Introduction
Neuroblastoma (NB) is an embryonic tumor derived
from sympathetic neural crest cells (1,2). It is
the most common type of extracranial solid tumor found in children
and the most common type of malignant tumor found in infants and
young children, with an incidence rate of 10.5 per million children
younger than 14 years old (3–5). The
site of the disease is concealed, and NB easily metastasizes
(5). Low-risk NB has the
characteristics of regression and inducing NB differentiation and
maturation in vitro, while high-risk NB has high malignancy
and can metastasize early (6). The
prognosis of patients with NB is extremely poor, which accounts for
~12–15% of all pediatric cancer-associated deaths (7,8). In
recent years, treatments for low- and medium-risk NB have improved;
however, the cure rate for patients with high-risk NB remains low
(9,10). It is therefore crucial to develop
novel treatments for patients with high-risk NB.
Enhancer of zeste homolog 2 (EZH2) protein is the
core catalytic element of polycomb repressor complex 2 (PRC2),
which regulates the transcription of target genes via promoting
histone H3 methylation (11–13). Previous studies have demonstrated
that high expression level of EZH2 is a poor prognostic factor in
various types of cancer (14,15). For
example, in pancreatic cancer, EZH2-mediated microRNA-139-5p
regulates epithelial-mesenchymal transition and lymph node
metastasis (16). Moreover, EZH2 and
EED directly regulate androgen receptor in advanced prostate cancer
(17). In NB, EZH2 is highly
expressed and can epigenetically silences NB tumor suppressor
genes, including CASZ1, CLU, RUNX3 and NGFR, suggesting that EZH2
may be an NB molecular target (18).
Long non-coding RNAs (lncRNAs) are a type of RNA of
>200 nucleotides in length that do not encode for proteins.
Following their discovery, lncRNAs were considered as ‘noises’ in
the genome transcription process (19,20).
However, subsequent studies demonstrated that lncRNAs regulate
genes at a number of levels, including epigenetic, genomic
transcription and post-transcriptional levels. lncRNAs participate
in cancer development and tumorigenesis by affecting tumor cell
proliferation, migration, invasion, apoptosis and the cell cycle,
and promoting angiogenesis (21,22).
lncRNAs directly bind to EZH2, recruiting it to the promoter region
of genes to repress their expression levels. lncRNAs also serve as
EZH2 effectors or regulators (23–25). The
present study investigated the association between lncRNAs and EZH2
using RNA immunoprecipitation (RIP)-, RNA- and chromatin
IP-sequencing (ChIP-seq).
Materials and methods
Cell culture
The SH-SY5Y cell line was obtained from the American
Type Culture Collection (cat. no. CRL-2266) and the 293T cell line
was obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Cells were cultured in Dulbecco's
Modified Eagle Medium supplemented with 10% FBS and 1%
penicillin-streptomycin solution (all from Biological Industries)
and placed in a humidified incubator with 5% CO2 at
37°C. Cell culture dishes were obtained from Hangzhou Xinyou
Biotechnology Co., Ltd.
Construction of short hairpin (sh)RNAs
and stable transfected cell lines
EZH2 shRNA plasmids [designed online (https://www.sigmaaldrich.com)] and pLKO1(Genomeditech,
Shanghai, China) were selected as vectors. The shRNA target
sequences were as follows: Forward shEZH2-F,
5′-CCGGCCCAACATAGATGGACCAAATCTCGAGATTTGGTCCATCTATGTTGGGTTTTTG-3′;
reverse shEZH2-R,
5′AATTCAAAAACCCAACATAGATGGACCAAATCTCGAGATTTGGTCCATCTATGTTGGG-3′;
forward shControl-F,
5′-CCGGTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAATTTTTG-3′;
reverse shControl-R,
5′-AATTCAAAAATTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAA.
Lentivirus with shEZH2 was packaged with 293T cells using
transfection reagent Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Stably transfected SH-SY5Y cells
with shRNA (12 µg) and control (12 µg) were acquired following
puromycin selection.
Western blotting
Cell were lysed using RIPA lysis buffer (Beyotime
Institute of Biotechnology) on ice, and western blotting was
performed as standard (26).
Following blocking with 8% skimmed milk for 1 h at room
temperature, membranes were incubated with primary antibodies
(EZH2; cat. no. 5246S; and β-tubulin; cat. no. 2146S; Cell
Signaling Technology, Inc.) on a decolorization shaker at 4°C
overnight. After washing with Tris-HCl + 0.05% Tween-20 TBST three
times, membranes were incubated with secondary antibody (goat
anti-rabbit IgG; 1:2,000; cat. no. CW0107; CoWin Biosciences) for 1
h at room temperature. Protein signals were detected via enhanced
chemiluminescence substrate (EMD Millipore).
RIP-seq
RNA Immunoprecipitation (RIP) was performed followed
as Gagliardi et al (27) In
simple terms, RNA was enriched with EZH2 antibody (1:100; cat. no.
5246S; Cell Signaling Technology, Inc) in SH-SY5Y cells. Enriched
RNA was broken into short fragments using fragmentation buffer
(cat. no. N402-VAHTS; Vazyme Biotech Co., Ltd.) at 94°C for 5 min.
Fragmented mRNA was used as a template to synthesize cDNA using
random hexamers, buffer, dNTPs and DNA polymerase I (VAHTS Stranded
mRNA-seq Library Prep kit for Illumina; cat. no. NR602-01; Vazyme
Biotech Co., Ltd.). After the synthesis of the double-stranded
cDNA, the double-stranded cDNA was purified. LC magnetic beads were
used for purification and target fragment binding. The EP tube was
placed on a magnetic stand (beads combined with cDNA), then the
supernatant was removed, and washed twice with 80% ethanol for 30
sec each time. End-repair and library preparation was performed by
the aforementioned kit (VAHTS Stranded mRNA-seq Library Prep kit
for Illumina). Target size selection was performed together with
magnetic purification. PCR amplification (using Amplification Mix;
cat. no. N611-01; Vazyme Biotech Co., Ltd.) was then performed as
following: Initial denaturation 95°C for 3 min, 12 cycles of
denaturation at 98°C for 20 sec, annealing at 55°C for 15 sec,
elongation at 72°C for 30 sec, and final extension 72°C for 5 min.
The primer sequences of PCR were as follows: Forward,
5′AATGATACGGCGACCACCGAGATCTACACACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′;
reverse,
5′-CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′.
Agarose electrophoresis was used for quality inspection of the
constructed library. Qubit 2.0 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to detect the concentration of the
library, and the loading concentration of the library was 42 ng/µl
with 20 µl. After library quality tests were passed, libraries were
sequenced using an Novaseq 6000 sequencer (Illumina, Inc.) with
PE150 model (double-ended 150 bp sequencing) according to effective
concentration and target data volume. RIP-seq and subsequent
bioinformatics Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) analysis was performed by Hangzhou Lianchuan
Biotechnology Co., Ltd with the DAVID website (https://david.ncifcrf.gov/).
RNA-seq and analysis
EZH2 protein was knocked down in SH-SY5Y cells using
shRNA. Total RNA was obtained from EZH2 knockdown and control
groups with TRIzol® (Takara Bio, Inc.). Each group
included three replicates. Illumina Paired End Sample Prep kits
(Illumina, Inc.) were used to prepare libraries. Each cDNA library
was sequenced using an Illumina Hiseq 4000 (cat. no. PE150;
Illumina, Inc.). Differential expression levels of lncRNA and mRNA
transcripts between the EZH2 knockdown and control groups were
measured. RNA-seq and subsequent GO and KEGG analysis was performed
by Hangzhou Lianchuan Biotechnology Co., Ltd following the previous
study (28).
ChIP-seq
Cells from one 10-cm dish of 80–90% confluence
cultures were sonicated 4 times for (30 sec on and 30 sec off) in
precooled conditions (Fisher Sonic Dismembrator; Thermo Fisher
Scientific, Inc.). DNA was disrupted into fragments of 200–1,000 bp
by nucleic acid gel. Anti-EZH2 (1:100; cat. no. 5246S; Cell
Signaling Technology, Inc) was used to capture chromatin fragments
from cell extracts, and libraries were constructed from
immunoprecipitated DNA. An Illumina sequencer was used for
high-throughput sequencing of lncRNA and mRNA. ChIP-seq and
subsequent GO and KEGG analysis were performed by Hangzhou
Lianchuan Biotechnology Co., Ltd. The ChIP protocol was conducted
as previously described by Kong et al (29).
Differential expression level
analysis
Gene expression levels were estimated using
fragments per kilobase of transcript per million mapped reads
(FPKM) values. Cuffdiff (v2.1.1; http://cole-trapnell-lab.github.io/cufflinks/) was
used to calculate FPKM values of lncRNAs and mRNAs.DAVID
(https://david.ncifcrf.gov/) was used to
perform GO and KEGG analysis. Official gene symbols of the
significantly different genes were enriched. We followed the
instructions on the website step by step until acquiring GO and
KEGG terms. Significantly differentially expressed genes were
obtained using P-value <0.05.
