A balance among cell proliferation, differentiation
and migration is required for proper tissue development.
Epidemiological studies investigating the association between
potential risk factors and an increased risk of cancer have
indicated that a number of factors, including diet, obesity,
hormones, immunosuppression, cancer-causing substances, chronic
inflammation, infectious agents and radiation, may increase the
risk of imbalance (1–3). Furthermore, genetic and/or epigenetic
changes or loss of function mutations in tumorigenesis-associated
genes and signaling pathways may disrupt this balance, resulting in
tumor progression and metastasis (4). Previous studies investigated gene
mutations of the RAS, WNT, MYC, ERK and TRK genes, that may result
in tumor initiation and progression, and may assist in
understanding the underlying mechanisms involved (5–8).
However, little is known about the mechanism and exact cause of
>100 types of human cancer. Current knowledge has revealed that
only 5–10% of cancer cases have a genetic component, which
indicates that gene mutation is not the sole cause of cancer
development (9–14). Therefore, investigating the
mechanisms underlying the alteration of protein expression profiles
may aid cancer study.
Atonal bHLH transcription factor 1 (ATOH1), an
evolutionarily conserved human ortholog of the Drosophila
proneural basic helix-loop-helix (bHLH) transcription factor
atonal, is involved in a variety of developmental processes. ATOH1
was cloned and identified as a proneural transcription factor based
on its sequence, structure and functional features (15). ATOH1 serves an important role in the
specification and regulation of skin mechanosensory cells and in
the development of the auditory system in the inner ear (16,17).
Furthermore, ATOH1 is required to establish the intestinal
epithelium secretory cell lineage and for the development of
rhombic lip derivatives, including respiratory rhythmogenesis and
the cerebellar external granule cell precursor layer (15,18–20).
ATOH1 positively regulates cell type specification and
differentiation, controls cell cycle arrest and maintains granule
neuron progenitors depending on the developmental context.
Therefore, ATOH1 plays an important role in neural development and
may serve as a tumor suppressor or an oncogene (21–27).
Similar to other proneural genes, including
achaete-scute complex like 1 and neurogenin 2, mutations that alter
the function or result in loss of function of ATOH1 are generally
lethal (28). Therefore, unlike the
classic oncogenes or tumor suppressor genes, ATOH1 loss of function
mutations are rarely found in tumor tissue and the majority of
tumors tend to exhibit abnormal increased or decreased expression
of ATOH1 (21,22,26,27,29,30).
Previous studies assessing the expression profile of ATOH1 in
various tumor tissues revealed an alteration of ATOH1 mRNA and
protein levels in brain, colon, thyroid, prostate and lung cancer
(21,22,26,27,29,30).
Several studies demonstrated that such alterations positively or
negatively regulate tumor initiation or progression via
tissue-specific mechanisms.
It is essential to identify novel molecular
biomarkers for the clinical diagnosis and molecular targeting of
cancer for clinical treatment. Considering the complexity of the
tumorigenic progress, drug resistance, the specificity of clinical
treatments and side effects, further developments are required in
the field of cancer therapy. ATOH1 regulates the expression of
several target genes, including BarH like homeobox 1 and hes family
bHLH transcription factor 6, and influences several important
signaling pathways, such as the sonic hedgehog (SHH) and notch
pathways (31,32). Therefore, further investigation into
the effects of ATOH1 alteration on tumorigenesis is required. The
present review investigated the role of ATOH1 in cancer, with a
particular emphasis on medulloblastoma (MB) and gastrointestinal
cancer. Furthermore, the present review aimed to develop a clearer
understanding of how alterations in ATOH1 expression and activation
affect tumor initiation, progression and metastasis. Additionally,
potential drug treatments for cancer therapy are discussed.
ATOH1, also referred to as Hath1 in humans, Math1 in
mice and Cath1 in chickens, encodes a class II bHLH transcription
factor. The functional bHLH domain consists of a basic DNA-binding
region and protein-binding region with two α-helices linked by a
variable loop region. The protein-binding region is required for
the formation of a heterodimer with a class I member of the bHLH
family protein E47/E12. ATOH1 shares ~70% homology with atonal in
the bHLH domain. However, the rest of the sequence exhibits much
less similarity and the positioning of the bHLH domain varies among
species (33,34). In vertebrates, protein sequence
comparisons have revealed >80% similarity in the serine-rich
region of the C-terminal (35).
Additionally, the N-terminus of the open reading frame exhibits a
high similarity among mammals (35).
Studies on atonal and its orthologs have revealed that the non-bHLH
domain of the protein serves an important role; for example, the
conserved serine residues are involved in post-translational
modifications which affect protein function (15,36).
Domain sweeping experiments have demonstrated that specific motifs
and their combinations are important for proper protein function
(36,37). Over the last few decades, research
has focused on identifying the downstream targets of ATOH1/atonal.
The majority of the target gene candidates identified are involved
in transcriptional regulation, chromosomal organization and cell
cycle control and are associated with Wnt, SHH, notch, transforming
growth factor-β signaling and Janus kinase (JNK)/mitogen-activated
protein kinase (MAPK) signaling pathways (26,38–46).
In vertebrates, MCs are derived from neuroendocrine
cells and are located in the basal layer of the epidermis (47). Clustered MCs consist of a touch
sensitive zone, which is in nervated by slowly adapting type I
mechanoreceptor nerves (48). These
epidermis-derived cells are required for light touch responses
(48). ATOH1 is expressed in MCs
during development and in adults (49,50).
ATOH1 may be required for MC progenitor differentiation, but its
expression is also maintained throughout development and in mature
MCs (51). ATOH1 null mice exhibited
a loss of type I mechanoreceptor nerve response and lacked MCs
(52). ATOH1 expression in MCs is
regulated by the transcription factor SRY-box transcription factor
2 (SOX2). The expression of SOX2 in MCs is controlled by the
polycomb repressor complex, which exhibits histone
methyltransferase activity (53),
suggesting the involvement of epigenetic regulation in cell lineage
development. However, to the best of the authors' knowledge, the
expression of ATOH1 in lung tissue during development has not been
documented.
MCC is a rare malignant skin cancer derived from
epithelial and neuroendocrine cell differentiation that carries a
very poor prognosis (54).
Approximately 80% of MCC cases are polyomavirus-positive. However,
the pathogenesis involved in polyomavirus-positive and negative MCC
is yet to be fully elucidated (55–57).
Therefore, the association between polyomavirus infection and the
development of MMC remains unclear.
Loss-of-function ATOH1 mutations or epigenetic
silencing via promoter methylation have been detected in a small
number of MCC cases (26). However,
a recent study with a larger number of MCC cases did not determine
a correlation between ATOH1 expression and MCC malignancy or an
association between ATOH1 mutations and MCC development (58). Interestingly, the same study revealed
a significant correlation between downregulated protein expression
levels of ATOH1 and MCC recurrence or mortality (58). Further studies are required to
determine the signaling pathways that interact with ATOH1 to
control the differentiation of epidermal progenitors into MCs and
to identify novel therapeutic agents for MCC.
Previous studies investigating ATOH1 function during
development and ATOH1 expression profiles in cancer have
demonstrated the tissue- and context-specific functions of ATOH1 in
physiological and pathological conditions (21–23,26,59–62).
Several gene expression analyses have determined that ATOH1 is
expressed in small cell lung carcinoma (SCLC), a neuroendocrine
tumor (63–66). Additionally, <20% adenocarcinoma
samples express ATOH1 and these tumors exhibit neuroendocrine
features (63). Cytoplasmic and
nuclear expression of ATOH1 has been detected in certain lung
squamous cell carcinoma (SCC) samples (30). However, ATOH1 is not known to be
expressed in lung tissue and has not been reported to be involved
in normal lung development (63). A
correlation analysis revealed that the expression of ATOH1 was
inversely correlated with lung cancer growth (25). Another study revealed that the
ectopic activation of ATOH1 occurred in lung cancer, resulting in a
poor patient prognosis (63).
However, the underlying mechanisms remain unclear and ATOH1
expression in lung cancer cells is poorly understood. The pathways
linking ATOH1 and lung cancer pathogenesis have not been fully
elucidated. However, the association between ATOH1 expression and
neuroendocrine tumors as well as the poor prognosis of these tumors
may unravel the underlying mechanisms. Future studies investigating
how ATOH1 expression alters protein expression profiles in lung
cancer cells are warranted and may shed light on the mechanisms
maintaining the balance among cell proliferation, differentiation
and migration.
The mouse ato orthologues Math1/ATOH1 mRNA is
detected in the dorsal neural tube and cranial nerve ganglia at
E9.5 (15). During embryonic brain
development, ATOH1 is expressed in the dorsal hindbrain neuro
epithelium, rhombic lip and the developing cerebellum (15,67).
