Introduction
Prostate cancer (PCa) is the second leading cause of
male morbidity and mortality in the world (1). In recent years, the incidence rate of
PCa has increased dramatically (1,2). In
China in 2015, ~603,000 men were estimated to be diagnosed with PCa
and 266,000 men died of it (3). One
of the most likely causes is that more reliable and effective
biomarkers for early diagnosis of PCa are still lacking. Although a
large number of studies revealed several key proteins in PCa
progression, including androgen receptor (AR), forkhead box A1
(FOXA1) and speckle-type BTB/POZ protein (SPOP) (4–7), the
pathogenesis and etiology of PCa is still not well-understood.
Therefore, here is an urgent need to identify novel regulators to
understand mechanisms underlying PCa carcinogenesis and to serve as
biomarkers.
Histone methylation is tightly controlled by histone
methyltransferases and histone demethylases, and it is one of the
most important types of chromatin post-translational modifications
(8). Emerging studies have revealed
that histone methylation plays a significant role in
transcriptional regulation, maintenance of genome integrity and
epigenetic inheritance (9–13). An imbalance between methylation and
demethylation is frequently observed in the pathogenesis of human
disorders, including cancer (14–17).
Lysine demethylase 5B (KDM5B) is a member of the jumonji/AT-rich
interaction domain-containing (ARID) family of histone
demethylases, and it is also known as JARID1B or PLU-1. KDM5B has
been found to be upregulated in squamous cell carcinoma of the head
and neck, breast cancer, hepatocellular carcinoma, gastric cancer
and glioma (18–24). Previous studies also revealed that
KDM5B is upregulated in PCa (25).
However, the functional roles of KDM5B in PCa remain largely
unknown.
In the present study, the prognostic value of KDM5B
in PCa was explored by analyzing 3 independent public datasets.
Moreover, experimental validation was performed by investigating
the effects of KDM5B on PCa cell proliferation, cell cycle
progression and migration. The current study may provide useful
information to explore potential candidate biomarkers for the
diagnosis of PCa and for predicting prognosis in patients.
Materials and methods
Cell culture
LNCaP, PC-3, 22Rv1, DU145, and WPMY-1 cells were
purchased from the American Type Culture Collection. The cell lines
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences) in an incubator at 37°C with 5%
CO2.
Cell transfection
Short-interfering RNAs (siRNAs) against KDM5B
(siKDM5B-546 and siKDM5B-943) and negative control siRNA (siNC)
were purchased from Shanghai GenePharma Co., Ltd. The sequences
were as follows: siKDM5B-546 sense, 5′-GCAGUUGUUUGCAAGGAUATT-3′ and
antisense, 5′-UAUCCUUGCAAACAACUGCTT-3′; siKDM5B-943 sense,
5′-GCAUCAAGCAAGAACCUAUTT-3′ and antisense,
5′-AUAGGUUCUUGCUUGAUGCTT-3′; and siNC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. Transfection with the siKDM5Bs or siNC
was performed using Lipofectamine 3000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.).
RNA extraction and RT-qPCR
Total RNA was extracted from the cells using the
Ultrapure RNA kit (CoWin Biosciences). RT was performed using the
SuperQuick RT MasterMix (CoWin Biosciences) according to the
manufacturer's protocol. RT-qPCR was performed using the AceQ qPCR
SYBR Green Master Mix (Vazyme) according to the manufacturer's
protocol. The cycling conditions were as follows: Initial
denaturation (2 min at 95°C) followed by 40 cycles of denaturation
(10 sec at 95°C), annealing (30 sec at 59°C), elongation (30 sec at
72°C) and a final extension (30 sec at 72°C). The PCR primers for
mature KDM5B and β-actin were as follows: KDM5B forward,
5′-AGCAGACTGGCATCTGTAAGG-3′ and reverse,
5′-GAAGTTTATCAACATCACATGCAA-3′; and β-actin forward,
5′-CCTCTCCCAAGTCCACACAG-3′ and reverse, 5′-GGGCACGAAGGCTCATCATT-3′.
β-actin was used as internal control. The 2−ΔΔCq method
was used to analyze the data (26).
Each sample was measured in triplicate.
Western blot analysis
The PCa cells were homogenized and sonicated using
Mammalian Protein Extraction Kit (CoWin Biosciences). Protein
concentrations were detected using a BCA Protein Quantification
kit, according to the manufacturer's protocol. The proteins (50 µg)
were separated by 10% SDS-PAGE and then transferred onto
polyvinylidene difluoride membranes. The membrane was blocked with
5% non-fat dry milk for 1 h at room temperature and incubated with
specific primary antibodies overnight at 4°C. The primary
antibodies used were as follows: KDM5B (1:1,000; Abnova), ACTB
(1:1,000, cat. no. ab8226; Abcam). The secondary antibodies were
Goat Anti-Mouse IgG H&L (1:1,000, cat. no. ab205719; Abcam) for
ACTB, and Goat Anti-Rabbit IgG H&L (1:1,000, cat. no. ab205719;
Abcam) for KDM5B. The blots were detected with an enhanced
chemiluminescence substrate kit (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The bands were scanned
and quantified by ImageJ v1.47 software (National Institutes of
Health).
Cell proliferation assay
Cell proliferation was detected using the Cell
Counting Kit-8 (CCK-8) assay (MedChemExpress) in 96-well plates.
