Introduction
Gastric cancer (GC) is one of the most common types
of cancer and is responsible for an estimated 783,000 deaths and
over 1,000,000 new cases in 2018 worldwide, making it the fifth
most frequently diagnosed cancer and the third leading cause of
cancer-associated mortality (1).
Surgical resection remains the most promising intervention towards
localized GC tumors; however, many patients with advanced-stage GC
present with inoperable disease at diagnosis or develop recurrent
disease following resection (2).
Patients with GC exhibit extremely poor quality of life and poor
prognosis with a five-year survival rate <15% (3). The development of sensitive and
specific biomarkers is therefore crucial and may improve current
management of GC, including early diagnosis, prognosis, recurrence
monitoring and treatment response evaluation.
Recent studies have focused on the function of
exosomes in local and systemic cell-to-cell communication in cancer
progression and on their applications in cancer screening and
diagnosis (3,4). Exosomes are lipid bilayer vesicles of
endocytic origin with a typical size of 30–100 nm, which are
detected in most body fluids, including serum, urine and saliva
(5). Exosomes are produced from
various cell types, and tumor cells secrete excessive amounts of
these vesicles compared with non-tumor normal cells (6). Furthermore, exosomes serve a pivotal
role in mediating signaling transduction between neighboring or
distant cells via delivering proteins, RNAs and DNAs (4). Comparing the contents of exosomes
derived from GC patients with the content of exosomes from healthy
individuals may provide a better understanding of the molecular
mechanisms of GC tumorigenesis and progression and may help the
identification of novel biomarkers for cancer development,
metastasis and prognosis.
Circular RNAs (circRNAs) are a type of non-coding
RNA (ncRNA) with a closed annular structure (7,8). As an
abundant cargo of exosomes, circRNAs can serve as biomarkers due to
the following advantages: i) circRNAs are not easily degraded by
RNase R due to their circular structure and may exist stably in
exosomes; ii) circRNAs are highly conserved and tissue-,
developmental stage- and tumor type-specific; and iii) some
circRNAs are involved in tumorigenesis and associated with
treatment response or patients' survival (9). A study of circRNAs expression in GC and
paracancerous tissues identified 467 significantly differentially
expressed circRNAs (DE circRNAs), including 214 upregulated and 253
downregulated ones (10). However,
the expression profile of circRNAs in circulating exosomes from
patients with GC remains unknown. In the present study, the RNA
sequencing (RNA-seq) technique was used to detect exosomal circRNAs
in the plasma from patients with GC and healthy donors and screen
DE circRNAs. The potential roles of these DE circRNAs and their
target miRNAs were predicted using bioinformatics methods.
Materials and methods
Patients and sample collection
In the present study, 5 patients with GC were
recruited from the First Hospital of Jilin University between
December 2018 and February 2019. All patients were pathologically
diagnosed with gastric adenocarcinoma and were free of any other
types of cancer. Patients who had received any type of anticancer
treatment were excluded from the present study. In addition, 5
healthy donors with no history of cancer were enrolled as healthy
controls (HC). The clinicopathological characteristics of patients
and volunteers are presented in Table
I. In total, 5 ml of peripheral blood samples were collected
intravenously from each individual in BD Vacutainer®
Venous Blood Collection Tubes containing EDTA, and plasma was
isolated and frozen immediately at −80°C until use. This study was
approved by the local independent Ethics Committee and the
Institutional Review Board of the First Hospital of Jilin
University (Changchun, China), and informed consent was obtained
from all patients and volunteers. Blood collection and experiments
were performed in accordance with the Declaration of Helsinki and
relevant guidelines and regulations.
 | Table I.Clinicopathological characteristics
of patients with GC and healthy volunteers. |
Table I.
Clinicopathological characteristics
of patients with GC and healthy volunteers.
| Sample ID | Age, years | Sex | Histological
type | Histological
grade | TNM stage |
|---|
| GC |
|
|
|
|
|
|
GC190121 | 71 | Male | Adenocarcinoma | Poorly | T4aN3aM0 IIIB |
|
GC190122 | 58 | Male | Adenocarcinoma | Poorly | T2N1M0 IIA |
|
GC190124 | 63 | Male | Adenocarcinoma | Poorly | T3N2MO IIIA |
|
GC190125-2 | 64 | Male | Adenocarcinoma | Poorly | T4aN3aM0 IIIB |
|
GC190128-2 | 65 | Male | Adenocarcinoma | Poorly | T3N3aMO IIIB |
| HC |
|
|
|
|
|
|
HC190128-1 | 33 | Male |
|
|
|
|
HC190128-2 | 43 | Male |
|
|
|
|
HC190128-3 | 29 | Male |
|
|
|
|
HC190128-4 | 26 | Male |
|
|
|
|
HC190128-5 | 37 | Male |
|
|
|
Exosome isolation and
identification
Exosomes were isolated from 1 ml of plasma samples
pre-filtered by passing through a 0.8 µm filter using an exoEasy
Maxi kit (Qiagen, Inc.) according to the manufacturers'
instructions and frozen immediately at −80°C until use.
Exosomes were visualized by transmission electron
microscope (TEM) as previously described (11). Briefly, following loading onto
carbon-coated formvar grids, and staining with 2% uranyl acetate
for 10 min at room temperature and lead citrate for 5 min at room
temperature, exosomes were visualized using TEM (FEI TF20, Thermo
Fisher Scientific, Inc.) with a magnification of ×75,000.
Western blotting was used to detect the typical
exosomal protein markers, CD9 and TSG101 (12). Exosomes were lysed on ice in
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology,) supplemented with 1% protease inhibitor cocktail
(Cell Signaling Technology). Following protein quantification using
a BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.),
30 µg exosomal proteins were separated by 12% SDS-PAGE and
transferred onto a polyvinylidene fluoride membrane. Membranes were
blocked with TBS-5% BSA at room temperature for 2 h and incubated
with the primary antibodies rabbit anti-CD9 (cat. no. D8O1A;
1:1,000; Cell Signaling Technology, Inc.), rabbit anti-tumor
susceptibility gene 101 (TSG101; cat. no. ab125011; 1:1,000; Abcam)
and rabbit anti-calnexin (cat. no. C5C9; 1:1,000, Cell Signaling
Technology, Inc.) at 4°C overnight. HRP-Goat Anti-Rabbit IgG (cat.
no. 111-035-003; Jackson ImmunoResearch Laboratories, Inc.) was
used as the secondary antibody at 1:2,000 dilution and incubated
for 2 h at room temperature. A EasyBlot ECL kit (cat. no.
