Introduction
Cervical cancer is the leading cause of
cancer-associated mortality among women worldwide and the estimated
age-standardized incidence of cervical cancer was 13.1 per 100,000
women (1). Several studies have
suggested that both genetic and epigenetic changes may serve
important roles in carcinogenesis (2,3);
however, the development of targeted molecular therapy for cervical
cancer is unsatisfactory. It is therefore crucial to improve the
therapeutic strategies by determining the underlying mechanisms
involved in cervical cancer.
The p21-activated kinase (PAK) family is a group of
serine/threonine kinases, which contain a Cdc42/Rac-interactive
binding domain and a Ste20-related kinase domain (4). Based on the sequence homology and
regulatory properties, PAKs have been classified into two groups:
Group I (PAK1-3) and Group II (PAK4-6) (5). Previous studies have revealed that the
PAK family was involved in numerous cellular functions, such as
cell motility, cell survival and gene regulation (6,7). The
overexpression or amplification of PAKs has been detected in
several types of human tumor, including breast cancer,
neurofibromatosis, colon cancer and lung cancer (8,9), and it
has been reported to be involved in several cancer signaling
pathways, including the NF-κB, Ras, AKT, Raf and p53 pathways
(10–13). Thus, the different biological roles
of PAKs have positioned them as attractive cancer therapeutic
targets.
PAK6, which was originally cloned as an androgen
receptor-interacting protein, is a unique member of the PAK family
(14,15). Although PAK6 expression levels were
reported to be overexpressed in primary and metastatic prostate
cancer (13), and hypothesized to be
a useful biomarker of adenocarcinoma (16), the current knowledge of the role of
PAK6 in the progression of other types of cancer remains
insufficient, with very little known about its underlying
mechanism. Thus, the present study aimed to determine the role of
PAK6 in the oncogenesis of cervical cancer, one of the most common
types of cancer to affect women, and to identify possible molecular
mechanisms.
Materials and methods
Cell culture
The human cervical cancer cell lines, HeLa and C33A,
and the 293T cell line were obtained from the China Center for Type
Culture Collection. All cells were cultured in DMEM (HyClone;
Cytiva), supplemented with 10% heat-inactivated FBS (Invitrogen;
Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 µg/ml
streptomycin, and placed at 37°C in a humidified incubator
containing 5% CO2.
Immunohistochemistry (IHC)
A total of 50 specimens with complete clinical
information on the clinical metastatic status of cervical cancer
were obtained from the CR501a tissue microarray (Alenabio). This
microarray included 40 cases of squamous cell carcinoma, 2 cases of
adenocarcinoma, 4 cases of adenosquamous carcinoma and 4
paracancerous tissues. Specimens were blocked using goat serum
(1:10; cat. no. AR1009, Boster Biological Technology) for 20 min at
room temperature, followed by incubation with PAK6 antibody (1:50;
cat. no. 13539–1-AP; ProteinTech Group, Inc.) at 4°C overnight.
Specimens were incubated with secondary antibody (goat anti-rabbit;
1:100; cat. no. S0001; Affinity Biosciences) at 37°C for 2 h.
Signal was detected using DAB detection kit (cat. no. AR1022;
Boster Biological Technology). The histological classifications and
clinical staging were performed in accordance with the
International Federation of Gynecology and Obstetrics (FIGO)
classification system (17). PAK6
expression levels were subsequently determined using IHC. All
tissue sections were examined qualitatively in five randomly
selected fields by two separate researchers using an Olympus CX31
light microscope (magnification, ×40; Olympus Corporation). The
scores were determined using an immunoreactivity score (18) (IRS; IRS=intensity score × extent
score). The intensity score for PAK6 was determined as follows: i)
0, negative; ii) 1, weak; iii) 2, moderate; and iv) 3, strong. The
extent scores were determined as follows: i) 0, <10%; ii) 1,
10–30%; iii) 2, 30–65%; and iv) 3, >65%, according to the
percentage of positively stained cells. The IHC results for PAK6
were assigned to one of four categories according to the IRS: i)
IRS of ≤1 was defined as (−); ii) 1<IRS ≤3 as (+); iii) 3<IRS
≤6 as (++); and iv) IRS>6 as (+++). The tissue microarray
testing was approved by the Medical Ethics Committee of Huazhong
University of Science and Technology (Wuhan, China) and all
patients provided written, informed consent.
Vector construction and
transfection
Short hairpin RNAs (shRNA/sh) targeting PAK6
(shPAK6) and the negative control (NC; scramble vector) were
obtained from Shanghai GeneChem Co., Ltd. (Table I). The plasmid was linearized by
restriction endonuclease double digestion with AgeI and
EcoR1, and the shPAK6 (GCAGGCTATTCCGAAGCAT) or shPAK6 NC
(TTCTCCGAACGTGTCACGT) sequences were inserted to construct
lentiviral vectors.
 | Table I.Sequences of p21-activated kinase 6
shRNA with AgeI and EcoR1 sites. |
Table I.
Sequences of p21-activated kinase 6
shRNA with AgeI and EcoR1 sites.
| shRNA | Sequence
(5′→3′) |
|---|
| Top strand |
5′–GAATTCGCAGGCTATTCCGAAGCATTTCAAGAGAATGCTTCGGAATAGCCTGCTTTTTT-3′ |
| Bottom strand |
3′-CGTCCGATAAGGCTTCGTAAAGTTCTCTTACGAAGCCTTATCGGACGAAAAAAGAGCTC−5′ |
To overexpress PAK6, a recombinant lentiviral vector
containing the full-length cDNA of PAK6 was used, and empty vector
was used as negative control. Briefly, the primer sequences were
designed and synthesized by Shanghai GeneChem Co., Ltd. (Table II). The PAK6 fragment, amplified by
reverse transcription-quantitative PCR using mRNA extracted from
HeLa cells, was subsequently cloned into the EcoR1 and
XhoI sites of the linearized pSico-eGFP-Flag plasmid
(FenghuiShengwu) to generate the recombinant lentiviral vector.
 | Table II.Primers for amplifying the full
length of p21-activated kinase 6 cDNA. |
Table II.
Primers for amplifying the full
length of p21-activated kinase 6 cDNA.
| Primer | Sequence
(5′→3′) |
|---|
| Forward |
5′-GGCCTCGAGATGTTCCGCAAGAAAAAGA-3′
(XhoI) |
| Reverse |
5′-CTGGAATTCCGCAGGTGGAGGTCTGCTTT-3′
(EcoR1) |
The lentiviral vectors were co-transfected with the
three packaging plasmids (pRSV-Rev, pMD2.G and pCMV–VSV-G, Hedgehog
Bio Science and Technology Ltd.) into 293T cells (1×106)
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) to produce the lentiviral particles. The
ratio of lentiviral vectors was as follows: pRSV-Rev: pMD2.G:
pCMV–VSV-G was 2:1:1. For a 60 mm dish, 3, 1.5, and 1.5 µg of those
plasmids was used individually. Following incubation at 37°C for 72
h, the lentiviruses were collected by centrifugation (1,200 × g,
4°C, 5 min) and used to infect the HeLa cells (1×106),
which were sub-cultured (with 3 µg/ml puromycin) for four
generations to obtain the stably transfected cell line. The changes
in the expression levels of PAK6, phosphorylated (p)-β-Catenin,
β-Catenin, glycogen synthase kinase (GSK)3β, p-GSK3β, cyclin D1 and
E-cadherin in the stably transfected cells were analyzed using
western blotting.