After transfection, 100 µl/well of cells were added to 96-well
plates. CCK-8 reagent was added to each well 2 h before the end of
the experiment, and the cells were incubated, and the absorbance
was then measured at 450 nm wavelength in a microplate reader.
Cell cycle analysis
Transfected LNCaP and PC-3 cells were collected 48 h
post-transfection. Triton X-100 (0.03%) and propidium iodide (50
ng/ml) were used to resuspend cells. After incubation at room
temperature for 10 min, the transfected cells were examined using a
flow cytometer (Beckman Coulter, Inc.). Each sample was measured in
triplicate.
Cell migration assay
Cells were treated with siRNAs, including siNC,
siKDM5B-546 and siKDM5B-943. Transwell plates were used for the
determination of migration ability. Briefly, 700 µl 1640 medium
supplemented with 10% FBS was added to the lower chamber of the
Transwells. A total of 2,000 cells were resuspended in 100 µl 1640
medium supplemented with 1% FBS, and then added to the upper
chamber of the system. After incubation in an incubator at 37°C for
3 days, the chambers were removed and unmigrated cells in the upper
chamber were wiped with a cotton swab. The migrated cells in the
upper chamber were washed twice with PBS and were then fixed with
700 µl methanol for 15 min. Next, the migrated cells were stained
with DAPI for 20 min, then washed 3 times with PBS. Cells were
imaged with a microscope. Each sample was measured in
triplicate.
Statistical analysis
The data are presented as the mean ± SD. Student's
t-test or Mann-Whitney U-test were used to compare the difference
between the 2 groups of data. Correlation analysis was performed
with Spearman's rank correlation. For multiple groups, Statistical
analyses were performed using a one-way analysis of variance with
the Bonferroni test for post hoc comparisons. Survival analysis was
based on the Kaplan-Meier method and the log-rank tests to compare
the differences between survival curves. P<0.05 was considered
to indicate a statistically significant difference.
Results
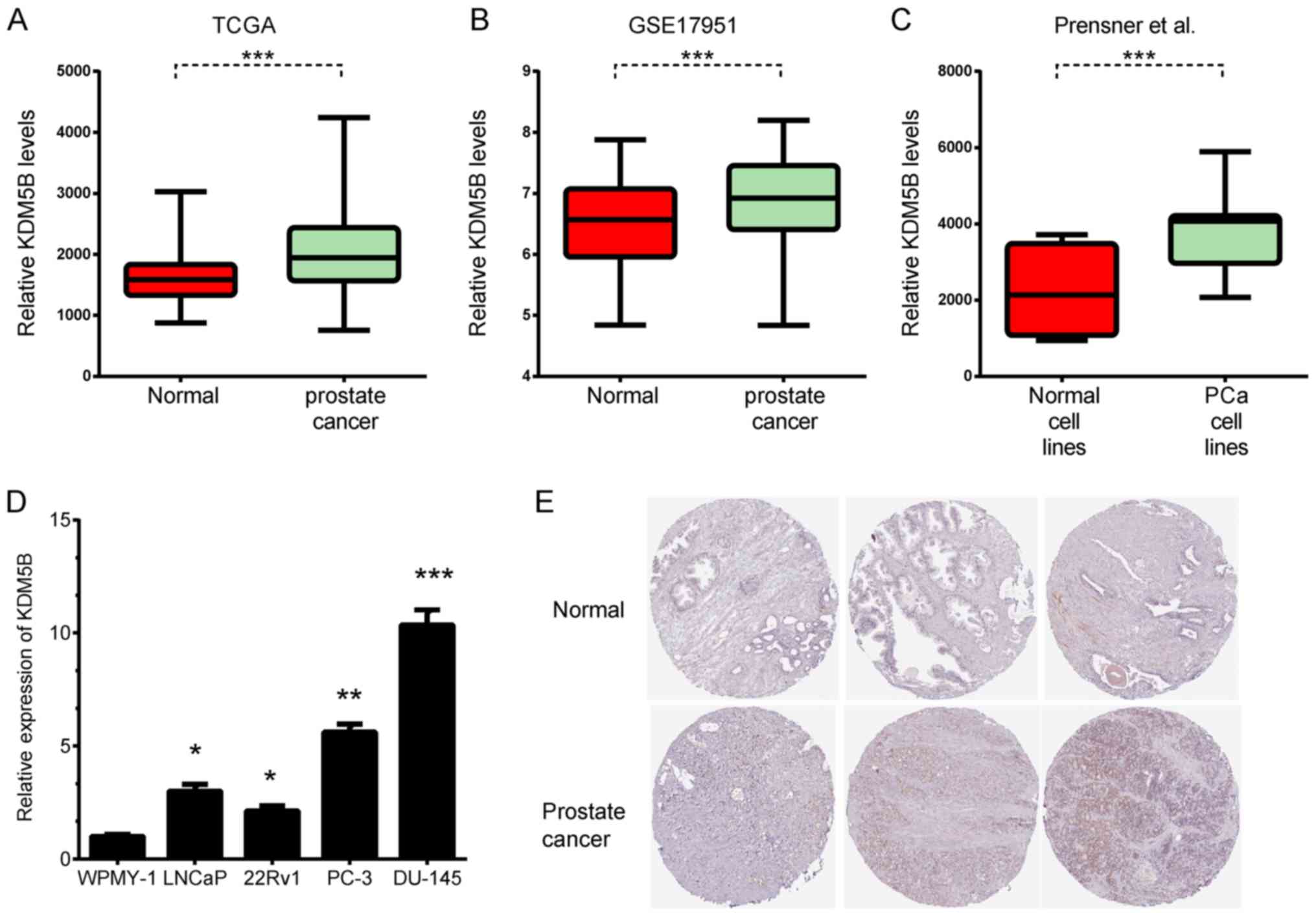
KDM5B is upregulated in PCa
The expression levels of KDM5B in PCa and normal
samples were previously unknown. In the present study, 3
independent datasets were analyzed, including TCGA, GSE17951
(27) and the Prensner datasets
(28). KDM5B was found to be
significantly upregulated in PCa tissues by analyzing TCGA
(Fig. 1A). To further validate this
result, two additional independent datasets, GSE17951 and Prensner
were analyzed, and consistent results were observed (Fig. 1B and C). To further compare KDM5B
protein levels in PCa and normal tissues, KDM5B protein expression
was analyzed using the Human Protein Atlas. KDM5B protein was
upregulated in PCa samples, however, KDM5B was not detected in
normal prostate glandular cells (Fig.
