Introduction
Skin cutaneous melanoma (SKCM) is the most
aggressive type of skin cancer, with an increasing number of cases
worldwide, potential for early metastasis and a high mortality rate
(1). The number of treatment options
available for human SKCM has increased, including radiotherapy,
photodynamic therapy, immunotherapy, chemotherapy and
biochemotherapy (2–6). Individualized treatment is difficult to
administer to patients with SKCM, as locally advanced SKCM confers
a major challenge in terms of surgical and medical management
(2). Therefore, effective prognostic
markers may provide an alternative therapeutic treatment strategy
for patients with SKCM. With the establishment of The Cancer Genome
Atlas (TCGA), bioinformatics research has enabled the
identification of differentially expressed and mutated genes in
different types of human cancer, which has led to the discovery of
novel biomarkers from the analysis of TCGA-SKCM samples that
display the potential to be used during the diagnosis, treatment
and prognosis of SKCM (7–10). However, further investigation into
the diagnostic and prognostic biomarkers identified in patients
with SKCM is required to determine their suitability, as well as
understand the molecular mechanisms and gene networks underlying
the development and prognosis of SKCM.
Cluster of differentiation 38 (CD38) is a versatile
membrane protein with various functions that was originally
identified as a cell surface differentiation marker in B
lymphocytes, and was subsequently characterized as a
multifunctional enzyme that is expressed ubiquitously (11). Increasing evidence has supported the
hypothesis that CD38 is involved during tumorigenesis, tumor growth
and metastasis (12–17). In particular, CD38 has been used as a
human multiple myeloma target for its combination therapy (12,15).
Morandi et al (18) also
confirmed that CD38 is expressed in human multiple myeloma cells.
Furthermore, several studies have revealed that CD38 serves an
important role during primary human melanoma metastasis and T cell
proliferation (13,18). The aforementioned studies suggested
that CD38 is overexpressed in multiple tumor types, and therefore,
may serve as a promising target for therapeutic antibodies and
drugs.
To determine the diagnostic and prognostic value of
CD38 in patients with SKCM, the publicly available TCGA-SKCM and
healthy samples were analyzed using University of Alabama Cancer
database (UALCAN) (19) and Gene
Expression Profiling Interactive Analysis 2 (GEPIA2) (20) online tools, as well as Search Tool
for the Retrieval of Interacting Genes/Proteins (STRING) (21), GeneMANIA (22), cBioPortal for Cancer Genomics
(23) and Metascape databases and
resources (24). In the present
study, the novel diagnostic and prognostic role of CD38 during SKCM
was identified. Therefore, the results of the present study might
advance the development of antagonist CD38 treatment strategies for
patients with SKCM.
Materials and methods
CD38 expression and mutations
analysis
To analyze the expression of CD38 in patients with
SKCM, the online software tools UALCAN (ualcan.path.uab.edu/analysis.html) and GEPIA2
(gepia2.cancer-pku.cn/#index) were used. The UALCAN online tool
analyzed CD38 expression levels in SKCM based on sample type,
individual cancer stage, and the sex, weight, age and race of the
patient. The cBioPortal for Cancer Genomics (www.cbioportal.org) was used to further evaluate the
gene expression levels and mutations of CD38 in SKCM samples. All
eight TCGA cutaneous melanoma categories were included. All
patients with CD38-positive status were included in the present
study, and both SKCM and healthy samples were analyzed.
Survival analysis
The Kaplan-Meier method was used to evaluate
survival analysis based on the expression of CD38 between various
groups. The primary endpoint was disease free survival (DFS), which
was defined as the time interval between the initiation of curative
treatment and the date of progression, the start date of a
second-line treatment or the date of death, whichever occurred
first. The secondary endpoint was overall survival (OS), which was
defined as the length of time between the date of diagnosis or
first therapy to the date of death or last follow-up. The follow-up
duration was calculated and presented using the Kaplan-Meier method
with 95% confidence intervals and the log-rank test was used to
identify significant differences among the various groups. The
effects of CD38 expression levels and the body weight, sex, race of
the patient on patient survival were determined using UALCAN. The
OS and DFS, based on the expression status of CD38 in patients with
SKCM, were determined using GEPIA2. In the total number of patients
with SKCM, 50% displayed high CD38 expression levels (n=229;
>median expression) and 50% displayed low expression levels of
CD38 (n=229; <median expression). Furthermore, the LOGpc
database (bioinfo.henu.edu.cn/DatabaseList.jsp) was used to
verify the identified OS, platinum-free interval (PFI) and
disease-specific survival (DSS) results.
Correlation genes analysis
To identify genes that were correlated with CD38
during SKCM, a correlation analysis was performed. The top 50 genes
that displayed a correlation with CD38 expression during SKCM were
obtained using the GEPIA2 online tool. The SKCM healthy and tumor
expression GDC TCGA Melanoma (SKCM) dataset (dataset ID:
TCGA-SKCM.cnv.tsv, http://gepia2.cancer-pku.cn/#dataset) was analyzed
from TCGA (http://gepia2.cancer-pku.cn/#similar). Pearson's
correlation coefficient (r) was used to screen the top 50
positively correlated genes (r≥0.75) with a similar expression
pattern to CD38 in SKCM tumor and normal tissues. Furthermore, the
UALCAN tool was used to determine the correlation between nine
highly correlated genes and CD38.
Expression and survival heatmap
analysis
The heatmap profile of nine correlated genes with
CD38 expression during SKCM and 32 additional TCGA cancer types,
including adrenocortical carcinoma, bladder urothelial carcinoma,
breast invasive carcinoma, cervical squamous cell carcinoma and
endocervical adenocarcinoma, cholangiocarcinoma, colon
adenocarcinoma, lymphoid neoplasm diffuse large B cell lymphoma,
esophageal carcinoma, glioblastoma multiforme, head and neck
squamous cell carcinoma, kidney chromophobe, kidney renal clear
cell carcinoma, kidney renal papillary cell carcinoma, acute
myeloid leukemia, brain lower grade glioma, liver hepatocellular
carcinoma, lung adenocarcinoma, lung squamous cell carcinoma,
mesothelioma, ovarian serous cystadenocarcinoma, pancreatic
adenocarcinoma, pheochromocytoma and paraganglioma, prostate
adenocarcinoma, rectum adenocarcinoma, sarcoma, stomach
adenocarcinoma, testicular germ cell tumors, thyroid carcinoma,
thymoma, uterine corpus endometrial carcinoma, uterine
carcinosarcoma, uveal melanoma, were generated using an interactive
heatmap and the multiple gene comparison tools in GEPIA2. Both
healthy and tumor data from TCGA were selected for the expression
heatmap analysis. To compare the survival contribution of the top
nine correlated genes with CD38 expression during SKCM and
additional TCGA cancer types, the survival map was calculated from
TCGA-tumor specimens using the Mantel-Cox test.
Occurrence of CD38 isoforms and
promoter methylation analysis
To verify the five structural isoforms of CD38 and
to compare the occurrence in SKCM, the TCGA-SKCM dataset in the
GEPIA2 online tool was used with the following parameters: Cancer,
X and isoform, Y. To analyze the CD38 promoter methylation levels
in patients with SKCM, the CD38 promoter methylation profile based
on individual cancer stage, and the sex, weight and age of the
patient was analyzed using the UALCAN online tool.
Protein-protein interaction (PPI)
networks and Gene Ontology (GO) enrichment analysis
The STRING database (string-db.org)
provides a critical evaluation and integration of PPI, including
physical and functional relevance (21). The PPI network of CD38 was produced
using STRING online tools (version 10.0). GeneMANIA (genemania.org) is a flexible user-friendly website for
generating hypotheses regarding gene function, analyzing gene lists
and prioritizing genes for functional assays. The Metascape tool
(metascape.org) provides a resource for biologists
for the analysis of systems-level datasets (24). Therefore, GeneMANIA and Metascape
were used to further analyze the related genes and functional
enrichment of CD38.