D601039-0050; Sangon Biotech Co., Ltd.) was used to visualize the
bands. CD9 and TSG101 are two of the most frequently identified
markers in exosomes, whereas calnexin is an endoplasmic reticulum
protein that is used as a negative control.
RNA isolation and quality control
Total RNA was extracted from exosomes using TRIzol
reagent (Life Technologies; Thermo Fisher Scientific, Inc.). The
quality control and quantification of the RNA samples were
performed using the NanoDrop ND 1000 spectrophotometer (Thermo
Fisher Scientific, Inc.). RNA integrity and DNA contamination were
assessed by electrophoresis on a 1% denaturing agarose gel. Only
DNA-free RNA samples with OD260/OD280 ratio
between 1.8 and 2.1 and RNA integrity number (RIN) ≥7 were used for
further experiments.
RNA library construction and circRNA
sequencing
High-throughput RNA-seq was performed by Cloud-Seq
Biotech, Inc. Briefly, the NEBNext® rRNA Depletion Kit
(New England BioLabs, Inc.) was used to remove ribosomal RNAs
(rRNAs) from total exosomal RNAs according to the
manufacturers' instructions. Subsequently, RNA libraries were
constructed from rRNA-depleted RNAs using the NEB
Next®Ultra™ II Directional RNA Library Prep Kit (New
England BioLabs, Inc.) according to the manufacturers' protocol.
The quality and quantity of the RNA libraries were evaluated using
the BioAnalyzer 2100 system (Agilent Technologies, Inc.). These
libraries were then sequenced on an Illumina HiSeq 4000 sequencer
(Illumina, Inc.) and 150 bp paired-end reads were generated.
Quality score of 30 (Q30) was used to determine sequencing quality.
Furthermore, cutadapt software (v1.9.2) (13) was used to trim 3′ adaptor sequences
and to remove low-quality reads to acquire clean reads, which were
then aligned to the reference genome/transcriptome via the Bowtie2
software (v2.2.4) (14). In
addition, circRNAs were identified with the find_circ software
(v1.2) (15). Finally, raw junction
reads for each sample were normalized to the number of total reads
and the expression levels of circRNAs were transformed into logCPM
values (log2 counts per million; normalized for library sizes).
Identification of DE circRNAs
Differential expression of circRNAs was calculated
using the edgeR package of the R software (v3.3.2). DE circRNAs
were identified through fold-change (FC ≥2.0; i.e.
|log2FC| >1) between GC cases and controls, and
t-tests (P<0.05). DE circRNAs with log2FC <0 and
log2FC >0 were considered as down- and upregulated
genes, respectively. Volcano plot visualization based on false
discovery rate (FDR)-adjusted P-values was performed using ggplot2
in R software (v3.5.1). Subsequently, a hierarchical heatmap was
constructed to reveal circRNAs expression pattern among samples via
the heatmap.2 function of the gplots package in R software
(v3.5.1).
Gene ontology (GO) and Kyoto
Encyclopedia of Gene and Genome (KEGG) pathway enrichment
analyses
DE circRNAs were submitted to The Database for
Annotation, Visualization and Integrated Discovery (DAVID:
http://david.abcc.ncifcrf.gov) to unveil
their potential roles (16) via GO
(17) and KEGG (18) enrichment analysis. The P-value
(−log10) scores indicated the significance of GO terms
and KEGG pathway correlations. The top 10 enriched GO terms,
including molecular function (MF), cellular component (CC) and
biological process (BP), and the top 10 KEGG pathways of
upregulated and downregulated circRNAs were presented via
enrichment scores. Finally, the pathway relation network was
established using the Cytoscape software (v3.6.1) based on the
significantly enriched KEGG pathways.
Reverse transcription quantitative PCR
(RT-qPCR)
RT-qPCR was performed to verify the expression
levels of the selected DE circRNAs in plasma exosomes derived from
patients with GC and healthy controls. Total RNA was reverse
transcribed to cDNA using SuperScriptTM III Reverse Transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturers' protocol. Subsequently, the cDNA library of each
sample was subjected to RT-qPCR reactions on a QuantStudio 5
Real-Time PCR System (Thermo Fisher Scientific, Inc.) using a qPCR
SYBR Green master mix (CloudSeq Biotech, Inc.). Primer sequences of
the randomly selected circRNAs and GAPDH (internal control) are
listed in Table II (Invitrogen;
Thermo Fisher Scientific, Inc.). The circRNA expression levels were
normalized using GAPDH as the reference gene and calculated by
applying the 2−ΔΔCq method (19). Each circRNA sample was analyzed in
triplicate.
 | Table II.Randomly selected circRNAs for
RT-qPCR and primer sequences. |
Table II.
Randomly selected circRNAs for
RT-qPCR and primer sequences.