1E). KDM5B expression was also detected in cell lines,
including LNCaP, PC-3, DU145, and 22Rv1 PCa cells and WPMY-1
noncancerous prostate cells. KDM5B was found to be upregulated in
PCa cell lines compared to WPMY-1 cells (Fig. 1D).
The knockdown of KDM5B inhibits PCa
cell proliferation
The roles of KDM5B on the proliferation of PCa cells
was then evaluated. siRNAs against KDM5B (siKDM5B-546 and
siKDM5B-943) were designed to knockdown KDM5B expression. LNCaP and
PC-3 cells were transfected with siNC or siKDM5B siRNAs. At 48 h
post-transfection, both the mRNA and protein levels of KDM5B were
significantly suppressed in both siKDM5B groups compared with the
siNC group (P<0.05 and P<0.01; Fig. 2A and C). Additionally, knockdown of
KDM5B inhibited proliferation of LNCaP and PC-3 cells (P<0.001
and P<0.001; Fig. 2B and D).
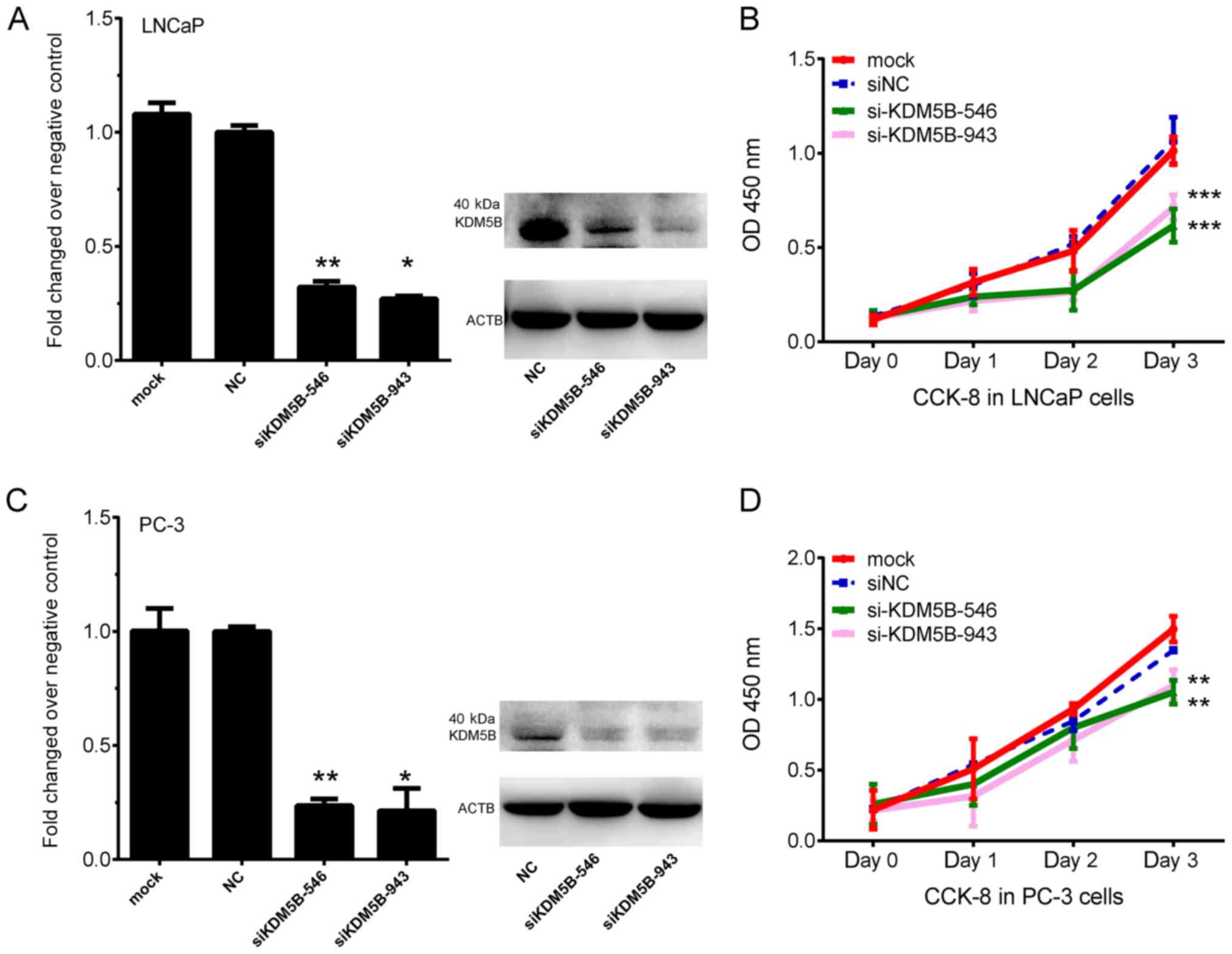 | Figure 2.Knockdown of KDM5B inhibits PCa cell
proliferation. (A) mRNA and protein expression of KDM5B after
transfection with mock, siNC, siKDM5B-546 and siKDM5B-943 in LNCaP
cells. (B) Knockdown of KDM5B inhibits LNCaP cell proliferation.
