Introduction
Osteosarcoma (OS) is the most common primary
malignancy of bone, which also has a high rate of metastasis to the
lungs (1). Currently, the 5-year
survival for patients with localized OS is 60–65%; however,
survival drops to 20–30% for patients with recurrent disease
(2). Despite various attempts, the
overall survival probabilities have not substantially improved in
the last 30 years (2). OS is a
heterogeneous tumor, with a wide diversity of genetic and
epigenetic alterations that are involved in its development and
progression (1,3). As such, the identification of these
genetic and epigenetic abnormalities may have a great impact on the
future therapies for OS.
Epigenetic regulations through post-translational
modifications (PTMs) of histone proteins by methylation have been
characterized as key factors in cell growth, differentiation and
DNA repair pathways (4,5). Aberrant histone methylation is a
universal feature of various types of cancer and most likely plays
a causal part in tumorigenesis (6,7).
However, due to the rarity of OS within the population, there are
limited studies investigating alterations of histone methylation in
OS. To the best of our knowledge, the present study is the first
showing aberrant histone methylation in OS.
Methylation of histone H4 lysine20 (H4K20) is
implicated genomic integrity, such as playing a role in the DNA
repair, DNA replication, chromatin compaction and transcriptional
regulation processes (8,9). Trimethylation at H4K20 (H4K20me3), a
marker of constitutive heterochromatin, is correlated with the
silencing of genes during the development of various types of
cancer (10,11). In particular, a global decrease of
H4K20me3 is considered a common hallmark of various types of human
cancers (11–14). It has been proposed that the loss of
H4K20me3 in repetitive DNA sequences is associated with the global
loss of DNA methylation in tumorigenesis (11). Furthermore, loss of H4K20me3 is
correlated with a poor prognosis in bladder, colon and breast
cancer, presenting H4K20me3 with a potential prognostic value for
various types of cancer (15–20).
Lysine methyltransferase 5B (SUV420H1) and lysine methyltransferase
5C (SUV420H2) are preferentially responsible for dimethylation of
H4K20 (H4K20me2) and trimethylation of H4K20 (H4K20me3) (21,22).
Suv420h-double-null (Suv420h1/Suv420h2 double-knockout) mice
present with perinatally death and lose nearly all H4K20me2 and
H4K20me3, as well as being accompanied with chromosomal aberrations
and deficiencies in DNA double-strand break repair (23). Abrogation of SUV420H2 or both SUV420H
HMTs but not SUV420H1 leads to decreased H4K20me3 in telomeric
chromatin that provides an increased tumorigenic potential,
indicating an important role for SUV420H HMTs in tumorgenicity
(24,25). In the present study, aberrations in
histone H4K20 trimethylation in OS tissues and cell lines were
observed, presumably associated with decreased SUV420H2 expression
levels, which is likely to be related to a poor survival in OS.
Additionally, a set of SUV420H2-regulated genes involved in
numerous signaling pathways was identified through RNA-sequencing
(RNA-seq) analysis. As such, the present findings indicated crucial
functions for H4K20me3 and its specific HMTs, SUV420H2, in OS.
Furthermore, it was illustrated that H4K20me3 and SUV420H2 may be
promising candidate biomarkers for the early detection of OS.
Materials and methods
Cell lines and cell culture
The human osteoblast cell line, hFOB1.19, and human
OS cell lines, HOS, U2OS and MG-63, were purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
Cells were grown in DMEM (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS (ScienCell Research Laboratories, Inc.)
and 1% antibiotic/antimycotic solution (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified incubator with 5%
CO2.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from the cells by using
Takara MiniBEST Universal RNA Extraction kit (Takara Bio, Inc.).
RNA was reverse transcribed to cDNA using the PrimeScript™ 1st
strand cDNA Synthesis kit (Takara Bio, Inc.) according to the
manufacturer's instruction. The cDNA synthesis reaction was
performed at 42°C for 45 min. Quantitative PCR was conducted using
TB Green™ Premix Ex Taq™ II (Takara Bio, Inc.) at 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec, in the
ABI StepOnePlus Real-time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Relative gene expression levels were
quantified relative to GAPDH levels using the 2−ΔΔCq
method (26). Each sample was
analyzed in triplicate. The primer sequences are listed in Table SI.
Small interfering RNA (siRNA)
transfection
siRNA oligonucleotide duplexes were synthesized by
Biolino Nucleic Acid Technology Co., Ltd. (www.biolino.cn), targeting the human SUV420H2
transcripts. siRNA targeting enhanced green fluorescent protein
[EGFP (siEGFP)] and negative control (siNC) were used as control
siRNAs. The siRNA sequences are described in Table SII. OS cancer cells were transfected
with siRNA duplexes (100 nM final concentration) using
Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.).
Lipofectamine® RNAiMAX and siRNA were separately diluted
in Opti-MEM® I Reduced Serum Medium (Gibco; Thermo
Fisher Scientific, Inc.). Two diluted reactions were mixed
together, incubated for 5 min at room temperature, and the
siRNA-lipid complex was added to cells. Transfected cells were
incubated continuously at 37°C for additional 96 h followed by
immediate RNA extraction or cell lysis.