Results
RIP-seq identifies EZH2-interacting
lncRNAs
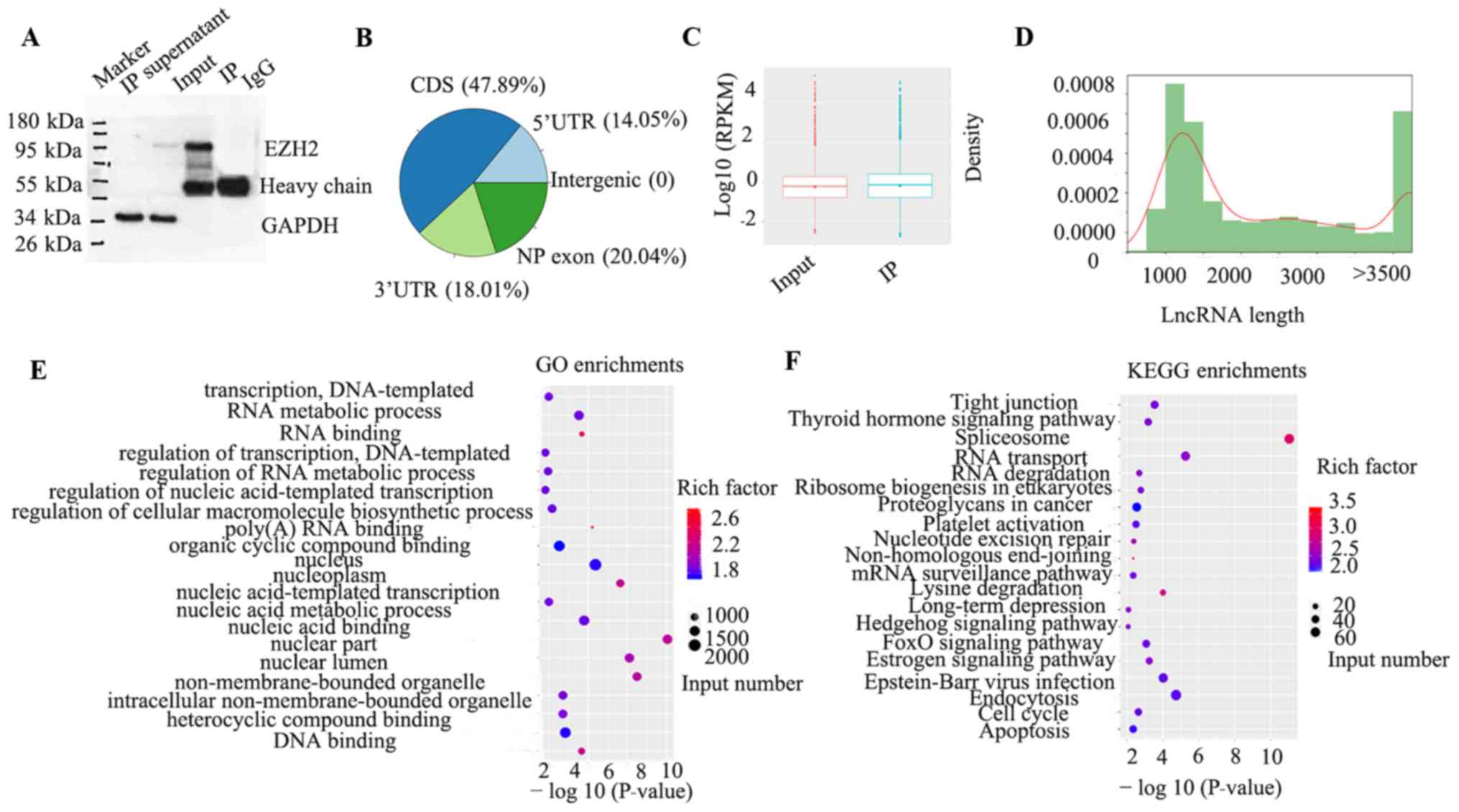
RNA Immunoprecipitation (RIP) was performed
using SH-SY5Y cells and western blotting was used to check IP
efficiency (Fig. 1A). Both IP
supernatants and inputs showed bands for the internal reference
gene GAPDH, demonstrating protein extraction and the western
blotting system is successful. Antibody heavy chain was detected in
the IP and IgG lanes, indicating that IP was successful.
Distribution statistics for peaks of each functional area of the
gene demonstrated that the coding region accounted for 50% and the
exon region of a non-coding gene was ~20% (Fig. 1B). By analyzing the distribution of
RPKM values, the gene expression level characteristics of the
sample were treated as a whole. If IP was significantly enriched
compared with the input group, the total expression level of all
genes in the IP group should have been higher than that in the
input group (Fig. 1C). lncRNAs are
>200 nucleotides in length. Length distribution statistics of
known lncRNAs were determined. The results demonstrated that
lncRNAs 500–1,000 and >3,500 bp in length were significantly
more common than lncRNAs of other lengths in SH-SY5Y cells
(Fig. 1D). A total of 2,595 lncRNAs
were identified by counting peaks associated with known lncRNAs.
Among these lncRNAs, 94 were identified via exclusion of processed
transcripts and retained introns (Table
I). GO analysis demonstrated that these lncRNAs were involved
in numerous biological processes, including cellular components and
molecular functions, such as metabolic process and transcription
regulation(Fig. 1E). Furthermore,
KEGG analysis indicated that ‘Hedgehog signaling pathway’ and ‘FoxO
signaling pathway’ were enriched in SH-SY5Y cells. ‘Tight
junction’, ‘apoptosis’ and ‘cell cycle’ may also be associated with
these lncRNAs (Fig. 1F).
 | Table I.lncRNAs determined by RNA
immunoprecipitation-sequencing. |
Table I.
lncRNAs determined by RNA
immunoprecipitation-sequencing.
| lncRNA name | lncRNA ID | lncRNA type | Chromosome |
|---|
| AF001548.2-201 |
ENST00000574212 | antisense_RNA | 16 |
| AC087280.2-201 |
ENST00000637205 | lincRNA | 11 |
| HOXA11-AS-205 |
ENST00000522863 | antisense_RNA | 7 |
| PCBP1-AS1-210 |
ENST00000416395 | antisense_RNA | 2 |
| AP002383.3-201 |
ENST00000545958 | antisense_RNA | 11 |
| CCDC18-AS1-205 |
ENST00000421202 | lincRNA | 1 |
| AL096701.3-202 |
ENST00000483736 | antisense_RNA | 22 |
| AC020661.4-201 |
ENST00000561388 | antisense_RNA | 15 |
| PITRM1-AS1-201 |
ENST00000430356 | antisense_RNA | 10 |
| AL121759.1-201 |
ENST00000434043 | antisense_RNA | 20 |
| AC244517.2-201 |
ENST00000607216 | antisense_RNA | 5 |
|
PPP1R26-AS1-202 |
ENST00000603624 | antisense_RNA | 9 |
| AC025188.1-201 |
ENST00000507514 | antisense_RNA | 5 |
| AC092115.3-201 |
ENST00000575838 | antisense_RNA | 16 |
| RAB11B-AS1-201 |
ENST00000593581 | antisense_RNA | 19 |
| AC069503.1-201 |
ENST00000538710 | lincRNA | 12 |
| LINC00843-201 |
ENST00000429104 | lincRNA | 10 |
| AC011477.3-201 |
ENST00000585571 | antisense_RNA | 19 |
| AC010320.4-201 |
ENST00000594379 | antisense_RNA | 19 |
| AL135787.1-201 |
ENST00000450154 | antisense_RNA | 9 |
| AC109587.1-201 |
ENST00000482368 | antisense_RNA | 3 |
| FAM201A-201 |
ENST00000377680 | antisense_RNA | 9 |
| SNHG7-201 |
ENST00000414282 | antisense_RNA | 9 |
| AC138150.1-201 |
ENST00000589950 | antisense_RNA | 17 |
| EMX2OS-206 |
ENST00000551288 | antisense_RNA | 10 |
| AC137932.1-201 |
ENST00000563087 | antisense_RNA | 16 |
| AC005920.2-201 |
ENST00000509833 | antisense_RNA | 17 |
| MZF1-AS1-202 |
ENST00000600534 | antisense_RNA | 19 |
| AC080112.2-201 |
ENST00000578774 | antisense_RNA | 17 |
| AC008741.1-201 |
ENST00000569456 | antisense_RNA | 16 |
| AC024563.1-201 |
ENST00000601860 | antisense_RNA | 19 |
| AC020911.1-201 |
ENST00000591038 | antisense_RNA | 19 |
| AC097467.3-216 |
ENST00000599555 | antisense_RNA | 4 |
| C5orf66-205 |
ENST00000555438 | antisense_RNA | 5 |
| SGO1-AS1-204 |
ENST00000634618 | lincRNA | 3 |
| AC011603.3-201 |
ENST00000549516 | antisense_RNA | 12 |
| ADIRF-AS1-203 |
ENST00000609111 | antisense_RNA | 10 |
| AC106864.1-201 |
ENST00000510655 | antisense_RNA | 4 |
| AC093110.1-202 |
ENST00000626206 | antisense_RNA | 2 |
| MCM3AP-AS1-201 |
ENST00000414659 | antisense_RNA | 21 |
| AC108449.1-202 |
ENST00000517632 | antisense_RNA | 8 |
| AC004923.4-201 |
ENST00000532296 | antisense_RNA | 11 |