(C) mRNA and protein expression of KDM5B after transfection with
mock, siNC, siKDM5B-546 and siKDM5B-943 in PC-3 cells. (D)
Knockdown of KDM5B inhibits PC-3 cell proliferation. *P<0.05,
**P<0.01, ***P<0.001 vs. NC. KDM5B, lysine demethylase 5B;
PCa, prostate cancer; si, small interfering; NC, negative control;
OD, optical density; CCK-8, Cell Counting Kit-8. |
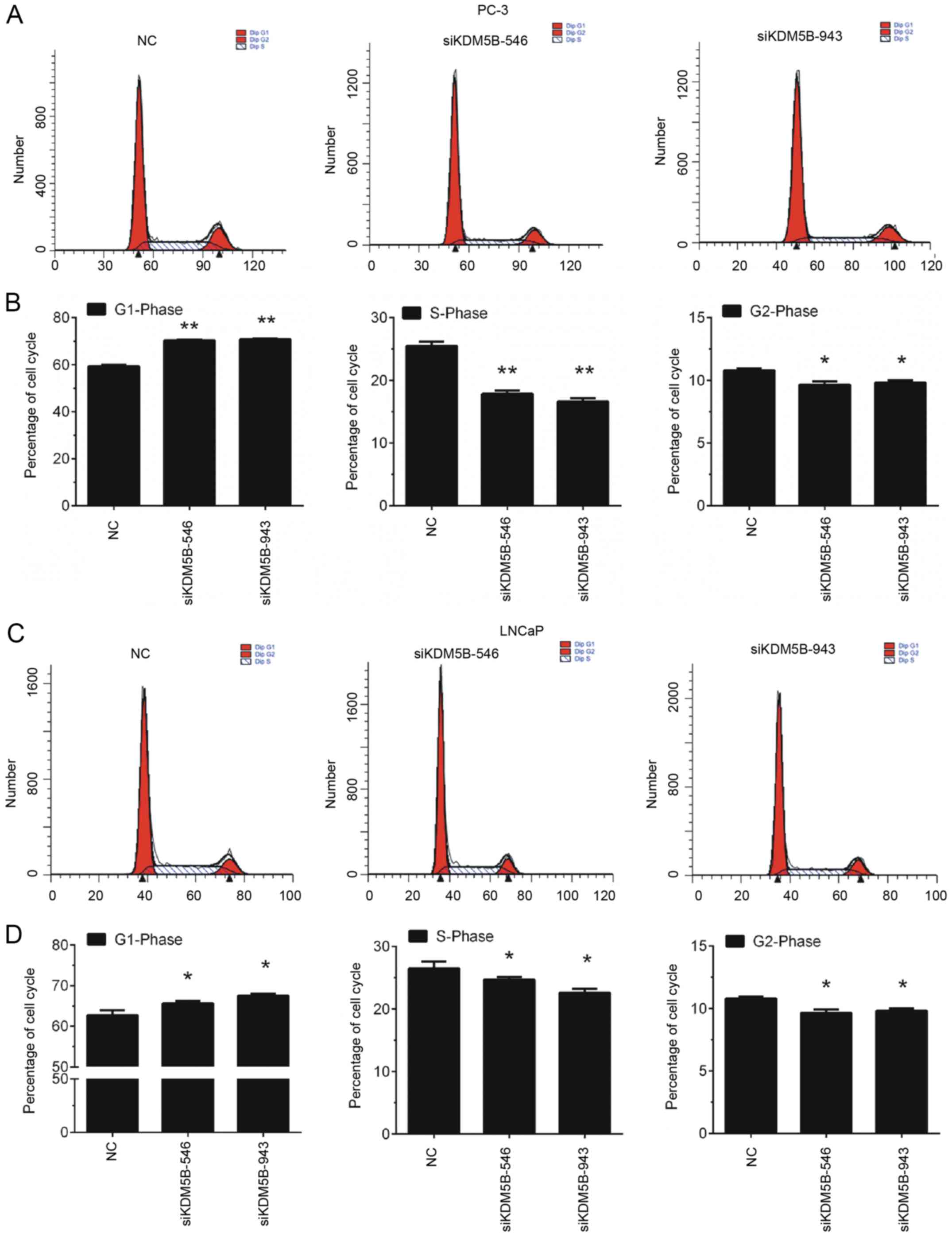
Knockdown of KDM5B induces PCa cell
cycle arrest in the G1 phase
Furthermore, the effects of KDM5B on cell cycle
progression of LNCaP and PC-3 cells were detected by flow
cytometry. The findings revealed that knockdown of KDM5B in LNCaP
and PC-3 cells induced a significant increase in the proportion of
cells in G1 phase, however, KDM5B knockdown decreased the
proportion of cells in S and G2/M phase (P<0.05 and P<0.01;
Fig. 3).