These ATOH1-dependent neurons are required for the generation of
dorsal commissural interneurons (68) and brainstem respiratory nuclei, and
the development of cerebellar granule cell lineages (18,69).
Unlike MCs, in which ATOH1 is expressed in both progenitor and
mature cells, ATOH1 expression persists during granule cell lineage
development and in cerebellar granule cell precursors, but
disappears during differentiation and migration from the external
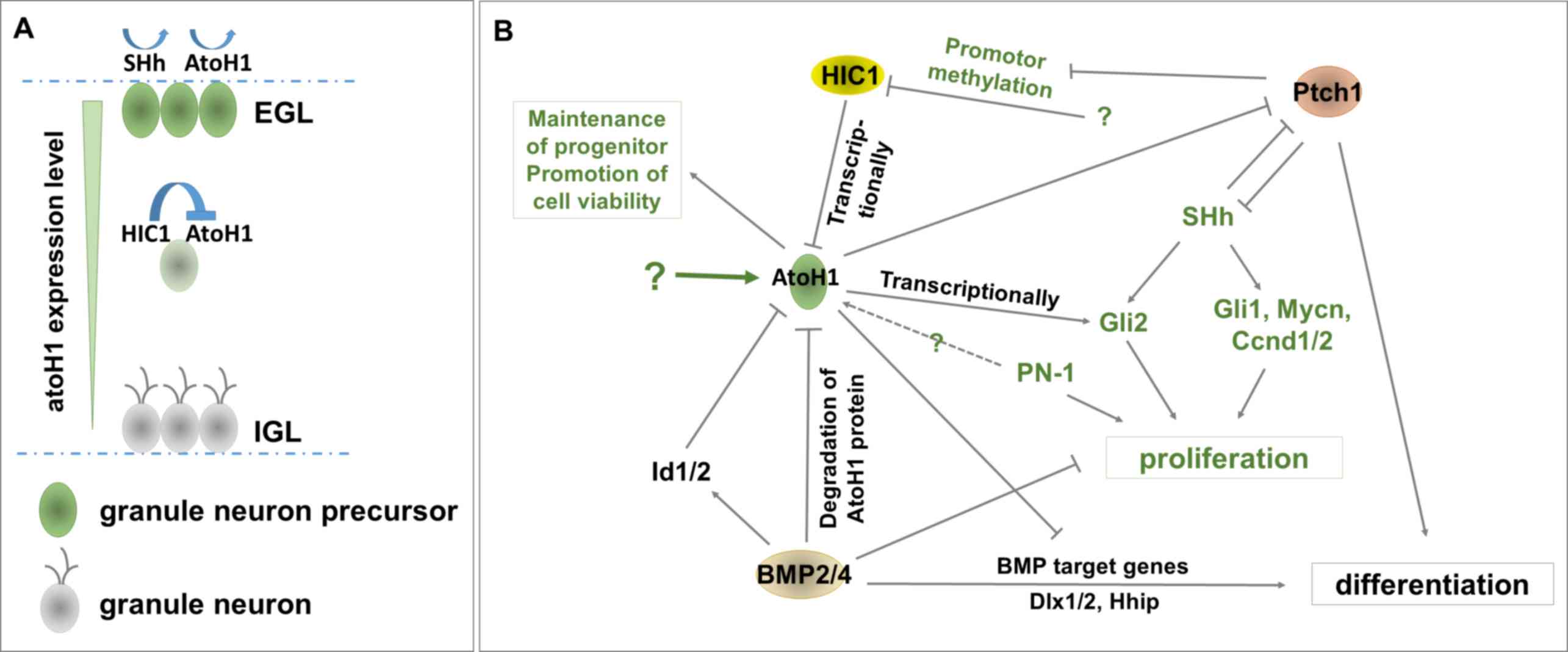
granule layer (EGL) to the internal granule layer (IGL; Fig. 1A) (18). ATOH1-null mice have a smaller
cerebellum compared with wild type or heterozygous mice, and lack
an EGL (18). The balance between
the protein activity of ATOH1 and signaling pathway activity of SHH
and Notch has been demonstrated to regulate granule cell
differentiation (40). These data
indicated that ATOH1 serves a crucial role in the development of
cerebellar granule cells.
Since ATOH1 is required for the regulation of
cerebellar granule neuron precursors during cerebellar development,
it is not surprising that ATOH1 exerts a crucial effect in the
malignant cerebellar tumor subgroup SHH-MB (70), which is closely associated with the
origin of granule cell progenitors (71,72). In
SHH-MB, ATOH1, as well as GLI family zinc finger (GLI) 1/2, MYCN
proto-oncogene, bHLH transcription factor (MYCN), cyclin D1/2
(CCND1/2) and protease nexin-1 (PN-1), are highly expressed when
compared with normal tissue (73,74) and
serve to control the proliferation of granule cell precursors
(21–23). However, the precise mechanism of
ATOH1 upregulation in MB is yet to be fully elucidated. Previous
studies revealed that ATOH1 was upregulated in cases of MB with
loss-of-function mutations of hypermethylated in cancer 1 (HIC1) (a
ZBTB transcriptional repressor) and super-activated SHH signaling
(22,59,75).
Furthermore, HIC1 is required for the transcriptional inhibition of
ATOH1, while cerebellar granule neuron precursors (GNPs) undergo
differentiation to granule neurons, migrating from the EGL to IGL
(Fig. 1A) (76). Another study revealed that the
phosphorylation of tyrosine 78 in ATOH1 was present only in human
colorectal cancer (CRC) and not normal tissues. This
phosphorylation served to both stabilize ATOH1 and increase its
transcriptional activity, hence promoting MB (77).
No evidence has demonstrated the direct activation
of ATOH1 by the SHH signaling pathway, although in the majority of
cases, the hyper activation of the SHH signaling pathway and high
levels of ATOH1 expression are observed in SHH-MB (23,59,78).
Additionally, the upregulation of ATOH1 alone does not initiate MB
(21), but an increased number of
MB-initiating cells were observed when both ATOH1 and Gli1 were
upregulated (21). Blocking ATOH1 in
GNPs limited the response of pre-proliferation genes to SHH
activity and accelerated differentiation (21). This indicates that the upregulation
of ATOH1 and the hyper activation of SHH may synergistically
interact with each in tumorigenesis. Furthermore, ATOH1 is required
for the maintenance of progenitor cells in MB progression rather
than MB initiation (21,59,79,80).
Gene expression data analysis has revealed a strong correlation
between ATOH1 gene expression and poor survival in patients with MB
(81). These data suggested that
ATOH1 is a tumor progression rather than a tumor initiation
oncogene in the SHH subgroup of MB.
Hyperactive SHH induces the expression of certain
downstream genes including GLI1/2, CCND1/2 and MYCN to drive the
proliferation of GNPs (82–84). In addition, a high expression of
ATOH1 transcriptionally induces GLI2 expression (Fig. 1B). It has been demonstrated that
ATOH1 regulates the SHH signaling pathway in GNPs (22). Furthermore, the overexpression of
ATOH1 in GNPs under active SHH signaling conditions accelerates MB
progression, however, proliferation was decreased in the absence of
SHH (21). ATOH1 may enhance GNP
proliferation by activating GLI2 and indirectly increasing the
activity of the SHH signaling pathway via the inhibition of the
transmembrane receptor patched 1 (PTCH1; Fig. 1B) (21,59,76).
It has been reported that <25% of MB cases are
associated with constantly active mutations of the SHH pathway
(85). Additionally, <20% of MB
cases carry a loss-of-function mutation in PTCH1 (85). Since PTCH1 and the SHH pathway have
been demonstrated to have an antagonistic relationship, hyperactive
SHH mutants and loss-of-function mutations in PTCH1 strongly
enhanced the progression of MB (Fig.
1B) (59). The markedly
increased methylation of the HIC1 promoter results in the silencing
of HIC1 expression in PTCH1 heterozygous mutant mice (21,76).
Furthermore, the loss of PTCH1 may cause HIC1 silencing (Fig. 1B), which in turn deregulates its
function to transcriptionally inhibit ATOH1 expression, potentially
causing an increased expression of ATOH1. In addition, Ptch1 not
only inhibits SHH to eliminate the enhancement function of SHH in
proliferation, but also induces GNP differentiation (22). Hence, ATOH1 may repress GNP cell
differentiation by downregulating PTCH1 (Fig. 1B). However, ATOH1 inhibits bone
morphogenetic protein (BMP) 2/4 induced GNP cell differentiation by
downregulating multiple BMP target genes. These genes, including
distal-less homeobox 1/2 and hedgehog interacting protein, are
required for GNP cell differentiation (Fig. 1B) (86–88).