| KDM4A-AS1-203 |
ENST00000434346 | antisense_RNA | 1 |
| FO393401.1-201 |
ENST00000453914 | antisense_RNA | 20 |
| AC078777.1-201 |
ENST00000425371 | antisense_RNA | 12 |
| PDZRN3-AS1-201 |
ENST00000478988 | antisense_RNA | 3 |
| AL139423.1-201 |
ENST00000606802 | antisense_RNA | 1 |
| AL118558.1-201 |
ENST00000557551 | antisense_RNA | 14 |
| FOXD1-AS1-201 |
ENST00000514661 | lincRNA | 5 |
| ZFPM2-AS1-207 |
ENST00000524045 | antisense_RNA | 8 |
| AC073167.1-201 |
ENST00000559303 | antisense_RNA | 15 |
| SH3BP5-AS1-202 |
ENST00000420195 | antisense_RNA | 3 |
| AC012170.2-201 |
ENST00000560380 | antisense_RNA | 15 |
| NEXN-AS1-201 |
ENST00000421331 | antisense_RNA | 1 |
| AC092329.1-201 |
ENST00000594653 | lincRNA | 19 |
| DNMBP-AS1-202 |
ENST00000434409 | antisense_RNA | 10 |
| AC013391.1-201 |
ENST00000560477 | antisense_RNA | 15 |
| PKP4-AS1-201 |
ENST00000342892 | antisense_RNA | 2 |
| LINC01089-202 |
ENST00000429892 | lincRNA | 12 |
| AC010978.1-202 |
ENST00000427050 | antisense_RNA | 2 |
| AL606534.3-201 |
ENST00000437499 | antisense_RNA | 1 |
| AL606534.1-201 |
ENST00000439562 | antisense_RNA | 1 |
| AL356599.1-205 |
ENST00000606388 | antisense_RNA | 6 |
| KTN1-AS1-202 |
ENST00000412224 | antisense_RNA | 14 |
| AC004893.2-201 |
ENST00000360902 | antisense_RNA | 7 |
| AC087286.3-201 |
ENST00000561409 | antisense_RNA | 15 |
| AL139021.2-201 |
ENST00000556390 | antisense_RNA | 14 |
| C1orf220-202 |
ENST00000521244 | lincRNA | 1 |
| H1FX-AS1-201 |
ENST00000383461 | antisense_RNA | 3 |
| SEC62-AS1-201 |
ENST00000479626 | antisense_RNA | 3 |
| AL161747.2-201 |
ENST00000535893 | antisense_RNA | 14 |
| DDN-AS1-202 |
ENST00000547866 | antisense_RNA | 12 |
| TTC3-AS1-201 |
ENST00000424733 | antisense_RNA | 1 |
| AC245452.1-201 |
ENST00000458178 | antisense_RNA | 22 |
| AC008676.1-201 |
ENST00000508443 | antisense_RNA | 5 |
| AC022395.1-201 |
ENST00000451610 | antisense_RNA | 10 |
| AC010976.1-203 |
ENST00000629005 | antisense_RNA | 2 |
| AP000229.1-201 |
ENST00000608591 | lincRNA | 21 |
| AC026427.1-201 |
ENST00000508993 | antisense_RNA | 5 |
| AC025043.1-201 |
ENST00000558047 | antisense_RNA | 15 |
|
SLC16A1-AS1-204 |
ENST00000428411 | antisense_RNA | 1 |
| ZNF337-AS1-201 |
ENST00000414393 | antisense_RNA | 20 |
| AL451047.1-201 |
ENST00000424451 | antisense_RNA | 1 |
| AC009185.1-201 |
ENST00000517634 | antisense_RNA | 5 |
| AC023790.2-201 |
ENST00000543321 | lincRNA | 12 |
| AC092143.3-201 |
ENST00000565150 | antisense_RNA | 16 |
| ASH1L-AS1-202 |
ENST00000456633 | antisense_RNA | 1 |
| SPG20-AS1-203 |
ENST00000493739 | antisense_RNA | 13 |
| ALKBH3-AS1-201 |
ENST00000499194 | antisense_RNA | 11 |
| SNHG22-201 |
ENST00000589499 | antisense_RNA | 18 |
| AC010624.3-201 |
ENST00000599914 | antisense_RNA | 19 |
| AL592166.1-202 |
ENST00000428791 | antisense_RNA | 1 |
RNA-seq for mRNAs and lncRNAs
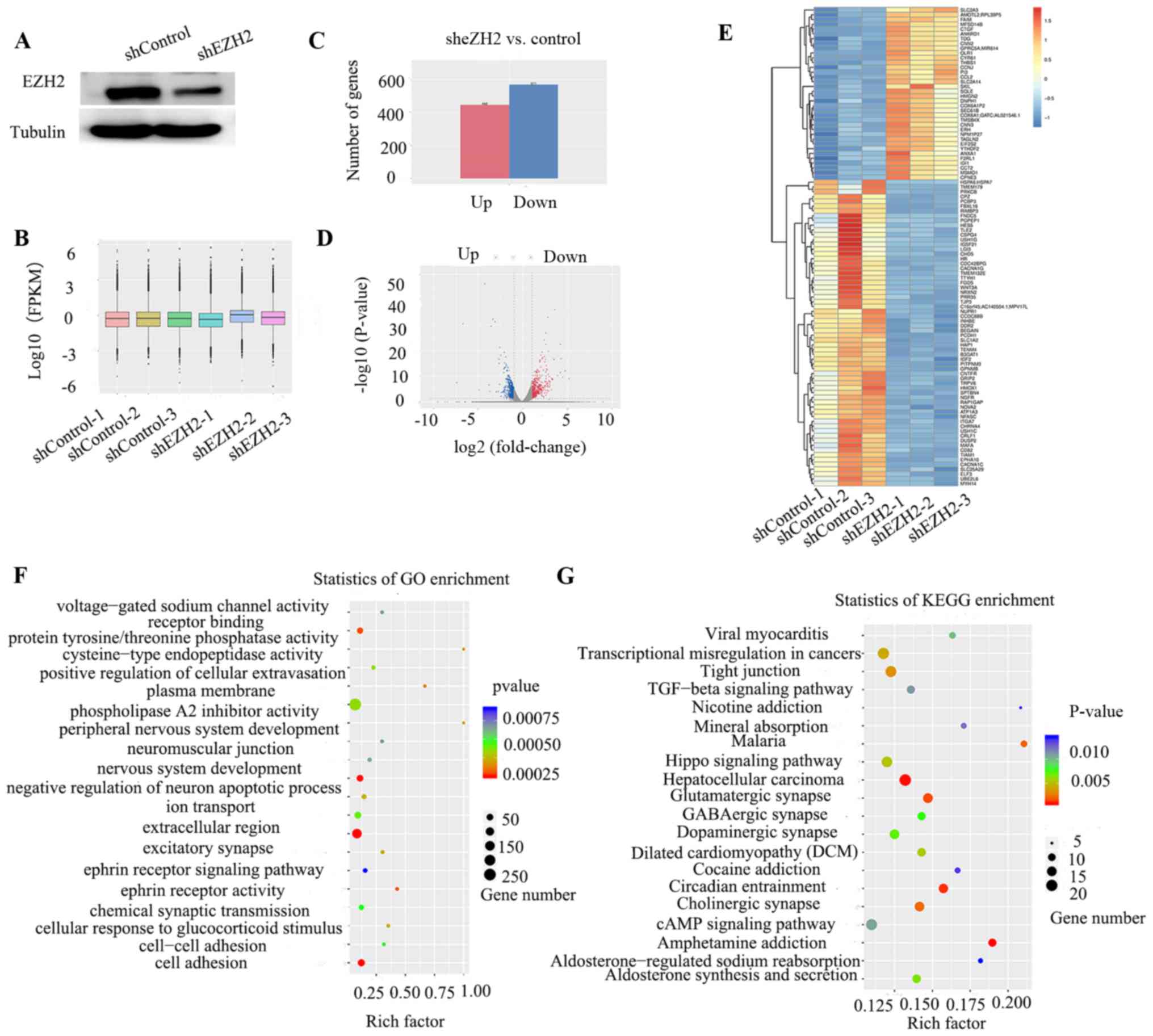
The results from western blotting demonstrated that
EZH2 was significantly downregulated following shRNA transfection
(Fig. 2A). By analyzing the
distribution of FPKM values, the gene expression level
characteristics of the sample were treated as a whole (Fig. 2B). Following EZH2 knockdown, 448 up-
and 571 downregulated genes were differentially expressed compared
with the normal control group (Fig. 2C
and D). A heatmap of the top 100 differentially expressed genes
was generated (Fig. 2E). GO analysis
demonstrated that these genes were associated with ‘negative
regulation of neuron apoptotic processes’, ‘nervous system
development’ and ‘peripheral nervous system development’. KEGG
analysis showed that enriched genes were primarily distributed in
the ‘TGF-β signaling pathway’, ‘Hippo signaling pathway’ and ‘cAMP
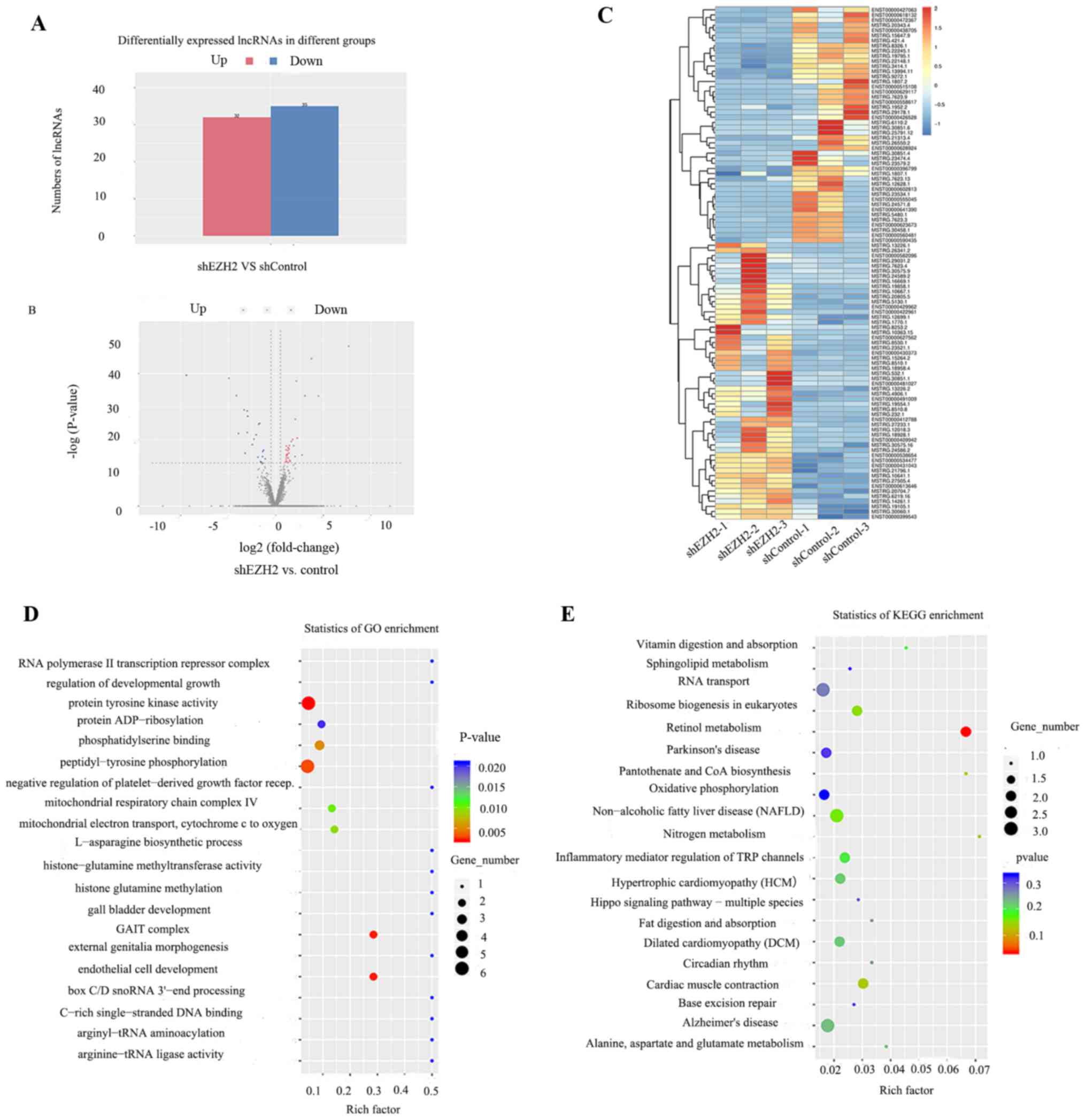
signaling pathway’. Compared with the normal control group, 32 up-
and 35 downregulated lncRNAs were differentially expressed in the
shEZH2 group (Fig. 3A and B;
Table II). A heatmap of the top 100
differentially expressed lncRNAs (including known and novel
lncRNAs) was generated (Fig. 3C). GO
analysis demonstrated that these lncRNAs were involved in numerous
biological processes, including ‘regulation of developmental
growth’, ‘peptidyl-tyrosine phosphorylation’ and ‘histone glutamine
methylation’ (Fig. 3D). KEGG
analysis demonstrated that ‘Hedgehog signaling pathway’,
‘Parkinson's disease’ and ‘Alzheimer's disease’ were associated
with these lncRNAs (Fig. 3E).