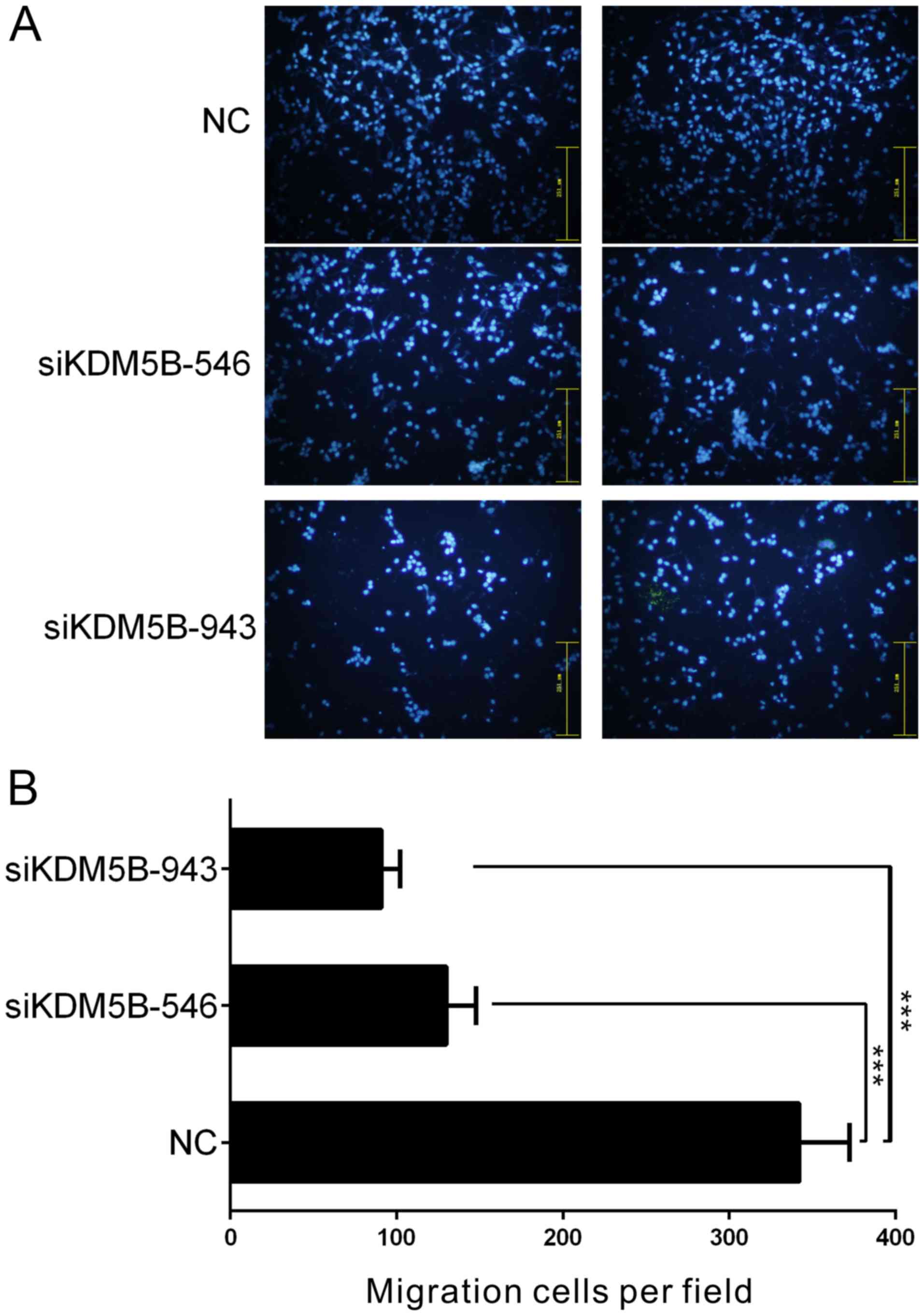
The knockdown of KDM5B inhibits the
migration of PCa cells in vitro
The number of PC-3 cells that migrated through the
filter of the Transwell chambers was used to estimate the migratory
ability of the cells (Fig. 4).
Compared with the scrambled siRNA-treated control group, the number
of migrating cells was decreased by 2.74- and 3.91-fold in PC-3
cells treated with siKDM5B-546 and siKDM5B-943, respectively
(P<0.001; Fig. 4).