Results
Elevated expression level and specific
mutations of CD38 in patients with SKCM from TCGA dataset
To determine the expression level of CD38 in
patients with SKCM, the online analytical UALCAN and GEPIA2 tools
were used. The expression level of CD38 was significantly higher in
SKCM tissues (n=461) compared with healthy tissues (n=558;
P<0.001; Fig. 1A). From the
UALCAN analysis, CD38 expression levels were significantly
increased in SKCM metastatic tumors compared with primary tumors
(P<0.001; Fig. 1B). In addition,
there was a significant difference in CD38 expression levels
between the sex (P<0.05; Fig.
1C), race (P<0.05; Caucasian vs. Asian; Fig. 1E) and individual cancer stage
(P<0.001; stage 1 vs. stage 2 and stage 2 vs. stage 3; Fig. 1G) of the patient, but there were no
significant differences in the expression levels of CD38 between
the weight (Fig. 1D) and age
(Fig. 1F) of the patient.
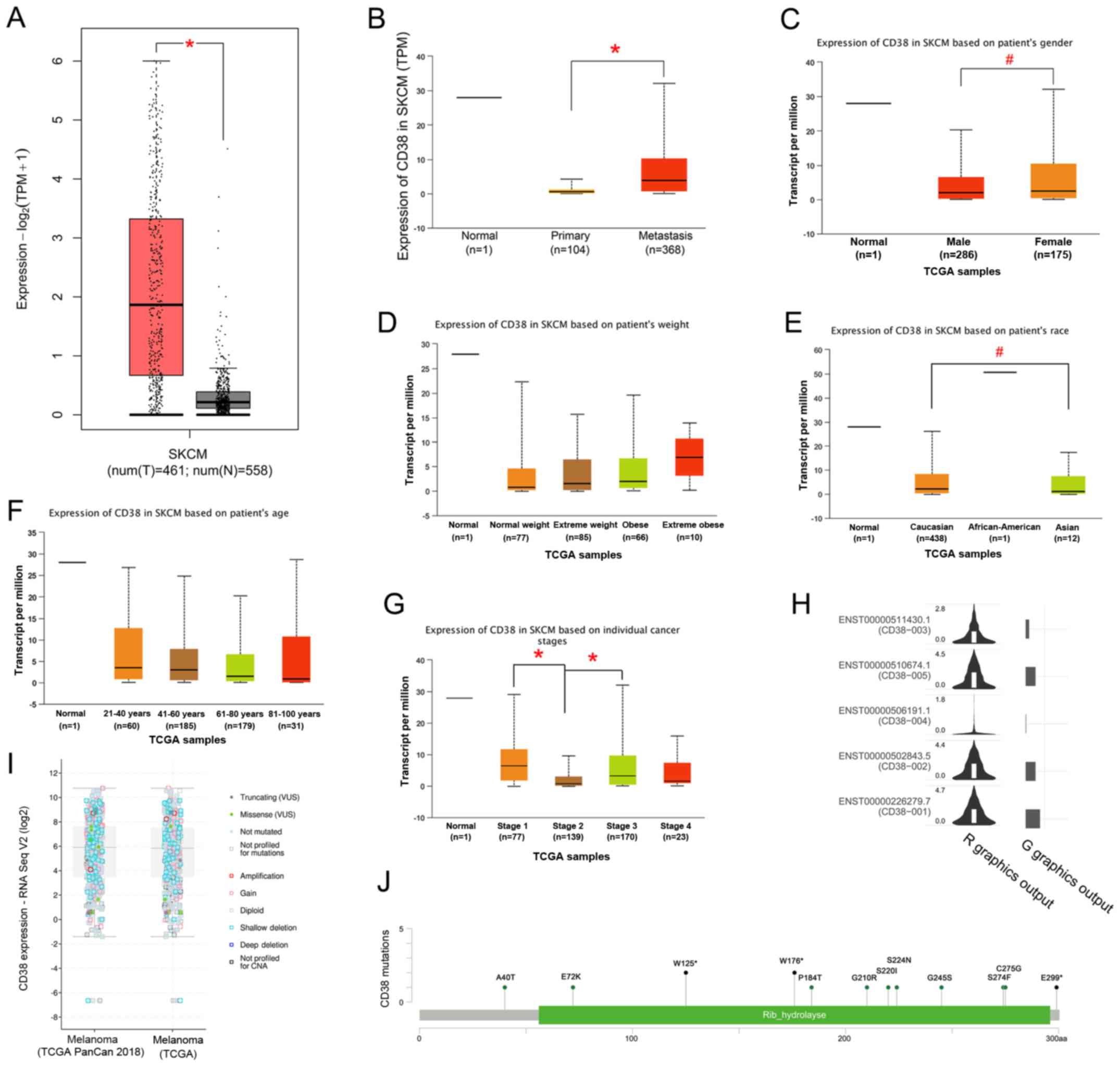 | Figure 1.CD38 expression and mutations in
patients with SKCM. (A) CD38 expression in SKCM using (A) the
GEPIA2 online tool and (B) UALCAN. CD38 expression in SKCM based on
(C) sex, (D) body weight, (E) race, (F) age and (G) individual
cancer stage of the patient. (H) Occurrence of CD38 isoforms in
patients from TGGA-SKCM dataset (X, cancer; Y, isoform). (I) CD38
expression and mutations analyzed using cBioPortal for Cancer
Genomic. (J) Location of specific CD38 mutations in patients with
SKCM. Green and black represent missense and truncating mutations,
respectively. #P<0.05; *P<0.001. CD38, cluster of
differentiation 38; SKCM, skin cutaneous melanoma; GEPIA2, Gene
Expression Profiling Interactive Analysis 2; UALCAN, University of
Alabama Cancer database; TCGA, The Cancer Genome Atlas; TPM,
Transcripts Per Million; T, tumor; N, normal. |
In addition, the GEPIA2 analysis indicated that the
occurrence of the two major isoforms, CD38-001 and CD38-005, are
more frequent in patients with SKCM compared with other isoforms
(Fig. 1H). The cBioPortal for Cancer
Genomics analysis suggested that there were a total of 22 mutations
in CD38 in TCGA-SKCM samples, including 14 missenses and 8
truncating mutations (Fig. 1I and J;
Table I). Among them, the W125
mutation affects CD38 enzyme activity, and the single nucleotide
polymorphism (SNP) at position (P184) is associated with autism
spectrum disorder and type II diabetes, and C275 is a disulfide
bond formation site for CD38 (11).
Collectively, the results from the present study suggested that
high expression levels and key mutations in CD38 might be involved
in the development of SKCM.
 | Table I.Mutations in CD38 gene in TCGA-SKCM
dataset. |
Table I.