In addition, pDsRed-N1 and pEGFP-N1 plasmids
(Shanghai Beinuo Biotech, Co., Ltd.) were introduced to construct
PAK6/pDsRed-N1 and GSK3β/pEGFP-N1 recombinants, based on the
designed primers (Table III)
following the steps described above. For a 6-well culture plate, 4
µg plasmid was co-transfected into 293T cells (2×105
cells/well) for 48 h using Lipofectamine® 2000 reagent.
Transfected cells were used for fluorescence colocalization
microscopy analysis.
 | Table III.Primers for amplifying PAK6 and
GSK3β. |
Table III.
Primers for amplifying PAK6 and
GSK3β.
| Gene | Primer sequence
(5′→3′) |
|---|
| PAK6 | F:
GGCCTCGAGATGTTCCGCAAGAAAAAGA
(XhoI) |
|
| R:
CTGGAATTCCGCAGGTGGAGGTCTGCTTT
(EcoRI) |
| GSK3β | F:
AGACTCGAGATGTCAGGGCGGCCCAGAA
(XhoI) |
|
| R:
GACGAATTCCGGTGGAGTTGGAAGCTGATG
(EcoRI) |
Reverse transcription-quantitative PCR
(RT-qPCR)
The mRNA level of PAK6 was evaluated by RT-qPCR.
Total RNA was extracted by TRIzol Reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and reversed transcribed into
single-strand cDNA using Superscript II reverse transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA
amplification was performed using SYBR Premix Ex Tap (Tli RNaseH
Plus; Takara) on the BIO-RAD CFX96 system. The following
thermocycling conditions were used: Initial denaturation at 95°C
for 3 min, followed by 40 cycles of 95°C for 15 sec and 95°C for 1
min, finally dissociation at 95°C for 10 min. β-actin was used as
an internal control. Fold-changes were calculated using the
relative quantification (2−ΔΔCq) method (19). The primers (Shanghai GeneChem Co.,
Ltd.) used are as follows: PAK6, forward
5′-GACTCCATCCTGCTGACCCTC-3′; reverse 5′-CACCTCAGTGGCATACAAAGACC-3′;
β-actin, forward 5′-ACCAGTTCGCCATGGATGAC-3′; reverse
5′-TGCCGGAGCCGTTGTC-3′.
Western blotting
Total protein was extracted from cells using a lysis
buffer (150 mM NaCl; 50 mM Tris-HCl, pH 7.4; 2 mM EDTA; 1% NP-40;
0.1% SDS), containing a protease inhibitor cocktail (cat. no.
ab65621; Abcam). Total protein was quantified using BCA (Beyotime
Institute of Biotechnology), and proteins (20 µg) were separated by
12% SDS-PAGE. The separated proteins were subsequently transferred
onto a PVDF membrane and blocked in 5% skim milk at room
temperature for 1 h. The membranes were then incubated at 4°C
overnight with the following primary antibodies: PAK6 (1:1,000;
cat. no. 13539-1-AP; ProteinTech Group Inc.), β-catenin (1:1,000;
cat. no. 51067-2-AP; ProteinTech Group Inc.), cyclinD1 (1:1,000;
cat. no. 60186-1-Ig; ProteinTech Group Inc.), E-cadherin (1:1,000;
cat. no. 20874-1-AP; ProteinTech Group Inc.) and GAPDH (1:10,000;
cat. no. 10494-1-AP; ProteinTech Group Inc.); GSK3β (1:1,000; cat.
no. AF7814; Affinity Biosciences), p-GSK3β (1:1,000; cat. no.
AF2016; Affinity Biosciences), p-β-catenin (1:2,000; cat. no.
DF2989; Affinity Biosciences). Following the primary antibody
incubation, the membranes were incubated with a horseradish
peroxidase-conjugated secondary antibodies (anti-mouse and
anti-rabbit; cat. nos. SA00001-1 and SA00001-2, respectively;
1:10,000; ProteinTech Group, Inc.) for 1 h at room temperature.
Bands were detected using enhanced chemiluminescence substrate
(Tanon Science and Technology Co., Ltd.). The expression levels
were semi-quantified using ImageJ 1.51K software (National
Institutes of Health).
Colony formation assay
A total of 200 stably transfected HeLa cells were
incubated in 5 ml DMEM (HyClone; Cytiva) supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml
streptomycin in a 6-cm dish at 37°C for 1 week. Once visible to the
naked eye, colonies were fixed with 4% paraformaldehyde for 30 min
at room temperature and stained with 0.1% crystal violet at room
temperature for 15 min. The colonies were then visualized using an
inverted microscope and counted. Data were analyzed using Image J
1.51 software.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8) assay (Beijing
Solarbio Science & Technology Co., Ltd.) was used to analyze
the cell proliferation, according to the manufacturer's protocol.
Briefly, 5×103 stably transfected HeLa cells/well were
seeded into 96-well plates. Following 0, 24, 48, 72 and 96 h of
incubation at 37°C, 10 µl CCK-8 solution was added/well at 37°C for
2 h. The absorbance of each well was measured at a wavelength of
450 nm to determine cell proliferation.
Cell migration and invasion
assays
The stably transfected HeLa cells (1×105)
were plated into a 24-well Transwell insert with DMEM containing
0.1% FBS (pore size, 8.0-µm), which was pre-coated with Matrigel
(Corning Inc.) at 37°C for 5 h for the invasion assay. A volume of
500 µl DMEM supplemented with 10% FBS was plated in the lower
chambers. Following incubation at 37°C for 48 h, the
non-migratory/invasive cells remaining on the top surface of the
membrane were removed by scraping, and the migratory/invasive cells
in the lower chamber were fixed with 4% paraformaldehyde at room
temperature for 30 min and stained with 0.1% crystal violet at room
temperature. The stained cells were counted in five randomly
selected visual fields using a light microscope (magnification,
×20).
Co-immunoprecipitation (Co-IP)
Cells were lysed in RIPA lysis buffer (Beyotime
Institute of Biotechnology), containing protease inhibitors (Roche
Diagnostics) for 30 min. Following centrifugation at 13,000 × g for
15 min 4°C, the 1/10 volume of supernatant was collected as input,
and half of the remaining supernatant was incubated with 20 µl/ml
protein A/G sepharose beads (Beyotime Institute of Biotechnology)
at 4°C for 1 h to remove non-specific hybrid proteins. Following
centrifugation (12,000 × g; 4°C; 5 min), the lysates were incubated
with 2 µg anti-PAK6 rabbit polyclonal antibody (cat. no.