Previous studies have revealed that ATOH1 expression
can be transcriptionally inhibited by HIC1 and
post-transcriptionally downregulated by BMP2/4 (79). Through the rapid proteasome-mediated
protein degradation of ATOH1, BMP2/4 is able to inhibit MB
progression (Fig. 1B) (21,79).
However, BMP2/4 may also inhibit ATOH1 by activating the ATOH1
inhibitors inhibitor of DNA binding 1 HLH protein (ID1) and
inhibitor of DNA binding 2 (ID2) (79,89).
ID1/2 are transcriptional repressors, which interact with ATOH1 to
prevent ATOH1-DNA binding (79,90).
BMPs, a subgroup of the transforming growth factor-β superfamily of
proteins, act as MB suppressors not only via the downregulation of
ATOH1 expression, but also via its regulatory effect on cell
differentiation and proliferation. It has been revealed that BMPs
antagonize SHH-dependent proliferation by activating Smad
phosphorylation and downregulating SHH-induced genes that are
required for the proliferation of GNPs in MB. In vitro and
in vivo data have also revealed that BMPs promote the
differentiation of GNPs in MB by increasing the expression of
multiple genes for cell differentiation (79).
Several genome-wide studies on gene expression
profile assessing the up and downregulation of ATOH1 in MB GNPs
have revealed that the outcome candidates are primarily involved in
two biological processes: Cell differentiation and proliferation.
Among these ATOH1 related genes, nearly two thirds are involved in
differentiation and less than one third are associated with cell
proliferation (79,91). Others are associated with cell
adhesion, cell migration, metabolism and chromosome modulation
(21,92–94). A
comparative study on the categorical differences in upregulated and
down regulated gene expression between the two MB groups induced by
the overexpression of ATOH1 or GLI1 revealed that genes involved in
neuronal differentiation, migration and adhesion, were
significantly enriched in ATOH1 overexpressing MB. However, genes
that regulated cell cycle progression were similarly expressed both
MB groups (21). The results
indicate that ATOH1 has more profound effects on controlling
neuronal differentiation to promote the viability of GNPs and
maintain the progenitor.
As an aggressive embryonic cerebellum tumor, MB is
the most common malignant pediatric brain tumor that exhibits a
high mortality. The continued analysis of the mechanism that
genetically and epigenetically regulates the relative gene
expression of MB has permitted a deeper elucidation of therapeutic
targets. The success of using Smoothened inhibitors, cyclopamine (a
plant steroid alkaloid) and HhAntag (a benzimidazole derivative) as
therapeutic drugs, has revealed their important regressional effect
on controlling the SHH signaling pathway in MB (95,96).
These agents not only decrease the proliferation of tumor cells,
but also induce apoptosis of MB cells (97). However, the efficiency of these
treatments are limited for MB with hyperactive SHH signaling and/or
a high expression of ATOH1, since ATOH1 directly activates GLI2
expression at the transcriptional level and high GLI1/2 expressions
may reduce drug efficiency. Therefore, targeting certain downstream
proteins, including GLI1/2, may be more efficient (98,99).
Recent reports using melanoma cell lines have demonstrated that GLI
inhibitor GANT61 treatment was able to repress melanoma cells
(100), indicating that additional
GANT61 treatment may be effective in multiple anti-MB targeted
therapy.
Although the downregulation of SHH signaling
activity does not affect ATOH1 expression and a high ATOH1
expression in SHH subgroup MB represents only 25–35% of all MB
cases (101), the expression of
ATOH1 should still be taken into account when SHH signaling is used
as a therapeutic target in MB. Despite transcription factors being
difficult targets for small molecule drug development, for SHH
subgroup MB, ATOH1 may still serve as a good potential therapeutic
target, due to its regulatory function on GNPs in MB.
Epigenetic therapy, which reverses DNA promoter
methylation in clinical treatment of myelodysplastic syndrome, has
been reported (21,96). This method may be utilized to restore
Hic1 function by demethylating the Hic1 promoter, thereby blocking
ATOH1 expression. Another possible way to downregulate ATOH1 is via
BMPs treatment. It has been demonstrated that BMPs reduce GNP
proliferation and promote differentiation, as well as induce
apoptosis in human MB cells (102).
BMPs may therefore be used in therapeutic interventions. However,
high levels of ATOH1 can override the neuronal differentiation
induced by BMP by inhibiting the expression of a multiple BMP
target genes (21). Therefore,
similar to SHH targeting therapy, ATOH1expression levels should be
considered when BMPs are used as treatment for MB.
As tumors are heterogeneous entities with different
causes of disease progression, previous studies have combined two
or more drugs to tackle multiple targets. For example, GNP-like MB
is inhibited via effective treatment with BMPs and cyclopamine
(79). Although there is no clear
evidence indicating the direct effect on ATOH1 by blocking PN-1,
loss of function studies on PN-1 in PTCH1+/− mice
revealed the loss of ATOH1 expression and the reduction of MB
formation (Fig. 1B) (74). Therefore, it may be worthwhile to
combine the smoothened frizzled class receptor inhibitor with the
PN-1 inhibitor to prevent relapse due to single drug resistance.
For clinic treatment, theoretical and practical caution should be
taken. Recent study that has performed multivariate Cox regression
analysis has proposed a multi-gene model for MB risk prediction
(81). They have also provided a
quantitative analysis method for the identification of molecular
markers and for the evaluation of these markers on influencing
clinical behavior. More effort is required to optimize the outcome
of therapeutic treatment.
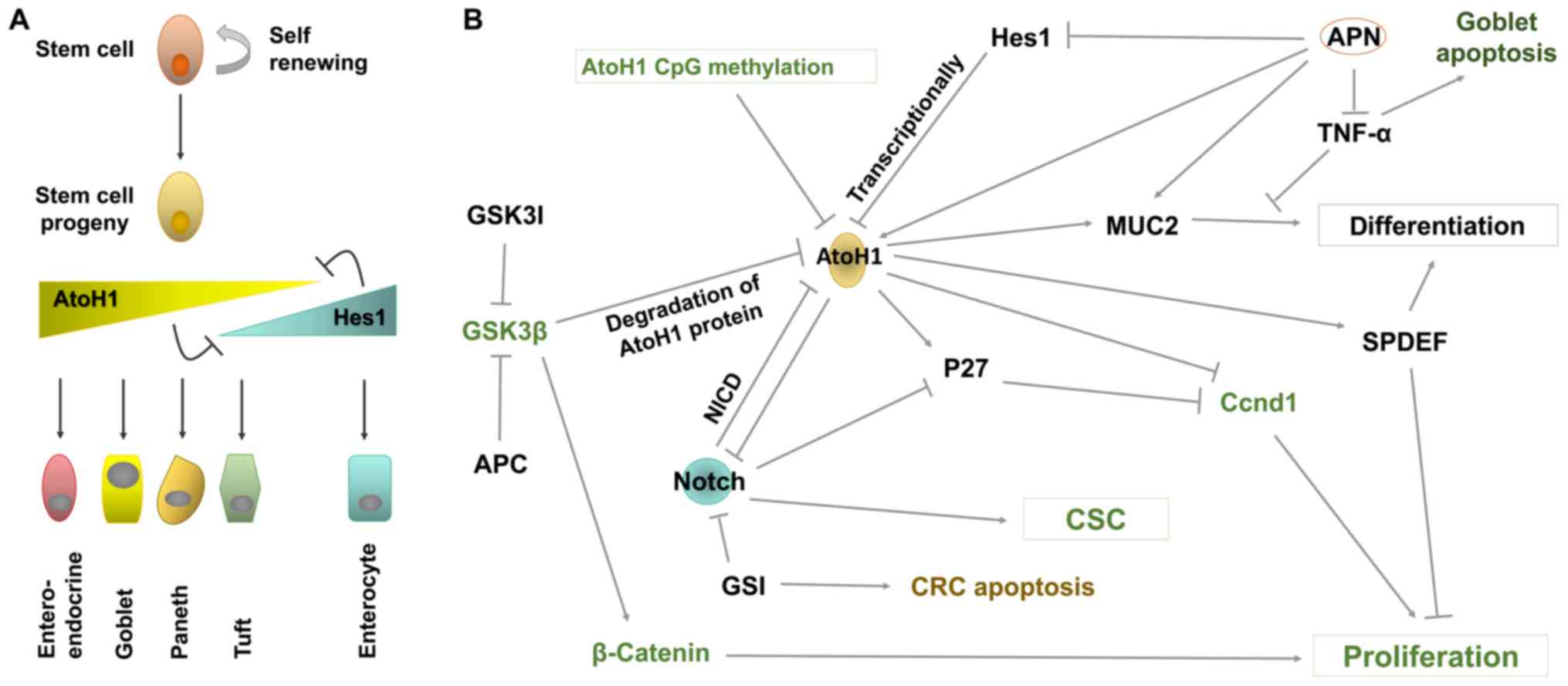
The intestinal epithelium is a self-renewing tissue
that is comprised of several cell lineages (Fig. 2A). Within the intestinal epithelium,
the major cell types can be classified as absorptive
(colonocytes/enterocytes) or secretory (goblet, Paneth and
enteroendocrine), based on their distinct genetic programs. In the
intestinal epithelium of mice, ATOH1/Math1 expression is maintained
throughout embryonic and adult phases (25). The expression of ATOH1 in the
intestinal epithelium is essential for the specification and
regulation of proliferation of the secretory cell lineage (25). The negative regulation of intestinal
epithelial cell proliferation by ATOH1 and the failure to develop
all types of secretory cells in ATOH1/Math1 null mice further
confirms the functions of ATOH1 in the intestinal epithelium
(103,104). In addition, interaction between the
ATOH1 and Notch signaling pathway regulates the lineage
differentiation of different types of intestinal cells. More
specifically, inhibition of ATOH1 via Hes, a Notch signaling
pathway downstream gene, promoted the absorptive vs. the secretory
fate (Fig. 2A) (105). Therefore, ATOH1 is required in
epidermal progenitors, in their progeny to specific MCs (106) and in their commitment to
neuroendocrine cells (107). These
results indicate that ATOH1 is involved in the general
neuroendocrine differentiation of epithelial cells.