| No. | CircRNA ID | Primer name | Primer
sequence |
|---|
| 1 |
chr2:61468695-61484485– | 2-Forward |
GCATGGAACCAGAGGAAGAA |
|
|
| 2-Reverse |
CAGAGGCTGCTTATCCATGG |
| 2 |
chr4:39915231-39927553– | 10-Forward |
GCAATGATGGAGAAGAGCGA |
|
|
| 10-Reverse |
TGCAATCTTCCAATTCAAAGCAG |
| 3 |
chr17:35797839-35800763+ | 7-Forward |
TGCTGAACCTGAAACAAGCA |
|
|
| 7-Reverse |
CCAAAGCCACAGTCCATCAC |
| Internal
standard | GAPDH | IS-Forward |
GGCCTCCAAGGAGTAAGACC |
|
|
| IS-Reverse |
AGGGGAGATTCAGTGTGGTG |
Prediction of circRNA-miRNA-mRNA
interaction
CircRNA-miRNA interaction was predicted using the
MiRanda software (August 2010 release) (20). Subsequently, top 5 putative target
miRNAs and their binding sites on DE circRNAs were identified. In
addition, the downstream target mRNAs of miRNAs were predicted
using TargetScan (http://www.targetscan.org) (21). Finally, the potential functional DE
circRNA-miRNA-mRNA network was visualized with Cytoscape software
(v3.6.1) (22).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.). Student's t-test was used to evaluate the
differences in circRNAs expression levels between patients with GC
and healthy controls. RT-qPCR data were expressed as the means ±
standard deviation of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of isolated
exosomes
Exosomes were isolated from the plasma of patients
with GC and healthy donors. The morphology and size of the obtained
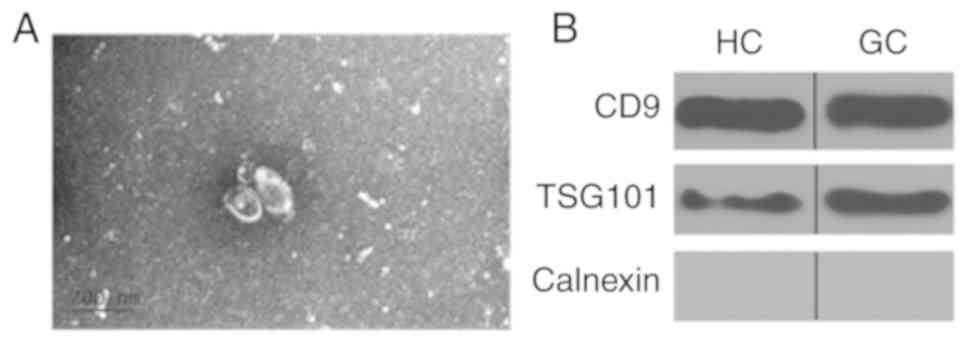
exosomes were identified using TEM. Exosomes ranged from 30 to 100
nm in size and exhibited a cup-shaped morphology with a clearly
defined and relatively intact membrane (Fig. 1A). The exosomal proteins CD9 and the
endosomal sorting complexes required for transport TSG101 were
detected in all samples by western blotting. These results
confirmed the exosomal characteristics of these extravesicles. In
addition, the endoplasmic reticulum marker calnexin was
undetectable in these exosomes, confirming the purity of the
isolation (Fig. 1B).
DE circRNAs in plasma exosomes from
patients with GC
The profiling of circRNAs expression in the
circulating exosomes derived from patients with GC and healthy
controls was performed using high-throughput RNA-seq. A total of
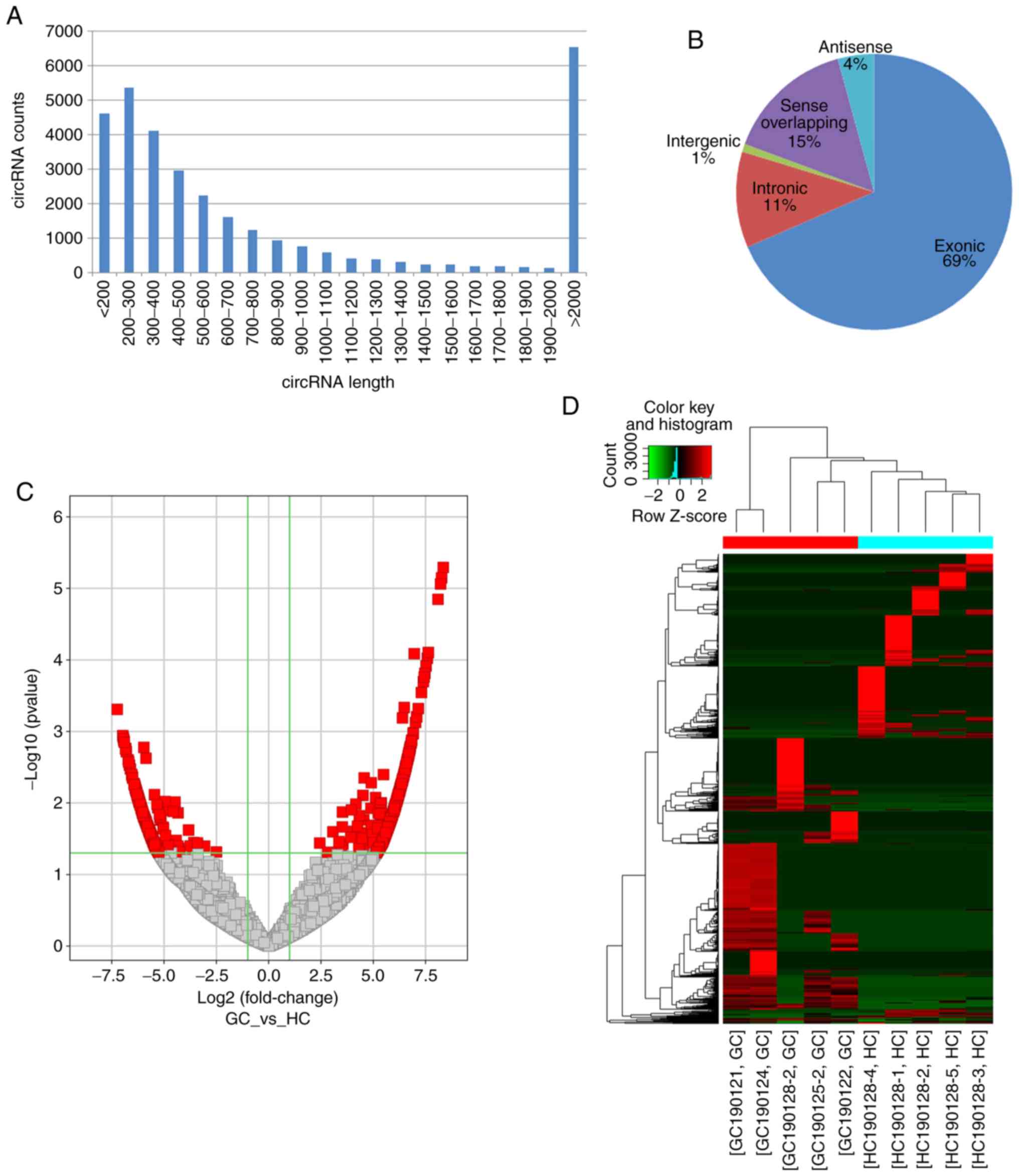
732,993,440 raw and 699,178,882 clean reads were obtained, and
67,880 circRNAs were identified in all samples. The number of
detected reads and circRNAs in each sample is listed in Table III. The predicted length of
circRNAs ranged between 53 and 99,588 nt with an average size of
4,176 nt. The length distribution of the circRNAs is presented in
Fig. 2A. According to their
association with protein-coding genes, circRNAs were classified
into the following five categories: 22,656 circRNAs were exonic;
3,728 circRNAs were intronic; 306 circRNAs were intergenic; 1,406
circRNAs were antisense; and 4,997 were sense overlapping (Fig. 2B). The raw read counts were
normalized and DE circRNAs were filtered using edgeR.