Immunohistochemistry (IHC), western
blotting (WB) and antibodies
Pre-fixed human OS tissue microarray containing 43
OS samples and 13 normal bone samples (from adjacent normal bone
tissue) was purchased from Alenabio. EliVision™ plus kit and DAB
kit (MXB Biotechnologies; http://maxim.com.cn/) were used for staining according
to the manufacturer's instruction. The sections were deparaffinized
with xylene and rehydrated through 100, 95, 85 and 70% ethanol for
5 min. Endogenous peroxidase activity was blocked by incubating
sections in 3% H2O2 solution in methanol at
room temperature for 10 min. After blocking with 10% goat serum
(Wuhan Boster Biological Technology, Ltd.) at room temperature for
20 min, the sections were sequentially incubated with rabbit
anti-H4K20me3 antibody (1:100) at 37°C for 2 h, signal enhancer
(from the EliVision™ Plus kit, MXB Biotechnologies) at room
temperature for 30 min and anti-rabbit IgG Fab-HRP (ready to use,
EliVision™ Plus kit) at 37°C for 30 min. Each incubation step was
followed by three washes in PBS for 5 min. DAB solution (DAB kit,
MXB Biotechnologies) was applied to reveal the color. After the
color development was stopped by washing with distilled water, the
slides were immersed into hematoxylin at room temperature for 10
min and washed with distilled water. The slides were dehydrated
through 4 changes of ethanol (70, 85, 95 and 100%) for 5 min each,
cleared with xylene, and mounted using Neutral balsam mounting
solution (Sinopharm Chemical Reagent Co., Ltd.). Finally, the
tissue slides were observed under a light microscope at 200× and
400× magnification (Olympus BX41). Two expert pathologists
performed semiquantitative analysis of H4K20me3 staining levels
using a 3-grade scale defined as: Mild grade, +1; moderate grade,
+2; and strong grade, +3. For WB, the cells were lysed using RIPA
lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% sodium
deoxycholate, 0.1% SDS, 1% Nonidet-P40, 0.1 mM PMSF] containing
Protease Inhibitor Cocktail (Roche Diagnostics), and protein
concentrations were determined using a BCA Protein assay kit (CoWin
Biosciences). In total, 20 µg proteins were loaded in each well
then subjected to 10% (for detection of SUV420H2 and β-actin) or
15% (for detection of H4K20me3 and H4) SDS-PAGE. The proteins were
transferred onto polyvinylidene fluoride (PVDF) membranes followed
by blocking with 5% milk in 0.1% TBST buffer for 1 h at room
temperature. Later, the blots were incubated with primary
antibodies at 4°C overnight, and subsequently incubated with
secondary antibodies for 1 h at room temperature. Finally, the
protein signals were detected by Tanon high-sig ECL western
blotting substrate (Tanon Science & Technology Co., Ltd.). The
relative density of the protein band of interest is qualified using
Tanon Image Software version 1.0 (Tanon Science & Technology
Co., Ltd.). The following antibodies were used: Anti-H4K20me3 (cat.
no. ab9053; dilution, 1:100 for IHC and 1:1,000 for WB; Abcam),
anti-H4 (cat. no. 16047-1-AP; dilution, 1:500; ProteinTech Group,
Inc.), anti-SUV420H2 antibody (cat. no. ab91224; dilution: 1:100;
Abcam), anti-β-actin (cat. no. sc-47778; dilution, 1:1,000; Santa
Cruz Biotechnology, Inc.), goat anti-rabbit IgG secondary antibody,
HRP conjugate (cat. no. KGAA35; dilution: 1:1,000; KeyGEN BioTECH)
and goat anti-mouse IgG secondary antibody, HRP conjugate (cat. no.
KGAA37; dilution, 1:1,000; KeyGEN BioTECH).
RNA-seq analysis
Indexed libraries from HOS cells incubated with
siEGFP or siSUV420H2 were subjected to RNA-seq using the Illumina
Hiseq2000 platform (Illumina, Inc.). Gene expression was quantified
using the fragments per kilobase of transcript per million mapped
reads (FPKM) normalization method (27) and Cufflinks version 2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/install/)
was used for FPKM quantification. Gene expression levels and
differential transcription between siEGFP and siSUV420H2 treated
cells (using the same methodology as aforementioned) were evaluated
using Cuffdiff (part of the Cufflinks software aforementioned). The
lists of significantly differentially expressed genes were obtained
using a threshold of P-value ≤0.05 and a fold-change ≥2. Gene
Ontology (GO) analysis was performed using the standard enrichment
computation method (28). KEGG
pathway analysis was performed by KOBAS and P<0.05 was set as
the cut-off criterion.
Statistical analysis
Statistical analyses were performed using SPSS
version 20.0 (IBM Corp.). Comparisons between two groups were
analyzed using an independent two-sample t-test (two-tailed), while
comparisons among multiple groups were analyzed using one-way
ANOVAs followed by Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference. Experiments
were performed in duplicate or triplicates and results are
presented as mean ± SD except for the RT-qPCR experiments. The mean
± SEM was used for RT-qPCR experiments. The status of overall
survival regarding the SUV420H2 expression was set using the median
SUV420H2 expression as the cut-off and was estimated with the
Kaplan-Meier method using GraphPad Prism v.6 software (GraphPad
Software, Inc.). The log-rank test was used to assess statistical
significance.