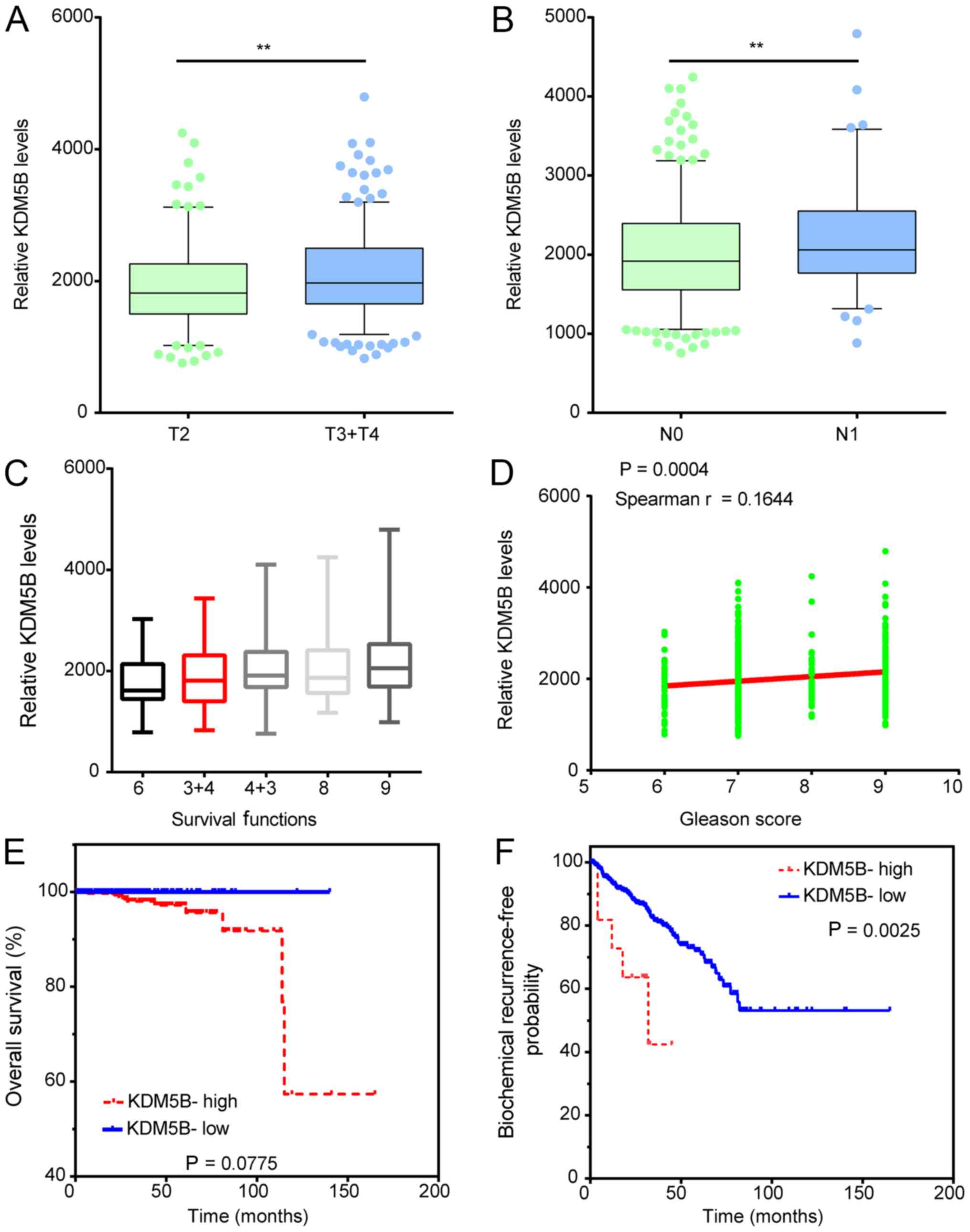
KDM5B expression is associated with
clinical variables in patients with PCa
The association between KDM5B expression and the
clinicopathological characteristics of patients with PCa was
investigated. As presented in Fig.
5, KDM5B expression was significantly upregulated in N1 stage
PCa samples compared to N0 samples (Fig.
5A), in T3/T4 PCa samples compared to T2 samples (Fig. 5B). Moreover, we analyzed the
correlation between KDM5B expression and Gleason score in patients
with PCa. The results showed that the higher expression levels of
KDM5B significantly correlated to the higher Gleason score in PCa
patients (Fig. 5C-D).
In addition, Kaplan-Meier analysis was performed to
determine whether KDM5B expression was associated with biochemical
recurrence (BCR)-free survival and overall survival in patients
with PCa by analyzing the TCGA dataset. In order to divide all PCa
samples into groups based on the high and low expression of KDM5B,
the cut-off value was calculated using the Cutoff Finder
(http://molpath.charite.de/cutoff/)
(29). In TCGA analysis, the
BCR-free survival and overall survival rates were higher in the
KDM5B-low compared with the KDM5B-high patients (Fig. 5E and F). These results indicate that
KDM5B expression may serve as a biomarker to predict the prognosis
of patients with PCa.
Discussion
In recent decades, studies focused on exploring the
functions of several key proteins, such as AR, SPOP, and FOXA1 in
PCa (4–7), however, the molecular mechanisms
underlying PCa progression remain largely unclear. Following the
application of high-throughput screening techniques like microarray
and small RNA sequencing, a series of studies identified genes
associated with PCa progression. For example, a study by Taylor
et al (30) and TCGA groups
performed integrative genomic analysis of human PCa. In the present
study, a comprehensive analysis of PCa-related genes was performed
by using 3 public datasets, TCGA and GSE17951, and KDM5B was found
to be upregulated in PCa samples.