13539-1-AP; ProteinTech Group, Inc.) or negative control rabbit IgG
(cat. no. A7016; Beyotime Institute of Biotechnology) at 4°C
overnight and then rotated at 4°C with a mixture of protein A/G
sepharose beads (20 µl/ml) for 4 h. The beads were then washed 3
times with RIPA buffer, and the bound proteins were boiled in 2X
Laemmli buffer and further analyzed using western blotting.
Fluorescence colocalization microscopy
analysis
The 293T cells transfected with PAK6/pDsRed-N1 and
GSK3β/pEGFP-N1 recombinants were cultured at 37°C for 48 h, and
then washed several times with PBS. Images of PAK6/pDsRed-N1 and
GSK3β/pEGFP-N1 positive cells were captured using Olympus
FV1000-IX81 microscope (FluoView 1000-IX81; Olympus
Corporation).
Statistical analysis
Data analysis was performed using GraphPad Prism 6.0
software (GraphPad Software, Inc.) and SPSS version 13 software
(SPSS, Inc.). The data are presented as the mean ± SD. A one-way
ANOVA was used to determine the statistical differences between the
groups presented in all figures. The data presented in Tables IV and V were analyzed using a Chi-squared test.
Each experiment was performed three times. P<0.05 was considered
to indicate a statistically significant difference.
 | Table IV.Expression levels of PAK6 in cervical
carcinoma and paracarcinoma tissues. |
Table IV.
Expression levels of PAK6 in cervical
carcinoma and paracarcinoma tissues.
|
|
| PAK6 expression
levels (n) |
|
|
|---|
|
|
|
|
|
|
|---|
| Histologic
type | Cases (n) | (−) | (+/++) | (+++) | Positive rate
(%) | P-value |
|---|
| Cervical
cancer | 46 | 8 | 27 | 11 | 82.61 | <0.01 |
| Paracarcinoma
tissue | 4 | 4 | 0 | 0 | 0.00 |
|
 | Table V.Association between PAK6 expression
levels and clinicopathological parameters in cervical cancer. |
Table V.
Association between PAK6 expression
levels and clinicopathological parameters in cervical cancer.
|
|
| PAK6 expression
levels (n) |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Cases (n) | (−) | (+/++) | (+++) | Positive rate
(%) | P-value |
|---|
| Age |
|
|
|
|
| >0.05 |
|
<45 | 21 | 3 | 13 | 5 | 85.71 |
|
|
≥45 | 25 | 5 | 14 | 6 | 80.00 |
|
| Pathological
type |
|
|
|
|
| >0.05 |
|
Adenocarcinoma | 2 | 1 | 1 | 0 | 50.00 |
|
|
Adenosquamous carcinoma | 4 | 1 | 2 | 1 | 75.00 |
|
|
Squamous cell carcinoma | 40 | 6 | 24 | 10 | 85.00 |
|
| International
federation of gynecology and obstetrics stage |
|
|
|
|
| >0.05 |
| I | 35 | 7 | 26 | 2 | 80.00 |
|
| II | 8 | 1 | 1 | 6 | 87.50 |
|
| III +
IV | 3 | 0 | 0 | 3 | 100.00 |
|
| Grade |
|
|
|
|
| <0.05 |
| 1 | 6 | 4 | 2 | 0 | 33.33 |
|
| 2 | 12 | 2 | 8 | 2 | 83.33 |
|
| 3 | 22 | 0 | 14 | 8 | 100.00 |
|
Results
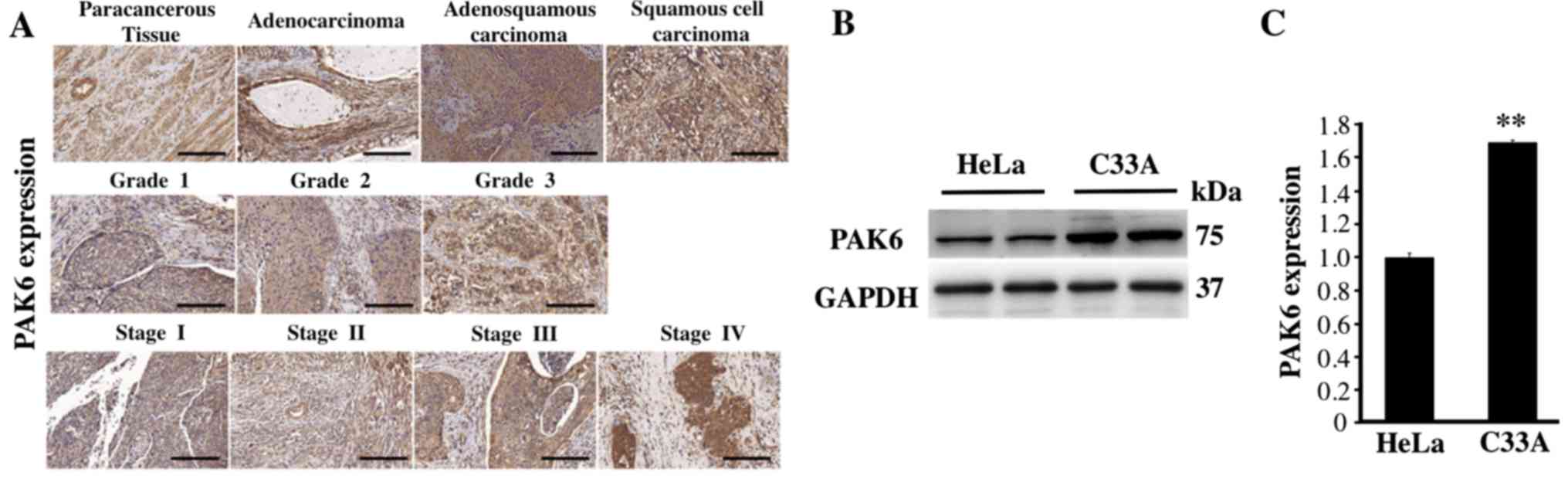
PAK6 expression levels are upregulated
in cervical cancer tissues and in C33A and HeLa cells
The expression levels of PAK6 were analyzed using
IHC and a commercial tissue microarray to investigate the function
of PAK6 in the development and progression of cervical carcinoma.
PAK6 staining was observed in both the cytoplasm and nuclei of the
positive cells in all of the different cervical tissue types, but
primarily in the cytoplasm (Fig.