Results
Aberrant pattern of H4K20me3 in
OS
The expression status of H4K20me3 was examined in
tissue samples from patients with OS using a tissue microarray
containing 43 OS samples and 13 normal bone samples (from adjacent
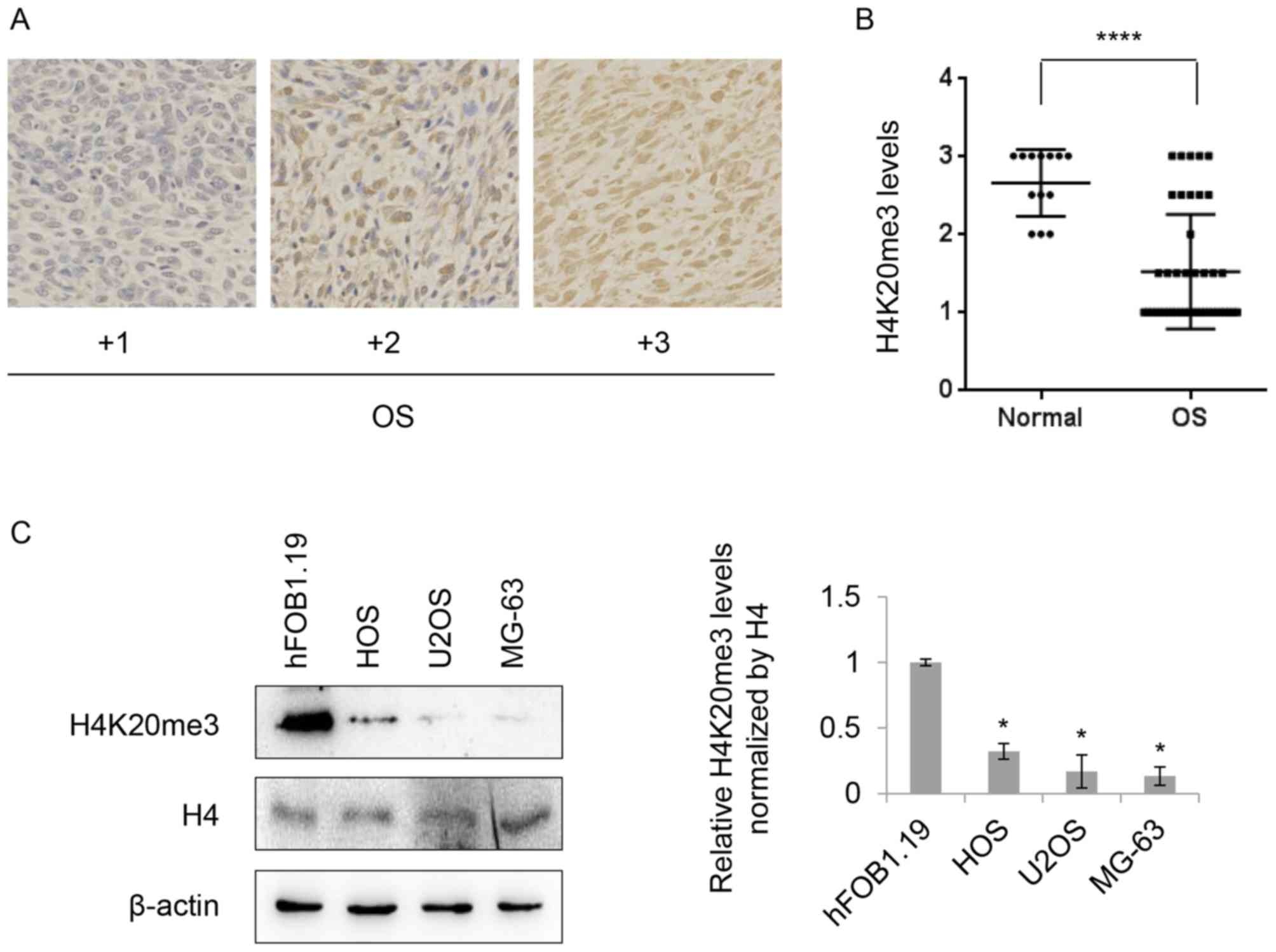
normal bone tissue) by IHC analysis. Fig. 1A presents representative results of
the IHC data, where scores were applied of +1 to +3. IHC analysis
revealed moderate or strong (≥+2) staining of H4K20me3 in all of
normal samples (13 in 13) compared with 25.58% (11 in 43) in the OS
cases. Approximately 55.81% (24 in 43) of OS cases expressed
H4K20me3 at mild levels (+1) and 18.6% (8 in 43) of OS cases
between mild to moderate levels (Fig.
1B), indicating decreased levels of H4K20me3 in OS tissues.
Subsequently, the levels of H4K20me3 in OS cell lines were
investigated. The levels of H4K20me3 were reduced in all of three
tested OS cell lines (HOS, U2OS and MG-63) compared with the normal
osteoblast cell line, hFOB1.19, whereas there was no significant
change in the expression of total histone H4 (Fig. 1C). Together, these results
illustrated that there was a loss of H4K20me3 in both the OS
tissues and cancer cell lines.
SUV420H2 expression is associated with
decreased H4K20me3 levels in OS
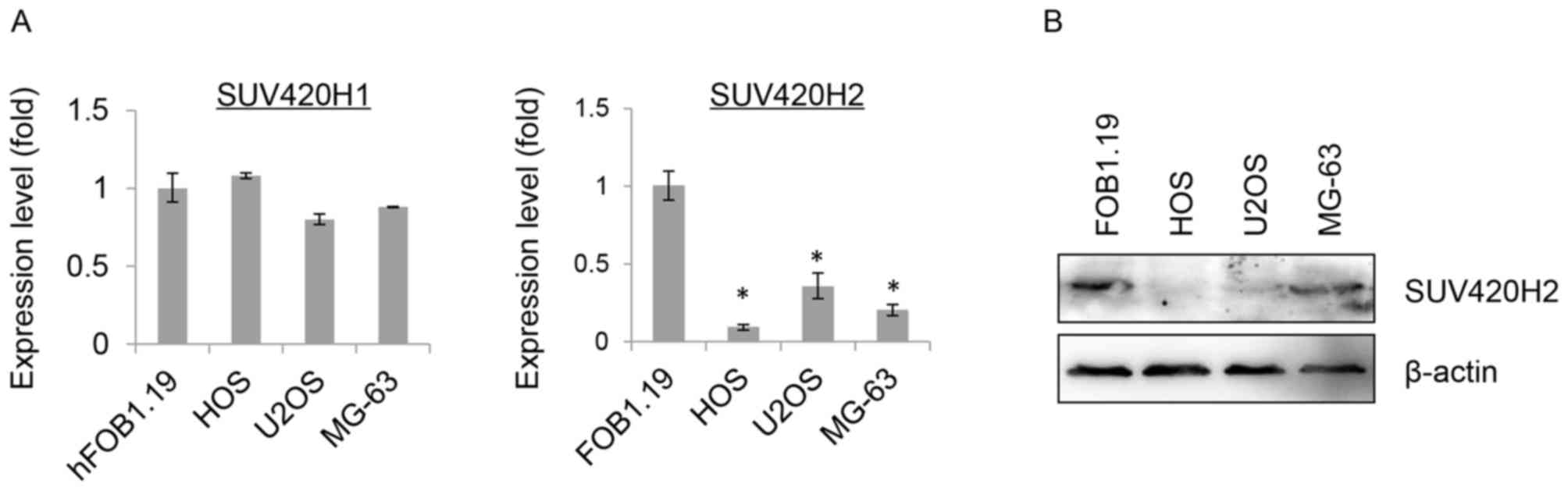
To identify the histone methyltransferase
responsible for the loss of H4K20me3, the mRNA expression levels of
both SUV420H1 and SUV420H2 were examined. Interestingly, only mRNA
expression levels of SUV420H2 were significantly reduced in all
three OS cell lines (HOS, U2OS and MG-63) compared to hFOB1.19
cells, but not mRNA expression levels of SUV420H1 (Fig. 2A). Similarly, a drastic reduction in
protein expression levels of SUV420H2 were observed in HOS, U2OS