Mutations in CD38 gene in TCGA-SKCM
dataset.
| Sample ID | Protein change | Mutation type | Copy number
variation | Number of
occurrences in COSMIC | Allele frequency
(total) | Number of mutations
in sample |
|---|
|
MEL-JWCI-WGS-12 | C275G | Missense | NA | NA | NA | 2703 |
|
TCGA-FS-A1ZK-06 | W176a | Nonsense | Gain | 2 | 0.16 | 1297 |
|
TCGA-EE-A2MR-06 | S274F | Missense | Diploid | 1 | 0.14 | 3944 |
|
TCGA-FW-A3R5-06 | W125a | Nonsense | Diploid | 1 | 0.18 | 15939 |
|
TCGA-GF-A6C9-06 | W125a | Nonsense | ShallowDel | 1 | 0.18 | 2762 |
|
TCGA-EB-A5UN-06 | G245S | Missense | Diploid | 1 | 0.19 | 625 |
|
TCGA-FW-A3R5-06 | E72K | Missense | Diploid | 1 | 0.37 | 15939 |
|
TCGA-BF-A5ER-01 | A40T | Missense | NA | NA | 0.38 | 165 |
|
TCGA-EB-A6QY-01 | S224N | Missense | NA | NA | 0.24 | 518 |
|
TCGA-FS-A1ZK-06 | W176a | Nonsense | Gain | 2 | 0.16 | 1459 |
|
TCGA-EE-A2MR-06 | S274F | Missense | Diploid | 1 | 0.14 | 2921 |
|
TCGA-FW-A3R5-06 | W125a | Nonsense | Diploid | 1 | 0.17 | 15832 |
|
TCGA-GF-A6C9-06 | W125a | Nonsense | ShallowDel | 1 | 0.15 | 2318 |
|
TCGA-EB-A5UN-06 | G245S | Missense | Diploid | 1 | 0.19 | 627 |
|
TCGA-FW-A3R5-06 | E72K | Missense | Diploid | 1 | 0.38 | 15832 |
|
TCGA-BF-A5ER-01 | A40T | Missense | NA | NA | 0.37 | 159 |
|
TCGA-EB-A6QY-01 | S224N | Missense | NA | NA | 0.24 | 506 |
|
TCGA-D3-A2JL-06 | P184T | Missense | Diploid | NA | 0.11 | 1202 |
|
TCGA-EE-A183-06 | S220I | Missense | Diploid | 1 | 0.12 | 2156 |
|
TCGA-FS-A1Z4-06 | E299a | Nonsense | ShallowDel | NA | 0.1 | 609 |
|
TCGA-Z2-A8RT-06 | G210R | Missense | ShallowDel | NA | 0.68 | 2422 |
| YUKLAB | W176a | Nonsense | NA | 2 | NA | 3656 |
Effect of CD38 expression level on
patient survival in TCGA-SKCM dataset
To evaluate the prognostic value of CD38 in patients
with SKCM, the effect of CD38 expression levels on survival time
was analyzed. Higher CD38 expression levels resulted in a
significantly higher survival probability compared with lower CD38
expression levels (P<0.0001; Fig.
2A). In addition, survival probability was also associated with
the body weight, sex and race of the patient. Low/medium CD38
expression levels in Asian patients resulted in a shorter survival
time compared with patients from other ethnicities (P<0.0001;
Fig. 2B). CD38 expression levels and
the sex (P=0.00029) and body weight (P=0.021) of the patients were
also significantly associated with the survival of patients with
SKCM (Fig. 2C and D). In particular,
female patients diagnosed with SKCM displayed a longer survival
time compared with male patients.
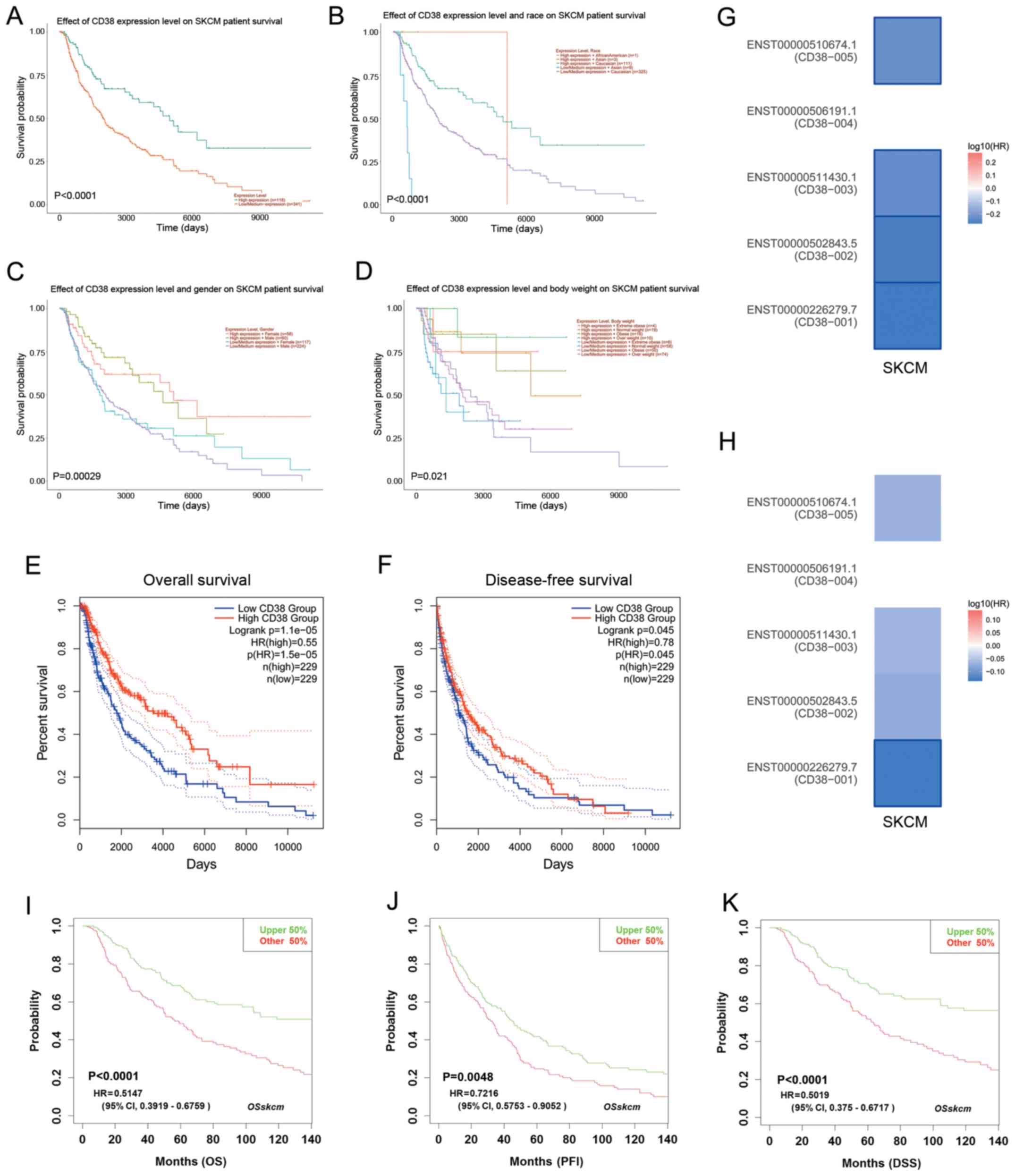 | Figure 2.Effect of CD38 expression on overall
survival in patients with SKCM. (A) Higher CD38 expression was
associated with improved survival probability in patients with
SKCM. Effect of CD38 expression and (B) race, (C) sex and (D) body
weight on survival in patients with SKCM. (E) OS and (F) DFS were
identified using the GEPIA2 online tool. (G) OS and (H) DFS
heatmaps of five CD38 isoforms in patients with SKCM. (I) OS, (J)
PFI and (K) DSS analysis using the LOGpc Database. CD38, cluster of
differentiation 38; SKCM, skin cutaneous melanoma; OS, overall
survival; DFS, disease-free survival; GEPIA2, Gene Expression
Profiling Interactive Analysis 2; PFI, platinum-free interval; DSS,
disease-specific survival; CI, confidence interval; HR, hazard
ratio. |
Moreover, the OS and DFS of CD38 expression levels
in patients with SKCM were analyzed using the GEPIA2 online tool.
Patients with high CD38 expression levels displayed a higher
survival probability for both OS and DFS compared with patients
with low CD38 expression levels (Fig. 2E
and F). The contribution of the CD38 isoforms to the survival
of patients with SKCM was determined. The survival map analysis
verified that major isoform, CD38-001, affected the survival of
patients with SKCM (Fig. 2G and
H).