 | Table II.Different lncRNAs determined by
RNA-sequencing following enhancer of zeste homolog 2 knockdown. |
Table II.
Different lncRNAs determined by
RNA-sequencing following enhancer of zeste homolog 2 knockdown.
| lncRNA name | lncRNA ID | Chromosome | Regulation |
|---|
| MALAT1 |
ENST00000618132 | 11 | Down |
| AL360012.1 |
ENST00000602813 | 1 | Down |
| TTN-AS1 |
ENST00000629117 | 2 | Down |
| AC108488.1 |
ENST00000422961 | 2 | Up |
| EXOC3-AS1 |
ENST00000623673 | 5 | Down |
| AC015813.1 |
ENST00000582096 | 17 | Up |
| AC025171.2 |
ENST00000515108 | 5 | Down |
| DHRS4-AS1 |
ENST00000555045 | 14 | Down |
| AC027237.3 |
ENST00000558617 | 15 | Down |
| SVIL-AS1 |
ENST00000427063 | 10 | Down |
| SNHG15 |
ENST00000438705 | 7 | Down |
| SNHG5 |
ENST00000431043 | 6 | Up |
| AC125494.1 |
ENST00000396799 | 12 | Down |
| AC079781.5 |
ENST00000641390 | 7 | Down |
| LINC00958 |
ENST00000534477 | 11 | Up |
| LRRC75A-AS1 |
ENST00000472367 | 17 | Down |
| H19 |
ENST00000412788 | 11 | Up |
| FAM212B-AS1 |
ENST00000430373 | 1 | Up |
| AC016757.1 |
ENST00000409942 | 2 | Up |
| AL590133.2 |
ENST00000560481 | 1 | Down |
| THAP7-AS1 |
ENST00000429962 | 22 | Up |
| EBLN3P |
ENST00000628924 | 9 | Down |
| AL139099.5 | MSTRG.8510.8 | 14 | Up |
| ARF4 | MSTRG.21313.4 | 3 | Down |
| GRAPL | MSTRG.12628.1 | 17 | Down |
| AC006064.4 | MSTRG.6110.2 | 12 | Down |
| RBFOX2 | MSTRG.20343.4 | 22 | Down |
| AL139099.5 | MSTRG.8510.1 | 14 | Up |
| LINC00854 | MSTRG.13226.1 | 17 | Up |
| AC092329.3 | MSTRG.15647.9 | 19 | Down |
| FIRRE | MSTRG.30851.6 | X | Down |
| AL627171.1 | MSTRG.8530.1 | 14 | Up |
| FAM182B | MSTRG.18928.1 | 20 | Up |
| RMRP | MSTRG.29178.1 | 9 | Down |
| LINC01021 | MSTRG.23474.4 | 5 | Down |
| AP002360.2 | MSTRG.5480.1 | 11 | Down |
| UBC | MSTRG.7623.9 | 12 | Down |
| MST1L | MSTRG.421.4 | 1 | Down |
| SNHG16 | MSTRG.13994.11 | 17 | Down |
| CCNG1 | MSTRG.24571.8 | 5 | Down |
| Z94721.1 | MSTRG.26341.2 | 6 | Up |
| NDUFA4 | MSTRG.26550.2 | 7 | Down |
| FBXL16 | MSTRG.10641.1 | 16 | Up |
| PANK3 | MSTRG.24589.2 | 5 | Up |
| LINC00854 | MSTRG.13226.2 | 17 | Up |
| C16orf74 | MSTRG.12018.3 | 16 | Up |
| MIR34AHG | MSTRG.232.1 | 1 | Up |
| MAP2K3 | MSTRG.12699.1 | 17 | Up |
| ASXL1 | MSTRG.18958.4 | 20 | Up |
| TJP1 | MSTRG.9272.1 | 15 | Down |
| MANBAL | MSTRG.19105.1 | 20 | Up |
| AL355075.4 | MSTRG.8253.2 | 14 | Up |
| XIST | MSTRG.30575.16 | X | Up |
| FIRRE | MSTRG.30851.1 | X | Up |
| ZNF436-AS1 | MSTRG.532.1 | 1 | Up |
| RPL37 | MSTRG.23579.2 | 5 | Down |
| FAM111B | MSTRG.4906.1 | 11 | Up |
| AC092821.3 | MSTRG.6219.16 | 12 | Up |
| NUp210 | MSTRG.20805.5 | 3 | Up |
| PARG | MSTRG.3414.1 | 10 | Down |
ChIP-seq for EZH2
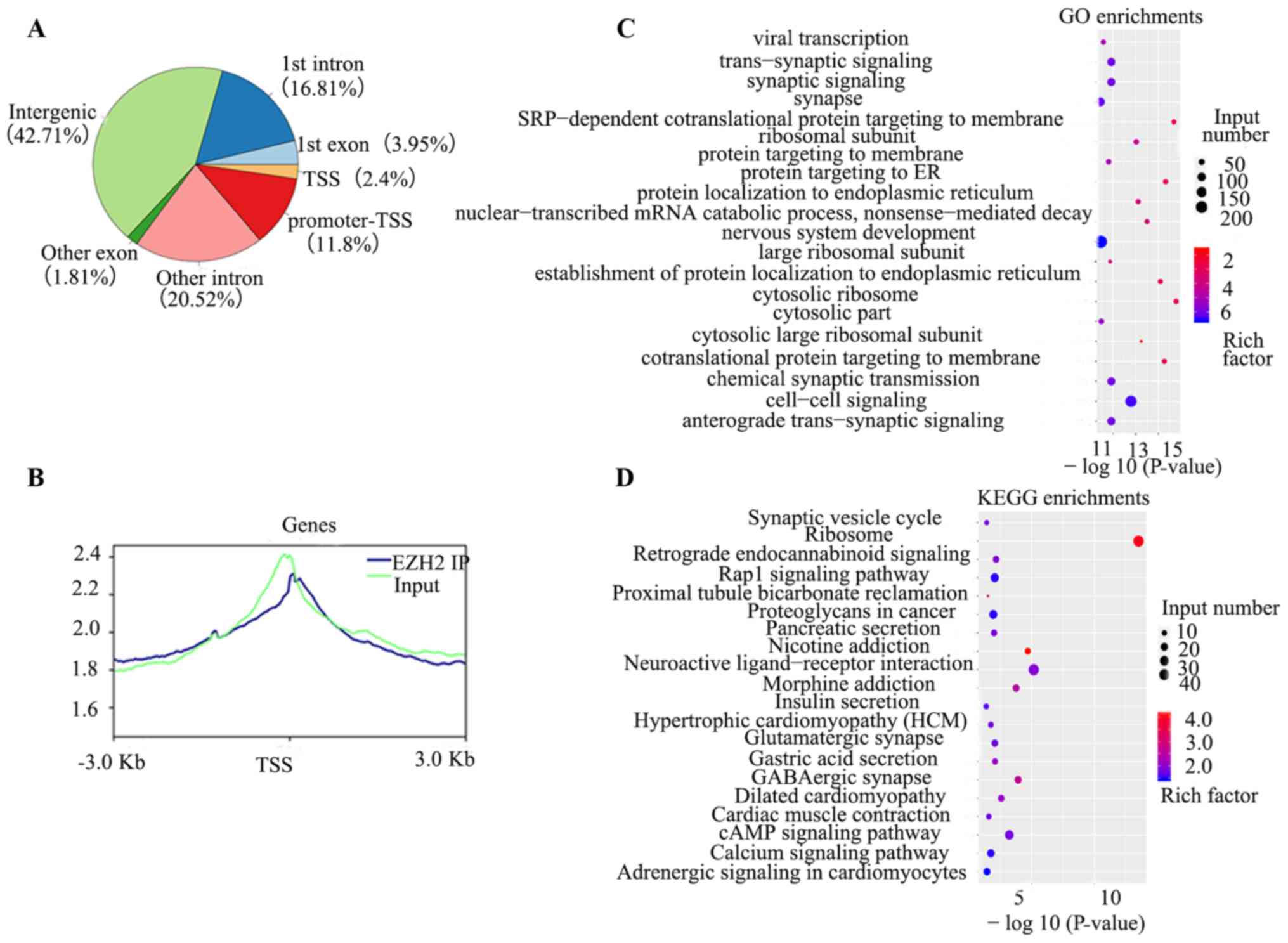
Due to the influence of chromosome conformation,
chromosome expression levels in the active region of gene
expression levels was more open. This resulted in input DNA reads
exhibiting greater abundance in promoter and gene body regions,
with a characteristic decrease near the transcription start site
(TSS). The distribution of IP DNA is associated with EZH2 proteins,
and apparent modifications such as transcription factors and
H3K27me3 were enriched in the promoter and gene body regions.