and MG-63 cells in contrast to hFOB1.19 cells, indicating that the
decreased SUV420H2 is associated with the global loss of H4K20me3
in OS (Fig. 2B). The present study
failed to detect the protein expression of SUV420H1 in cell lines
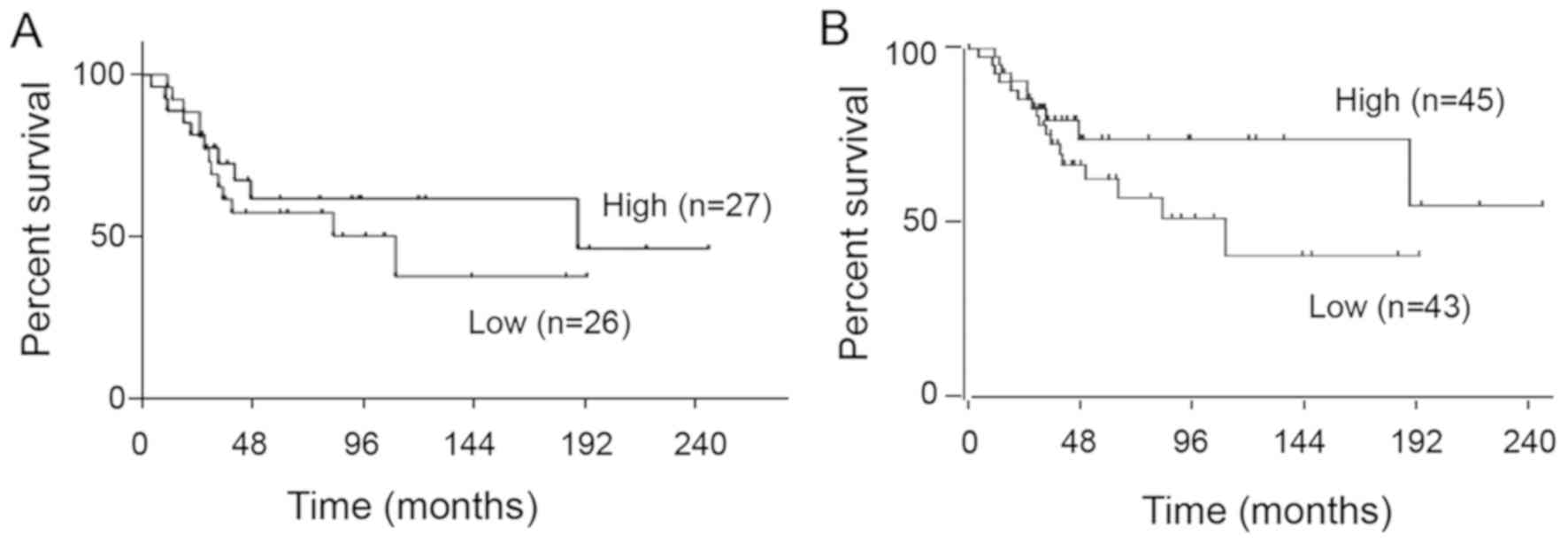
due to the antibody issue. Next, using the expression profile
dataset GSE21257, which included 53 OS samples, and the GSE42352
dataset, which included 88 OS samples, the prognostic significance
of SUV420H2 expression was investigated. As shown in Fig. 3A and B, no prognostic value for
SUV420H2 expression was observed (P=0.47 in GSE21257; P=0.11 in
GSE42352). Nevertheless, there was a tendency towards a poorer
prognosis in low SUV420H2 expression group even though the
association was not significant.
Identification of downstream genes of
SUV420H2
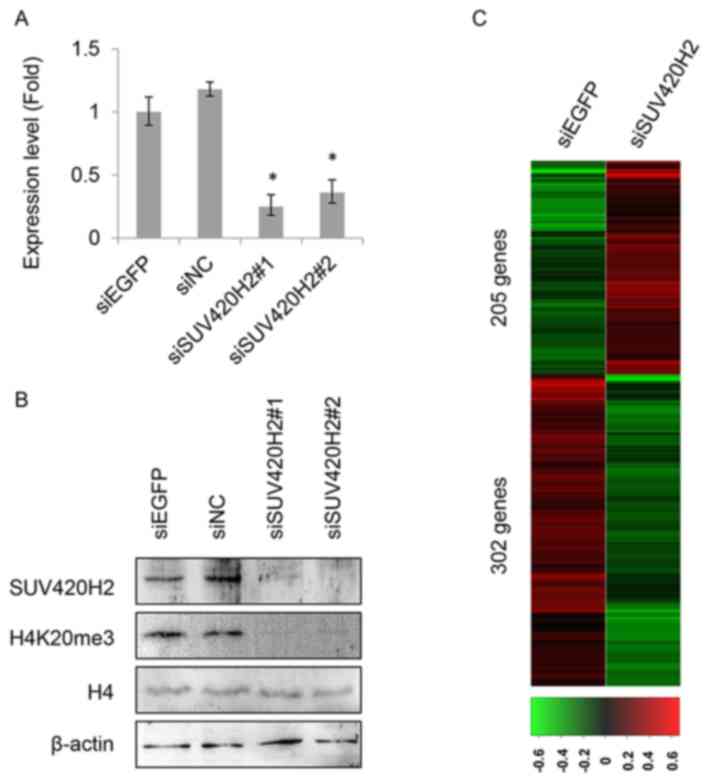
To identify the involvement of SUV420H2 in OS,
RNA-seq analysis was performed to find SUV420H2-regulated genes in
HOS cells which were expressed at higher levels with H4K20me3. The
knockdown efficiency of SUV420H2 was validated both at the mRNA and
protein levels, as shown in Fig. 4A and
B. The expression levels of SUV420H2 were substantially reduced
in the siSUV420H2 treated cells. Subsequently, RNA sequencing of
the gene expression analysis was performed in HOS cells treated
with siSUV420H2#1 or siEGFP as a control. A variety of genes were
identified, including 205 upregulated genes and 302 downregulated
genes in SUV420H2-depleted HOS cells, with a threshold of P≤0.05
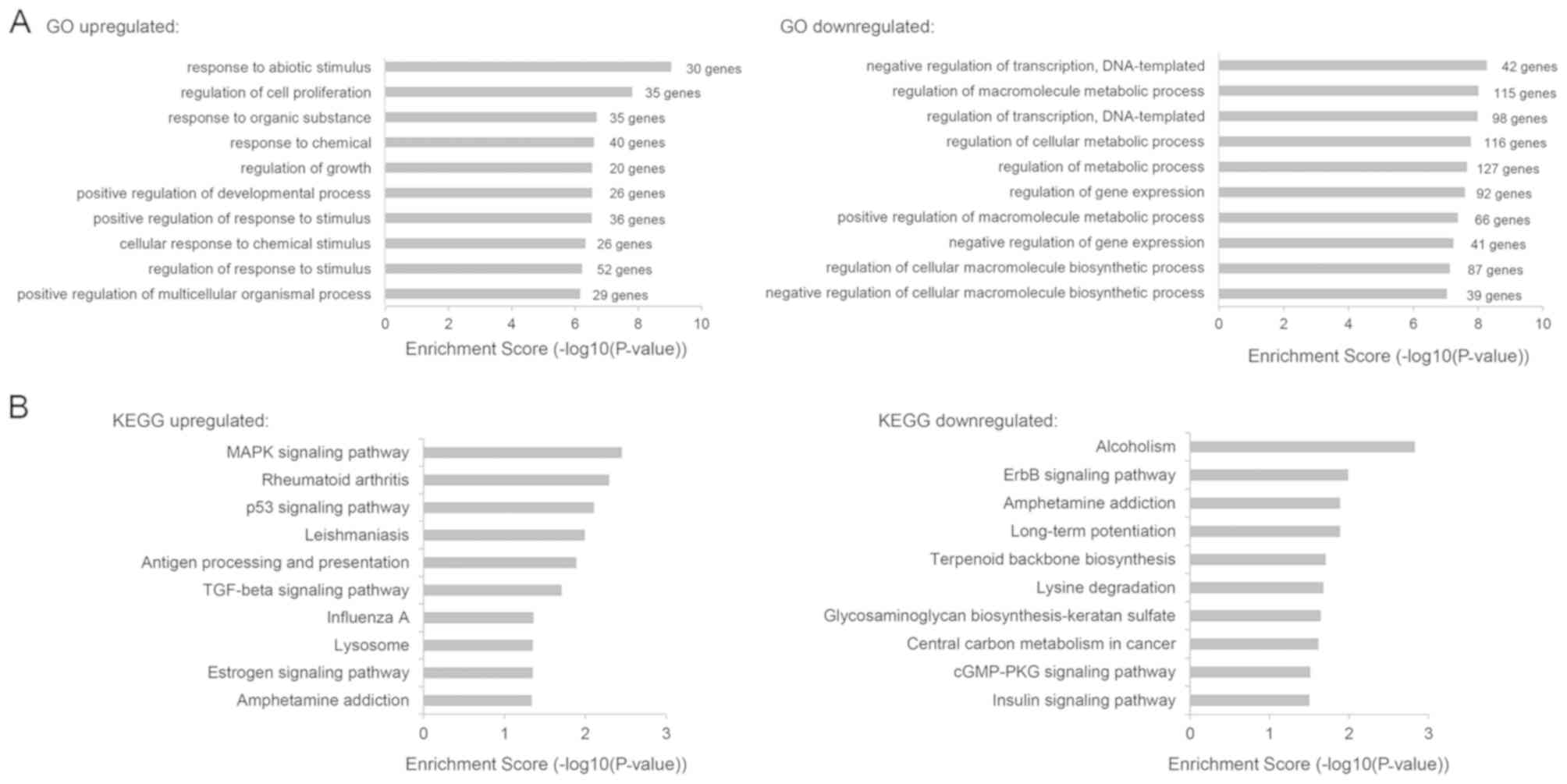
and a fold-change ≥2 as compared with the control (Fig. 4C). GO term enrichment analysis of the
biological process category revealed that upregulated genes after
SUV420H2 depletion were involved in various biological processes,