It is well known that ATOH1 and Notch signaling
inhibit one and another, and are involved in the development of the
secretory and absorptive cell lineages. It has been demonstrated in
several previous studies that intestinal ATOH1 regulates cell cycle
arrest and represses proliferation, hence promoting differentiation
or stimulating apoptosis (26,41). A
significant decrease in ATOH1 mRNA levels has been detected in ~70%
of CRC cases (24,26). In addition, the Notch signaling
pathway has been determined to be associated with human colon
adenocarcinomas (108). In addition
to inhibiting ATOH1, Notch signaling functions to maintain stem
cells in an uncommitted state (Fig.
2B) (109). Active Notch1/2 and
its transcriptional target HES1 have been detected in human colon
adenocarcinomas and CRC cell lines and were enriched in adenomas
from adenomatous polyposis coli (APC) mutant mice (27,110–113).
Previous studies have revealed that at least one
copy deletion of ATOH1 is present in ~50% of tumors and ATOH1 CpG
methylation has been detected in ~70% of tumors (Fig. 2B) (26). Other evidence obtained from the
analysis of 48 patients with colon cancer has revealed that ATOH1
mRNA levels drop ~20-fold and goblet cell populations are markedly
reduced in colon adenocarcinomas (114). These data indicate that genetic and
epigenetic mechanisms may be involved in the silencing of ATOH1
expression in CRC, and that ATOH1 is required for goblet lineage
development.
The cell cycle inhibitor p27 has been determined to
be activated by ATOH1 and repressed by Notch signaling via Hes1
(Fig. 2B) (114,133).
ATOH1 induces cell cycle exit by inhibiting the cell cycle marker
gene CCND1 either directly or via p27, blocking the proliferation
of intestinal progenitors (103).
ATOH1 therefore serves as a tumor suppressor, which promotes
differentiation and inhibits the proliferation of intestinal
progenitors.
Active Notch signaling represses p27 and ATOH1 via
Hes1 activation, thereby indirectly enhancing cell proliferation
and maintaining cancer stem cells (130,133,134).
Similarly, the aberrant activation of Wnt/GSK3 signaling promotes
cell proliferation indirectly by activating the β-catenin protein
(117), whilst indirectly
inhibiting cell differentiation and enhancing cell proliferation by
degrading ATOH1 (115).
It has been demonstrated that a high Notch activity
and an accumulation of β-catenin protein as a result of aberrant
Wnt/GSK3 signaling, together with the inactivation of ATOH1,
induces carcinogenesis and maintains the undifferentiated state of
colon cancer (135). Therefore,
treatment with GSK3 inhibitors (GSK3I), such as APC, and Notch
signaling inhibitors (such as Notch-targeting antibodies or
γ-secretase) may represent a novel therapeutic approach (109).
GSK is considered to be a key enzyme in various
biological processes. Therefore, the development of a drug
targeting Wnt/GSK signaling is difficult due to its complex
protein-protein interactions and various functions in different
cell types (113). The risk of
treatment and pathological safety should becarefully evaluated. One
particular GSK3 inhibitor, lithium chloride, serves a protective
effect against colon cancer and exerts no detectable pathological
changes in other major organs while being used to treat bipolar
disorder treatment (136). Previous
studies assessing the molecular pathways and mechanisms involved in
the cancer suppressive effect of GSK3I were performed in cell lines
and rodent model systems (137–139).
In addition, inactivation of the Wnt/GSK signaling pathway via the
overexpression of a full-length APC gene in colon cancer cells
revealed the stabilization of ATOH1 and that the degradation of
β-catenin (Fig. 2B) results in cell
differentiation (115). Combination
with other treatments may provide a sufficient effect when compared
with single GSK3I therapies for CRC (140). For instance, the overexpression of
ATOH1 or the stabilization of ATOH1 protein in combination with
GSK3I treatment may serve as a potential cancer therapy (135). However, detailed analyses and
evaluations are required to optimize drug dosages, multiple
treatments and tissue specificity. Certain cases of CRC exhibit an
undifferentiated proliferative phenotype caused by constitutively
activated Notch signaling (111,112,141).
Maintaining a negative regulative Notch activity is initiated via
the release and entry of the active domain of Notch into the
nucleus. The release of the Notch signaling receptor active domain
and intracellular domain requires γ-secretase activity.
Theoretically, antibody-mediated Notch inhibition and γ-secretase
inhibitors (GSIs) would be good candidates to inhibit colonic
cancer. However, antibody-mediated Notch inhibition exerts a weak
effect on blocking CRC growth (142). A previous study on γ-secretase
inhibitor treatment in colon cancer cell lines and primary human
CRC cell cultures revealed that GSI treatment upregulates ATOH1
expression and increases Muc2 and p27, resulting in the reduction
of anchorage independent cellular growth and increasing Muc2
positive cells in ATOH1 positive CRC, but no detectable effect in
ATOH1 negative CRC (134). In
addition, a significant proapoptotic effect on CRC cell lines has
been observed in GSI treatment (111). These data indicate that ATOH1 is
crucial to the tumorigenesis regulatory network of CRC.
Undifferentiated CRC represents only a fraction of colonic cancers.
There are also moderately and well-differentiated classes of CRC
(143). Furthermore, the hyper
activation of β-catenin signaling overrides the forced
differentiation induced by GSI treatment (27). These data indicate that multiple
treatments or the combination of other drugs is required to obtain
improved outcomes in patients.
Currently, certain cytotoxic drugs, including
taxanes or platinum compounds, have been studied in CRC cell lines
(144,145). However, more detailed analyses are
required for further clarification. Assessing the possibility and
optimizing the proper conditions of GSK3I and GSI co-treatment in
CRC should be considered. Another gene, SPDEF, has been revealed to
regulate the terminal differentiation of intestinal goblet cells.
In breast and prostate cancer, SPDEF acts as tumor suppressor by
inhibiting invasion and metastasis (26,116)
and/or tumor growth and survival (24). In CRC, SPDEF serves as a key mediator
of ATOH1 for tumor suppressive activity (130). Future studies are therefore
worthwhile to assess whether SPDEF upregulating treatment alone or
in combination with GSK3I and/or GSI is sufficient to activate
goblet cell differentiation and block proliferation in ATOH1-null
CRC cells. However, since ATOH1 is a potential anti-tumor gene in
CRC, whether it is sufficient to block tumor growth by elevating
ATOH1 levels or by force expressing ATOH1, should be elucidated. A
previous study assessing the role of adiponectin (APN) in the
prevention of goblet cell apoptosis and the differentiation of
epithelial cells to goblet cells revealed that APN may block goblet
cell apoptosis by inhibiting TNF-α and promoting differentiation by
upregulating ATOH1 and Muc2, and downregulating Hes1 (74) (Fig.
2B). Therefore, APN may serve as a potential clinical target
candidate for CRC.
The effect of ATOH1on the tumorigenesis of
different tissue organs and the possibility of targeted clinical
therapy indicated that variations in the mechanisms of
tumorigenesis existed in different type of tumors. ATOH1 serves as
a tumor suppressor in MCC and CRC but is an oncogene in MC and
SCLC/SCC. The expression profile of the ATOH1 protein also exhibits
a differential change in different types of tumors. In MCC and CRC,
ATOH1 expressions are downregulated. By contrast, ATOH1 expressions
were upregulated in MC and SCLC/SCC. However, regardless of these
specificities, ATOH1 displayed a common feature that ATOH1
influenced tumorigenesis by promoting the transcription of its
target genes for cell proliferation and differentiation. In the
case of MCC and CRC, these genes are required for enhancing
differentiation and inhibiting proliferation. The opposite is
observed in MC and SCLS/SCC. The abnormal expression of ATOH1
results in an unpaired balance between differentiation and
proliferation, which promotes the progression of cancer.