To verify the aforementioned results, the LOGpc
database was used. Patients with high CD38 expression levels
displayed higher OS (P<0.0001), PFI (P<0.0001) and DSS
(P<0.005) probabilities compared with patients with low CD38
expression levels (Fig. 2I-K).
Correlated genes with CD38 in patients
with SKCM from TCGA dataset
To identify genes correlated with CD38 in SKCM, a
correlation analysis was conducted. The top 50 correlated genes
with CD38 expression in SKCM were investigated using GEPIA2
(Table II; Pearson CC≥0.75). A PPI
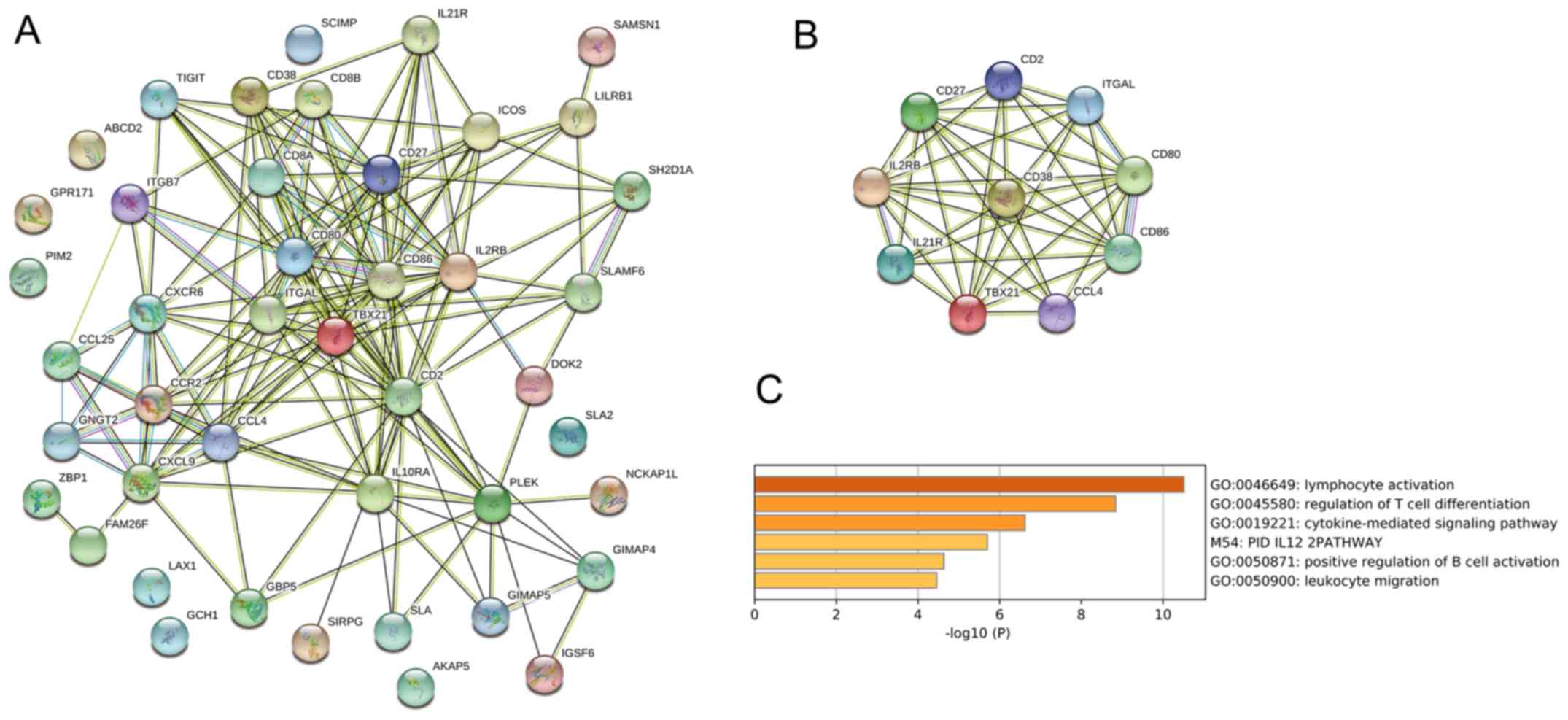
network of these genes was created using the STRING tool (Fig. 3A). The identified genes were from
diverse functional groups, such as cell adhesion molecules (CD80,
CD86, CD8A, CD8B, CD2, ICOS, ITGAL, ITGB7 and TIGIT), chemokine
signaling pathway molecules (CCL4, CCL25, CXCR6, CXCL9, CCR2 and
GNGT2) and intestinal immune network for IgA production molecules
(CD80, CD86, ICOS, ITGB7 and CCL25). A total of nine correlated
genes that interacted with CD38 were further predicted using the
STRING database (Fig. 3B), which
included CD86, CD80, CD27, CD2, IL21R, IL2RB, ITGAL, CCL4 and
TBX21. Functional enrichment analysis from Metascape revealed that
the nine correlated genes with CD38 in patients with SKCM might be
involved with lymphocyte activation and regulation of T cell
differentiation (Fig. 3C).
 | Table II.Top 50 ranked CD38 correlated genes
in TCGA-SKCM dataset. |
Table II.
Top 50 ranked CD38 correlated genes
in TCGA-SKCM dataset.
| Genenumber | Gene symbol | Gene ID | r |
|---|
| 1 | TIGIT |
ENSG00000181847.11 | 0.82 |
| 2 | TBX21 |
ENSG00000073861.2 | 0.81 |
| 3 | PLEK |
ENSG00000115956.9 | 0.81 |
| 4 | ZBP1 |
ENSG00000124256.14 | 0.81 |
| 5 | CCR2 |
ENSG00000121807.5 | 0.81 |
| 6 | LAX1 |
ENSG00000122188.12 | 0.80 |
| 7 | SLA |
ENSG00000155926.13 | 0.80 |
| 8 | SAMSN1 |
ENSG00000155307.17 | 0.80 |
| 9 | CXCR6 |
ENSG00000172215.5 | 0.79 |
| 10 | PIM2 |
ENSG00000102096.9 | 0.79 |
| 11 | SIRPG |
ENSG00000089012.14 | 0.79 |
| 12 | GBP5 |
ENSG00000154451.14 | 0.78 |
| 13 | GNGT2 |
ENSG00000167083.6 | 0.78 |
| 14 | SLA2 |
ENSG00000101082.13 | 0.78 |
| 15 | SLAMF6 |
ENSG00000162739.13 | 0.78 |
| 16 | IL2RB |
ENSG00000100385.13 | 0.78 |
| 17 | SH2D1A |
ENSG00000183918.14 | 0.78 |
| 18 | AKAP5 |
ENSG00000179841.8 | 0.77 |
| 19 | GCH1 |
ENSG00000131979.18 | 0.77 |
| 20 | ITGAL |
ENSG00000005844.17 | 0.77 |
| 21 | CD2 |
ENSG00000116824.4 | 0.77 |
| 22 | SCIMP |
ENSG00000161929.14 | 0.76 |
| 23 | USP30-AS1 |
ENSG00000256262.1 | 0.76 |
| 24 | ICOS |
ENSG00000163600.12 | 0.76 |
| 25 | IL10RA |
ENSG00000110324.9 | 0.76 |
| 26 | CD86 |
ENSG00000114013.15 | 0.76 |
| 27 | IL21R |
ENSG00000103522.15 | 0.76 |
| 28 | GIMAP5 |
ENSG00000196329.10 | 0.76 |
| 29 | GIMAP4 |
ENSG00000133574.9 | 0.76 |
| 30 | IGSF6 |
ENSG00000140749.8 | 0.76 |
| 31 | NCKAP1L |
ENSG00000123338.12 | 0.76 |
| 32 | TRGV10 |
ENSG00000211694.2 | 0.76 |
| 33 | CD8B |
ENSG00000172116.21 | 0.76 |
| 34 | CXCL9 |
ENSG00000138755.5 | 0.76 |
| 35 | CXCR2P1 |
ENSG00000229754.1 | 0.76 |
| 36 | TRGC2 |
ENSG00000227191.6 | 0.76 |
| 37 | FAM26F |
ENSG00000188820.12 | 0.75 |
| 38 | ITGB7 |
ENSG00000139626.15 | 0.75 |
| 39 | GPR171 |
ENSG00000174946.6 | 0.75 |
| 40 | CD8A |
ENSG00000153563.15 | 0.75 |
| 41 | TRGV2 |
ENSG00000233306.2 | 0.75 |
| 42 | CD80 |
ENSG00000121594.11 | 0.75 |
| 43 | ABCD2 |
ENSG00000173208.3 | 0.75 |
| 44 | CD27 |
ENSG00000139193.3 | 0.75 |
| 45 | RP11-493L12.5 |
ENSG00000257924.1 | 0.75 |
| 46 | DOK2 |
ENSG00000147443.12 | 0.75 |
| 47 | LILRB1 |
ENSG00000104972.14 | 0.75 |
| 48 | CCL4 |
ENSG00000275302.1 | 0.75 |
| 49 | CCL25 |
ENSG00000131142.13 | 0.75 |
| 50 | AC104820.2 |
ENSG00000234663.5 | 0.75 |
In addition, the correlation between CD38 and the
nine aforementioned correlated genes was further illustrated in
Fig. 4A. To determine the similarity
of the correlated genes, expression and survival heatmaps across
TCGA tumors were generated. The survival heatmaps of the nine
aforementioned CD38-correlated genes displayed a positive
correlation with survival in patients with SKCM, which was
consistent with the results of CD38, but there was no correlation
with other types of tumor. Moreover, the expression heatmaps in
patients with SKCM and healthy tissues of the nine aforementioned
genes was similar to the expression heatmap of CD38 (Fig. 4C). However, further studies are
required to validate the correlated genes and to explore underlying
functional mechanisms.