including regulation of cell proliferation (GO:0042127, 35
upregulated DEGs were included), regulation of growth (GO:0040008,
20 upregulated DEGs were included) and the response to chemicals
(GO:0042221, 40 upregulated DEGs were included), whereas
downregulated genes after SUV420H2 depletion were associated with
the negative regulation of transcription (GO:0045892, 42
downregulated DEGs were included), regulation of metabolic
processes (GO:0019222, 127 downregulated DEGs were included) and
the negative regulation of gene expression (GO:0010629, 41
downregulated DEGs were included) (Fig.
5A), indicating that SUV420H2 expression is involved with a
complicated of functions in vivo. Additionally, the top ten
KEGG pathways were analyzed and presented in Fig. 5B. These results demonstrated that
changes in SUV420H2 expression may influence a variety of key
signaling pathways in carcinogenesis including the
mitogen-activated protein kinase (MAPK), p53 and ErbB signaling
pathways, indicating substantial contributions of SUV420H2 to
carcinogenesis. To validate the findings of the RNA-seq, the top 20
upregulated and downregulated candidate genes were selected for
further analysis using RT-qPCR. Two SUV420H2 specific siRNAs
(siSUV420H2#1 and siSUV420H2#2), targeting different sections of
SUV420H2, were used to exclude any off-target candidate genes. As a
result, 7 upregulated and 13 downregulated genes, as listed in
Fig. 6A and B, were identified from
the depletion of SUV420H2. Fos proto-oncogene (FOS), which is
implicated as a regulator of cell proliferation and differentiation
(29) and was upregulated with the
reduction in SUV420H2 expression. Growth differentiation factor 15,
which is involved in transforming growth factor (TGF) signaling
(30), was downregulated with the
silencing of SUV420H2.