1A). The positive rate of PAK6 expression in 46 cases of
cervical cancer was 82.61%, which was significantly increased
compared with the expression levels observed in the 4 paracancerous
tissues (P<0.01; Fig. 1A;
Table IV). The positive rates of
PAK6 in cervical adenocarcinoma, adenosquamous carcinoma and
squamous cell carcinoma were 50.00, 75.00 and 85.00%, respectively,
with no significant differences observed between the three groups
(P>0.05; Table V). In addition,
the positive rates of PAK6 expression in stages I, II and III + IV
of cervical cancer (according to the clinical classification of
FIGO staging) were 80.00, 87.50 and 100.00%, respectively. Finally,
the positive rates of PAK6 expression in grade 1, 2 and 3 of
cervical cancer were 33.33, 83.33 and 100.00%, respectively
(Fig. 1A; Table V). The expression levels of PAK6 were
also analyzed in both C33A and HeLa cells; significantly
upregulated expression levels of PAK6 were identified in the C33A
cells compared with the HeLa cells (P<0.01; Fig. 1B and C). These results suggested that
PAK6 may serve an important role in cervical cancer.
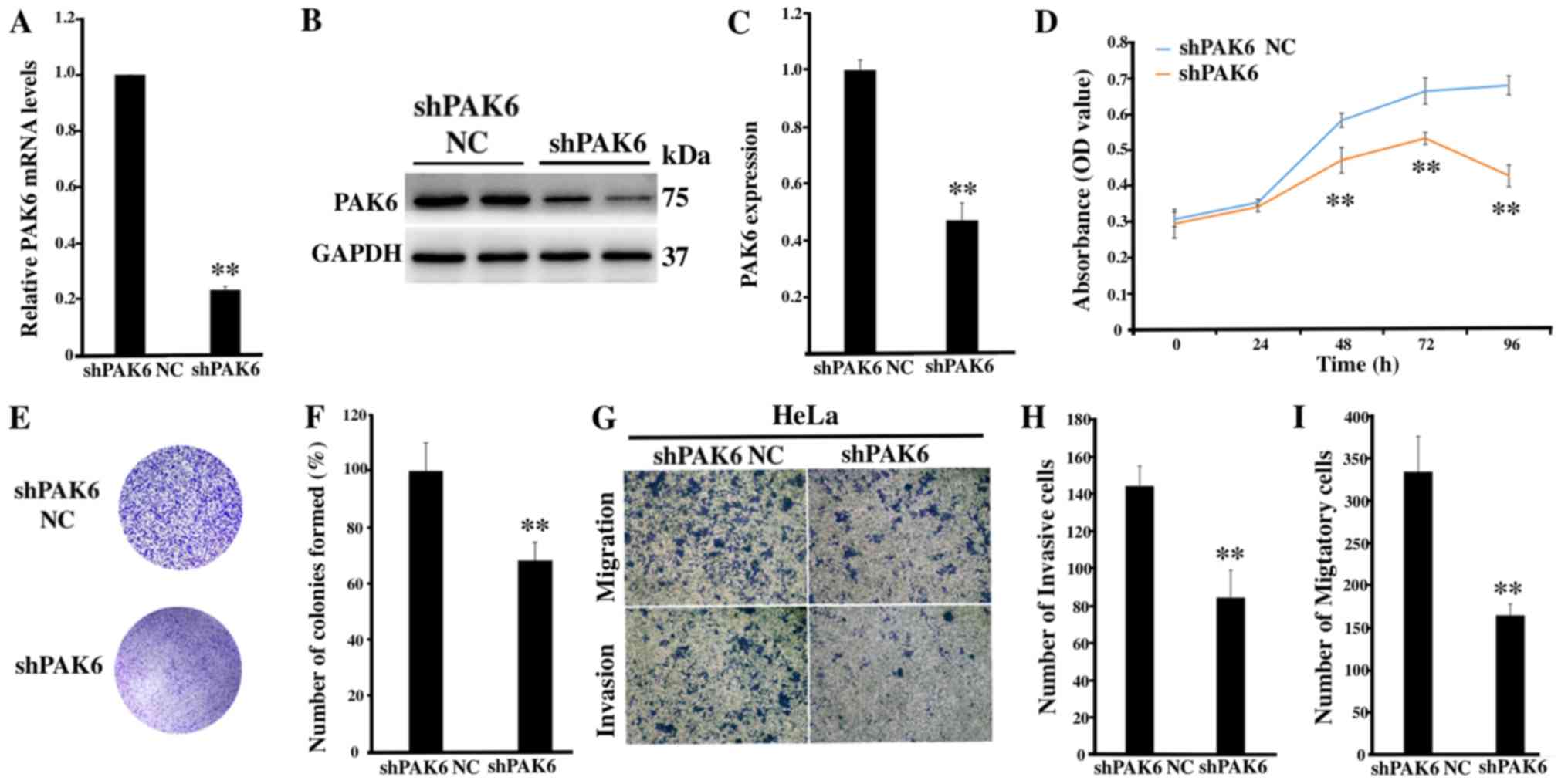
Inhibition of PAK6 attenuates the
proliferation, migration and invasion of HeLa cells
To determine the role of PAK6 in cervical
carcinogenesis, lentiviral-mediated RNA interference targeting PAK6
was used to investigate any biological changes in the HeLa cell
line. The transfection efficiency of shPAK6 in HeLa cells was
subsequently determined; the mRNA and protein expression levels of
PAK6 were significantly downregulated in the shPAK6-transfected
cells compared with the shPAK6 NC group (both P<0.01; Fig. 2A-C). Notably, the proliferation rate
(P<0.01; Fig. 2D) and the number
of cell colonies formed (P<0.01; Fig.
2E and F) were significantly reduced in the shPAK6-transfected
cells compared with the shPAK6 NC-transfected cells. In addition,
the invasive and migratory abilities of HeLa cells following PAK6
knockdown were investigated. The cells transfected with shPAK6 were
revealed to have a significantly reduced invasive and migratory
capacity compared with the cells transfected with the shPAK6 NC
(Fig. 2G-I). These results suggested
that inhibiting PAK6 expression levels may affect a number of
hallmarks of cancer, including the cell proliferative, migratory
and invasive abilities of HeLa cells.