Through the distribution of reads in the intervals of these genes,
the success of ChIP-seq experiments was verified (Fig. 4B). Analysis of peak distribution in
the genomic functional area indicated that intergenic and
promoter-TSS were the most frequent areas (Fig. 4A). ChIP-seq identified 634 genes
located in the promoter region, including 138 long intervening
non-coding RNAs (lincRNA; Table
III). GO analysis demonstrated enrichment of ‘nervous system
development’, ‘chemical synaptic transmission’ and ‘trans-synaptic
signaling’ (Fig. 4C). KEGG analysis
indicated that ‘Rap1 signaling pathway’, ‘cAMP signaling pathway’
and ‘retrograde endocannabinoid signaling’ were enriched (Fig. 4D).
 | Table III.Promoter of long non-coding RNA
determined by chromatin immunoprecipitation-sequencing. |
Table III.
Promoter of long non-coding RNA
determined by chromatin immunoprecipitation-sequencing.
| Gene ID | Chromosome | Start | End | Strand |
Annotated_Transcript | logFC | P-value |
|---|
| RP11-672L10.3 | 18 | 907879 | 908507 | − |
ENST00000582554 | 3.129033173 |
1.33492×10−12 |
| KIRREL3 | 11 | 127003233 | 127004161 | − |
ENST00000533026 | 3.003238319 |
1.50761×10−06 |
| KIRREL3 | 11 | 127000973 | 127001187 | − |
ENST00000547738 | 3.003238319 |
1.50761×10−06 |
| IGF2-AS | 11 | 2139621 | 2141215 | + |
ENST00000381361 | 2.877117623 |
1.14607×10−17 |
| AP000282.2 | 21 | 33070946 | 33071173 | − |
ENST00000454622 | 2.813201993 |
4.67603×10−05 |
| RP11-88H9.2 | 1 | 111989337 | 111991186 | + |
ENST00000438293 | 2.775316437 |
2.86341×10−10 |
| TBX5-AS1 | 12 | 114407959 | 114408321 | + |
ENST00000531202 | 2.734420323 |
9.25093×10−07 |
| MIR1-1HG | 20 | 62549839 | 62550053 | + |
ENST00000624914 | 2.522145791 |
9.65857×10−10 |
| FLJ16779 | 20 | 63253277 | 63253737 | + |
ENST00000612722 | 2.510159699 |
1.96314×10−09 |
| RP11-92C4.6 | 9 | 98942992 | 98944517 | − |
ENST00000605631 | 2.391645807 |
1.12556×10−08 |
| RP11-672L10.2 | 18 | 904551 | 904765 | − |
ENST00000582921 | 2.372166725 |
1.71009×1007 |
| RP11-672L10.2 | 18 | 904176 | 904390 | − |
ENST00000582921 | 2.372166725 |
1.71009×10−07 |
| RP11-672L10.2 | 18 | 906549 | 907654 | − |
ENST00000581719 | 2.372166725 |
1.71009×10−07 |
| RP3-525N10.2 | 6 | 68634166 | 68635846 | − |
ENST00000604392 | 2.366348664 |
2.47756×10−10 |
| AC099754.1 | 3 | 26622451 | 26622769 | − |
ENST00000435884 | 2.28090657 |
4.94043×10−05 |
| RP11-436F23.1 | 4 | 46389763 | 46390468 | + |
ENST00000502455 | 2.248486391 |
1.13494×10−08 |
| CTC-235G5.3 | 5 | 76084156 | 76085290 | − |
ENST00000503652 | 2.221930293 |
2.35777×10−09 |
| RP11-274G22.1 | X | 21373658 | 21374071 | − |
ENST00000636317 | 2.211953324 |
1.02635×10−11 |
| RP11-343J3.2 | 10 | 69574708 | 69575542 | + |
ENST00000428753 | 2.207584122 |
1.94744×10−06 |
| AP002856.5 | 11 | 131252716 | 131253002 | + |
ENST00000416553 | 2.203360843 | 0.001177692 |
| RP11-452C13.1 | 7 | 157867952 | 157868291 | + |
ENST00000608596 | 2.19401377 | 0.001695242 |
| GPR50-AS1 | X | 151177247 | 151178372 | − |
ENST00000454196 | 2.152759945 |
3.44626×10−06 |
| LINC00200 | 10 | 1159259 | 1159473 | + |
ENST00000425630 | 2.151520464 | 0.000313333 |
| RNF219-AS1 | 13 | 77918846 | 77919422 | + |
ENST00000607862 | 2.145410121 | 0.000124562 |
| C8orf34-AS1 | 8 | 68331425 | 68331669 | − |
ENST00000512294 | 2.129407806 | 0.00016937 |
| RP11-17E2.2 | 4 | 21948016 | 21948230 | + |
ENST00000510705 | 2.129407806 | 0.00016937 |
| RP1-269M15.3 | 20 | 43189349 | 43189646 | + |
ENST00000611791 | 2.129407806 | 0.00016937 |
| RP6-24A23.3 | X | 108735146 | 108736079 | + |
ENST00000608811 | 2.103500759 |
2.23745×10−05 |
| RP4-683L5.1 | 11 | 35418158 | 35420102 | + |
ENST00000534165 | 2.073937887 |
6.79163×10−10 |
| PTPRD-AS2 | 9 | 10612459 | 10613283 | + |
ENST00000429581 | 2.04619868 | 0.000273436 |
| LINC01210 | 3 | 137771274 | 137772805 | + |
ENST00000478772 | 2.039638671 |
5.14222×10−08 |
| RP11-563K23.1 | 7 | 143363541 | 143363755 | + |
ENST00000609674 | 2.008614264 |
1.33265×10−05 |
| RP11-588H7.1 | 3 | 14601566 | 14601780 | + |
ENST00000635800 | 1.989744589 | 0.00057344 |
| RP11-343P9.1 | 8 | 135456592 | 135457331 | − |
ENST00000518674 | 1.980487281 |
5.20702×10−06 |
| FGF14-AS1 | 13 | 102368126 | 102368468 | + |
ENST00000451630 | 1.970094904 | 0.004327089 |
| CTD-2523D13.2 | 11 | 119729856 | 119730070 | + |
ENST00000533253 | 1.954654555 | 0.000134587 |
| RP11-901H12.1 | 3 | 21227123 | 21227337 | − |
ENST00000634947 | 1.950333985 | 0.006647239 |
| CASC16 | 16 | 52606660 | 52606874 | − |
ENST00000510238 | 1.943876617 | 0.010319298 |
| ADGRA1-AS1 | 10 | 133088588 | 133089156 | − |
ENST00000366099 | 1.898488801 |
8.76189×10−06 |
| RP11-231C18.1 | 4 | 54331878 | 54332092 | + |
ENST00000511634 | 1.883283382 | 0.001289255 |
| MIR519A1 | 19 | 53751934 | 53752148 | + |
ENST00000385257 | 1.874499442 | 0.025042572 |
| MIR129-2 | 11 | 43580825 | 43581855 | + |
ENST00000362207 | 1.872185866 |
1.61125×10−07 |
| CD1E | 1 | 158352934 | 158353309 | + |
ENST00000368167 | 1.847765953 | 0.00285251 |
| RP11-415C15.1 | 4 | 27140713 | 27140939 | − |
ENST00000506878 | 1.82461652 | 0.001888542 |
| RP11-21C4.1 | 8 | 64578103 | 64578381 | − |
ENST00000520834 | 1.797854867 |
1.29304×10−05 |
| AL022344.7 | 10 | 42751909 | 42752386 | − |
ENST00000568976 | 1.796142677 | 0.000430003 |
| HAR1B | 20 | 63102600 | 63103193 | − |
ENST00000447910 | 1.784741755 | 0.000108551 |
| RP1-273N12.4 | 6 | 98832858 | 98833303 | − |
ENST00000635423 | 1.782829544 | 0.000111871 |
| RP11-461O7.1 | 16 | 56190697 | 56191236 | − |
ENST00000501259 | 1.742859438 | 0.000158433 |
| MAPT-AS1 | 17 | 45895359 | 45895627 | − |
ENST00000579599 | 1.733234586 |
9.03861×10−06 |
| RP1-90G24.10 | 22 | 32204199 | 32204413 | + |
ENST00000434942 | 1.705684675 | 0.000158433 |
| HAR1A | 20 | 63102059 | 63102459 | + |
ENST00000433161 | 1.705257532 | 0.000218753 |
|
LL22NC03-121E8.3 | 22 | 48141487 | 48141701 | + |
ENST00000446364 | 1.699601022 | 0.004015236 |
| CTD-2194D22.3 | 5 | 1882806 | 1883957 | + |
ENST00000506335 | 1.67006809 |
4.45899×10−07 |
| FOXC2-AS1 | 16 | 86567608 | 86568108 | − |
ENST00000563280 | 1.667099473 | 0.000418716 |
| MIR124-2HG | 8 | 64376996 | 64377210 | + |
ENST00000524060 | 1.656091826 | 0.009625718 |
| RP11-876N24.2 | 16 | 10888696 | 10889194 | − |
ENST00000572017 | 1.645368552 | 0.001847602 |
| AC010729.1 | 2 | 5695877 | 5696091 | + |
ENST00000455579 | 1.634091046 | 0.000299766 |
| RP11-146I2.1 | 6 | 15089927 | 15090141 | − |
ENST00000437648 | 1.632781249 | 0.005823769 |
| CTD-2089N3.1 | 5 | 50967242 | 50967513 | − |
ENST00000510349 | 1.624569379 | 0.009038249 |
| RP11-386B13.3 | 4 | 185051181 | 185051395 | + |
ENST00000509017 | 1.614500015 | 0.009625718 |
| RP11-1263C18.2 | 4 | 576322 | 576536 | + |
ENST00000637674 | 1.611131906 | 0.001648005 |
| RP11-486P11.1 | 7 | 20095996 | 20096210 | + |
ENST00000590912 | 1.608559926 | 0.014306174 |
| CTA-125H2.2 | 22 | 25898897 | 25899111 | − |
ENST00000595102 | 1.601862196 | 0.031052599 |
| CTA-125H2.2 | 22 | 25903547 | 25903761 | − |
ENST00000608257 | 1.601862196 | 0.031052599 |
| RP11-140A10.3 | 10 | 132185784 | 132186004 | − |