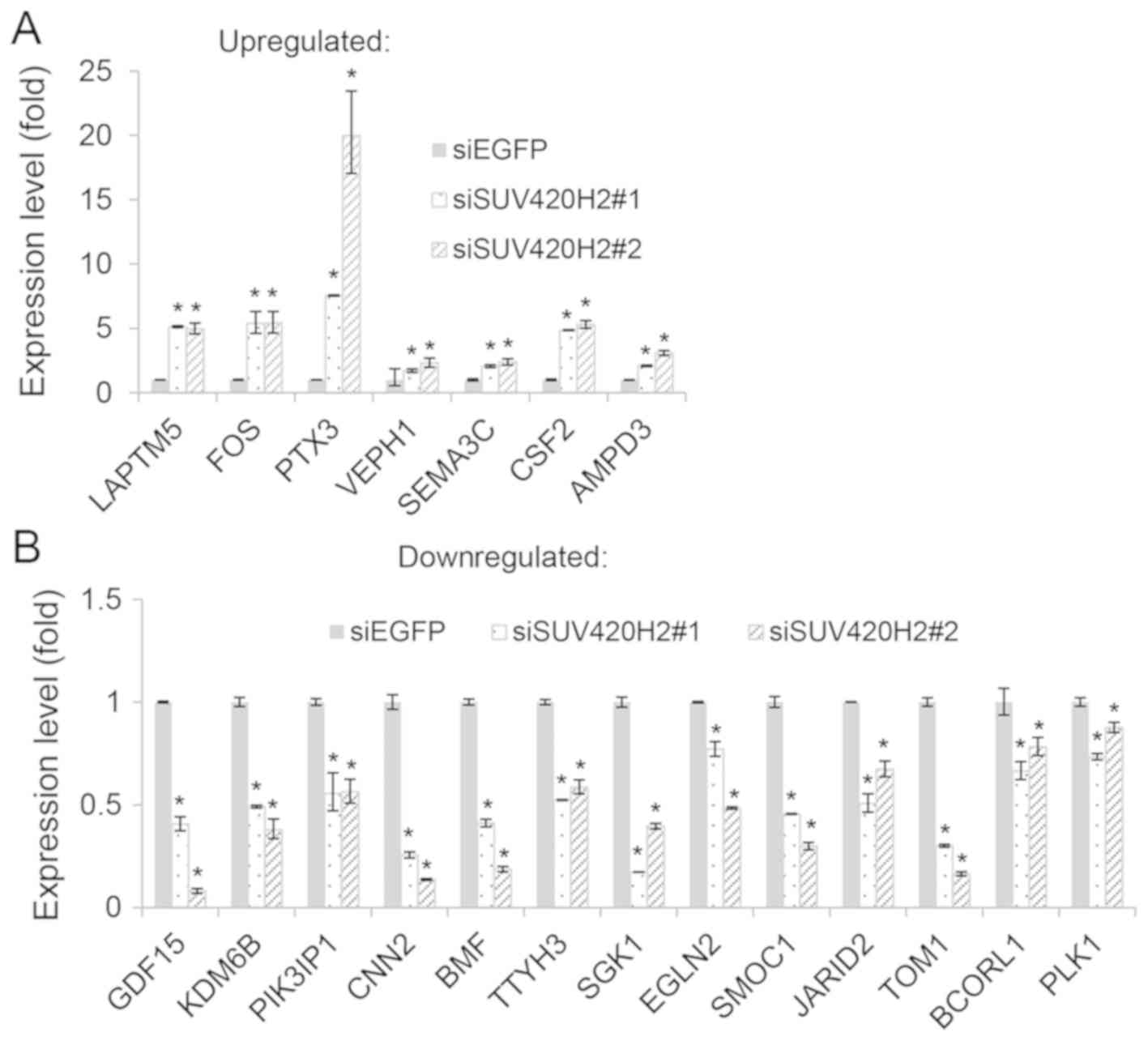 | Figure 6.Validation of RNA-sequencing results
using RT-qPCR. Expression of selected SUV420H2-associated genes,
including (A) upregulated and (B) downregulated genes upon SUV420H2
depletion, was validated in control (siEGFP) and SUV420H2-depleted
(siSUV420H2#1 and siSUV420H2#2) HOS cells using RT-qPCR analysis.
The relative mRNA expression levels (normalized to GAPDH) in
siSUV420H2#1 and siSUV420H2#2 samples were compared to that in the
control siEGFP sample. Data are presented as the mean ± SEM of
three experimental repeats; *P<0.05, siSUV420H2 vs. siEGFP.
RT-qPCR, reverse transcription-quantitative PCR; si, small
interfering RNA; LAPTM5, lysosomal protein transmembrane 5; FOS,
Fos proto-oncogene; PTX3, pentraxin 3; VEPH1, ventricular zone
expressed PH domain containing 1; SEMA3C, semaphorin 3C; CSF2,
colony stimulating factor 2; AMPD3, adenosine monophosphate
deaminase 3; GDF15, growth differentiation factor 15; KDM6B, lysine
demethylase 6B; PIK3IP1, phosphoinositide-3-kinase interacting
protein 1; CNN2, calponin 2; BMF, Bcl2 modifying factor; TTYH3,
tweety family member 3; SGK1, serum/glucocorticoid regulated kinase
1; EGLN2, egl-9 family hypoxia inducible factor 2; SMOC1, SPARC
related modular calcium binding 1; JARID2, jumonji and AT-rich
interaction domain containing 2; TOM1, target of myb1 membrane
trafficking protein; BCORL1, BCL6 corepressor like 1; PLK1,
polo-like kinase 1. |
Discussion
The nucleosome is comprised of histone proteins
containing a 147-bp DNA segment and is a fundamental unit of
chromatin structures (31). All four
core histone proteins, including H3, H4, H2A and H2B undergo
diverse PTMs such as acetylation, phosphorylation, methylation,
ubiquitination, SUMOylation and ADP-ribosylation (32). Through dynamic regulations of
chromatin structure, these histone modifications impact on genomic
integrity and a set of physiological functions, including
transcriptional regulation and DNA replication, as well as DNA
damage and repair (4,5). Among these histone modifications,
aberrant histone methylation events are likely to play a causal
role in tumorigenesis (6). H4K20me3,
a hallmark of silenced heterochromatic regions, is closely related
with various types of cancer (10).