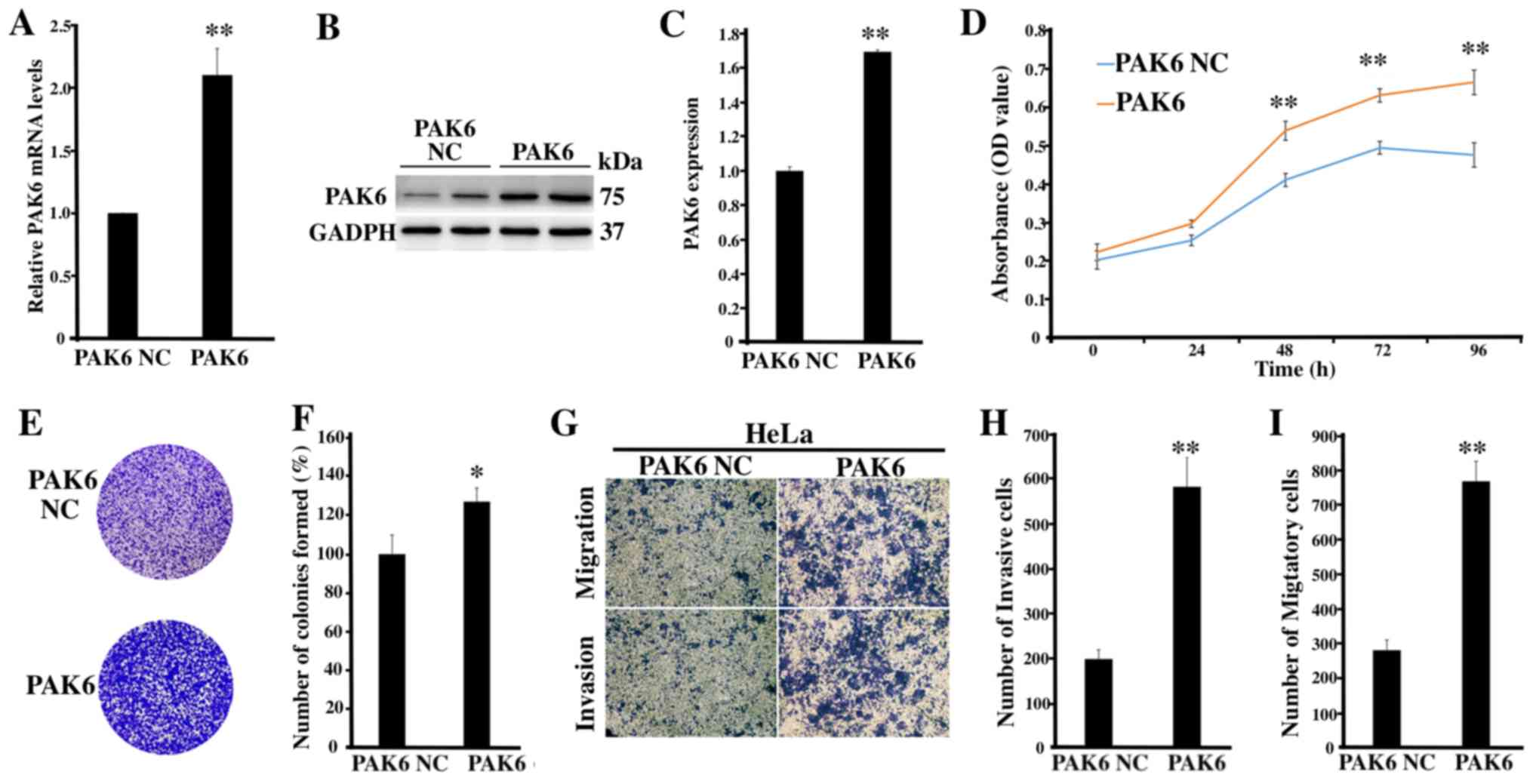
Overexpression of PAK6 promotes the
proliferation, migration and invasion of HeLa cells
To further determine the role of PAK6 in cervical
cancer, PAK6 was overexpressed to investigate the biological
changes of HeLa cells, of which the transfection was identified as
being successful (P<0.01; Fig.
3A-C). The proliferative rate (P<0.01; Fig. 3D) and the number of cell colonies
formed (P<0.05; Fig. 3E and F) in
the PAK6 overexpression group were significantly increased compared
with the PAK6 NC group. Moreover, according to the results of the
Transwell and Matrigel assays, the number of invasive and migratory
cells in the PAK6 overexpression group were significantly increased
compared with the PAK6 NC group (both P<0.01; Fig. 3G-I). Collectively, these findings
suggested that PAK6 overexpression may promote the cell
proliferative, migratory and invasive abilities of HeLa cells.
Effect of PAK6 knockdown or
overexpression on the Wnt/β-catenin signaling pathway in HeLa
cells
According to the western blotting results, the ratio
p-GSK3β/GSK3β and the downstream Cyclin D1 proteins were
significantly downregulated in shPAK6-transfected HeLa cells
compared with the shPAK6 NC-transfected cells (all P<0.05;
Fig. 4A-C). However, the ratio
p-β-catenin/β-catenin (P<0.01) and E-cadherin (P<0.05) were
significantly upregulated in PAK6 knockdown cells compared with the
shPAK6 NC group (Fig. 4A-C). In
contrast, the ratio p-GSK3β/GSK3β (P<0.01) and the expression of
Cyclin D1 (P<0.05) in stable PAK6 overexpressing HeLa cells were
all significantly upregulated (Fig.
4D-F), while the ratio p-β-catenin/β-catenin (P<0.01) and
E-cadherin levels (P<0.05) were significantly downregulated,
compared with the PAK6 NC group (Fig.
4D-F). Taken together, these findings suggested that PAK6 may
have a promoting role in the progression of cervical cancer by
activating the Wnt/β-catenin signaling pathway.