Subsrequently, 1,060 significantly DE circRNAs with FC≥2.0 and
P<0.05 were identified (Fig. 2C),
including 620 upregulated (Table
SI) and 440 downregulated ones (Table SII). The top 5 up- and downregulated
circRNAs in patients with are listed in Table IV. Eventually, the heatmap of
circRNA expression profiles in all samples revealed a remarkably
distinguishable exosomal circRNAs expression pattern in GC cases
compared with HC (Fig. 2D).
 | Table III.Number of detected reads and circRNAs
in each sample. |
Table III.
Number of detected reads and circRNAs
in each sample.
| Sample | Raw reads | Q30a (%) | Clean reads | Clean ratio
(%) | CircRNA number |
|---|
| GC190121 | 79,522,714 | 85.51 | 78,394,480 | 98.58 | 8,585 |
| GC190122 | 70,645,332 | 83.05 | 69,090,516 | 97.80 | 6,210 |
| GC190124 | 81,482,080 | 80.57 | 71,190,454 | 87.37 | 7,893 |
| GC190125-2 | 74,364,070 | 82.04 | 72,423,264 | 97.39 | 8,994 |
| GC190128-2 | 74,726,070 | 82.57 | 70,623,532 | 94.51 | 1,690 |
| HC190128-1 | 76,588,480 | 82.48 | 73,503,134 | 95.97 | 50,075,766 |
| HC190128-2 | 72,454,750 | 81.14 | 69,163,734 | 95.46 | 46,694,522 |
| HC190128-3 | 75,169,572 | 83.23 | 73,402,884 | 97.65 | 51,661,182 |
| HC190128-4 | 70,163,844 | 86.56 | 61,296,786 | 87.36 | 38,213,918 |
| HC190128-5 | 70,567,646 | 82.58 | 68,197,198 | 96.64 | 47,517,420 |
 | Table IV.The top 5 up- and downregulated
circRNAs in patients with gastric cancer. |
Table IV.
The top 5 up- and downregulated
circRNAs in patients with gastric cancer.
| CircRNAID | Regulation | logFC | logCPM | P-value | circBaseID | Source |
Best_transcript | Gene name |
Cataloga | Length |
|---|
|
chr10:27356086-27368090– | Up | 8.3486443 | 8.161152 | 5.099E-06 |
hsa_circ_0093425 | circBase | NM_014915 | ANKRD26 | Exonic | 467 |
|
chr2:153535866-153537850– | Up | 8.2675039 | 8.0871901 | 7.155E-06 |
| Novelb | NM_017892 | PRPF40A | Exonic | 232 |
|
chr1:235582788-235594119+ | Up | 8.2199474 | 8.043562 | 8.684E-06 |
hsa_circ_0007247 | circBase | NM_003193 | TBCE | Exonic | 289 |
|
chr9:72895675-72901187+ | Up | 8.0946435 | 7.9307965 | 1.425E-05 |
| Novelb | NM_015110 | SMC5 | Exonic | 375 |
|
chr13:25839957-25848325– | Up | 7.6344005 | 7.5242861 | 7.897E-05 |
hsa_circ_0029780 | circBase | NM_004685 | MTMR6 | Exonic | 567 |
|
chr6:106649847-106665476– | Down | −7.232753 | 7.1855505 | 0.0004927 |
| Novelb | NM_004849 | ATG5 | Sense
overlapping | 15630 |
|
chrX:17040246-17047746+ | Down | −6.970749 | 6.9710703 | 0.0011441 |
hsa_circ_0139983 | circBase | NM_004726 | REPS2 | Exonic | 374 |
|
chr13:36886282-36886614– | Down | −6.952648 | 6.9556143 | 0.0011978 |
hsa_circ_0001989 | circBase | NM_015087 | SPG20 | Exonic | 250 |
|
chr1:28337458-28343750– | Down | −6.934458 | 6.9415945 | 0.0012714 |
hsa_circ_0011115 | circBase | NM_001990 | EYA3 | Exonic | 410 |
|
chr10:103567487-103567658– | Down | −6.923018 | 6.932443 | 0.001315 |
hsa_circ_0019618 | circBase | NM_012215 | MGEA5 | Exonic | 172 |
Validation of DE circRNAs expression
by RT-qPCR
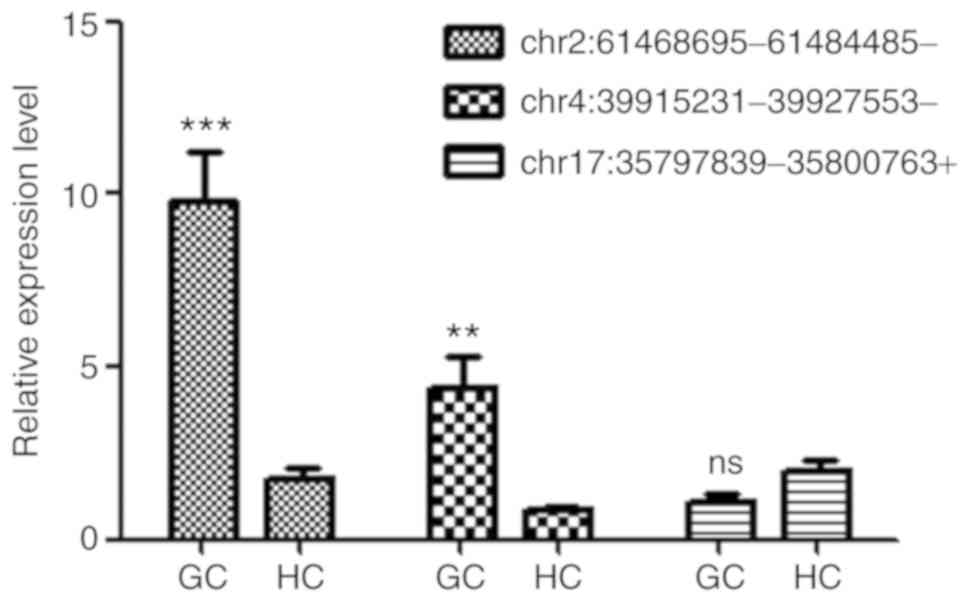
RT-qPCR was performed to verify the differential
expression of circRNAs. Among the high-throughput RNA-seq results,
two upregulated (chr2: 61468695-61484485- and chr4:
39915231-39927553-) and one unaffected circRNA (chr17:
35797839-35800763+) were randomly selected for validation. The
circRNA expression levels were normalized using GAPDH as the
internal control. The results demonstrated that the expression
levels of chr2: 61468695-61484485- and chr4: 39915231-39927553-
were significantly upregulated in samples from patients with GC
compared with HC. In addition, the expression of chr17:
35797839-35800763+ was comparable between the two groups (Fig. 3), suggesting that the observed
circRNA expression profiles were reliable.