Crucial proteins are more often deregulated in
multiple ways at different levels, such as at the transcriptional,
translational and post translational level and during mRNA and/or
protein stability. The present discussion of ATOH1 in MB and CRC
indicated that the temporal/spatial regulation of protein and
protein functions with tissue/context specificity are observed not
only during development but also in cancer progression. Clinical
drug treatments should therefore address the specificity and
regulatory aspects of their targets. The cross-interaction of
proteins causes single drug therapeutic treatments to be
ineffective. Optimal target selection or a combined treatment
approach for cancer therapy should therefore consider this
cross-effect. Multiple targeted treatments have revealed a more
efficient effect during cancer therapy. An improved understanding
of the mechanisms of tumorigenesis and cancer regulatory networks
would improve clinical approaches. The core reason for the
formation of cancer is the unpaired balance of biological systems
to a certain extent. Instead of assessing genes or pathways as
clinical targets, understanding how this balance is disturbed and
subsequently elucidating an approach for adjusting this would be
another direction of cancer therapy.
The authors would like to thank Professor Bassem A.
Hassan (Laboratory of Brain Development, Institut du Cerveau et de
la Moelle Épinière, Hôpital Pitié-Salpêtrière, Paris) for his
comments on the manuscript.
The present review was supported by the National
Nature Science Foundation (grant no. 81772943).
Not applicable.
YF and SSY wrote the manuscript. LJZ wrote the
manuscript and edited the figures. ZLJ and XJQ critically evaluated
and revised the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Kompella P and Vasquez KM: Obesity and
cancer: A mechanistic overview of metabolic changes in obesity that
impact genetic instability. Mol Carcinog. 58:1531–1550. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murata M: Inflammation and cancer. Environ
Health Prev Med. 23:502018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Axelrad JE, Lichtiger S and Yajnik V:
Inflammatory bowel disease and cancer: The role of inflammation,
immunosuppression, and cancer treatment. World J Gastroenterol.
22:4794–4801. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raskov H, Soby JH, Troelsen J, Bojesen RD
and Gogenur I: Driver gene mutations and epigenetics in colorectal
Cancer. Ann Surg. 271:75–85. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Todd R and Wong DT: Oncogenes. Anticancer
Res. 19:4729–4746. 1999.PubMed/NCBI
|
|
6
|
Bos JL: Ras oncogenes in human cancer: A
review. Cancer Res. 49:4682–4689. 1989.PubMed/NCBI
|
|
7
|
Felsher DW and Bishop JM: Reversible
tumorigenesis by MYC in hematopoietic lineages. Mol Cell.
4:199–207. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hisada M, Garber JE, Fung CY, Fraumeni JF
and Li FP: Multiple primary cancers in families with Li-Fraumeni
syndrome. J Natl Cancer Inst. 90:606–611. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
National Cancer Institute, . Genetic
Testing for Hereditary Cancer Syndromes. June
21–, 2016
|
|
11
|
National Cancer Institute, . Physician
Data Query (PDQ). Cancer Genetics Overview. 2016. June
21–2016
|
|
12
|
National Cancer Institute, . Physician
Data Query (PDQ). Genetics of Breast and Ovarian Cancer. 2016.
June
21–2016
|
|
13
|
Chang-Claude J: Inherited genetic
susceptibility to breast cancer. IARC Sci Publ. 154:177–190.
2001.PubMed/NCBI
|
|
14
|
National Cancer Institute, . Physician
Data Query (PDQ). Genetics of Colorectal Cancer. 2016. June
21–2016
|
|
15
|
Akazawa C, Ishibashi M, Shimizu C,
Nakanishi S and Kageyama R: A mammalian helix-loop-helix factor
structurally related to the product of Drosophila proneural gene
atonal is a positive transcriptional regulator expressed in the
developing nervous system. J Biol Chem. 270:8730–8738. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cai T and Groves AK: The role of atonal
factors in mechanosensory cell specification and function. Mol
Neurobiol. 52:1315–1329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chonko KT, Jahan I, Stone J, Wright MC,
Fujiyama T, Hoshino M, Fritzsch B and Maricich SM: Atoh1 directs
hair cell differentiation and survival in the late embryonic mouse
inner ear. Dev Biolo. 381:401–410. 2013. View Article : Google Scholar
|
|
18
|
Ben-Arie N, Bellen HJ, Armstrong DL,
McCall AE, Gordadze PR, Guo Q, Matzuk MM and Zoghbi HY: Math1 is
essential for genesis of cerebellar granule neurons. Nature.
390:169–172. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Machold R and Fishell G: Math1 is
expressed in temporally discrete pools of cerebellar rhombic-lip
neural progenitors. Neuron. 48:17–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rose MF, Ren J, Ahmad KA, Chao HT, Klisch
TJ, Flora A, Greer JJ and Zoghbi HY: Math1 is essential for the
development of hindbrain neurons critical for perinatal breathing.
Neuron. 64:341–354. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ayrault O, Zhao H, Zindy F, Qu C, Sherr CJ
and Roussel MF: Atoh1 inhibits neuronal differentiation and
collaborates with Gli1 to generate medulloblastoma-initiating
cells. Cancer Res. 70:5618–5627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Flora A, Klisch TJ, Schuster G and Zoghbi
HY: Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the
developing cerebellum and prevents medulloblastoma. Science.
326:1424–1427. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang ZJ, Ellis T, Markant SL, Read TA,
Kessler JD, Bourboulas M, Schüller U, Machold R, Fishell G, Rowitch
DH, et al: Medulloblastoma can be initiated by deletion of Patched
in lineage-restricted progenitors or stem cells. Cancer Cell.
14:135–145. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leow CC, Romero MS, Ross S, Polakis P and
Gao WQ: Hath1, down-regulated in colon adenocarcinomas, inhibits
proliferation and tumorigenesis of colon cancer cells. Cancer Res.
64:6050–6057. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Q, Bermingham NA, Finegold MJ and
Zoghbi HY: Requirement of Math1 for secretory cell lineage
commitment in the mouse intestine. Science. 294:2155–2158. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bossuyt W, Kazanjian A, De Geest N, Van
Kelst S, De Hertogh G, Geboes K, Boivin GP, Luciani J, Fuks F,
Chuah M, et al: Atonal homolog 1 is a tumor suppressor gene. PLoS
Biol. 7:e392009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peignon G, Durand A, Cacheux W, Ayrault O,
Terris B, Laurent-Puig P, Shroyer NF, Van Seuningen I, Honjo T,
Perret C, et al: Complex interplay between beta-catenin signalling
and Notch effectors in intestinal tumorigenesis. Gut. 60:166–176.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang C, Chan JA and Schuurmans C:
Proneural bHLH genes in development and disease. Curr Top Dev Biol.
110:75–127. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Leonard JH, Cook AL, Van Gele M, Boyle GM,
Inglis KJ, Speleman F and Sturm RA: Proneural and proneuroendocrine
transcription factor expression in cutaneous mechanoreceptor
(Merkel) cells and Merkel cell carcinoma. Int J Cancer.