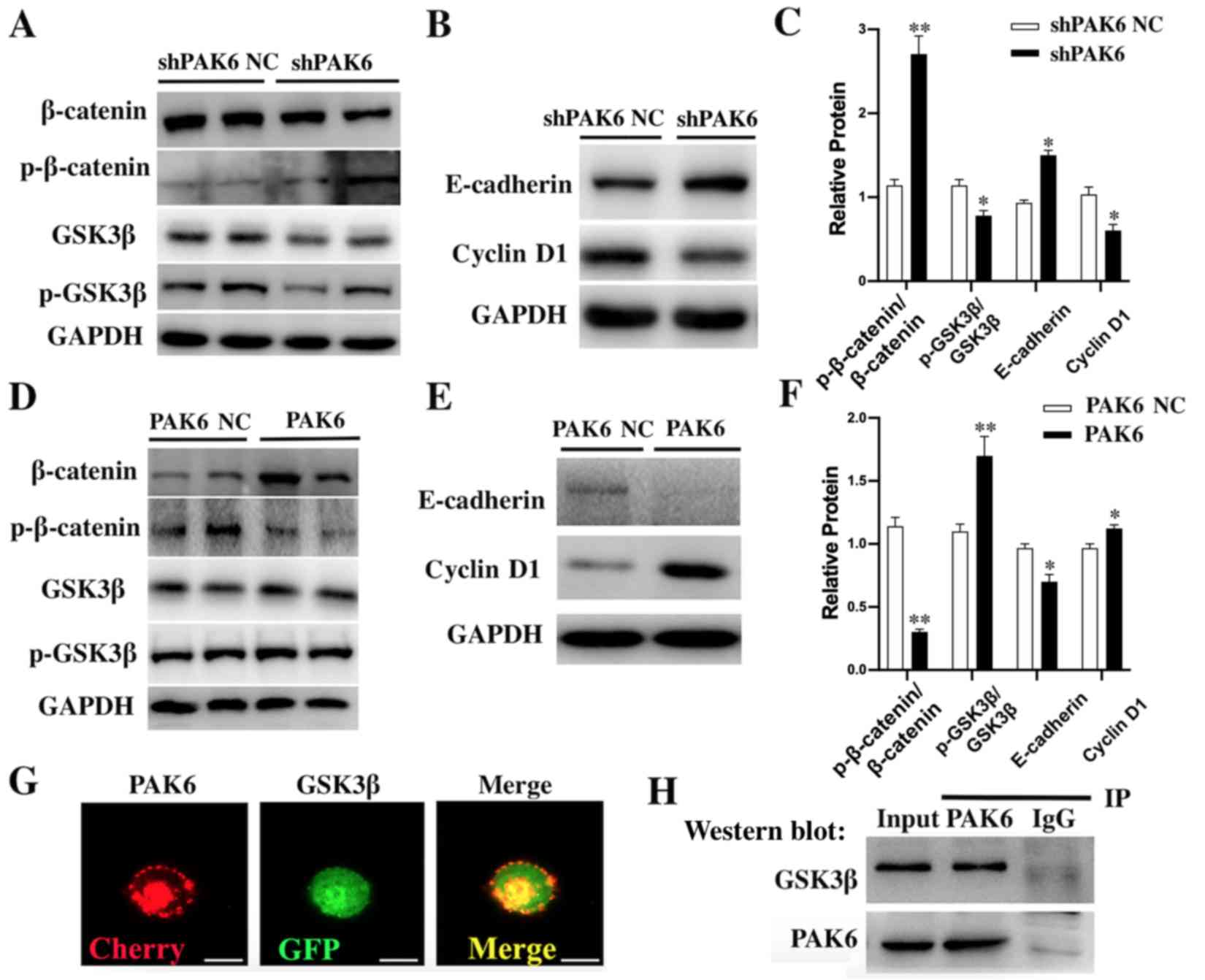 | Figure 4.Effect of PAK6 knockdown or
overexpression on the Wnt/β-catenin signaling pathway in HeLa
cells. Western blotting was used to analyze the expression levels
of (A) β-catenin, p-β-catenin, GSK3β and p-GSK3β, and (B)
E-cadherin and Cyclin D1 in stably shPAK6-transfected HeLa cells.
(C) Semi-quantification of the expression levels of proteins in
parts (A) and (B). *P<0.05, **P<0.01 vs. shPAK6 NC. Western
blotting was used to analyze the expression levels of (D)
β-catenin, p-β-catenin, GSK3β and p-GSK3β, and (E) E-cadherin and
cyclin D1 in stable PAK6 overexpressing HeLa cells. (F)
Semi-quantification of the expression levels of proteins in parts
(D) and (E). *P<0.05, **P<0.01 vs. PAK6 NC. (G)
Immunofluorescence was used to demonstrate the co-localization of
PAK6 and GSK3β. Scale bars, 10 µm. (H) Co-IP was used to analyze
the interaction between PAK6 and GSK3β. PAK6, p21-activated kinase
6; sh, short hairpin RNA; NC, negative control; p-, phosphorylated;
GSK3β, glycogen synthase kinase 3β; IP, immunoprecipitation. |
To investigate the mechanism underlying the effects
of PAK6 on the biological characteristics of cervical cancer cells,
the interaction between PAK6 and GSK3β was investigated using
fluorescence microscopy and Co-IP. PAK6 and GSK3β were observed to
be highly expressed in the cell membrane, cytoplasm and nucleus,
indicated by the red and green fluorescence, respectively (Fig. 4G). In the merged slice, the markedly
high levels of yellow fluorescence suggested that PAK6 and GSK3β
were co-localized in the cells. The results of the Co-IP further
confirmed their interaction; following PAK6 immunoprecipitation, a
positive band was detected after probing for the anti-GSK3β
antibody (Fig. 4H).
Discussion
Manser et al (20) first identified PAKs as molecules that
interacted with small Rho-like G proteins in 1994. PAKs have been
discovered to serve important roles in numerous cellular biological
processes, including cell cycle regulation, cell polarity,
cytoskeletal reorganization, gene transcription and translation
(21). More importantly, PAKs were
reported to be overexpressed in numerous types of human malignancy,
such as colon and breast cancers (22–24). The
present study discovered that the expression levels of PAK6 were
upregulated in cervical cancer tissues, and that the downregulation
of PAK6 expression levels by shRNA decreased cell growth and
proliferation, and inhibited the migratory and invasive abilities
of HeLa cells. Also, the expression levels of proteins related to
the Wnt/β-catenin signaling pathway, including β-catenin, p-GSK3β
and cyclin D1, were all downregulated following PAK6 knockdown. In
contrast, following the overexpression of PAK6, the expression
levels of β-catenin, p-GSK3β and cyclin D1 were all upregulated.
Further analysis by fluorescence microscopy and Co-IP suggested
that PAK6 may interact with GSK3β. Thus, these results indicated
that PAK6 may serve a role in promoting cervical cancer by
activating the Wnt/β-catenin signaling pathway.
Although the six PAKs in mammals are divided into
two groups, each member in Group 1 (PAK1-3) shares >90% homology
in its kinase domain, while Group 2 members (PAK4-6) share 50%
homology in the kinase domain, suggesting that there may be some
overlapping functions between PAKs (25). However, PAKs also present significant
differences in their tissue distribution and subcellular
localization, which may partly explain the organ-specific effects
of these molecules (26). A previous
study reported that transfected PAK6 was primarily localized in the
cytoplasm and on the plasma membrane in HeLa cells; however, the
presence of lower levels of nuclear PAK6 could not be ruled out,
indicating that PAK6 may be mostly cytoplasmic, but a small
fraction might be nuclear (27).
Therefore, specific antibodies should be used to evaluate the
cellular and tissue distribution of the endogenous PAK6 protein in
future studies. In addition, IHC analysis demonstrated that the
expression levels of PAK6 in cervical cancer tissues were not only
significantly upregulated compared with the paracancerous tissues,
but were also associated with the FIGO stage and degree of
malignancy of the cancers, suggesting that PAK6 expression level
may be increased at a higher grade and that PAK6 may be involved in
the occurrence and development of cervical cancer and may be a
potential therapeutic target.
Numerous studies have revealed that PAK4 and PAK5
serve crucial roles in cell processes involved in tumorigenesis and
tumor development, including anchorage-independent growth,
apoptosis and survival, migration and invasion. During mid-cell
division, spindle positioning and anchoring have been discovered to
require the involvement of PAK4 (28). In addition, PAK4 attenuated the
stability of the p57Kip2 protein through the ubiquitin-proteasome
pathway, resulting in the increased proliferation of breast cancer
cells (29). PAK4 suppression
downregulated the expression levels of cyclin A1, D1 and E1, and
upregulated the expression levels of p27 and p21, leading to G1
arrest in pancreatic cancer cells (30). Moreover, PAK4 knockdown reduced the
activation of several pro-survival pathways, including the NF-κB,
ERK and JNK signaling pathways (31,32).