GO and KEGG enrichment analyses of DE
circRNAs
To reveal the biological properties and functions of
DE circRNAs, GO and KEGG analyses were carried out. Furthermore,
DAVID website was used to visualize the DE circRNAs enrichment for
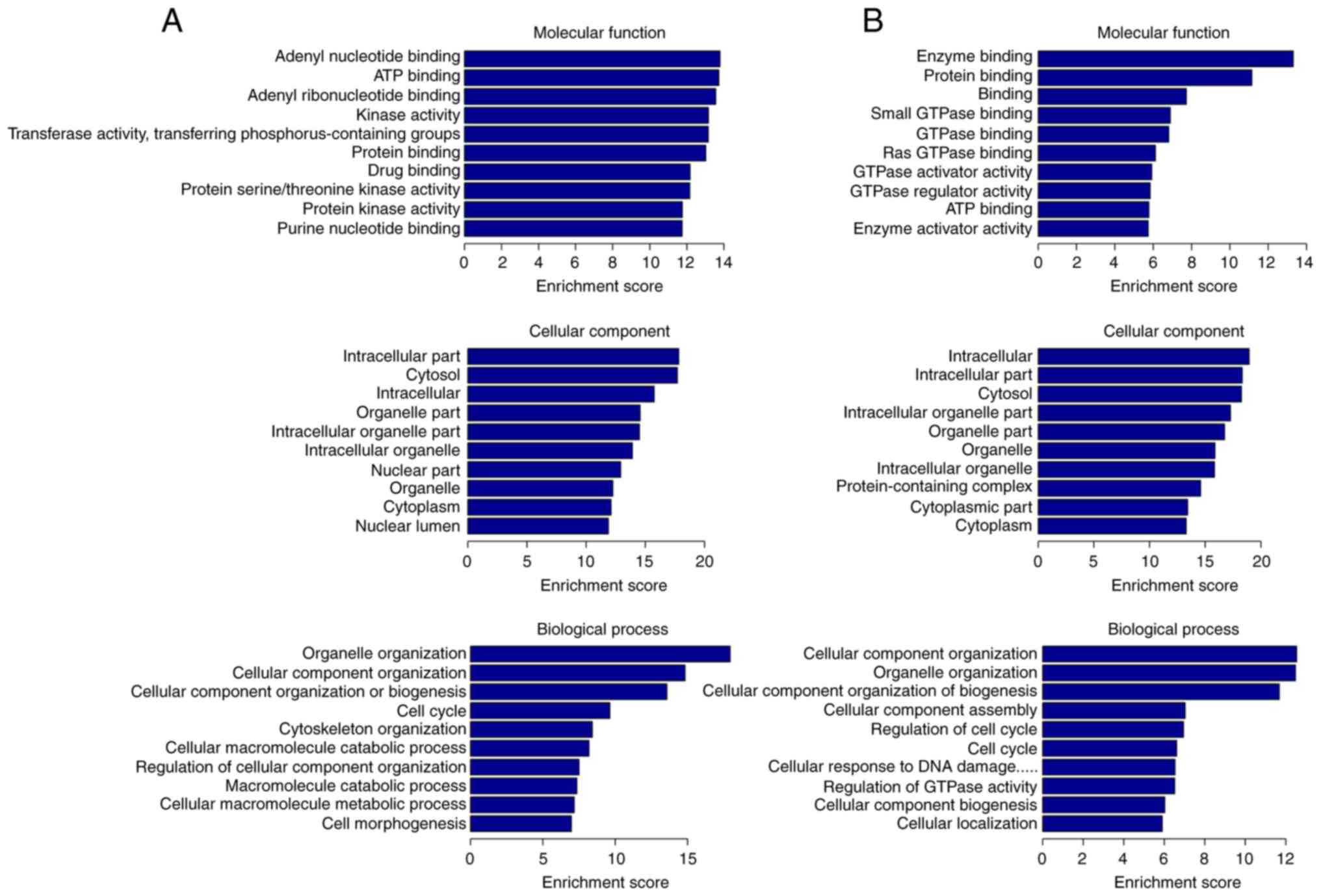
the associated GO terms and KEGG pathways. Regarding the MF,
upregulated DE circRNAs were associated with ‘purine nucleotide
binding’, especially with ‘adenyl nucleotide binding’, ‘ATP
binding’, ‘protein binding’, ‘drug binding’, ‘transferase activity,
transferring phosphorus-containing groups’ and ‘protein kinase
activity’, while the downregulated ones were associated with
‘enzyme binding’, especially ‘GTPase binding’, ‘protein binding’,
‘ATP binding’ and ‘GTPase regulator activity’. For GO CC,
upregulated DE circRNAs were significantly enriched in
‘intracellular part’, including ‘organelle’, ‘cytoplasm’ and
‘nucleus part’, whereas downregulated ones were also enriched in
‘intracellular part’, including ‘organelle’, ‘cytoplasm’ and
‘protein-containing complex’. For BP, upregulated DE circRNAs were
particularly involved in ‘cellular component organization or
biogenesis’, ‘cell cycle’, ‘cytoskeleton organization’, ‘cellular
macromolecule catabolic process’ and ‘cell morphogenesis’. Finally,
downregulated DE circRNAs participated in ‘cellular component
organization’, ‘cellular component organization or biogenesis’,
‘cellular component assembly’, ‘regulation of cell cycle’,
‘cellular response to DNA damage and regulation of GTPase activity’
(Fig. 4).