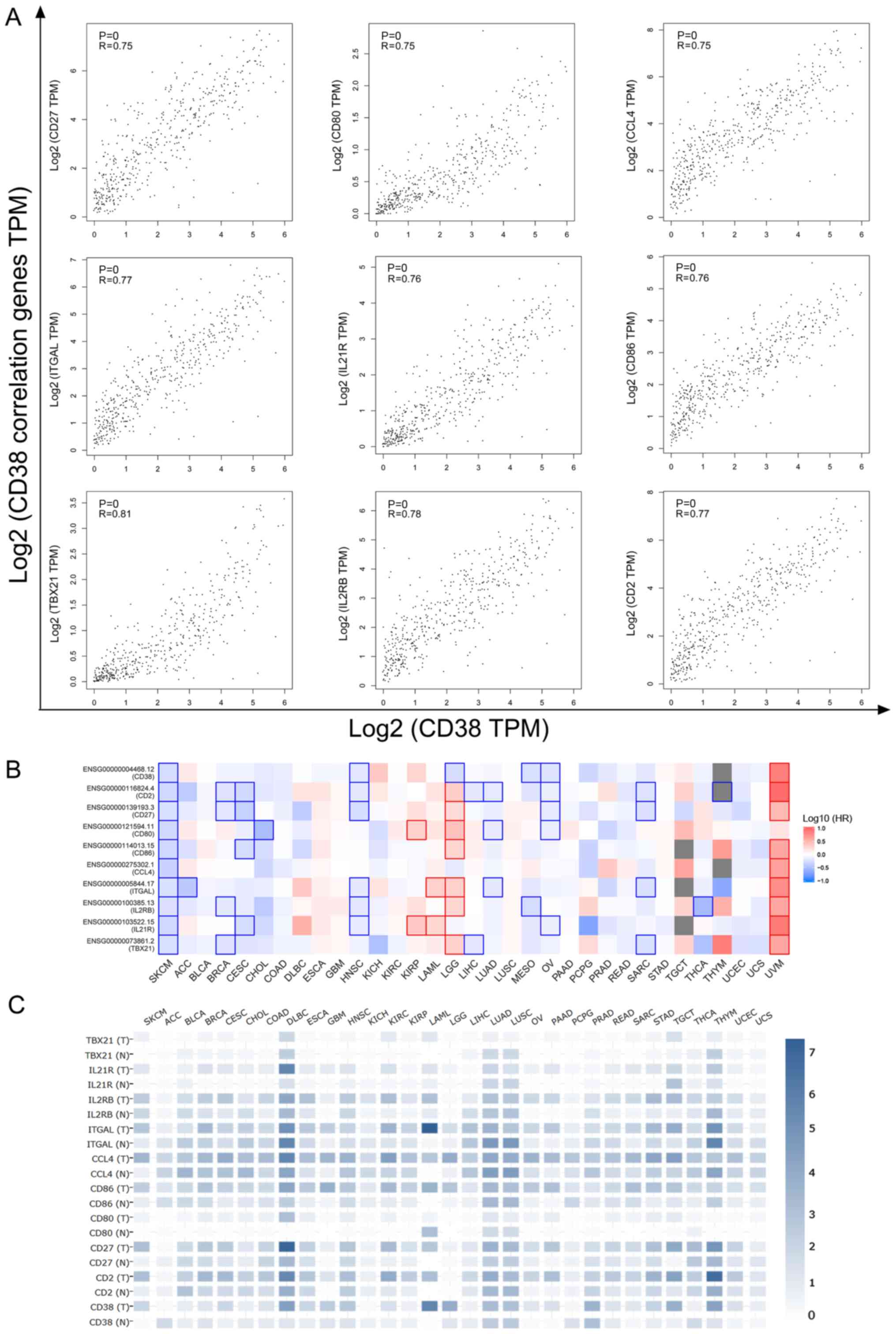 | Figure 4.Correlation between CD38 and nine
correlated genes. (A) Correlation analysis of CD86, CD80, CD27,
CD2, IL21R, IL2RB, ITGAL, CCL4 and TBX21 with CD38 in patients with
SKCM. (B) Survival heatmap of nine correlated genes and CD38 in
SKCM and other TCGA tumors. (C) Expression heatmap of nine
correlated genes and CD38 across TCGA tumors. The shades of blue
represent the expression levels of these genes in different tissues
(normal and tumor). Log2 (TPM + 1) transformed
expression data were chosen for plotting. CD38, cluster of
differentiation 38; SKCM, skin cutaneous melanoma; ACC,
adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA,
breast invasive carcinoma; CESC, cervical squamous cell carcinoma
and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD,
colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG,
brain lower grade glioma; LIHC, liver hepatocellular carcinoma;
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma;
MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma; TGCT,
testicular germ cell tumors; THCA, thyroid carcinoma; THYM,
thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine
carcinosarcoma; UVM, uveal melanoma; TCGA, The Cancer Genome Atlas;
TPM, Transcripts Per Million; HR, hazard ratio. |
Promoter methylation levels of CD38 in
patients with SKCM from TCGA dataset
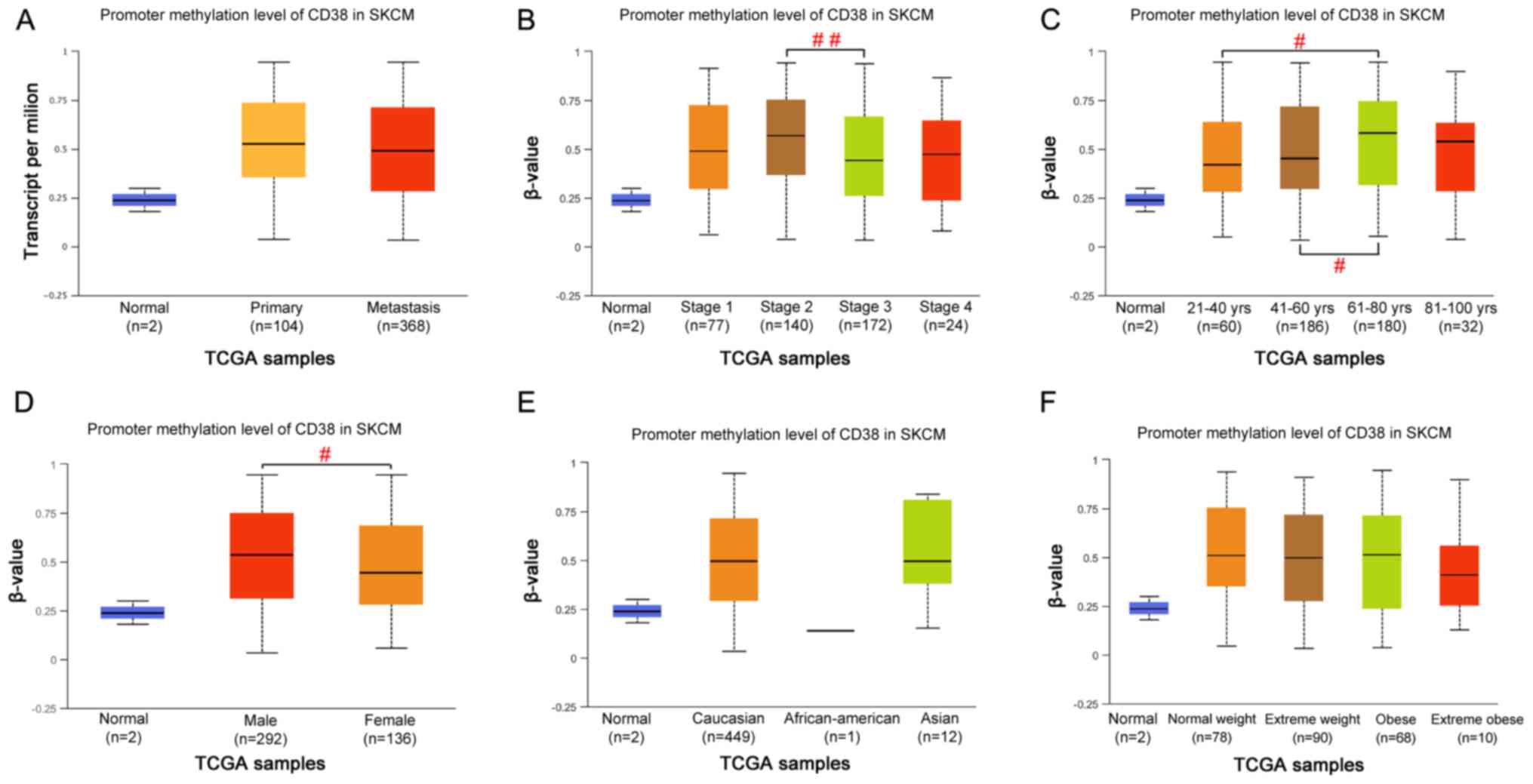