101:103–110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu HT, Xie XM, Li QC, Liu SL, Dai SD, Liu
Y and Wang EH: Atonal homolog 1 expression in lung cancer
correlates with inhibitors of the Wnt pathway as well as the
differentiation and primary tumor stage. APMIS. 121:111–119. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hou K, Jiang H, Karim MR, Zhong C, Xu Z,
Liu L, Guan M, Shao J and Huang X: A Critical E-box in
Barhl1 3′ enhancer is essential for auditory hair cell
differentiation. Cells. 8(pii): E4582019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scheffer D, Sage C, Corey DP and Pingault
V: Gene expression profiling identifies Hes6 as a transcriptional
target of ATOH1 in cochlear hair cells. FEBS Lett. 581:4651–4666.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jarman AP, Grau Y, Jan LY and Jan YN:
Atonal is a proneural gene that directs chordotonal organ formation
in the Drosophila peripheral nervous system. Cell. 73:1307–1321.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jarman AP, Grell EH, Ackerman L, Jan LY
and Jan YN: Atonal is the proneural gene for Drosophila
photoreceptors. Nature. 369:398–400. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mulvaney J and Dabdoub A: Atoh1, an
essential transcription factor in neurogenesis and intestinal and
inner ear development: Function, regulation, and context
dependency. J Assoc Res Otolaryngol. 13:281–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weinberger S, Topping MP, Yan J, Claeys A,
Geest N, Ozbay D, Hassan T, He X, Albert JT, Hassan BA and
Ramaekers A: Evolutionary changes in transcription factor coding
sequence quantitatively alter sensory organ development and
function. Elife. 6(pii): e264022017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Quan XJ, Denayer T, Yan J, Jafar-Nejad H,
Philippi A, Lichtarge O, Vleminckx K and Hassan BA: Evolution of
neural precursor selection: Functional divergence of proneural
proteins. Development. 131:1679–1689. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aerts S, Quan XJ, Claeys A, Naval Sanchez
M, Tate P, Yan J and Hassan BA: Robust target gene discovery
through transcriptome perturbations and genome-wide enhancer
predictions in Drosophila uncovers a regulatory basis for sensory
specification. PLoS Biol. 8:e10004352010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Klisch TJ, Xi Y, Flora A, Wang L, Li W and
Zoghbi HY: In vivo Atoh1 targetome reveals how a proneural
transcription factor regulates cerebellar development. Proc Natl
Acad Sci USA. 108:3288–3293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gazit R, Krizhanovsky V and Ben-Arie N:
Math1 controls cerebellar granule cell differentiation by
regulating multiple components of the Notch signaling pathway.
Development. 131:903–913. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
VanDussen KL and Samuelson LC: Mouse
atonal homolog 1 directs intestinal progenitors to secretory cell
rather than absorptive cell fate. Dev Biol. 346:215–223. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peignon G, Durand A, Cacheux W, Ayrault O,
Terris B, Laurent-Puig P, Shroyer NF, Van Seuningen I, Honjo T,
Perret C and Romagnolo B: Complex interplay between β-catenin
signalling and Notch effectors in intestinal tumorigenesis. Gut.
60:166–176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Flora A, Garcia JJ, Thaller C and Zoghbi
HY: The E-protein Tcf4 interacts with Math1 to regulate
differentiation of a specific subset of neuronal progenitors. Proc
Natl Acad Sci USA. 104:15382–15387. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee KJ, Dietrich P and Jessell TM: Genetic
ablation reveals that the roof plate is essential for dorsal
interneuron specification. Nature. 403:734–740. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao H, Ayrault O, Zindy F, Kim JH and
Roussel MF: Post-transcriptional down-regulation of Atoh1/Math1 by
bone morphogenic proteins suppresses medulloblastoma development.
Genes Dev. 22:722–727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hu X, Huang J, Feng L, Fukudome S,
Hamajima Y and Lin J: Sonic hedgehog (SHH) promotes the
differentiation of mouse cochlear neural progenitors via the
Math1-Brn3.1 signaling pathway in vitro. J Neurosci Res.
88:927–935. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Moll I, Roessler M, Brandner JM, Eispert
AC, Houdek P and Moll R: Human Merkel cells-aspects of cell
biology, distribution and functions. Eur J Cell Biol. 84:259–271.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wellnitz SA, Lesniak DR, Gerling GJ and
Lumpkin EA: The regularity of sustained firing reveals two
populations of slowly adapting touch receptors in mouse hairy skin.
J Neurophysiol. 103:3378–3388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ben-Arie N, Hassan BA, Bermingham NA,
Malicki DM, Armstrong D, Matzuk M, Bellen HJ and Zoghbi HY:
Functional conservation of atonal and Math1 in the CNS and PNS.
Development. 127:1039–1048. 2000.PubMed/NCBI
|
|
50
|
Haeberle H, Fujiwara M, Chuang J, Medina
MM, Panditrao MV, Bechstedt S, Howard J and Lumpkin EA: Molecular
profiling reveals synaptic release machinery in Merkel cells. Proc
Natl Acad Sci USA. 101:14503–14508. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wright MC, Reed-Geaghan EG, Bolock AM,
Fujiyama T, Hoshino M and Maricich SM: Unipotent, ATOH1+
progenitors maintain the Merkel cell population in embryonic and
adult mice. J Cell Biol. 208:367–379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Maricich SM, Wellnitz SA, Nelson AM,
Lesniak DR, Gerling GJ, Lumpkin EA and Zoghbi HY: Merkel cells are
essential for light-touch responses. Science. 324:1580–1582. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bardot ES, Valdes VJ, Zhang J, Perdigoto
CN, Nicolis S, Hearn SA, Silva JM and Ezhkova E: Polycomb subunits
Ezh1 and Ezh2 regulate the Merkel cell differentiation program in
skin stem cells. EMBO J. 32:1990–2000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Coggshall K, Tello TL, North JP and Yu SS:
Merkel cell carcinoma: An update and review: Pathogenesis,
diagnosis, and staging. J Am Acad Dermatol. 78:433–442. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tello TL, Coggshall K, Yom SS and Yu SS:
Merkel cell carcinoma: An update and review: Current and future
therapy. J Am Acad Dermatol. 78:445–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu W, MacDonald M and You J: Merkel cell
polyomavirus infection and Merkel cell carcinoma. Curr Opin Virol.
20:20–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Schadendorf D, Lebbé C, Zur Hausen A,
Avril MF, Hariharan S, Bharmal M and Becker JC: Merkel cell
carcinoma: Epidemiology, prognosis, therapy and unmet medical
needs. Eur J Cancer. 71:53–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gambichler T, Mohtezebsade S, Wieland U,
Silling S, Hoh AK, Dreissigacker M, Schaller J, Schulze HJ, Oellig
F, Kreuter A, et al: Prognostic relevance of high atonal homolog-1
expression in Merkel cell carcinoma. J Cancer Res Clin Oncol.
143:43–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Briggs KJ, Corcoran-Schwartz IM, Zhang W,
Harcke T, Devereux WL, Baylin SB, Eberhart CG and Watkins DN:
Cooperation between the Hic1 and Ptch1 tumor suppressors in
medulloblastoma. Genes Dev. 22:770–785. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Schüller U, Heine VM, Mao J, Kho AT,
Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al:
Acquisition of granule neuron precursor identity is a critical
determinant of progenitor cell competence to form Shh-induced
medulloblastoma. Cancer Cell. 14:123–134. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shroyer NF, Helmrath MA, Wang VY, Antalffy
B, Henning SJ and Zoghbi HY: Intestine-specific ablation of mouse
atonal homolog 1 (Math1) reveals a role in cellular homeostasis.
Gastroenterology. 132:2478–2488. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Peignon G, Durand A, Cacheux W, Ayrault O,
Terris B, Laurent-Puig P, Shroyer NF, Van Seuningen I, Honjo T,
Perret C, et al: Complex interplay between β-catenin signalling and
Notch effectors in intestinal tumorigenesis. Gut. 60:166–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Westerman BA, Breuer RH, Poutsma A,
Chhatta A, Noorduyn LA, Koolen MG, Postmus PE, Blankenstein MA and
Oudejans CB: Basic helix-loop-helix transcription factor profiling
of lung tumors shows aberrant expression of the proneural gene
atonal homolog 1 (ATOH1, HATH1, MATH1) in neuroendocrine tumors.
Int J Biol Markers. 22:114–123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hiroshima K, Iyoda A, Shibuya K, Toyozaki
T, Haga Y, Fujisawa T and Ohwada H: Prognostic significance of
neuroendocrine differentiation in adenocarcinoma of the lung. Ann
Thorac Surg. 73:1732–1735. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Berendsen HH, de Leij L, Poppema S,
Postmus PE, Boes A, Sluiter HJ and The H: Clinical characterization
of non-small-cell lung cancer tumors showing neuroendocrine
differentiation features. J Clin Oncol. 7:1614–1620. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ben-Arie N, McCall AE, Berkman S, Eichele
G, Bellen HJ and Zoghbi HY: Evolutionary conservation of sequence
and expression of the bHLH protein Atonal suggests a conserved role
in neurogenesis. Hum Mol Genet. 5:1207–1216. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bermingham NA, Hassan BA, Wang VY,
Fernandez M, Banfi S, Bellen HJ, Fritzsch B and Zoghbi HY:
Proprioceptor pathway development is dependent on Math1. Neuron.
30:411–422. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Iulianella A, Wingate RJ, Moens CB and
Capaldo E: The generation of granule cells during the development
and evolution of the cerebellum. Dev Dyn. 248:506–513. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Thompson MC, Fuller C, Hogg TL, Dalton J,
Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM,
Kellie SJ, et al: Genomics identifies medulloblastoma subgroups
that are enriched for specific genetic alterations. J Clin Oncol.
24:1924–1931. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gibson P, Tong Y, Robinson G, Thompson MC,
Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, et
al: Subtypes of medulloblastoma have distinct developmental
origins. Nature. 468:1095–1099. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gilbertson RJ and Ellison DW: The origins
of medulloblastoma subtypes. Ann Rev Pathol. 3:341–365. 2008.