Similar to PAK4, PAK5 knockdown delayed the G0/G1 phase of the
human gastric cancer, liver cancer and glioma cell cycle, thereby
inhibiting cell proliferation (33).
Furthermore, PAK5 was reported to promote the migration and
invasion of glioma and breast cancer cells via the PAK5/early
growth response protein1/matrix metalloproteinase 2 signaling
pathway (34). However, compared
with PAK4 and PAK5, the role of PAK6 in the development of cancer
remains unclear. To investigate the functional role of PAK6 in the
occurrence and development of cervical cancer, the present study
used knockdown and overexpression experiments. Following the
knockdown of the PAK6 gene in HeLa cells, the number of cell
colonies formed was significantly reduced compared with the control
group, in addition to the proliferation rate. The migratory and
invasive abilities were also significantly decreased. In contrast,
following the overexpression of PAK6 in HeLa cells, the number of
cell colonies formed were significantly increased compared with the
PAK6 NC group and the proliferation rate was increased. In
addition, the migratory and invasive abilities were significantly
increased. These results indicated that PAK6 may have the ability
to promote cell growth, proliferation, migration and invasion in
the onset and development of cervical cancer.
Previous studies have revealed that the
Wnt/β-catenin signaling pathway, which is essential for the
maintenance of cervical cancer cells and epithelial-mesenchymal
transition-associated stem cell-like features, was prominently
involved in the development and invasion of cervical cancer
(35,36). In the present study, the expression
levels of PAK6 protein were closely related to Wnt-related
proteins. PAK6 knockdown significantly downregulated the ratio
p-GSK3β/GSK3β and the downstream Cyclin D1 proteins in HeLa cells,
while the ratio p-β-catenin/β-catenin and E-cadherin were
significantly upregulated, indicating that the Wnt/β-catenin
signaling pathway may be inhibited. Moreover, following the
overexpression of PAK6, the ratio p-GSK3β/GSK3β and the expression
of Cyclin D1 were significantly upregulated in HeLa cells, while
the ratio p-β-catenin/β-catenin and E-cadherin levels were
downregulated, suggesting that the Wnt/β-catenin signaling pathway
may be activated.
Further investigations using immunofluorescence and
Co-IP identified that PAK6 may interact with GSK3β, which may be
the trigger that activates the Wnt/β-catenin signaling pathway.
GSK3β is considered to be an important molecule in the canonical
Wnt signaling pathway, negatively regulating the phosphorylation
and degradation of β-catenin (37).
In the present study, following the knockdown of PAK6, the
expression levels of p-GSK3β and β-catenin were discovered to be
significantly downregulated in HeLa cells. In contrast, the
overexpression of PAK6 resulted in the upregulated expression
levels of β-catenin and p-GSK3β in HeLa cells, activating the Wnt
signaling pathway in the cytoplasm. These results indicated that
GSK3β regulated β-catenin positively, instead of negatively, in the
Wnt signaling pathway in HeLa cells. A previous study reported that
GSK3β served a positive regulatory role in Wnt signaling, by
phosphorylating the LDL-receptor related protein 6, which is the
co-receptor of the Wnt protein (38). In the current study, the results of
Co-IP results revealed that PAK6 and GSK3β were expressed in the
cell membrane, cytoplasm and nucleus, and were co-localized in the
cell, providing a spatial basis for the interaction between the two
proteins. It is therefore hypothesized that PAK6 may serve as a Wnt
signal to interact with GSK3β on the cell or nuclear membrane in
HeLa cells, thereby affecting the entry and exit of β-catenin into
the nucleus, and positively regulating β-catenin in the Wnt
signaling pathway. However, the mechanism underlying the
interactions between PAK6 and GSK3β remain unclear and further
study is required.
In conclusion, the findings of the present study
indicated that PAK6 may promote the growth, proliferation,
migration and invasion of HeLa cells through the Wnt/β-catenin
signaling pathway. The functional and mechanistic investigations
suggested that the interactions between PAK6 and GSK3β, which may
positively regulate β-catenin, may be the trigger for the
activation of the Wnt/β-catenin signaling pathway in the
pathogenesis of cervical cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QY, GLL and GW designed the experiments; QY, YCZ,
YSC and AH performed the experiments; QY, GW, TX and YC analyzed
the data; and QY, GLL and GW prepared and revised the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Huazhong University of Science and Technology (Wuhan,
China) and all patients provided written, informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arbyn M, Weiderpass E, Bruni L, de Sanjosé
S, Saraiya M, Ferlay J and Bray F: Estimates of incidence and
mortality of cervical cancer in 2018: A worldwide analysis. Lancet
Glob Health. 8:e191–e203. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Whitham HK, Hawes SE, Chu H, Oakes JM,
Lifson AR, Kiviat NB, Sow PS, Gottlieb GS, Ba S, Sy MP and
Kulasingam SL: A comparison of the natural history of hpv infection
and cervical abnormalities among HIV-positive and HIV-negative
women in Senegal, Africa. Cancer Epidemiol Biomarkers Prev.
26:886–894. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gomih A, Smith JS, North KE, Hudgens MG,
Brewster WR, Huang Z, Skaar D, Valea F, Bentley RC, Vidal AC, et
al: DNA methylation of imprinted gene control regions in the
regression of low-grade cervical lesions. Int J Cancer.
143:552–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu X, Busby J, John C, Wei J, Yuan X and
Lu ML: Direct interaction between AR and PAK6 in
androgen-stimulated PAK6 activation. PLoS One. 8:e773672013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang K, Baldwin GS, Nikfarjam M and He H:
p21-activated kinase signalling in pancreatic cancer: New insights
into tumour biology and immune modulation. World J Gastroenterol.
24:3709–3723. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chow HY, Dong B, Valencia CA, Zeng CT,
Koch JN, Prudnikova TY and Chernoff J: Group I Paks are essential
for epithelial-mesenchymal transition in an Apc-driven model of
colorectal cancer. Nat Commun. 9:34732018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu H, Liu S, Zhang G, Bin Wu, Zhu Y,
Frederick DT, Hu Y, Zhong W, Randell S, Sadek N, et al: PAK
signalling drives acquired drug resistance to MAPK inhibitors in
BRAF-mutant melanomas. Nature. 550:133–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han K, Zhou Y, Tseng KF, Hu H, Li K, Wang
Y, Gan Z, Lin S, Sun Y and Min D: PAK5 overexpression is associated
with lung metastasis in osteosarcoma. Oncol Lett. 15:2202–2210.
2018.PubMed/NCBI
|
|
9
|
Siekmann IK, Dierck K, Prall S, Klokow M,
Strauss J, Buhs S, Wrzeszcz A, Bockmayr M, Beck F, Trochimiuk M, et
al: Combined inhibition of receptor tyrosine and p21-activated
kinases as a therapeutic strategy in childhood ALL. Blood Adv.