KDM5B was included in this hub-network. The
functional roles of KDM5B in PCa remain largely unknown. Previous
studies had observed that dysregulation of KDM5B was associated
with cancer progression. KDM5B was found to be upregulated in
squamous cell carcinoma of the head and neck, breast cancer,
hepatocellular carcinoma, gastric cancer and glioma (18–24).
These studies suggested that KDM5B may serve as a diagnostic and
therapeutic target for cancers. In the present study, the function
of KDM5B was explored in PCa cells. KDM5B was found to act as an
oncogene in PCa cells, as knockdown of KDM5B significantly
inhibited cell proliferation, cell cycle progression, and
migration. To the best of our knowledge, this is the first study to
reveal the effects of KDM5B on the biological functions of PCa
cells.
Prostate-specific antigen testing is the most widely
used biomarker for patients with PCa, but its efficacy is limited
by low specificity. Of note, several recent studies revealed that
Low serum total testosterone level and Body mass index could serve
as a predictor of upstaging and upgrading in low-risk prostate
cancer patients. For example, Ferro et al reported that low
serum total testosterone levels as a predictor of upstaging and
upgrading in low-risk prostate cancer patients meeting the
inclusion criteria for active surveillance (31). And de Cobelli et al revealed
that Body mass index was associated with upstaging and upgrading in
patients with low-risk prostate cancer who met the inclusion
criteria for active surveillance (32). Moreover, the expression levels of
multiple protein coding genes or non-protein coding genes were also
revealed to be associated with the progression and prognosis of
patients with PCa, such as PHI, PCA3, sarcosine, and Urotensin II
receptor. For example, de Cobelli et al showed Urotensin II
receptor on preoperative biopsy is associated with upstaging and
upgrading in prostate cancer (33).
Sreekumar and his colleges found sarcosine levels in PCa samples
were associated with the progression of cancer (34). However, lacking reliable and
effective biomarkers for PCa diagnosis remained to be one of the
biggest challenges in PCa treatment was lacking reliable and
effective biomarkers for PCa diagnosis. In the current study, KDM5B
was evaluated as a potential biomarker for PCa. By analyzing public
datasets, KDM5B was found to be upregulated in PCa compared with
normal samples, in T3/T4 PCa samples compared with T2 samples, in
N1 stage samples compared to N0 samples and in Gleason score, ≥8
samples compared to Gleason score ≤7 samples. Moreover, overall
survival rates were higher in patients with low expression of KDM5B
compared with those with high expression. These results indicate
that KDM5B expression may serve as a biomarker of PCa. We also
realized that the combined analysis of KDM5B levels and other
potential biomarkers, such as low serum total testosterone level,
Body mass index, PHI, PCA3, sarcosine, and Urotensin II receptor
levels, in PCa samples using clinical samples could strength the
clinical importance of KDM5B in PCa.
In conclusion, 3 public datasets were analyzed to
identify differentially expressed genes in PCa. A total of 3,834
genes were found to be dysregulated in PCa. Bioinformatic analysis
revealed that these DEGs were associated with cell cycle,
translation, and metabolic pathways. PPI network analysis revealed
that KDM5B was a key regulator in PCa progression. Knockdown of
KDM5B in PCa cells significantly inhibited proliferation, cell
cycle progression and migration. In addition, KDM5B was upregulated
in PCa tissues and associated with PCa clinical variables. High
expression of KDM5B was associated with worse prognosis in patients
with PCa. Given these results, KDM5B may be a potential therapeutic
target for PCa.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
Conception and design of the study was conducted by
ZY and JXX. Development of methodology was conducted by ZY, JXX,
DPF and JK. ZY, JXX, DPF and JK performed the analysis and
interpretation of data, and wrote, reviewed and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PCa
|
prostate cancer
|
|
siNC
|
small interfering RNAs against
negative control
|
|
KDM5B
|
lysine demethylase 5B
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Antonarakis ES, Lu C, Wang H, Luber B,
Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et
al: AR-V7 and resistance to enzalutamide and abiraterone in
prostate cancer. N Engl J Med. 371:1028–1038. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sahu B, Laakso M, Ovaska K, Mirtti T,
Lundin J, Rannikko A, Sankila A, Turunen JP, Lundin M, Konsti J, et
al: Dual role of FoxA1 in androgen receptor binding to chromatin,
androgen signalling and prostate cancer. EMBO J. 30:3962–3976.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barbieri CE, Baca SC, Lawrence MS,
Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van
Allen E, Stransky N, et al: Exome sequencing identifies recurrent
SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet.
44:685–689. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geng C, Rajapakshe K, Shah SS, Shou J,
Eedunuri VK, Foley C, Fiskus W, Rajendran M, Chew SA, Zimmermann M,
et al: Androgen receptor is the key transcriptional mediator of the
tumor suppressor SPOP in prostate cancer. Cancer Res. 74:5631–5643.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kouzarides T: Histone methylation in
transcriptional control. Curr Opin Genet Dev. 12:198–209. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Berger SL: Histone modifications in
transcriptional regulation. Curr Opin Genet Dev. 12:142–148. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kulis M, Queirós AC, Beekman R and
Martín-Subero JI: Intragenic DNA methylation in transcriptional
regulation, normal differentiation and cancer. Biochim Biophys
Acta. 1829:1161–1174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sharif J, Muto M, Takebayashi S, Suetake
I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T,
Okamura K, et al: The SRA protein Np95 mediates epigenetic
inheritance by recruiting Dnmt1 to methylated DNA. Nature.