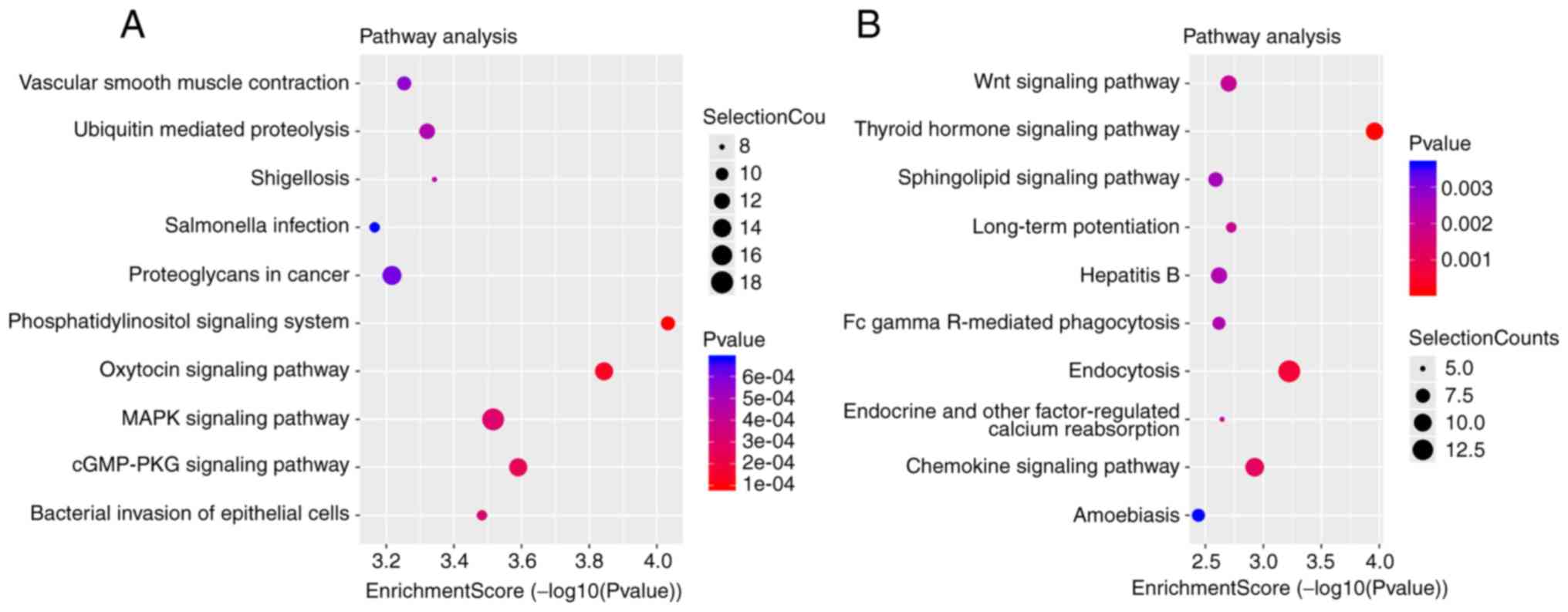
KEGG analysis displayed the involvement of these DE
circRNAs in different biological pathways. Top 10 significantly
enriched pathways for upregulated and downregulated circRNAs are
presented in Fig. 5A and B,
respectively. For upregulated circRNAs, the top 10 significantly
enriched pathways were ‘phosphatidylinositol signaling system’,
‘oxytocin signaling system’, ‘cGMP/PKG signaling system’ ‘MAPK
signaling pathway’, ‘bacterial invasion of epithelial cells’,
‘shigellosis’, ‘ubiquitin mediated proteolysis’, ‘vascular smooth
muscle contraction’, ‘proteoglycans in cancer’ and ‘salmonella
infection’ (Fig. 5A). Finally, for
the downregulated circRNAs, the top 10 pathways were ‘thyroid
hormone signaling system’, ‘chemokine signaling system’, ‘Wnt
signaling system’, ‘sphingolipid signaling pathway’, ‘endocytosis’,
‘long-term potentiation’, ‘endocrine and other factor-regulated
calcium reabsorption’, ‘Fc gamma R-mediated phagocytosis’,
‘hepatitis B’ and ‘amoebiasis’ (Fig.
5B).
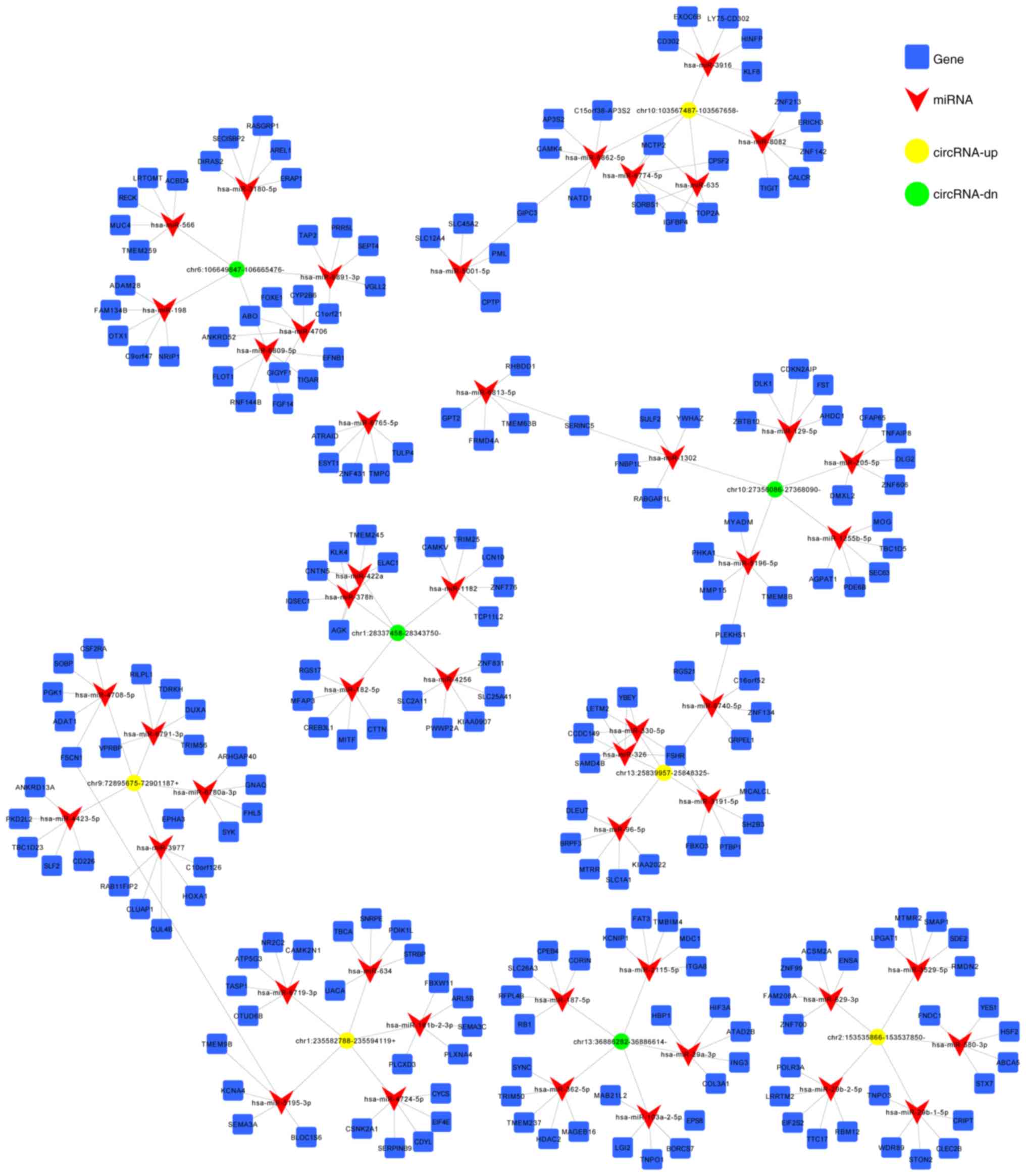
Construction of circRNA-miRNA-mRNA
network
CircRNAs affect tumorigenesis by regulating gene
expression via interaction with miRNAs and acting as an efficient
miRNA sponge (23). In addition,
miRNAs are involved in post-transcriptional gene silencing by base
pairing with mRNAs to mediate their degradation and/or
translational repression (24).