ENST00000443922 | 1.597660222 | 0.000116767 |
| LHX5-AS1 | 12 | 113470956 | 113471262 | + |
ENST00000551357 | 1.571644358 | 0.001181247 |
| RP11-923I11.6 | 12 | 51816816 | 51817177 | + |
ENST00000562343 | 1.562378841 | 0.0022933 |
| RP11-255H23.4 | 19 | 23875050 | 23875264 | − |
ENST00000599944 | 1.558300834 | 0.031052599 |
| RP11-636O21.2 | 18 | 39841267 | 39841481 | + |
ENST00000637369 | 1.546964313 | 0.013197567 |
| RP13-977J11.9 | 12 | 132168767 | 132169107 | + |
ENST00000619983 | 1.525547657 | 0.005463975 |
| CTA-299D3.8 | 22 | 48547918 | 48548151 | − |
ENST00000626321 | 1.50297288 | 0.045280597 |
| LINC01497 | 17 | 69960846 | 69961199 | + |
ENST00000455460 | 1.481003517 | 0.007180229 |
| FGF12-AS2 | 3 | 192514512 | 192515287 | + |
ENST00000443165 | 1.467946667 | 0.004402766 |
| AC145110.1 | 8 | 29748261 | 29748499 | + |
ENST00000517491 | 1.464946227 | 0.019169253 |
| DOCK4-AS1 | 7 | 111808080 | 111808294 | + |
ENST00000452714 | 1.464946227 | 0.019169253 |
| AC011513.4 | 19 | 41786996 | 41787210 | − |
ENST00000601409 | 1.459633143 | 0.006071182 |
| RP11-347C12.11 | 16 | 30359204 | 30359692 | + |
ENST00000611264 | 1.442748682 | 0.002976471 |
| GRM7-AS1 | 3 | 7560233 | 7560551 | − |
ENST00000427273 | 1.438859533 | 0.015604312 |
| RP11-164C12.2 | 15 | 93423281 | 93423680 | − |
ENST00000556708 | 1.438859533 | 0.015604312 |
| RP11-655C2.3 | 11 | 58505612 | 58505826 | − |
ENST00000527054 | 1.438859533 | 0.015604312 |
| RP11-476M19.2 | 12 | 3367415 | 3367629 | + |
ENST00000542449 | 1.429070956 | 0.002593418 |
| RP11-583F24.7 | 11 | 18864950 | 18865166 | − |
ENST00000524957 | 1.412555095 | 0.003778445 |
| FGF10-AS1 | 5 | 44388123 | 44388744 | + |
ENST00000502457 | 1.397829443 | 0.000213665 |
| LINC00342 | 2 | 95816770 | 95817393 | − |
ENST00000448494 | 1.39486642 | 0.011048418 |
| RP11-31E13.2 | 10 | 78696881 | 78697101 | − |
ENST00000455498 | 1.39486642 | 0.011048418 |
| CTC-490G23.2 | 19 | 43331306 | 43331853 | − |
ENST00000595748 | 1.393500649 | 0.001691433 |
| RP11-715J22.3 | 16 | 2453190 | 2453657 | − |
ENST00000561653 | 1.382523434 | 0.009165782 |
| CTC-293G12.1 | 5 | 100658715 | 100658963 | − |
ENST00000511592 | 1.377982806 | 0.027678904 |
| RP11-1151B14.2 | 18 | 58415921 | 58416146 | + |
ENST00000585470 | 1.372986331 | 0.006071182 |
| LINC00905 | 19 | 16034867 | 16035081 | + |
ENST00000589071 | 1.349005167 | 0.011418275 |
| RP11-417J1.1 | 5 | 8524277 | 8524621 | + |
ENST00000505784 | 1.346034828 | 0.001397979 |
| RP11-339N8.1 | 9 | 108703360 | 108703574 | − |
ENST00000415465 | 1.329449778 | 0.034844927 |
| RP11-100E13.1 | 1 | 224615665 | 224615973 | − |
ENST00000437416 | 1.313540747 | 0.003978429 |
| CTD-2369P2.4 | 19 | 10260054 | 10260664 | − |
ENST00000587088 | 1.291980197 | 0.008432619 |
| RP11-120A1.1 | 4 | 21304232 | 21304532 | + |
ENST00000515680 | 1.290820753 | 0.018753813 |
| CTC-529I10.2 | 17 | 16040460 | 16040674 | + |
ENST00000442828 | 1.289168246 | 0.019530136 |
| RP11-404O13.1 | 1 | 158147337 | 158147621 | − |
ENST00000635685 | 1.289168246 | 0.019530136 |
| RP11-554D14.6 | 12 | 107864072 | 107864415 | − |
ENST00000547452 | 1.279468004 | 0.006918611 |
| CH17-125A10.2 | 1 | 144642430 | 144642671 | − |
ENST00000615763 | 1.278798153 | 0.017419713 |
| CH17-125A10.2 | 1 | 144643049 | 144643263 | − |
ENST00000615763 | 1.278798153 | 0.017419713 |
| RP11-308B16.2 | 5 | 12573811 | 12574025 | − |
ENST00000502209 | 1.271835066 | 0.005263785 |
| RP5-1159O4.2 | 7 | 7553201 | 7553548 | − |
ENST00000608770 | 1.262358355 | 0.018753813 |
| SATB1-AS1 | 3 | 18444817 | 18445100 | + |
ENST00000414198 | 1.243227131 | 0.011232888 |
| SATB1-AS1 | 3 | 18692681 | 18693359 | + |
ENST00000425799 | 1.243227131 | 0.011232888 |
| CTD-2278I10.1 | 19 | 17360281 | 17360495 | + |
ENST00000597592 | 1.242486357 | 0.048953034 |
| LINC01252 | 12 | 11547257 | 11547497 | + |
ENST00000499291 | 1.242486357 | 0.048953034 |
| LINC01194 | 5 | 12574630 | 12574871 | + |
ENST00000505196 | 1.210837745 | 0.008262948 |
| AC002539.1 | 17 | 70074000 | 70074236 | − |
ENST00000587325 | 1.203691494 | 0.02513009 |
| RP11-30J20.1 | 8 | 136529857 | 136530169 | + |
ENST00000524346 | 1.203691494 | 0.02513009 |
| RP11-30J20.1 | 8 | 136536567 | 136536781 | + |
ENST00000517345 | 1.203691494 | 0.02513009 |
| RP4-555D20.3 | 3 | 43996414 | 43996646 | + |
ENST00000605537 | 1.192768756 | 0.014908014 |
| GFOD1-AS1 | 6 | 13486482 | 13486696 | + |
ENST00000446001 | 1.18462265 | 0.009134047 |
| U91319.1 | 16 | 13245463 | 13245712 | + |
ENST00000571619 | 1.165322808 | 0.03830181 |
| AC009501.4 | 2 | 63048680 | 63049151 | − |
ENST00000437346 | 1.160170784 | 0.004406531 |
| AC000403.4 | 13 | 76887415 | 76887795 | + |
ENST00000613696 | 1.151662451 | 0.003423805 |
| AC009404.2 | 2 | 117833865 | 117834191 | + |
ENST00000420330 | 1.14253746 | 0.044449271 |
| CTD-3224K15.2 | 5 | 139649694 | 139650142 | − |
ENST00000514287 | 1.1368449 | 0.009213888 |
| RP11-234B24.2 | 12 | 4719653 | 4719867 | − |
ENST00000527518 | 1.121955081 | 0.02513009 |
| LINC00694 | 3 | 44439800 | 44440014 | − |
ENST00000636468 | 1.120622631 | 0.043957484 |
| RP1-35C21.1 | 1 | 177350670 | 177350941 | + |
ENST00000451341 | 1.120622631 | 0.043957484 |
| RP1-90J20.2 | 6 | 2916876 | 2917111 | + |
ENST00000437718 | 1.106734529 | 0.017533495 |
| PABPC5-AS1 | X | 91435542 | 91435757 | − |
ENST00000456187 | 1.103269093 | 0.029430716 |
| MMP25-AS1 | 16 | 3059866 | 3060087 | − |
ENST00000572574 | 1.099462156 | 0.011720469 |
| RP11-331F9.4 | 9 | 35645503 | 35645717 | + |
ENST00000428948 | 1.070621547 | 0.038802283 |
| RP11-307E17.8 | 9 | 94331494 | 94331776 | + |
ENST00000454869 | 1.069513076 | 0.044449271 |
| RP11-353N14.1 | 17 | 79800162 | 79800376 | + |
ENST00000570512 | 1.069080411 | 0.004834217 |
| RP11-297M9.1 | 16 | 9677330 | 9677544 | − |
ENST00000561538 | 1.060801047 | 0.033509944 |
| RP11-637O11.2 | 3 | 168287466 | 168287707 | + |
ENST00000496247 | 1.060801047 | 0.033509944 |
| RP1-253P7.4 | 17 | 17857479 | 17857693 | + |
ENST00000354746 | 1.060801047 | 0.033509944 |
| CTB-118N6.2 | 5 | 116573419 | 116573633 | + |
ENST00000508766 | 1.041943716 | 0.018538748 |
| CTD-2516F10.2 | 11 | 7435808 | 7436022 | − |
ENST00000530846 | 1.041943716 | 0.018538748 |
| CASC15 | 6 | 21664562 | 21664824 | + |
ENST00000606336 | 1.014892605 | 0.030066952 |
| RP11-410N8.3 | 20 | 32560238 | 32560452 | + |
ENST00000413983 | 1.008861783 | 0.030000331 |
| RP11-805F19.1 | 18 | 15165032 | 15165460 | − |
ENST00000581690 | 0.992961131 | 0.043825627 |
| RP11-452I5.2 | 17 | 74749000 | 74749350 | − |
ENST00000585285 | 0.958163776 | 0.023648425 |
| AC009133.12 | 16 | 29821695 | 29821963 | − |
ENST00000564980 | 0.896472053 | 0.046484806 |
| RP11-680H20.2 | 11 | 94237505 | 94238448 | + |
ENST00000506309 | 0.678965838 | 0.003627177 |
Discussion
Although numerous treatments for NB currently exist,
patients with NB have only 40% survival rate (2,30). A
novel treatment is therefore needed to improve survival rate. EZH2
is a member of the polycomb group protein family that is
upregulated in various types of cancer, including NB (31,32). Li
et al (32) demonstrated that
EZH2 knockdown significantly inhibits NB differentiation.