450:908–912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukuda A and Hisatake K: Histone
methylation and transcriptional regulation. Seikagaku. 79:362–365.
2007.(In Japanese). PubMed/NCBI
|
|
13
|
An W: Histone acetylation and methylation:
Combinatorial players for transcriptional regulation. Subcell
Biochem. 41:351–369. 2007.PubMed/NCBI
|
|
14
|
Schlesinger Y, Straussman R, Keshet I,
Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E,
Reubinoff BE, et al: Polycomb-mediated methylation on Lys27 of
histone H3 pre-marks genes for de novo methylation in cancer. Nat
Genet. 39:232–236. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
de Sousa E Melo F, Colak S, Buikhuisen J,
Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR,
Fessler E, van den Bergh SP, et al: Methylation of
cancer-stem-cell-associated Wnt target genes predicts poor
prognosis in colorectal cancer patients. Cell Stem Cell. 9:476–485.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Parrella P, Poeta ML, Gallo AP, Prencipe
M, Scintu M, Apicella A, Rossiello R, Liguoro G, Seripa D, Gravina
C, et al: Nonrandom distribution of aberrant promoter methylation
of cancer-related genes in sporadic breast tumors. Clin Cancer Res.
10:5349–5354. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayami S, Yoshimatsu M, Veerakumarasivam
A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue
H, et al: Overexpression of the JmjC histone demethylase KDM5B in
human carcinogenesis: Involvement in the proliferation of cancer
cells through the E2F/RB pathway. Mol Cancer. 9:592010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barrett A, Santangelo S, Tan K, Catchpole
S, Roberts K, Spencer-Dene B, Hall D, Scibetta A, Burchell J,
Verdin E, et al: Breast cancer associated transcriptional repressor
PLU-1/JARID1B interacts directly with histone deacetylases. Int J
Cancer. 121:265–275. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang D, Han S, Peng R, Jiao C, Wang X,
Yang X, Yang R and Li X: Depletion of histone demethylase KDM5B
inhibits cell proliferation of hepatocellular carcinoma by
regulation of cell cycle checkpoint proteins p15 and p27. J Exp
Clin Cancer Res. 35:372016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Tang F, Qi G, Yuan S, Zhang G,
Tang B and He S: KDM5B is overexpressed in gastric cancer and is
required for gastric cancer cell proliferation and metastasis. Am J
Cancer Res. 5:87–100. 2015.PubMed/NCBI
|
|
22
|
Dai B, Hu Z, Huang H, Zhu G, Xiao Z, Wan
W, Zhang P, Jia W and Zhang L: Overexpressed KDM5B is associated
with the progression of glioma and promotes glioma cell growth via
downregulating p21. Biochem Biophys Res Commun. 454:221–227. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bamodu OA, Huang WC, Lee WH, Wu A, Wang
LS, Hsiao M, Yeh CT and Chao TY: Aberrant KDM5B expression promotes
aggressive breast cancer through MALAT1 overexpression and
downregulation of hsa-miR-448. BMC Cancer. 16:1602016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Catchpole S, Spencer-Dene B, Hall D,
Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG,
Burchell JM and Taylor-Papadimitriou J: PLU-1/JARID1B/KDM5B is
required for embryonic survival and contributes to cell
proliferation in the mammary gland and in ER+ breast cancer cells.
Int J Oncol. 38:1267–1277. 2011.PubMed/NCBI
|
|
25
|
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z,
Ma Y, Yu Y, Lin H, Chen AP and Chen CD: JARID1B is a histone H3
lysine 4 demethylase up-regulated in prostate cancer. Proc Natl
Acad Sci USA. 104:19226–19231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Xia XQ, Jia Z, Sawyers A, Yao H,
Wang-Rodriquez J, Mercola D and McClelland M: In silico estimates
of tissue components in surgical samples based on expression
profiling data. Cancer Res. 70:6448–6455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff Finder: A
comprehensive and straightforward Web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E Reva B,
et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ferro M, Lucarelli G, Bruzzese D, Di
Lorenzo G, Perdonà S, Autorino R, Cantiello F, La Rocca R, Busetto
GM, Cimmino A, et al: Low serum total testosterone level as a
predictor of upstaging and upgrading in low-risk prostate cancer
patients meeting the inclusion criteria for active surveillance.
Oncotarget. 8:18424–18434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
de Cobelli O, Terracciano D, Tagliabue E,
Raimondi S, Galasso G, Cioffi A, Cordima G, Musi G, Damiano R,
Cantiello F, et al: Body mass index was associated with upstaging
and upgrading in patients with low-risk prostate cancer who met the
inclusion criteria for active surveillance. Urol Oncol.
33:201.e1–e8. 2015. View Article : Google Scholar
|
|
33
|
de Cobelli O, Buonerba C, Terracciano D,
Bottero D, Lucarelli G, Bove P, Altieri V, Coman I, Perdonà S,
Facchini G, et al: Urotensin II receptor on preoperative biopsy is
associated with upstaging and upgrading in prostate cancer. Future
Oncol. 11:3091–3098. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sreekumar A, Poisson LM, Rajendiran TM,
Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al:
Metabolomic profiles delineate potential role for sarcosine in
prostate cancer progression. Nature. 457:910–914. 2009. View Article : Google Scholar : PubMed/NCBI
|