Therefore, the target miRNAs and their binding sites on DE circRNAs
were predicted using MiRanda software. The first 5 predicted miRNA
targets of each DE circRNA are presented in Table SIII. Furthermore, the top 5 up- and
downregulated circRNAs as listed in Table IV and were used to construct the
circRNA-miRNA network (Fig. 6).
Discussion
GC remains one of the most common and lethal types
of cancer worldwide. Despite the declining incidence rate and
advances in diagnosis, GC causes >700,000 deaths annually
(1). Clarifying the underlying
mechanisms of GC pathogenesis and developing sensitive and specific
biomarkers are therefore required to improve GC management.
Exosomes represent an important group of
extracellular vesicles, which have crucial roles in the initiation,
progression and metastasis of several types of cancer, including GC
(3). In addition, exosomes are
commonly found in various body fluids and may also exhibit a
potential role in early diagnosis and evaluation of therapeutic
effects of drugs in patients with GC (25). As a key class of exosomal cargos,
circRNAs mediate tumor and stromal communication and are considered
as emerging biomarkers for cancer diagnosis (5). In the present study, the expression
profiles of exosomal circRNAs were compared between GC patients and
HC using high-throughput RNA-seq. The findings from the present
study may contribute to the identification of novel biomarkers for
GC and may provide insights for future studies to investigate the
mechanisms of GC pathogenesis.
A total of 1,060 DE circRNAs between GC patients and
HC were identified, including 620 upregulated and 440 downregulated
ones. The selected circRNAs were further evaluated using RT-qPCR
and the results confirmed the reliability of the RNA-seq analysis.
Furthermore, GO and KEGG enrichment analyses were performed to gain
insights into the potential functions of theses DE circRNAs in GC
oncogenesis.
The results from GO analysis revealed that DE
circRNAs were mainly located in the intracellular cytoplasm and
organelle and were associated with several important biological
processes involved in tumorigenesis, including cell cycle,
cytoskeleton organization, cellular response to DNA damage and
regulation of GTPase activity. The members of small GTPase family,
including RhoA and RhoC, have been associated with the progression
of GC via regulation of cell proliferation, invasion and metastasis
of GC cells. They regulate the proliferation, invasion and
metastasis of GC and has potential clinical significance (26–28).
The majority of signaling pathways enriched in KEGG
analysis, including phosphatidylinositol (29–31),
MAPK (32), thyroid hormone
(33,34), chemokine (6,35,36) and
Wnt (37,38) signaling pathways, are also associated
with GC progression and chemotherapy resistance.
The phosphatase and tensin homolog
(PTEN)/phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt)
signaling pathway serves a critical role in several cellular
processes, including cell proliferation, apoptosis and invasion
(29). Furthermore, PTEN/PI3K/Akt is
the second most commonly altered pathway in human cancer following
p53 pathway, with a frequency of 30–60% among different tumor types
(30). It has been reported that GC
harbors one of the highest rates of oncogenic alterations in PI3K,
which confers a therapeutic potential to the PI3K pathway (31).
MAPK signaling pathway has been implicated in the
invasion and metastasis of GC cells (32), whereas Wnt signaling pathway serves a
key role in cell proliferation during both normal and cancerous gut
development (37,38). It has also been reported that both
pathways, as well as PI3K, are activated following Helicobacter
pylori infection, resulting in increased expression of
inflammatory cytokines and changes in apoptosis, proliferation and
differentiation. These changes ultimately lead to the
transformation of normal epithelial cells into oncogenic ones
(39).
Thyroid hormone signaling has been characterized as
a major effector of growth and homeostasis of the digestive system.
Based on this finding, several studies have demonstrated that the
occurrence of GC is associated with alterations in the protein
levels of thyroid hormone receptor TRα, autoimmune thyroid disease
and goiter, suggesting a potential role of thyroid hormone
signaling pathway in GC (33,34).
Several chemokines secreted by cancer cells and
tumor-associated stromal cells are involved in metastatic tumor
microenvironment (6). In addition,
the chemokine signaling pathway has been shown to be associated
with the survival, proliferation, angiogenesis and metastasis of
tumor cells. Therefore, therapeutic strategies blocking the
chemokine signaling pathway, including C-C chemokine ligand 5/C-C
chemokine receptor type 5 and C-X-C motif chemokine ligand 12/C-X-C
chemokine receptor type 4 axes, have been considered as effective
strategies in the treatment of GC (35,36).
CircRNAs may control GC progression by regulating
the expression of the key components of the aforementioned
signaling pathways. As suggested by a previous study that screened
GC tissues and adjacent tissues for differences in mRNAs and
circRNAs expression using Agilent microarray technology, DE
circRNAs had corresponding miRNA binding sites, and these circRNAs
regulated the expression of target genes through interactions with
miRNAs (10). Therefore the present
study identified the putative miRNA targets of DE circRNAs and
constructed the circRNA-miRNA-mRNA network to improve the general
understanding on the role of circRNAs in regulating the expression
of specific genes.