To investigate the CD38 promoter methylation profile
in patients with SKCM based on the cancer type, individual cancer
stage, age, sex, race and weight of the patient, the UALCAN online
tool was used. The results suggested that the individual cancer
stage (P<0.05; stage 2 vs. stage 3; Fig. 5B), age (P<0.05; 41–60 vs.
21–40/61–80 years; Fig. 5C) and sex
(P<0.05; male vs. female; Fig.
5D) of the patient might contribute to the promoter methylation
level of CD38 in patients with SKCM. However, there were no
significant alterations in the promoter methylation levels of CD38
in the TCGA-SKCM dataset based on the cancer type (Fig. 5A), race (Fig. 5E) or weight (Fig. 5F) of the patient.
PPI network and GO enrichment analysis
of CD38
The functional interactions between proteins can
provide important information of the molecular mechanism involved.
The PPI network of CD38 was determined using the STRING database
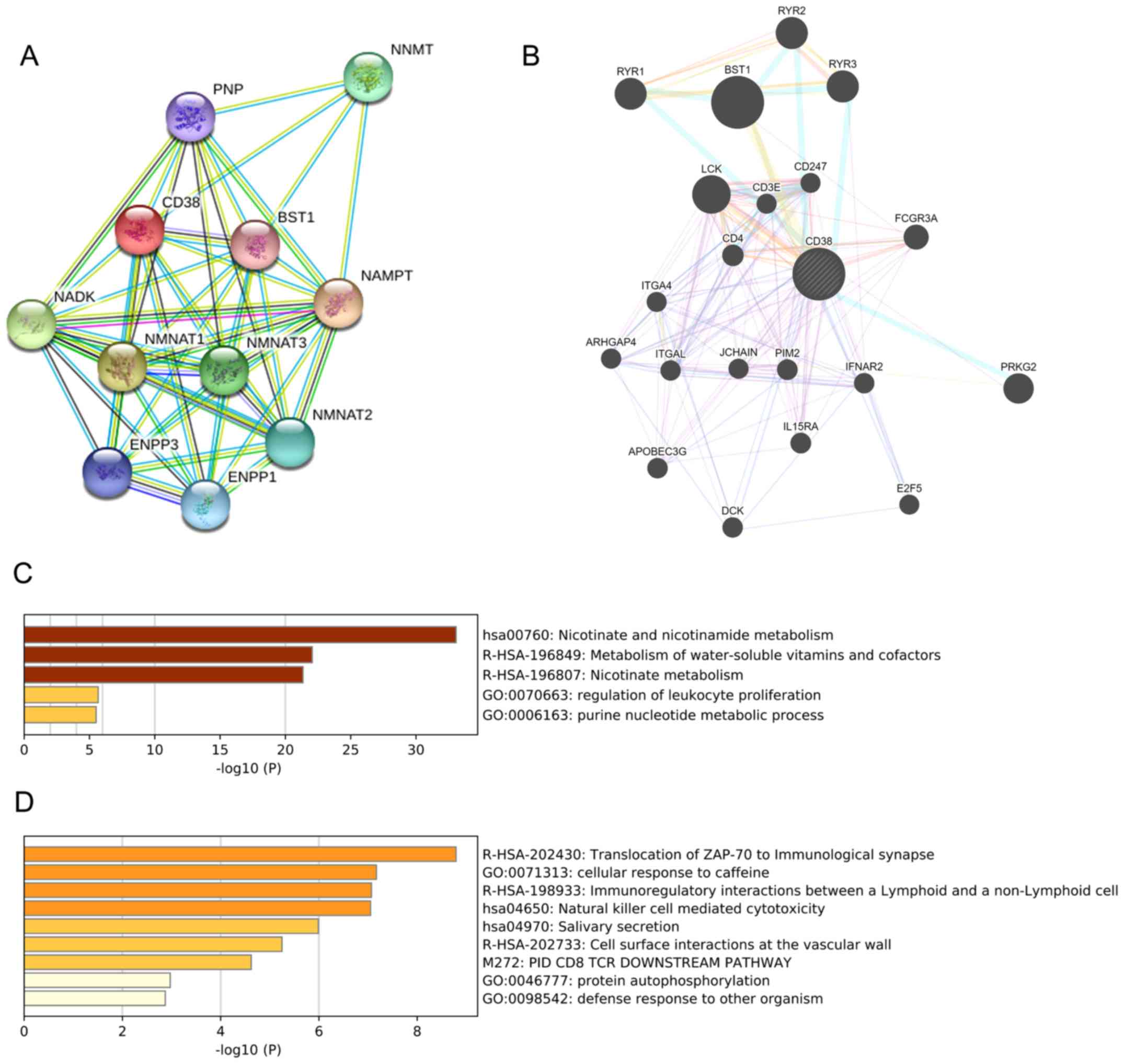
(Fig. 6A). The results indicated
that CD38 interacted with 10 functional genes, including NMNAT1,
NMNAT2, NMNAT3, NAMPT, ENPP1, ENPP3, NADK, BST1, PNP and NNMT. From
the STRING functional enrichments, the 10 associated genes were
involved in the nucleotide metabolic process and nine of the genes
were involved in the nicotinamide adenine dinucleotide (NAD)
metabolic process. A further 20 genes interacting with CD38 were
identified using GeneMANIA (Fig.