View Article : Google Scholar
|
|
73
|
Lee Y, Miller HL, Jensen P, Hernan R,
Connelly M, Wetmore C, Zindy F, Roussel MF, Curran T, Gilbertson RJ
and McKinnon PJ: A molecular fingerprint for medulloblastoma.
Cancer Res. 63:5428–5437. 2003.PubMed/NCBI
|
|
74
|
Vaillant C, Valdivieso P, Nuciforo S, Kool
M, Schwarzentruber-Schauerte A, Mereau H, Cabuy E, Lobrinus JA,
Pfister S, Zuniga A, et al: Serpine2/PN-1 is required for
proliferative expansion of pre-neoplastic lesions and malignant
progression to medulloblastoma. PLoS One. 10:e01248702015.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Blaess S, Corrales JD and Joyner AL: Sonic
hedgehog regulates Gli activator and repressor functions with
spatial and temporal precision in the mid/hindbrain region
Development 133. 1799–1809. 2006.PubMed/NCBI
|
|
76
|
Briggs KJ, Eberhart CG and Watkins DN:
Just say no to ATOH: How HIC1 methylation might predispose
medulloblastoma to lineage addiction. Cancer Res. 68:8654–8666.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Klisch TJ, Vainshtein A, Patel AJ and
Zoghbi HY: Jak2-mediated phosphorylation of ATOH1 is critical for
medulloblastoma growth. Elife. 6(pii): e311812017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Salsano E, Pollo B, Eoli M, Giordana MT
and Finocchiaro G: Expression of MATH1, a marker of cerebellar
granule cell progenitors, identifies different medulloblastoma
sub-types. Neurosci Lett. 370:180–185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhao H, Ayrault O, Zindy F, Kim JH and
Roussel MF: Post-transcriptional down-regulation of ATOH1/Math1 by
bone morphogenic proteins suppresses medulloblastoma development.
Genes Dev. 22:722–727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Garraway LA, Weir BA, Zhao X, Widlund H,
Beroukhim R, Berger A, Rimm D, Rubin MA, Fisher DE, Meyerson ML and
Sellers WR: ‘Lineage addiction’ in human cancer: Lessons from
integrated genomics. Cold Spring Harb Symp Quant Biol. 70:25–34.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zakrzewska M, Gresner SM, Zakrzewski K,
Zalewska-Szewczyk B and Liberski PP: Novel gene expression model
for outcome prediction in paediatric medulloblastoma. J Mol
Neurosci. 51:371–379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lum L and Beachy PA: The Hedgehog response
network: Sensors, switches, and routers. Science. 304:1755–1759.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Katoh Y and Katoh M: Integrative genomic
analyses on GLI1: Positive regulation of GLI1 by Hedgehog-GLI,
TGFbeta-Smads, and RTK-PI3K-AKT signals, and negative regulation of
GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol.
35:187–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Browd SR, Kenney AM, Gottfried ON, Yoon
JW, Walterhouse D, Pedone CA and Fults DW: N-myc can substitute for
insulin-like growth factor signaling in a mouse model of sonic
hedgehog-induced medulloblastoma. Cancer Res. 66:2666–2672. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Eberhart CG: Medulloblastoma in mice
lacking p53 and PARP: All roads lead to Gli. Am J Pathol. 162:7–10.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chiba S, Takeshita K, Imai Y, Kumano K,
Kurokawa M, Masuda S, Shimizu K, Nakamura S, Ruddle FH and Hirai H:
Homeoprotein DLX-1 interacts with Smad4 and blocks a signaling
pathway from activin A in hematopoietic cells. Proc Natl Acad Sci
USA. 100:15577–15582. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Harris SE, Guo D, Harris MA, Krishnaswamy
A and Lichtler A: Transcriptional regulation of BMP-2 activated
genes in osteoblasts using gene expression microarray analysis:
Role of Dlx2 and Dlx5 transcription factors. Front Biosci.
8:s1249–s1265. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
88
|
Katoh Y and Katoh M: Comparative genomics
on HHIP family orthologs. Int J Mol Med. 17:391–395.
2006.PubMed/NCBI
|
|
89
|
Kamaid A, Neves J and Giraldez F: Id gene
regulation and function in the prosensory domains of the chicken
inner ear: A link between Bmp signaling and ATOH1. J Neurosci.
30:11426–11434. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Angley C, Kumar M, Dinsio KJ, Hall AK and
Siegel RE: Signaling by bone morphogenetic proteins and Smad1
modulates the postnatal differentiation of cerebellar cells. J
Neurosci. 23:260–268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Killeen MT and Sybingco SS: Netrin, Slit
and Wnt receptors allow axons to choose the axis of migration. Dev
Biol. 323:143–151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zou YR, Kottmann AH, Kuroda M, Taniuchi I
and Littman DR: Function of the chemokine receptor CXCR4 in
haematopoiesis and in cerebellar development. Nature. 393:595–599.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Adamson DC, Shi Q, Wortham M, Northcott
PA, Di C, Duncan CG, Li J, McLendon RE, Bigner DD, Taylor MD and
Yan H: OTX2 is critical for the maintenance and progression of
Shh-independent medulloblastomas. Cancer Res. 70:181–191. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Berman DM, Karhadkar SS, Hallahan AR,
Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale
J, Olson JM and Beachy PA: Medulloblastoma growth inhibition by
hedgehog pathway blockade. Science. 297:1559–1561. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Romer JT, Kimura H, Magdaleno S, Sasai K,
Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL and
Curran T: Suppression of the Shh pathway using a small molecule
inhibitor eliminates medulloblastoma in Ptc1(+/-)p53(−/-) mice.
Cancer Cell. 6:229–240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hallahan AR, Pritchard JI, Hansen S,
Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein
ID, Beachy PA and Olson JM: The SmoA1 mouse model reveals that
notch signaling is critical for the growth and survival of sonic
hedgehog-induced medulloblastomas. Cancer Res. 64:7794–7800. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Northcott PA, Nakahara Y, Wu X, Feuk L,
Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, et
al: Multiple recurrent genetic events converge on control of
histone lysine methylation in medulloblastoma. Nat Genet.
41:465–472. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
99
|
Stanton BZ and Peng LF: Small-molecule
modulators of the Sonic Hedgehog signaling pathway. Mol Biosyst.
6:44–54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Vlckova K, Reda J, Ondrusova L, Krayem M,
Ghanem G and Vachtenheim J: GLI inhibitor GANT61 kills melanoma
cells and acts in synergy with obatoclax. Int J Oncol. 49:953–960.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Northcott PA, Rutka JT and Taylor MD:
Genomics of medulloblastoma: From Giemsa-banding to next-generation
sequencing in 20 years. Neurosurg Focus. 28:E62010. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Hallahan AR, Pritchard JI, Chandraratna
RA, Ellenbogen RG, Geyer JR, Overland RP, Strand AD, Tapscott SJ
and Olson JM: BMP-2 mediates retinoid-induced apoptosis in
medulloblastoma cells through a paracrine effect. Nat Med.
9:1033–1038. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
103
|
Kazanjian A, Noah T, Brown D, Burkart J
and Shroyer NF: Atonal homolog 1 is required for growth and
differentiation effects of notch/gamma-secretase inhibitors on
normal and cancerous intestinal epithelial cells. Gastroenterology.
139:918–928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Kim TH and Shivdasani RA: Genetic evidence
that intestinal Notch functions vary regionally and operate through
a common mechanism of Math1 repression. J Biol Chem.
286:11427–11433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Noah TK and Shroyer NF: Notch in the
intestine: Regulation of homeostasis and pathogenesis. Annu Rev
Physiol. 75:263–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Van Keymeulen A, Mascre G, Youseff KK,
Harel I, Michaux C, De Geest N, Szpalski C, Achouri Y, Bloch W,
Hassan BA and Blanpain C: Epidermal progenitors give rise to Merkel
cells during embryonic development and adult homeostasis. J Cell
Biol. 187:91–100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Barker N, van Es JH, Kuipers J, Kujala P,
van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H,
Peters PJ and Clevers H: Identification of stem cells in small
intestine and colon by marker gene Lgr5. Nature. 449:1003–1307.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Reedijk M, Odorcic S, Zhang H, Chetty R,
Tennert C, Dickson BC, Lockwood G, Gallinger S and Egan SE:
Activation of Notch signaling in human colon adenocarcinoma. Int J
Oncol. 33:1223–1229. 2008.PubMed/NCBI
|
|
109
|
van Es JH, de Geest N, van de Born M,
Clevers H and Hassan BA: Intestinal stem cells lacking the Math1
tumour suppressor are refractory to Notch inhibitors. Nat Commun.