Aberrant H4K20me3 has been reported in breast and lung cancers
(11,13,14).
However, there is no reported study regarding the status of
H4K20me3 in OS. As such, the present study firstly examined the
levels of H4K20me3 in OS tissue samples and cancer cell lines. The
results found decreased levels of H4K20me3 in OS tissue samples and
cell lines compared with normal samples. These findings indicated
that the loss of H4K20me3 is likely to be involved in the
development and progression of OS.
SUV420H proteins, including SUV420H1 and SUV420H2,
mediate the vast majority of H4K20me3 modifications (33). In SUV420H1-null primary mouse
embryonic fibroblasts, H4K20me2, but not H4K20me3, is reduced. In
contrast, SUV420H2-null cells exhibit selective loss of H4K20me3,
but maintain H4K20me2, indicating that SUV420H2 has a preference to
induce H4K20me3 (23). In order to
further elucidate which histone methyltransferase are responsible
for the loss of H4K20me3 in OS, the expression levels of SUV420H1
and SUV420H2 were investigated in OS cell lines. Notably, there was
a pronounced decrease in SUV420H2 expression levels in OS cells,
whereas no significant changes were detected in the expression
levels of SUV420H1. These findings suggested that aberrant SUV420H2
expression was largely associated with the loss of H4K20me3 in OS
cells. However, the protein expression levels of SUV420H1 were
unable to be examined in the present study due to specificity
issues with the available antibodies. Similarly, reduced expression
levels of SUV420H2 are also observed in breast cancer, lung cancer,
liver cancer and colon cancer (12,13,34). In
addition, the expression of SUV420H2 was found to have no
prognostic value for patients with OS; however, this may be due to
the limited small sample size in the present study. Despite a lack
of significance being observed in the present study, there was some
tendency for SUV420H2 expression to be associated with a poorer
prognosis, implying that SUV420H2 and H4K20me3 may have value as
prognostic factors in OS. To clarify this, the expression levels of
SUV420H2 and H4K20me3 in OS tissue samples should be examined with
a larger sample size. Unfortunately, due to issues with
antibody-specificity for targeting SUV420H2, the present study was
unable to examine the expression levels of SUV420H2 in OS tissue
samples.
Transient depletion of SUV420H2 results in a
decreased expression of several osteoblastic markers, such as
integrin binding sialoprotein and alkaline phosphatase,
biomineralization associated, as well as osteoblastic transcription
factors, such as Sp7 transcription factor (35). To the best of our knowledge, there is
limited information regarding the functions of SUV420H2 as well as
H4K20me3 in OS. To explore the involvement of SUV420H2 in the
development and progression of OS, SUV420H2 regulated genes were
identified through RNA-seq analysis. GO term enrichment analysis
using SUV420H2 regulated genes revealed that as well as regulation
of cell proliferation, transcription and gene expression, SUV420H2
was also likely to be involved in the response to abiotic stimulus
(GO:0009628), cellular response to chemical stimulus (GO:0070887),
regulation of response to stimulus (GO:0048583), and metabolic
processes (GO:0019222, regulation of metabolic process; GO:0031323,
regulation of cellular metabolic process; GO:0080090, regulation of
primary metabolic process). Furthermore, SUV420H2 expression was
found to be associated with MAPK, P53, TGF and ErbB signaling
pathway through regulations of the lysosomal protein transmembrane
5, FOS and phosphoinositide-3-kinase interacting protein 1
genes. Further studies should focus on confirming that there has
been accumulation of H4K20me3 within these targeted genes in
SUV420H2-overexpressing OS cells.
In conclusion, the present study revealed that
aberrations in SUV420H2 expression and the resultant loss of
H4K20me3 in OS may be important factors for further understanding
the biological significance of SUV420H2 and H4K20me3 in OS.
However, further studies are required to elucidate the underlying
mechanisms of action behind the involvement of SUV420H2 and
H4K20me3 in OS through the regulation of the aforementioned
genes.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Shan Chang
(Institute of Bioinformatics and Medical Engineering, Jiangsu
University of Technology) for their support.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81903661) and
Changzhou Sci&Tech Program (grant no. 20180170).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contribution
LP and XY participated in the whole project and
performed the majority of the experiments. LW, XX and MZ performed
immunohistochemistry-related experiments. JL and RK analyzed
RNA-seq data. ZL was involved in the conception of the study and
also supervised the quality of all the work throughout the entire
process. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there is no competing
interest associated with the manuscript.
References
|
1
|
Morrow JJ and Khanna C: Osteosarcoma
genetics and epigenetics: Emerging biology and candidate therapies.
Crit Rev Oncog. 20:173–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hattinger CM, Fanelli M, Tavanti E, Vella
S, Ferrari S, Picci P and Serra M: Advances in emerging drugs for
osteosarcoma. Expert Opin Emerg Drugs. 20:495–514. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martin JW, Squire JA and Zielenska M: The
genetics of osteosarcoma. Sarcoma. 2012:6272542012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luco RF, Pan Q, Tominaga K, Blencowe BJ,
Pereira-Smith OM and Misteli T: Regulation of alternative splicing
by histone modifications. Science. 327:996–1000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huertas D, Sendra R and Muñoz P: Chromatin
dynamics coupled to DNA repair. Epigenetics. 4:31–42. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hamamoto R, Saloura V and Nakamura Y:
Critical roles of non-histone protein lysine methylation in human
tumorigenesis. Nat Rev Cancer. 15:110–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balakrishnan L and Milavetz B: Decoding
the histone H4 lysine 20 methylation mark. Crit Rev Biochem Mol
Biol. 45:440–452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jorgensen S, Schotta G and Sorensen CS:
Histone H4 lysine 20 methylation: Key player in epigenetic
regulation of genomic integrity. Nucleic Acids Res. 41:2797–2806.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwon MJ, Kim SS, Choi YL, Jung HS, Balch
C, Kim SH, Song YS, Marquez VE, Nephew KP and Shin YK: Derepression
of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with
loss of repressive histone modifications. Carcinogenesis.