6B). Among them, LCK, CD3E, CD247 CD4 and FCGR3A were
identified by physical interactions with CD38. In particular, the
functional enrichment analysis revealed that the proteins were
involved in the immune response activating cell surface receptor
signaling pathway. RYR1, RYR2 and RYR3 were also implicated in
calcium release channel activity and intracellular ligand-gated
calcium channel activity process. Consistent with the
aforementioned correlation analysis, ITGAL belonged to the group of
nine correlated genes with CD38 in patients with SKCM.
To further determine the potential function of CD38
in SKCM, GO enrichment analysis of CD38 and the genes it interacts
with was performed using Metascape. The results suggested that CD38
and the 10 genes it interacted with were enriched in nicotinate and
nicotinamide metabolism, metabolism of water-soluble vitamins and
cofactors, and nicotinate metabolism processes (−log10
(P)>20; Fig. 6C). A total of 20
CD38-correlated genes identified using GeneMANIA were significantly
enriched in translocation of ZAP-70 to immunological synapse,
cellular response to caffeine, immunoregulatory interactions
between a lymphoid and a non-lymphoid cell, and natural killer cell
mediated cytotoxicity (−log10 (P)>6; Fig. 6D).
Discussion
Over the past few years, several reports have
concentrated on differentially expressed genes and mutations in
patients with SKCM, identifying diagnostic and prognostic
biomarkers for SKCM (8,25–27).
However, the current curative biomarkers identified for the therapy
of patients with SKCM are not adequate (28). CD38 has emerged as an effective
target for therapeutic antibodies and drugs in human multiple
myeloma and neuroblastoma (14–16,29). The
present study indicated that higher CD38 expression levels and
specific CD38 mutations were favorable diagnostic factors in SKCM.
Both UALCAN and GEPIA2 online tools revealed that the expression
level of CD38 was significantly higher in patients with SKCM
compared with healthy tissues. Furthermore, the results indicated
that high expression of CD38 may serve as a biomarker for SKCM
metastasis. In addition, several specific mutations of CD38 were
identified in patients with SKCM. In particular, W125 is one of the
key residues, which is responsible for nucleosidase activity of
CD38 (30), P184 is a common SNP
(11), and the cysteine residues
between C275 and C254 form a disulfide bond within CD38 (11). The aforementioned mutations or
polymorphisms might increase the risk of developing SKCM. In
addition, a positive correlation between CD38 expression levels and
survival probability in patients with SKCM was identified. The
results suggested that higher CD38 expression levels resulted in
improved survival probability, which could affect the sensitization
of chemotherapy drugs or antibodies.
To the best of our knowledge, the present study also
screened the top 50 correlated genes with CD38 in patients with
SKCM for the first time, which might be implicated in the prognosis
of SKCM. However, further studies are required to identify the
molecular mechanism involved and the possible applications of the
correlated genes in SKCM. The PPI network generated using the
STRING online tool also identified nine CD38-interacting genes for
correlation analysis in SKCM. The GO enrichment analysis revealed
that the nine correlated genes (CD86, CD80, CD27, CD2, IL21R,
IL2RB, ITGAL, CCL4 and TBX21) were involved in the regulation of T
cell differentiation and lymphocyte activation. In particular, a
previous study also identified CD2 as a correlated gene, indicating
that CD2 might be an independent predictor of disease recurrence
and OS in patients with primary cutaneous melanoma (27).
From the PPI network analysis, STRING and GeneMANIA
identified 10- and 20-associated genes, respectively. Among the
genes identified using STRING were genes from diverse functional
groups, such as nicotinate-nucleotide adenylyltransferase activity
(NMNAT1, NMNAT2 and NMNAT3), NAD+ nucleotidase and
cyclic ADP-ribose generating [bone marrow stromal cell antigen-1
(BST1)] and pyrimidine metabolism (PNP, ENPP1 and ENPP3) genes.
Several genes, such as NMNAT1, NMNAT2 and NMNAT3, have been
previously identified to be involved in catalyzing NAD+
synthesis in the nicotinate and nicotinamide metabolism pathways
(31). The genes predicted using
GeneMANIA were from various functional enrichments, such as
response to virus (LCK, CD247, CD4, PIM2 and IFNAR2) and cellular
response to alkaloid (RYR1, RYR2 and RYR3). BST1 was predicted as a
CD38-associated protein by both databases. It has been reported
that some SNPs in BST1 serve as predictors for Parkinson's disease
(32). Moreover, a previous study
revealed that a deletion involving CD38 and BST1 facilitates a
fusion transcript in a patient with autism and asthma (33). GeneMANIA also revealed that ITGAL was
associated with CD38, which is consistent with the correlation
analysis from GEPIA2, and further supports the findings of a
previous study that indicated that ITGAL might serve as a
prognostic factor in patients with R0 resected Dukes stage B and C
colorectal cancer (34). A previous
study also investigated ITGAL as a prognostic factor in the
survival of men with castration-resistant prostate cancer (CRPC),
and SNPs in ITGAL may be associated with the risk of death in men
with CRPC (35).
Nevertheless, further investigation is required to
determine the potential mechanism and application value of the
aforementioned CD38-associated genes in SKCM, including CD2, BST1
and ITGAL. In a previous study, photodynamic therapy was
implemented for human melanoma (5,6). Further
studies developed related photodynamic therapy schemes and
identified possible mechanisms (36,37).
Recent reports also demonstrate that targeting CD38 enhances
antileukemic activity (38,39). Therefore, CD38-targeted photodynamic
therapy for SKCM requires further investigation.
In summary, the present study identified that CD38
was highly expressed in patients with SKCM and was associated with
SKCM metastasis. Moreover, elevated expression of CD38 was
associated with OS in patients with SKCM. The present study might
aid the identification of the mechanism underlying SKCM progression
and advance the development of antagonist CD38 strategies for
patients with SKCM. However, a key limitation of the present study
was that the data obtained from TCGA-SKCM lacked analysis of
patients with CD38 positive and negative expression.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 31700736), China
Scholarship Council (grant no. 201908420102), Hubei Medical Youth
Tip-Top Talent, Leading Talent Program of Yangtze Talent Project
and the College Students Innovative Entrepreneurial Training
Program in Yangtze University (grant nos. 2018184 and 2019372).
Availability of data and materials
The dataset GDC TCGA Melanoma (SKCM) generated
and/or analyzed during the present study are available in The
Cancer Genome Atlas repository (https://portal.gdc.cancer.gov/).
Authors' contributions
XW and SH designed the study. XW and PW drafted the
manuscript. XW, PW, LG, JW and SMASN performed the research and
analyzed the data. XW, PW and LG revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved and consented by the
Ethics Committee of the Second Affiliated Hospital of Yangtze
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khunger A, Buchwald ZS, Lowe M, Khan MK,
Delman KA and Tarhini AA: Neoadjuvant therapy of locally/regionally
advanced melanoma. Ther Adv Med Oncol. 11:17588359198669592019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krayem M, Sabbah M, Najem A, Wouters A,
Lardon F, Simon S, Sales F, Journe F, Awada A, Ghanem GE and Van
Gestel D: The benefit of reactivating p53 under MAPK inhibition on
the efficacy of radiotherapy in melanoma. Cancers (Basel).