In summary, the present study identified a series of
DE circRNAs in exosomes isolated from the plasma of patients with
GC and HC by using high-throughput RNA-seq analysis. Furthermore,
the potential functions of DE circRNAs were predicted using
bioinformatics analysis. The results of the present study may
contribute to uncover the underlying mechanisms of GC oncogenesis
and help the development of targeted therapies and predictive
biomarkers for GC diagnosis.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This research was supported by the Research Fund of
the First Hospital of Jilin University (grant no. 20170142) and the
Natural Science Foundation of Science and Technology Department of
Jilin Province (20200201496JC).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PG and MR conceived and designed the experiments,
and wrote the article. MR, YZ and LQ performed the experiments. MR,
FH and PG analyzed the data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All participants in this study gave informed consent
according to the Helsinki Declaration, and all experimental
protocols were approved by the Ethics Committee of the First
Hospital of Jilin University (Changchun, China) (approval no.
2019-070).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang T, Liu C, Huang S, Ma Y, Fang J and
Chen Y: A down modulated MicroRNA profiling in patients with
gastric cancer. Gastroenterol Res Pract. 2017:15269812017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan Y, Fu G, Ye Y and Ming L: Exosomes
participate in the carcinogenesis and the malignant behavior of
gastric cancer. Scand J Gastroenterol. 52:499–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Liu Y, Sun W, Zhang Q, Gu T and Li
G: Plasma exosomes as novel biomarker for the early diagnosis of
gastric cancer. Cancer Biomark. 21:805–812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hou J, Jiang, Zhu L, Zhong S, Zhang H, Li
J, Zhou S, Yang S, He Y, Wang D, et al: Circular RNAs and exosomes
in cancer: A mysterious connection. Clin Transl Oncol.
20:1109–1116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kharaziha P, Ceder S, Li Q and Panaretakis
T: Tumor cell-derived exosomes: A message in a bottle. Biochim
Biophys Acta. 1826:103–111. 2012.PubMed/NCBI
|
|
7
|
Gao Y, Wang J, Zheng Y, Zhang J, Chen S
and Zhao F: Comprehensive identification of internal structure and
alternative splicing events in circular RNAs. Nat Commun.
7:120602016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y and Wang Z: Efficient back splicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia L, Song M, Sun M, Wang F and Yang C:
Circular RNAs as biomarkers for cancer. Adv Exp Med Biol.
1087:171–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J,
Liu H, Liu F and Dai L: Circular RNA and gene expression profiles
in gastric cancer based on microarray chip technology. Oncol Rep.
37:1804–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar D, Gupta D, Shankar S and Srivastava
RK: Biomolecular characterization of exosomes released from cancer
stem cells: Possible implications for biomarker and treatment of
cancer. Oncotarget. 6:3280–3291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J. 17:2011.
View Article : Google Scholar
|
|
14
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Yang F, Wei S and Xu G:
Identification of key genes affecting results of hyperthermia in
osteosarcoma based on integrative ChIP-Seq/TargetScan analysis. Med
Sci Monit. 23:2042–2048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vishnoi A and Rani S: MiRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boukouris S and Mathivanan S: Exosomes in
bodily fluids are a highly stable resource of disease biomarkers.
Proteomics Clin Appl. 9:358–367. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nam S, Kim JH and Lee DH: RHOA in gastric
cancer: Functional roles and therapeutic potential. Front Genet.
10:4382019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu N, Zhang G, Bi F, Pan Y, Xue Y, Shi Y,
Yao L, Zhao L, Zheng Y and Fan D: RhoC is essential for the
metastasis of gastric cancer. J Mol Med (Berl). 85:1149–1156. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan Y, Bi F, Liu N, Xue Y, Yao X, Zheng Y
and Fan D: Expression of seven main Rho family members in gastric
carcinoma. Biochem Biophys Res Commun. 315:686–691. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu M, Zhu S, Xiong S, Xue X and Zhou X:
MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review).
Oncol Rep. 41:1439–1454. 2019.PubMed/NCBI
|
|
30
|
Klempner SJ, Myers AP and Cantley LC: What
a tangled web we weave: Emerging resistance mechanisms to
inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov.
3:1345–1354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tran P, Nguyen C and Klempner SJ:
Targeting the phosphatidylinositol-3-kinase pathway in gastric
cancer: Can omics improve outcomes? Int Neurourol J. 20 (Supp
2):S131–S140. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang M and Huang CZ: Mitogen-activated
protein kinase signaling pathway and invasion and metastasis of
gastric cancer. World J Gastroenterol. 21:11673–11679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brown AR, Simmen RC and Simmen FA: The
role of thyroid hormone signaling in the prevention of digestive
system cancers. Int J Mol Sci. 14:16240–16257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kandemir EG, Yonem A and Narin Y: Gastric
carcinoma and thyroid status. J Int Med Res. 33:222–227. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aldinucci D and Casagrande N: Inhibition
of the CCL5/CCR5 axis against the progression of gastric cancer.
Int J Mol Sci. 19:14772018. View Article : Google Scholar
|
|
36
|
Xue LJ, Mao XB, Ren LL and Chu XY:
Inhibition of CXCL12/CXCR4 axis as a potential targeted therapy of
advanced gastric carcinoma. Cancer Med. 6:1424–1436. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ooi CH, Ivanova T, Wu J, Lee M, Tan IB,
Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al: Oncogenic
pathway combinations predict clinical prognosis in gastric cancer.
PLoS Genet. 5:e10006762009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Korourian A, Roudi R, Shariftabrizi A,
Kalantari E, Sotoodeh K and Madjd Z: Differential role of wnt
signaling and base excision repair pathways in gastric
adenocarcinoma aggressiveness. Clin Exp Med. 17:505–517. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yousefi B, Mohammadlou M, Abdollahi M,
Salek Farrokhi A, Karbalaei M, Keikha M, Kokhaei P, Valizadeh S,
Rezaiemanesh A, Arabkari V and Eslami M: Epigenetic changes in
gastric cancer induction by Helicobacter pylori. J Cell
Physiol. 234:21770–21784. 2019. View Article : Google Scholar : PubMed/NCBI
|