Transcriptome sequencing has demonstrated that neurotrophic
receptor tyrosine kinase 1 may be a target of EZH2. Chen et
al (31) reported that the MYCN
gene binds to the EZH2 promoter, directly promoting EZH2 expression
and EZH2 inhibition of neuronal differentiation in a PRC2-dependent
manner (33). Tsubota et al
(34) demonstrated that EZH2
inhibitors significantly repress the growth of tyrosine
hydroxylase-MYCN NB mice, and that MYCN and PRC2 targets are
positively correlated in NB. EZH2 may therefore be considered as a
novel target for NB treatment. Bate-Eya et al (35) demonstrated that high expression of
EZH2 has a survival function independent of its methyltransferase
activity in NB. Although inhibitors of EZH2 are at pre-clinical
stage in many cancers, their efficacy and underlying mechanism in
NB remain unknown.
In a previous study, certain lncRNAs were
demonstrated to serve key roles in NB. For example, FOXD3-antisense
RNA (AS) 1 is downregulated in NB tissues and cell lines; this is
an independent prognostic marker for favorable outcomes for
patients with NB. FOXD3-AS1 inhibits the progression of NB via
repressing poly-ADP ribose polymerase 1-mediated CCCTC-binding
factor activation (36). The lncRNA
pancEts-1 is upregulated and is an independent prognostic factor
for unfavorable NB outcomes. In addition, pancEts-1 directly
interacts with heterogeneous nuclear ribonucleoprotein K to
increase its interaction with β-catenin, resulting in stabilization
and transactivation of β-catenin and promotion of the growth and
metastasis of NB both in vitro and in vivo (37). EZH2 is a transcriptional repressor
associated with lncRNA. Numerous lncRNAs are associated with EZH2
with positive or negative correlation (38,39).
Since the interacting gene product enhances the co-expressed gene,
positively correlated lncRNA is a potential ligand for EZH2 or has
the same transcriptional machinery as EZH2 (40). Knocking down EZH2 using small
interfering RNA has previously confirmed that lncRNA is negatively
correlated with EZH2 expression and is inhibited by EZH2 (41). The present study demonstrated that
numerous lncRNAs were associated with EZH2. RIP-seq identified 94
lncRNAs that may bind to EZH2 directly. Among lncRNAs, Chi et
al (42) reported that small
nucleolar host gene (SNHG) 7 facilitates NB progression via the
microRNA (miR)-653-5p/signal transducer and activator of
transcription 2 pathway, providing a novel therapeutic target and
prognostic biomarker for NB. The lncRNA family with sequence
similarity 201A may affect the radiosensitivity of esophageal
squamous cell cancer by regulating ataxia telangiectasia mutated
(ATM) and mTOR expression via miR-101 (43). In the present study, RNA-seq
demonstrated that 32 up- and 35 downregulated lncRNAs were
differentially expressed in the shEZH2 group compared with the
control group. Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) (44), H19 (45) and X-inactive specific transcript
(XIST) (46) were some of the first
reported lncRNAs associated with NB development. Koshimizu et
al (47) demonstrated that
expression level of the tumor marker MALAT1 is sensitive to cell
surface receptor activation by oxytocin in an NB cell line. In
addition, a six-center case-control study identified three single
nucleotide polymorphisms (SNPs; rs2839698 G>A, rs3024270 C>G
and rs217727 G>A) from the H19 gene in a Chinese population (700
people with NB and 1,516 controls) and investigated the effect of
individual and combined SNPs on NB risk (48). Zhang et al (49) demonstrated that XIST downregulates
the Dickkopf Wnt signaling pathway inhibitor 1 by promoting H3
histone methylation via EZH2, inhibiting proliferation, migration
and invasion of NB cells and limiting tumor development. In
addition, SNHG family members, SNHG5, is upregulated while SNHG15
and SNHG16 is downregulated in NB. SNHG16 is reported to facilitate
proliferation, migration, invasion and autophagy of NB cells via
sponging miR-542-3p and upregulating autophagy-related 5 expression
levels (50). However, the
involvement of these lncRNAs in NB remains unknown. Among the 138
lincRNAs identified by EZH2 ChIP-seq, cancer susceptibility 15 was
identified as a tumor suppressor that can regulate numerous genes
involved in neural crest development (51). GO analysis demonstrated that EZH2
participated in a number of biological processes, such as ‘nervous
system development’, ‘regulation of developmental growth’ and
‘histone glutamine methylation’. KEGG analysis showed that
‘Hedgehog signaling pathway’ was enriched in both RIP-seq and
RNA-seq, indicating that the pathway may be important in
EZH2-associated lincRNAs.
In conclusion, the present study demonstrated that
numerous lincRNAs could directly bind to EZH2. Certain lincRNAs may
regulate or be regulated by EZH2. Certain lncRNAs were associated
with N6-methyladenosine and may potentially encode functional
polypeptides. In addition, the difficulty of EZH2-targeted drug
research may be associated with these lincRNAs. These lincRNAs may
provide a novel option for EZH2-centered molecular target
therapy.
Acknowledgements
Not applicable.
Funding
This study received financial support from Shanghai
Key Disciplines (grant no. 2017ZZ02022), National Natural Science
Foundation of China (grant nos. 81771633 and 81572324) and Science
Foundation of Shanghai (grant nos. 17411960600 and
15ZR1404200).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
KD, DM and MY designed the study. MY, LX and JZ
collected the data and performed experiments. BL, XL and JH
analyzed and interpreted the data. DM and KD were involved in
critical reviewing of the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PRC2
|
polycomb repressor complex 2
|
|
RNA-seq
|
RNA sequencing
|
|
RIP-seq
|
RNA immunoprecipitation sequencing
|
|
ChIP-seq
|
chromatin immunoprecipitation
sequencing
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Alam MW, Boreas M, Lind DE,
Cervantes-Madrid D, Umapathy G, Palmer RH and Hallberg B:
Alectinib, an anaplastic lymphoma kinase inhibitor, abolishes ALK
activity and growth in alk-positive neuroblastoma cells. Front
Oncol. 9:5792019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calvo C, Storey C, Morcrette G, Akl P,
Freneaux P, Pierron G, Trang H, Aerts I, Schleiermacher G,
Philippe-Chomette P, et al: Metastatic neuroblastoma in a patient
with ROHHAD: A new alert regarding the risk of aggressive
malignancies in this rare condition. Pediatr Blood Cancer.
66:e279062019.PubMed/NCBI
|
|
3
|
Garcia M, Rodriguez-Hernandez CJ,
Mateo-Lozano S, Perez-Jaume S, Goncalves-Alves E, Lavarino C, Mora
J and de Torres C: Parathyroid hormone-like hormone plays a dual
role in neuroblastoma depending on PTH1R expression. Mol Oncol.
13:1959–1975. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grasso S, Cangelosi D, Chapelle J, Alzona
M, Centonze G, Lamolinara A, Salemme V, Angelini C, Morellato A,
Saglietto A, et al: The SRCIN1/p140Cap adaptor protein negatively
regulates the aggressiveness of neuroblastoma. Cell Death Differ.
27:790–807. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhoopathi P, Pradhan AK, Bacolod MD, Emdad
L, Sarkar D, Das SK and Fisher PB: Regulation of neuroblastoma
migration, invasion, and in vivo metastasis by genetic and
pharmacological manipulation of MDA-9/syntenin. Oncogene.
38:6781–6793. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pacenta HL and Macy ME: Entrectinib and
other ALK/TRK inhibitors for the treatment of neuroblastoma. Drug
Des Devel Ther. 12:3549–3561. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Campbell K, Shyr D, Bagatell R, Fischer M,
Nakagawara A, Nieto AC, Brodeur GM, Matthay KK, London WB and
DuBois SG: Comprehensive evaluation of context dependence of the
prognostic impact of MYCN amplification in neuroblastoma: A report
from the International Neuroblastoma Risk Group (INRG) project.
Pediatr Blood Cancer. 66:e278192019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu Y, Chen F, Yang Y, Jin Y, Shi J, Han S,
Chu P, Lu J, Tai J, Wang S, et al: lncRNA SNHG16 is associated with
proliferation and poor prognosis of pediatric neuroblastoma. Int J
Oncol. 55:93–102. 2019.PubMed/NCBI
|
|
9
|
Avitabile M, Lasorsa VA, Cantalupo S,
Cardinale A, Cimmino F, Montella A, Capasso D, Haupt R, Amoroso L,
Garaventa A, et al: Association of PARP1 polymorphisms with
response to chemotherapy in patients with high-risk neuroblastoma.
J Cell Mol Med. 24:4072–4081. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye M, Ma J, Liu B, Liu X, Ma D and Dong K:
Linc01105 acts as an oncogene in the development of neuroblastoma.
