of metastatic colorectal
cancer with anti-EGFR monoclonal antibodies: A meta-analysis. J
Hematol Oncol. 5:522012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Brudvik KW, Kopetz SE, Li L, Conrad C,
Aloia TA and Vauthey JN: Meta-analysis of KRAS mutations and
survival after resection of colorectal liver metastases. Br J Surg.
102:1175–1183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shindoh J, Nishioka Y, Yoshioka R,
Sugawara T, Sakamoto Y, Hasegawa K, Hashimoto M and Kokudo N: KRAS
mutation status predicts Site-Specific recurrence and survival
after resection of colorectal liver metastases irrespective of
location of the primary lesion. Ann Surg Oncol. 23:1890–1896. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yaeger R, Cowell E, Chou JF, Gewirtz AN,
Borsu L, Vakiani E, Solit DB, Rosen N, Capanu M, Ladanyi M and
Kemeny N: RAS mutations affect pattern of metastatic spread and
increase propensity for brain metastasis in colorectal cancer.
Cancer. 121:1195–1203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nagakubo Y, Hirotsu Y, Amemiya K, Oyama T,
Mochizuki H and Omata M: Accurate detection of KRAS, NRAS and BRAF
mutations in metastatic colorectal cancers by bridged nucleic
acid-clamp real-time PCR. BMC Med Genomics. 12:1622019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gilson P, Franczak C, Dubouis L, Husson M,
Rouyer M, Demange J, Perceau M, Leroux A, Merlin JL, Harlé A, et
al: Evaluation of KRAS, NRAS and BRAF hotspot mutations detection
for patients with metastatic colorectal cancer using direct DNA
pipetting in a fully-automated platform and next-generation
sequencing for laboratory workflow optimisation. PLoS One.
14:e02192042019. View Article : Google Scholar : PubMed/NCBI
|