1:182010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Guilmeau S, Flandez M, Mariadason JM and
Augenlicht LH: Heterogeneity of Jagged1 expression in human and
mouse intestinal tumors: Implications for targeting Notch
signaling. Oncogene. 29:992–1002. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Sikandar SS, Pate KT, Anderson S, Dizon D,
Edwards RA, Waterman ML and Lipkin SM: NOTCH signaling is required
for formation and self-renewal of tumor-initiating cells and for
repression of secretory cell differentiation in colon cancer.
Cancer Res. 70:1469–1478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Fre S, Pallavi SK, Huyghe M, Lae M,
Janssen KP, Robine S, Artavanis-Tsakonas S and Louvard D: Notch and
Wnt signals cooperatively control cell proliferation and
tumorigenesis in the intestine. Proc Natl Acad Sci USA.
106:6309–6314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
van Es JH, van Gijn ME, Riccio O, van den
Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ,
Radtke F and Clevers H: Notch/gamma-secretase inhibition turns
proliferative cells in intestinal crypts and adenomas into goblet
cells. Nature. 435:959–963. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhu DH, Niu BL, Du HM, Ren K, Sun JM and
Gong JP: Hath1 inhibits proliferation of colon cancer cells
probably through up-regulating expression of Muc2 and p27 and
down-regulating expression of cyclin D1. Asian Pac J Cancer Prev.
13:6349–6355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tsuchiya K, Nakamura T, Okamoto R, Kanai T
and Watanabe M: Reciprocal targeting of Hath1 and beta-catenin by
Wnt glycogen synthase kinase 3beta in human colon cancer.
Gastroenterology. 132:208–220. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Park ET, Oh HK, Gum JR Jr, Crawley SC,
Kakar S, Engel J, Leow CC, Gao WQ and Kim YS: HATH1 expression in
mucinous cancers of the colorectum and related lesions. Clin Cancer
Res. 12:5403–5410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Aragaki M, Tsuchiya K, Okamoto R, Yoshioka
S, Nakamura T, Sakamoto N, Kanai T and Watanabe M: Proteasomal
degradation of ATOH1 by aberrant Wnt signaling maintains the
undifferentiated state of colon cancer. Biochem Biophys Res Commun.
368:923–929. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.PubMed/NCBI
|
|
119
|
Polakis P: The oncogenic activation of
beta-catenin. Curr Opin Genet Dev. 9:15–21. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Quan XJ, Yuan L, Tiberi L, Claeys A, De
Geest N, Yan J, van der Kant R, Xie WR, Klisch TJ, Shymkowitz J, et
al: Post-translational control of the temporal dynamics of
transcription factor activity regulates neurogenesis. Cell.
164:460–475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Tomic G, Morrissey E, Kozar S, Ben-Moshe
S, Hoyle A, Azzarelli R, Kemp R, Chilamakuri CSR, Itzkovitz S,
Philpott A and Winton DJ: Phospho-regulation of ATOH1 is required
for plasticity of secretory progenitors and tissue regeneration.
Cell Stem Cell. 23:436–43.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yang X, Zhang L, Song X, He W, Zhang D, Lu
Q, Wu J, Wu C and Jiang J: MicroRNA-613 promotes colon cancer cell
proliferation, invasion and migration by targeting ATOH1. Biochem
Biophys Res Commun. 504:827–833. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ponz de Leon M and Di Gregorio C:
Pathology of colorectal cancer. Digestive and liver disease:
Official journal of the Italian Society of Gastroenterology and the
Italian Association for the Study of the Liver. 33:372–388. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Hooper LV and Macpherson AJ: Immune
adaptations that maintain homeostasis with the intestinal
microbiota. Nat Rev Immunol. 10:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Hollingsworth MA and Swanson BJ: Mucins in
cancer: Protection and control of the cell surface. Nat Rev Cancer.
4:45–60. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sheng XZ, Xu GJ, Tang XQ and Zhan WB:
Monoclonal antibodies recognizing mucus immunoglobulin and surface
immunoglobulin-positive cells of flounder (Paralichthys olivaceus).
Vet Immunol Immunopathol. 145:143–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Corfield AP, Carroll D, Myerscough N and
Probert CS: Mucins in the gastrointestinal tract in health and
disease. Front Biosci. 6:D1321–D1357. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Velcich A, Yang W, Heyer J, Fragale A,
Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K and
Augenlicht L: Colorectal cancer in mice genetically deficient in
the mucin Muc2. Science. 295:1726–1729. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Byrd JC and Bresalier RS: Mucins and mucin
binding proteins in colorectal cancer. Cancer Metastasis Rev.
23:77–99. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Noah TK, Kazanjian A, Whitsett J and
Shroyer NF: SAM pointed domain ETS factor (SPDEF) regulates
terminal differentiation and maturation of intestinal goblet cells.
Exp Cell Res. 316:452–265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lo YH, Chung E, Li Z, Wan YW, Mahe MM,
Chen MS, Noah TK, Bell KN, Yalamanchili HK, Klisch TJ, et al:
Transcriptional regulation by ATOH1 and its Target SPDEF in the
intestine. Cell Mol Gastroenterol Hepatol. 3:51–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Noah TK, Lo YH, Price A, Chen G, King E,
Washington MK, Aronow BJ and Shroyer NF: SPDEF functions as a
colorectal tumor suppressor by inhibiting β-catenin activity.
Gastroenterology. 144:1012–1023.e6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Riccio O, van Gijn ME, Bezdek AC,
Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T,
Clevers H and Radtke F: Loss of intestinal crypt progenitor cells
owing to inactivation of both Notch1 and Notch2 is accompanied by
derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep.
9:377–383. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Souaze F, Bou-Hanna C, Kandel C, Leclair
F, Devalliere J, Charreau B, Bézieau S, Mosnier JF and Laboisse CL:
Differential roles of Hath1, MUC2 and P27Kip1 in relation with
gamma-secretase inhibition in human colonic carcinomas: A
translational study. PLoS One. 8:e559042013. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Aragaki M, Tsuchiya K, Okamoto R, Yoshioka
S, Nakamura T, Sakamoto N, Kanai T and Watanabe M: Proteasomal
degradation of Atoh1 by aberrant Wnt signaling maintains the
undifferentiated state of colon cancer. Biochem Biophys Res Commun.
368:923–929. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Cohen Y, Chetrit A, Cohen Y, Sirota P and
Modan B: Cancer morbidity in psychiatric patients: Influence of
lithium carbonate treatment. Med Oncol. 15:32–36. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Gould TD, Gray NA and Manji HK: Effects of
a glycogen synthase kinase-3 inhibitor, lithium, in adenomatous
polyposis coli mutant mice. Pharmacol Res. 48:49–53.
2003.PubMed/NCBI
|
|
138
|
Shakoori A, Mai W, Miyashita K, Yasumoto
K, Takahashi Y, Ooi A, Kawakami K and Minamoto T: Inhibition of
GSK-3 beta activity attenuates proliferation of human colon cancer
cells in rodents. Cancer Sci. 98:1388–1393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Tan J, Zhuang L, Leong HS, Iyer NG, Liu ET
and Yu Q: Pharmacologic modulation of glycogen synthase
kinase-3beta promotes p53-dependent apoptosis through a direct
Bax-mediated mitochondrial pathway in colorectal cancer cells.
Cancer Res. 65:9012–9020. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ring DB, Johnson KW, Henriksen EJ, Nuss
JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, et
al: Selective glycogen synthase kinase 3 inhibitors potentiate
insulin activation of glucose transport and utilization in vitro
and in vivo. Diabetes. 52:588–595. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Veenendaal LM, Kranenburg O, Smakman N,
Klomp A, Borel Rinkes IH and van Diest PJ: Differential Notch and
TGFbeta signaling in primary colorectal tumors and their
corresponding metastases. Cell Oncol. 30:1–11. 2008.PubMed/NCBI
|
|
142
|
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de
Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, et al:
Therapeutic antibody targeting of individual Notch receptors.
Nature. 464:1052–1057. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Kim JW, Shin MK and Kim BC:
Clinicopathologic impacts of poorly differentiated cluster-based
grading system in colorectal carcinoma. J Korean Med Sci. 30:16–23.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Akiyoshi T, Nakamura M, Yanai K, Nagai S,
Wada J, Koga K, Nakashima H, Sato N, Tanaka M and Katano M:
Gamma-secretase inhibitors enhance taxane-induced mitotic arrest
and apoptosis in colon cancer cells. Gastroenterology. 134:131–144.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Aleksic T and Feller SM: Gamma-secretase
inhibition combined with platinum compounds enhances cell death in
a large subset of colorectal cancer cells. Cell Commun Signal.
6:82008. View Article : Google Scholar : PubMed/NCBI
|