Oncol Rep. 2019.(Epub ahead of print). View Article : Google Scholar
|
|
11
|
Liu Z, Yang L, Zhong C and Zhou L: EZH2
regulates H2B phosphorylation and elevates colon cancer cell
autophagy. J Cell Physiol. 235:1494–1503. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kosalai ST, Morsy M, Papakonstantinou N,
Mansouri L, Stavroyianni N, Kanduri C, Stamatopoulos K, Rosenquist
R and Kanduri M: EZH2 upregulates the PI3K/AKT pathway through
IGF1R and MYC in clinically aggressive chronic lymphocytic
leukaemia. Epigeneticst. 14:1125–1140. 2019. View Article : Google Scholar
|
|
13
|
Pediconi N, Salerno D, Lupacchini L,
Angrisani A, Peruzzi G, De Smaele E, Levrero M and Belloni L: EZH2,
JMJD3, and UTX epigenetically regulate hepatic plasticity inducing
retro-differentiation and proliferation of liver cells. Cell Death
Dis. 10:5182019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim KH and Roberts CW: Targeting EZH2 in
cancer. Nat Med. 22:128–134. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan KS, Lin CY, Liao TW, Peng CM, Lee SC,
Liu YJ, Chan WP and Chou RH: EZH2 in cancer progression and
potential application in cancer therapy: A friend or foe? Int J Mol
Sci. 18:11722017. View Article : Google Scholar
|
|
16
|
Ma J, Zhang J, Weng YC and Wang JC:
EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal
transition and lymph node metastasis of pancreatic cancer. Mol
Cells. 41:868–880. 2018.PubMed/NCBI
|
|
17
|
Liu Q, Wang G, Li Q, Jiang W, Kim JS, Wang
R, Zhu S, Wang X, Yan L, Yi Y, et al: Polycomb group proteins EZH2
and EED directly regulate androgen receptor in advanced prostate
cancer. Int J Cancer. 145:415–426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei
JS, Marquez VE, Bates SE, Jin Q, Khan J, et al: EZH2 mediates
epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU,
RUNX3, and NGFR. Cancer Res. 72:315–324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Delas MJ, Jackson BT, Kovacevic T,
Vangelisti S, Munera ME, Wild SA, Stork EM, Erard N, Knott S and
Hannon GJ: lncRNA spehd regulates hematopoietic stem and progenitor
cells and is required for multilineage differentiation. Cell Rep.
27:719–729.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhong Y, Wang J, Lv W, Xu J, Mei S and
Shan A: lncRNA TTN-AS1 drives invasion and migration of lung
adenocarcinoma cells via modulation of miR-4677-3p/ZEB1 axis. J
Cell Biochem. 120:17131–17141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cairns J, Ingle JN, Kalari KR, Shepherd
LE, Kubo M, Goetz MP, Weinshilboum RM and Wang L: The lncRNA
MIR2052HG regulates ERalpha levels and aromatase inhibitor
resistance through LMTK3 by recruiting EGR1. Breast Cancer Res.
21:472019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmidt K, Carroll JS, Yee E, Thomas DD,
Wert-Lamas L, Neier SC, Sheynkman G, Ritz J and Novina CD: The
lncRNA SLNCR recruits the androgen receptor to EGR1-bound genes in
melanoma and inhibits expression of tumor suppressor p21. Cell Rep.
27:2493–2507.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin L, Cai Q, Wang S, Wang S, Mondal T,
Wang J and Quan Z: Long noncoding RNA MEG3 regulates LATS2 by
promoting the ubiquitination of EZH2 and inhibits proliferation and
invasion in gallbladder cancer. Cell Death Dis. 9:10172018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Xie Y, Li L, He Y, Zheng D, Yu P,
Yu L, Tang L, Wang Y and Wang Z: EZH2 RIP-seq identifies
tissue-specific long non-coding RNAs. Curr Gene Ther. 18:275–285.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Su M, Xiao Y, Tang J, Wu J, Ma J, Tian B,
Zhou Y, Wang H, Yang D, Liao QJ and Wang W: Role of lncRNA and EZH2
interaction/regulatory network in lung cancer. J Cancer.
9:4156–4165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim B: Western blot techniques. Methods
Mol Biol. 1606:133–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gagliardi M and Matarazzo MR: RIP: RNA
immunoprecipitation. Methods Mol Biol. 1480:73–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia W, Hu J, Ma J, Huang J, Jing T, Deng
L, Zhang J, Jiang N, Ma D and Ma Z: Mutations in TOP2B cause
autosomal-dominant hereditary hearing loss via inhibition of the
PI3K-Akt signalling pathway. Febs Lett. 593:2008–2018. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong L, Tan L, Lv R, Shi Z, Xiong L, Wu F,
Rabidou K, Smith M, He C, Zhang L, et al: A primary role of TET
proteins in establishment and maintenance of De Novo bivalency at
CpG islands. Nucleic Acids Res. 44:8682–8692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ozcelik D and Pezacki JP: Small molecule
inhibition of protein disulfide isomerase in neuroblastoma cells
induces oxidative stress response and apoptosis pathways. Acs Chem
Neurosci. 10:4068–4075. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen L, Alexe G, Dharia NV, Ross L,
Iniguez AB, Conway AS, Wang EJ, Veschi V, Lam N, Qi J, et al:
CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma
dependency on EZH2. J Clin Invest. 128:446–462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Takenobu H, Setyawati AN, Akita N,
Haruta M, Satoh S, Shinno Y, Chikaraishi K, Mukae K, Akter J, et
al: EZH2 regulates neuroblastoma cell differentiation via NTRK1
promoter epigenetic modifications. Oncogene. 37:2714–2727. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bellamy J, Szemes M, Melegh Z, Dallosso A,
Kollareddy M, Catchpoole D and Malik K: Increased efficacy of
histone methyltransferase G9a inhibitors against MYCN-amplified
neuroblastoma. Front Oncol. 10:8182020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsubota S, Kishida S, Shimamura T, Ohira
M, Yamashita S, Cao D, Kiyonari S, Ushijima T and Kadomatsu K:
PRC2-mediated transcriptomic alterations at the embryonic stage
govern tumorigenesis and clinical outcome in MYCN-driven
neuroblastoma. Cancer Res. 77:5259–5271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bate-Eya LT, Gierman HJ, Ebus ME, Koster
J, Caron HN, Versteeg R, Dolman M and Molenaar JJ: Enhancer of
zeste homologue 2 plays an important role in neuroblastoma cell
survival independent of its histone methyltransferase activity. Eur
J Cancer. 75:63–72. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao X, Li D, Huang D, Song H, Mei H, Fang
E, Wang X, Yang F, Zheng L, Huang K and Tong Q: Risk-associated
long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by
repressing PARP1-mediated activation of CTCF. Mol Ther. 26:755–773.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li D, Wang X, Mei H, Fang E, Ye L, Song H,
Yang F, Li H, Huang K, Zheng L and Tong Q: Long noncoding RNA
pancEts-1 promotes neuroblastoma progression through
hnRNPK-mediated β-catenin stabilization. Cancer Res. 78:1169–1183.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng
W, Shi M and Wang Y: Long non-coding RNA CASC15 regulates gastric
cancer cell proliferation, migration and epithelial mesenchymal
transition by targeting CDKN1A and ZEB1. Mol Oncol. 12:799–813.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sanli I, Lalevee S, Cammisa M, Perrin A,
Rage F, Lleres D, Riccio A, Bertrand E and Feil R: Meg3 non-coding
RNA expression controls imprinting by preventing transcriptional
upregulation in cis. Cell Rep. 23:337–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang M, Guo C, Wang L, Luo G, Huang C, Li
Y, Liu D, Zeng F, Jiang G and Xiao X: Long noncoding RNA GAS5
promotes bladder cancer cells apoptosis through inhibiting EZH2
transcription. Cell Death Dis. 9:2382018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng W and Yu A: EZH2-mediated
suppression of lncRNA-LET promotes cell apoptosis and inhibits the
proliferation of post-burn skin fibroblasts. Int J Mol Med.
41:1949–1957. 2018.PubMed/NCBI
|
|
42
|
Chi R, Chen X, Liu M, Zhang H, Li F, Fan
X, Wang W and Lu H: Role of SNHG7-miR-653-5p-STAT2 feedback loop in
regulating neuroblastoma progression. J Cell Physiol.
234:13403–13412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen M, Liu P, Chen Y, Chen Z, Shen M, Liu
X, Li X, Li A, Lin Y, Yang R, et al: Long noncoding RNA FAM201A
mediates the radiosensitivity of esophageal squamous cell cancer by
regulating ATM and mTOR expression via miR-101. Front Genet.
9:6112018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dugimont T, Curgy JJ, Wernert N, Delobelle
A, Raes MB, Joubel A, Stehelin D and Coll J: The H19 gene is
expressed within both epithelial and stromal components of human
invasive adenocarcinomas. Biol Cell. 85:117–124. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kelley RL and Kuroda MI: Noncoding RNA
genes in dosage compensation and imprinting. Cell. 103:9–12. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Koshimizu TA, Fujiwara Y, Sakai N, Shibata
K and Tsuchiya H: Oxytocin stimulates expression of a noncoding RNA
tumor marker in a human neuroblastoma cell line. Life Sci.
86:455–460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Y, Zhuo ZJ, Zhou H, Liu J, Zhang J,
Cheng J, Zhou H, Li S, Li M, He J and Xiao Y: H19 gene
polymorphisms and neuroblastoma susceptibility in Chinese children:
A six-center case-control study. J Cancer. 10:6358–6363. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang J, Li WY, Yang Y, Yan LZ, Zhang SY,
He J and Wang JX: LncRNA XIST facilitates cell growth, migration
and invasion via modulating H3 histone methylation of DKK1 in
neuroblastoma. Cell Cycle. 18:1882–1892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wen Y, Gong X, Dong Y and Tang C: Long non
coding RNA SNHG16 facilitates proliferation, migration, invasion
and autophagy of neuroblastoma cells via sponging miR-542-3p and
upregulating ATG5 expression. Onco Targets Ther. 13:263–275. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Russell MR, Penikis A, Oldridge DA,
Alvarez-Dominguez JR, McDaniel L, Diamond M, Padovan O, Raman P, Li
Y, Wei JS, et al: CASC15-S is a tumor suppressor lncRNA at the 6p22
neuroblastoma susceptibility locus. Cancer Res. 75:3155–3166. 2015.
View Article : Google Scholar : PubMed/NCBI
|