31:974–983. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fraga MF, Ballestar E, Villar-Garea A,
Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S,
Petrie K, et al: Loss of acetylation at Lys16 and trimethylation at
Lys20 of histone H4 is a common hallmark of human cancer. Nat
Genet. 37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pogribny IP, Ross SA, Tryndyak VP,
Pogribna M, Poirier LA and Karpinets TV: Histone H3 lysine 9 and H4
lysine 20 trimethylation and the expression of Suv4-20h2 and
Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced
by methyl deficiency in rats. Carcinogenesis. 27:1180–1186. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tryndyak VP, Kovalchuk O and Pogribny IP:
Loss of DNA methylation and histone H4 lysine 20 trimethylation in
human breast cancer cells is associated with aberrant expression of
DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and
methyl-binding proteins. Cancer Biol Ther. 5:65–70. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Van Den Broeck A, Brambilla E,
Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B and Gazzeri S:
Loss of histone H4K20 trimethylation occurs in preneoplasia and
influences prognosis of non-small cell lung cancer. Clin Cancer
Res. 14:7237–7245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schneider AC, Heukamp LC, Rogenhofer S,
Fechner G, Bastian PJ, von Ruecker A, Müller SC and Ellinger J:
Global histone H4K20 trimethylation predicts cancer-specific
survival in patients with muscle-invasive bladder cancer. BJU Int.
108:E290–E296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benard A, Goossens-Beumer IJ, van Hoesel
AQ, de Graaf W, Horati H, Putter H, Zeestraten EC, van de Velde CJ
and Kuppen PJ: Histone trimethylation at H3K4, H3K9 and H4K20
correlates with patient survival and tumor recurrence in
early-stage colon cancer. BMC Cancer. 14:5312014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yokoyama Y, Matsumoto A, Hieda M, Shinchi
Y, Ogihara E, Hamada M, Nishioka Y, Kimura H, Yoshidome K,
Tsujimoto M and Matsuura N: Loss of histone H4K20 trimethylation
predicts poor prognosis in breast cancer and is associated with
invasive activity. Breast Cancer Res. 16:R662014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Paydar P, Asadikaram G, Nejad HZ, Akbari
H, Abolhassani M, Moazed V, Nematollahi MH, Ebrahimi G and Fallah
H: Epigenetic modulation of BRCA-1 and MGMT genes, and histones H4
and H3 are associated with breast tumors. J Cell Biochem.
120:13726–13736. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y, Shi W, Tang T, Wang Y, Yin X, Chen
Y, Zhang Y, Xing Y, Shen Y, Xia T, et al: miR-29a contributes to
breast cancer cells epithelial-mesenchymal transition, migration,
and invasion via down-regulating histone H4K20 trimethylation
through directly targeting SUV420H2. Cell Death Dis. 10:1762019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Özgür E, Keskin M, Yörüker EE,
Holdenrieder S and Gezer U: Plasma histone H4 and H4K20
trimethylation levels differ between colon cancer and precancerous
polyps. In vivo. 33:1653–1658. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang H, Pesavento JJ, Starnes TW,
Cryderman DE, Wallrath LL, Kelleher NL and Mizzen CA: Preferential
dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem.
283:12085–12092. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakaguchi A, Karachentsev D, Seth-Pasricha
M, Druzhinina M and Steward R: Functional characterization of the
Drosophila Hmt4-20/Suv4-20 histone methyltransferase. Genetics.
179:317–322. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schotta G, Sengupta R, Kubicek S, Malin S,
Kauer M, Callén E, Celeste A, Pagani M, Opravil S, De La
Rosa-Velazquez IA, et al: A chromatin-wide transition to H4K20
monomethylation impairs genome integrity and programmed DNA
rearrangements in the mouse. Genes Dev. 22:2048–2061. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benetti R, Gonzalo S, Jaco I, Schotta G,
Klatt P, Jenuwein T and Blasco MA: Suv4-20h deficiency results in
telomere elongation and derepression of telomere recombination. J
Cell Biol. 178:925–936. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marion RM, Schotta G, Ortega S and Blasco
MA: Suv4-20h abrogation enhances telomere elongation during
reprogramming and confers a higher tumorigenic potential to iPS
cells. PLoS One. 6:e256802011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian L, Greenberg SA, Kong SW, Altschuler
J, Kohane IS and Park PJ: Discovering statistically significant
pathways in expression profiling studies. Proc Natl Acad Sci USA.
102:13544–13549. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eferl R and Wagner EF: AP-1: A
double-edged sword in tumorigenesis. Nat Rev Cancer. 3:859–868.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Strelau J, Bottner M, Lingor P,
Suter-Crazzolara C, Galter D, Jaszai J, Sullivan A, Schober A,
Krieglstein K and Unsicker K: GDF-15/MIC-1 a novel member of the
TGF-beta superfamily. Journal of neural transmission. J Neural
Transm Suppl. 273–276. 2000.PubMed/NCBI
|
|
31
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schotta G, Lachner M, Sarma K, Ebert A,
Sengupta R, Reuter G, Reinberg D and Jenuwein T: A silencing
pathway to induce H3-K9 and H4-K20 trimethylation at constitutive
heterochromatin. Genes Dev. 18:1251–1262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shinchi Y, Hieda M, Nishioka Y, Matsumoto
A, Yokoyama Y, Kimura H, Matsuura S and Matsuura N: SUV420H2
suppresses breast cancer cell invasion through down regulation of
the SH2 domain-containing focal adhesion protein tensin-3. Exp Cell
Res. 334:90–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Khani F, Thaler R, Paradise CR, Deyle DR,
Kruijthof-de Julio M, Galindo M, Gordon JA, Stein GS, Dudakovic A
and van Wijnen AJ: Histone H4 methyltransferase Suv420h2 maintains
fidelity of osteoblast differentiation. J Cell Biochem.
118:1262–1272. 2017. View Article : Google Scholar : PubMed/NCBI
|