2:2554–2567. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sabra H, Brunner M, Mandati V,
Wehrle-Haller B, Lallemand D, Ribba AS, Chevalier G, Guardiola P,
Block MR and Bouvard D: β1 integrin-dependent Rac/group I PAK
signaling mediates YAP activation of Yes-associated protein 1
(YAP1) via NF2/merlin. J Biol Chem. 292:19179–19197. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Raghavan S, Venkatraman G and Rayala SK:
Cloning and functional characterization of human Pak1 promoter by
steroid hormones. Gene. 646:120–128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Si W, Liu X, He L, Ren J, Yang Z,
Yang J, Li W, Liu S, Pei F, et al: JMJD6 promotes melanoma
carcinogenesis through regulation of the alternative splicing of
PAK1, a key MAPK signaling component. Mol Cancer. 16:1752017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rane CK and Minden A: P21 activated kinase
signaling in cancer. Semin Cancer Biol. 54:40–49. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar R, Sanawar R, Li X and Li F:
Structure, biochemistry, and biology of PAK kinases. Gene.
605:20–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang M, Siedow M, Saia G and Chakravarti
A: Inhibition of p21-activated kinase 6 (PAK6) increases
radiosensitivity of prostate cancer cells. Prostate. 70:807–816.
2010.PubMed/NCBI
|
|
16
|
Lee EJ, McClelland M, Wang YP, Long F,
Choi SH and Lee JH: Distinct DNA methylation profiles between
adenocarcinoma and squamous cell carcinoma of human uterine cervix.
Oncol Res. 18:401–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bhatla N, Aoki D, Sharma DN and
Sankaranarayanan R: Cancer of the cervix uteri. Int J Gynaecol
Obstet. 143 (Suppl 2):S2–S36. 2018. View Article : Google Scholar
|
|
18
|
Lee DW, Ryu HS, Jin MS, Lee KH, Suh KJ,
Youk J, Kim JY, Min A, Lee HB, Moon HG, et al: Immune recurrence
score using 7 immunoregulatory protein expressions can predict
recurrence in stage I–III breast cancer patients. Br J Cancer.
121:230–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Manser E, Leung T, Salihuddin H, Zhao ZS
and Lim L: A brain serine/threonine protein kinase activated by
Cdc42 and Rac1. Nature. 367:40–46. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao ZS and Manser E: PAK family kinases:
Physiological roles and regulation. Cell Logist. 2:59–68. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu G, Li X, Guo B, Ke Q, Dong M and Li F:
PAK5-mediated E47 phosphorylation promotes epithelial-mesenchymal
transition and metastasis of colon cancer. Oncogene. 35:1943–1954.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramos-Alvarez I and Jensen RT:
P21-activated kinase 4 in pancreatic acinar cells is activated by
numerous gastrointestinal hormones/neurotransmitters and growth
factors by novel signaling, and its activation stimulates
secretory/growth cascades. Am J Physiol Gastrointest Liver Physiol.
315:G302–G317. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dou Q, Chen HN, Wang K, Yuan K, Lei Y, Li
K, Lan J, Chen Y, Huang Z, Xie N, et al: Ivermectin induces
cytostatic autophagy by blocking the PAK1/Akt axis in breast
cancer. Cancer Res. 76:4457–4469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aburatani T, Inokuchi M, Takagi Y,
Ishikawa T, Okuno K, Gokita K, Tomii C, Tanioka T, Murase H, Otsuki
S, et al: High expression of P21-activated kinase 5 protein is
associated with poor survival in gastric cancer. Oncol Lett.
14:404–410. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang F, Li X, Sharma M, Zarnegar M, Lim B
and Sun Z: Androgen receptor specifically interacts with a novel
p21-activated kinase, PAK6. J Biol Chem. 276:15345–15353. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee SR, Ramos SM, Ko A, Masiello D,
Swanson KD, Lu ML and Balk SP: AR and ER interaction with a
p21-activated kinase (PAK6). Mol Endocrinol. 16:85–99. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bompard G, Rabeharivelo G, Cau J, Abrieu
A, Delsert C and Morin N: P21-activated kinase 4 (PAK4) is required
for metaphase spindle positioning and anchoring. Oncogene.
32:910–919. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Wang D, Zhang H, Wang C, Dai W,
Cheng Z, Wang G and Li F: P21-activated kinase 4 regulates the
cyclin-dependent kinase inhibitor p57(kip2) in human breast cancer.
Anat Rec (Hoboken). 296:1561–1567. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tyagi N, Bhardwaj A, Singh AP, McClellan
S, Carter JE and Singh S: p-21 activated kinase 4 promotes
proliferation and survival of pancreatic cancer cells through AKT-
and ERK-dependent activation of NF-κB pathway. Oncotarget.
5:8778–8789. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He W, Zhao Z, Anees A, Li Y, Ashraf U,
Chen Z, Song Y, Chen H, Cao S and Ye J: p21-activated kinase 4
signaling promotes japanese encephalitis virus-mediated
inflammation in astrocytes. Front Cell Infect Microbiol. 7:2712017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu X, Feng J, Zeng D, Ding Y, Yu C and
Yang B: PAK4 confers cisplatin resistance in gastric cancer cells
via PI3K/Akt- and MEK/ERK-dependent pathways. Biosci Rep.
34:e000942014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang ZP, Jiang BG, Gu XF, Zhao B, Ge RL
and Zhang FB: P21-activated kinase 5 plays essential roles in the
proliferation and tumorigenicity of human hepatocellular carcinoma.
Acta Pharmacol Sin. 35:82–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Han ZX, Wang XX, Zhang SN, Wu JX, Qian HY,
Wen YY, Tian H, Pei DS and Zheng JN: Downregulation of PAK5
inhibits glioma cell migration and invasion potentially through the
PAK5-Egr1-MMP2 signaling pathway. Brain Tumor Pathol. 31:234–241.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun X and Liu Y: Activation of the
Wnt/β-catenin signaling pathway may contribute to cervical cancer
pathogenesis via upregulation of twist. Oncol Lett. 14:4841–4844.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang YH, Chu TY and Ding DC:
WNT/β-Catenin signaling pathway regulates non-tumorigenesis of
human embryonic stem cells co-cultured with human umbilical cord
mesenchymal stem cells. Sci Rep. 7:419132017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mao XW, Xiao JQ, Li ZY, Zheng YC and Zhang
N: Effects of microRNA-135a on the epithelial-mesenchymal
transition, migration and invasion of bladder cancer cells by
targeting GSK3β through the Wnt/β-catenin signaling pathway. Exp
Mol Med. 50:e4292018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taelman VF, Dobrowolski R, Plouhinec JL,
Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD and De Robertis
EM: Wnt signaling requires sequestration of glycogen synthase
kinase 3 inside multivesicular endosomes. Cell. 143:1136–1148.
2010. View Article : Google Scholar : PubMed/NCBI
|