11:10932019. View Article : Google Scholar
|
|
4
|
Dummer R, Siano M, Hunger RE, Lindenblatt
N, Braun R, Michielin O, Mihic-Probst D, von Moos R, Najafi Y,
Guckenberger M and Arnold A: The updated Swiss guidelines 2016 for
the treatment and follow-up of cutaneous melanoma. Swiss Med Wkly.
146:w142792016.PubMed/NCBI
|
|
5
|
Lee EH, Lim SJ and Lee MK: Chitosan-coated
liposomes to stabilize and enhance transdermal delivery of
indocyanine green for photodynamic therapy of melanoma. Carbohydr
Polym. 224:1151432019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi L, Liu P, Wu J, Ma L, Zheng H, Antosh
MP, Zhang H, Wang B, Chen W and Wang X: The effectiveness and
safety of X-PDT for cutaneous squamous cell carcinoma and melanoma.
Nanomedicine (Lond). 14:2027–2043. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Network, . Genomic
classification of cutaneous melanoma. Cell. 161:1681–1696. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Q, Pan F, Li S, Huang R, Wang X, Wang
S, Liao X, Li D and Zhang L: The prognostic value of the proteasome
activator subunit gene family in skin cutaneous melanoma. J Cancer.
10:2205–2219. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Q, Wang X, Liang Q, Wang S, Xiwen L,
Pan F, Chen H and Li D: Distinct prognostic value of mRNA
expression of guanylate-binding protein genes in skin cutaneous
melanoma. Oncol Lett. 15:7914–7922. 2018.PubMed/NCBI
|
|
10
|
Xiong J, Bing Z and Guo S: Observed
survival interval: A supplement to TCGA pan-cancer clinical data
resource. Cancers (Basel). 11:2802019. View Article : Google Scholar
|
|
11
|
Wang X, Song J, Wu Z, Fan B and Mode X:
Dual roles of CD38 in autophagy. Yangtze Medicine. 01:8–19. 2017.
View Article : Google Scholar
|
|
12
|
O'Steen S, Comstock ML, Orozco JJ, Hamlin
DK, Wilbur DSS, Jones JC, Kenoyer A, Nartea ME, Lin Y, Miller BW,
et al: The α-Emitter astatine-211 targeted to CD38 can eradicate
multiple myeloma in a disseminated disease model. Blood.
134:1247–1256. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ben Baruch B, Blacher E, Mantsur E,
Schwartz H, Vaknine H, Erez N and Stein R: Stromal CD38 regulates
outgrowth of primary melanoma and generation of spontaneous
metastasis. Oncotarget. 9:31797–31811. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morandi F, Marimpietri D, Horenstein AL,
Corrias MV and Malavasi F: Microvesicles expressing adenosinergic
ectoenzymes and their potential role in modulating bone marrow
infiltration by neuroblastoma cells. Oncoimmunology.
8:e15741982019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schutze K, Petry K, Hambach J, Schuster N,
Fumey W, Schriewer L, Rockendorf J, Menzel S, Albrecht B, Haag F,
et al: CD38-Specific biparatopic heavy chain antibodies display
potent complement-dependent cytotoxicity against multiple myeloma
cells. Front Immunol. 9:25532018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu T, Qiao C, Lv M and Tang L: Novel
anti-CD38 humanized mAb SG003 possessed enhanced cytotoxicity in
lymphoma than Daratumumab via antibody-dependent cell-mediated
cytotoxicity. BMC Biotechnol. 19:282019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Peng X, Zhang X, Xu H, Lu C, Liu
L, Song J and Zhang Y: The emerging roles of CIB1 in cancer. Cell
Physiol Biochem. 43:1413–1424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morandi F, Morandi B, Horenstein AL,
Chillemi A, Quarona V, Zaccarello G, Carrega P, Ferlazzo G, Mingari
MC, Moretta L, et al: A non-canonical adenosinergic pathway led by
CD38 in human melanoma cells induces suppression of T cell
proliferation. Oncotarget. 6:25602–25618. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Franz M, Rodriguez H, Lopes C, Zuberi K,
Montojo J, Bader GD and Morris Q: GeneMANIA update 2018. Nucleic
Acids Res. 46(W1): W60–W64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Selitsky SR, Mose LE, Smith CC, Chai S,
Hoadley KA, Dittmer DP, Moschos SJ, Parker JS and Vincent BG:
Prognostic value of B cells in cutaneous melanoma. Genome Med.
11:362019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guan J, Gupta R and Filipp FV: Cancer
systems biology of TCGA SKCM: Efficient detection of genomic
drivers in melanoma. Sci Rep. 5:78572015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harcharik S, Bernardo S, Moskalenko M, Pan
M, Sivendran M, Bell H, Hall LD, Castillo-Martin M, Fox K,
Cordon-Cardo C, et al: Defining the role of CD2 in disease
progression and overall survival among patients with completely
resected stage-II to -III cutaneous melanoma. J Am Acad Dermatol.
70:1036–1044. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hyams DM, Cook RW and Buzaid AC:
Identification of risk in cutaneous melanoma patients: Prognostic
and predictive markers. J Surg Oncol. 119:175–186. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
van de Donk NWCJ, Richardson PG and
Malavasi F: CD38 antibodies in multiple myeloma: Back to the
future. Blood. 131:13–29. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Q, Graeff R, Kriksunov IA, Lam CM, Lee
HC and Hao Q: Conformational closure of the catalytic site of human
CD38 induced by calcium. Biochemistry. 47:13966–13973. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brazill JM, Li C, Zhu Y and Zhai RG:
NMNAT: It's an NAD+ synthase. It's a chaperone. It's a
neuroprotector. Curr Opin Genet Dev. 44:156–162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shen YT, Wang JW, Wang M, Zhi Y, Li JY,
Yuan YS, Wang XX, Zhang H and Zhang KZ: BST1 rs4698412 allelic
variant increases the risk of gait or balance deficits in patients
with Parkinson's disease. CNS Neurosci Ther. 25:422–429. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ceroni F, Sagar A, Simpson NH, Gawthrope
AJ, Newbury DF, Pinto D, Francis SM, Tessman DC, Cook EH, Monaco
AP, et al: A deletion involving CD38 and BST1 results in a fusion
transcript in a patient with autism and asthma. Autism Res.
7:254–263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vendrell E, Ribas M, Valls J, Sole X, Grau
M, Moreno V, Capella G and Peinado MA: Genomic and transcriptomic
prognostic factors in R0 Dukes B and C colorectal cancer patients.
Int J Oncol. 30:1099–1107. 2007.PubMed/NCBI
|
|
35
|
Xie W, Stopsack KH, Drouin SJ, Fu H,
Pomerantz MM, Mucci LA, Lee GM and Kantoff PW: Association of
genetic variation of the six gene prognostic model for
castration-resistant prostate cancer with survival. Prostate.
79:73–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Xu Y, Peng X, Huang J, Yang M and
Wang X: A novel photosensitizer Znln2S4
mediated photodynamic therapy Induced-HepG2 cell apoptosis. Radiat
Res. 192:422–430. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang X, He X, Hu S, Sun A and Lu C:
Involvement of Bim in Photofrin-mediated photodynamically induced
apoptosis. Cell Physiol Biochem. 35:1527–1536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Roepcke S, Plock N, Yuan J, Fedyk ER, Lahu
G, Zhao L and Smithson G: Pharmacokinetics and pharmacodynamics of
the cytolytic anti-CD38 human monoclonal antibody TAK-079 in
monkey-model assisted preparation for the first in human trial.
Pharmacol Res Perspect. 6:e004022018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Manna A, Aulakh S, Jani P, Ahmed S, Akhtar
S, Coignet M, Heckman M, Meghji Z, Bhatia K, Sharma A, et al:
Targeting CD38 enhances the antileukemic activity of ibrutinib in
chronic lymphocytic leukemia. Clin Cancer Res. 25:3974–3985. 2019.
View Article : Google Scholar : PubMed/NCBI
|