Introduction
Primary brain and central nervous system (CNS)
tumors have a mortality of 7% in patients <70 years old, and an
estimated 270,000 non-malignant and 120,000 malignant tumors were
reported between 2011 and 2015 in the USA (1,2). In
2017, ~80,000 new cases and 17,000 primary brain tumor-related
mortalities were reported (3).
Furthermore, one-third of the cases were diagnosed as malignant
tumors (4). Glioma is a broad term
that encompasses several types of malignant tumor derived from
neuroepithelial cells, such as glial cells and other supporting
cells of the CNS (1,5,6). Gliomas
account for ~80% of primary brain tumors in adults and can be
categorized as astrocytomas, oligodendrogliomas, ependymomas, mixed
gliomas and other rare types, including brain stem and optic nerve
gliomas (7). According to the World
Health Organization criteria, gliomas vary histopathologically and
may include benign ependymomas, as well as the most aggressive
grade-IV glioblastoma (GBM) (5).
Surgical removal is the primary therapy for intracranial tumors.
However, tumors located in inoperable or sensitive regions of the
brain pose clinical challenges (8).
In addition, limited effectiveness of systemic chemotherapy can be
attributed to the blood-brain barrier, which protects the brain
from harmful compounds but also restricts the entry of
chemotherapeutic drugs (9). The
molecular mechanisms underlying the development of gliomas remain
largely unknown. Therefore, elucidating the associated mechanisms
may accelerate the development of targeted therapies for the
disease.
The competing endogenous RNA (ceRNA) hypothesis,
proposed by Salmena et al (10), has provided insight for RNA
regulatory networks. According to this hypothesis, mRNA, long
non-coding RNA (lncRNA), pseudogenes and other molecules can
competitively bind to the same microRNA (miRNA/miR) response
element and modulate miRNA function, forming an RNA regulatory
network (10).
lncRNAs are RNAs >200 bp in length that do not
encode proteins (11). Previous
studies suggested that lncRNAs are a by-product of RNA
transcription with biological functions that include modulation of
the nervous system at the epigenetic, transcriptional and
post-transcriptional levels (12–14).
Additionally, lncRNAs serve as ceRNAs and participate in regulating
gene expression and encoding miRNAs (10,15).
Furthermore, lncRNAs were demonstrated to serve an important role
in oncogenesis and tumor progression (16).
The present study constructed a ceRNA network in
order to investigate and to identify potential biomarkers for
glioma, and to determine the molecular mechanisms of glioma
pathogenesis, using Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG). Additionally, the prognostic value of the
identified mRNAs, miRNAs and lncRNAs was analyzed by survival
analysis.
Materials and methods
Data acquisition and processing
RNA sequencing and the corresponding clinical data
of patients with glioma (low-grade glioma and glioblastoma) were
obtained from The Cancer Genome Atlas (TCGA) data portal
(https://tcga-data.nci.nih.gov/tcga/)
(17). The lncRNA, mRNA and miRNA
sequence data were derived from the Illumina HiSeq platform
(Illumina, Inc.). A total of 598 glioma tissues and 5 normal brain
tissues were included in the present study. DESeq package in R
software was confirmed to identify significant DEGs in the study
(https://bioconductor.org/packages/release/bioc/html/DESeq.html)
(18). The present study was
conducted in accordance with the publication guidelines provided by
TCGA (http://cancergenome.nih.gov/publications/publicationguidelines).
Therefore, further approval from the local ethics committee was not
required.
Identification of differentially
expressed (DE) RNA
DElncRNAs and DEmRNAs were defined and encoded based
on the annotations from the Ensembl database (http://www.ensembl.org/index.html). Using the
edgeR package version.3.53 in R language to further analyze the
data, the DElncRNAs, DEmRNAs and DEmiRNAs were identified
(https://www.r-project.org).
|log2 fold change (FC)|>1.5 and false discovery rate
(FDR) adjusted to P<0.01 were set as the thresholds. In
addition, heat maps and volcano maps of the DE RNAs were generated,
using the gplots and heatmap R packages v3.53 (https://www.r-project.org).
Construction of the ceRNA and
cytohubba networks
The miRcode (http://www.mircode.org/) database and the Perl program
(http://www.perl.org) were used to predict
lncRNA-miRNA interactions, and the miRNAs sequences were identified
by using StarBase version.2.0 database (http://starbase.sysu.edu.cn/). miRNA-targeted mRNAs
were retrieved from the miRDB (http://mirdb.org/), miRTarBase version.7.0 (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and
TargetScan databases version.7.2 (http://www.targetscan.org/vert_72/) (19–21).
miRTarBase, miRDB, and TargetScan were used to identify the target
genes of miRNAs. Only mRNAs recognized by all three databases were
considered as candidate mRNAs and intersected with the DEmRNAs to
screen out the DEmRNAs targeted by the DEmiRNAs. A co-expression
network of DE genes (DEGs) was then constructed, based on
DEmiRNA-DElncRNA and DEmiRNA-DEmRNA interactions, which were
visualized using Cytoscape version.3.61 (National Institutes of
Health). The cytohubba plugin (22)
was used to evaluate the top-10 network in the network based on the
degree of association between RNAs, which was termed as
closeness.
Functional enrichment analysis
To examine the underlying biological mechanisms of
DEmRNAs in the ceRNA crosstalk network, GO annotation and KEGG
pathway analyses were conducted using the DAVID version.6.7)
(https://david.ncifcrf.gov/) online tool
(23) and cluster Profiler
(https://www.rdocumentation.org/packages/clusterProfiler/versions/3.0.4),
an R package for functional classification and enrichment of gene
clusters using hypergeometric distribution. The GO plot package of
R software was utilized to display the results of the GO and KEGG
analyses (http://wencke.github.io/). GO and
KEGG enrichment analysis was based on the threshold of
P<0.05.
Survival analysis
To assess the prognostic value of DElncRNAs in
patients with glioma, survival analysis for these DERNAs in the
ceRNA network was conducted using the survival package in R
(https://www.rdocumentation.org/packages/survival/versions/2.42-3).
Survival curves were generated using the Kaplan-Meier method and
the log-rank test was used to compare the difference between the
groups. Univariate Cox regression analyses were performed to
independently identify the effects of DEmRNAs, DElncRNAs and
DEmiRNAs on overall survival (OS). P<0.05 was considered to
indicate a statistically significant difference.
Results
DElncRNAs, DEmiRNAs and DEmRNAs in
glioma
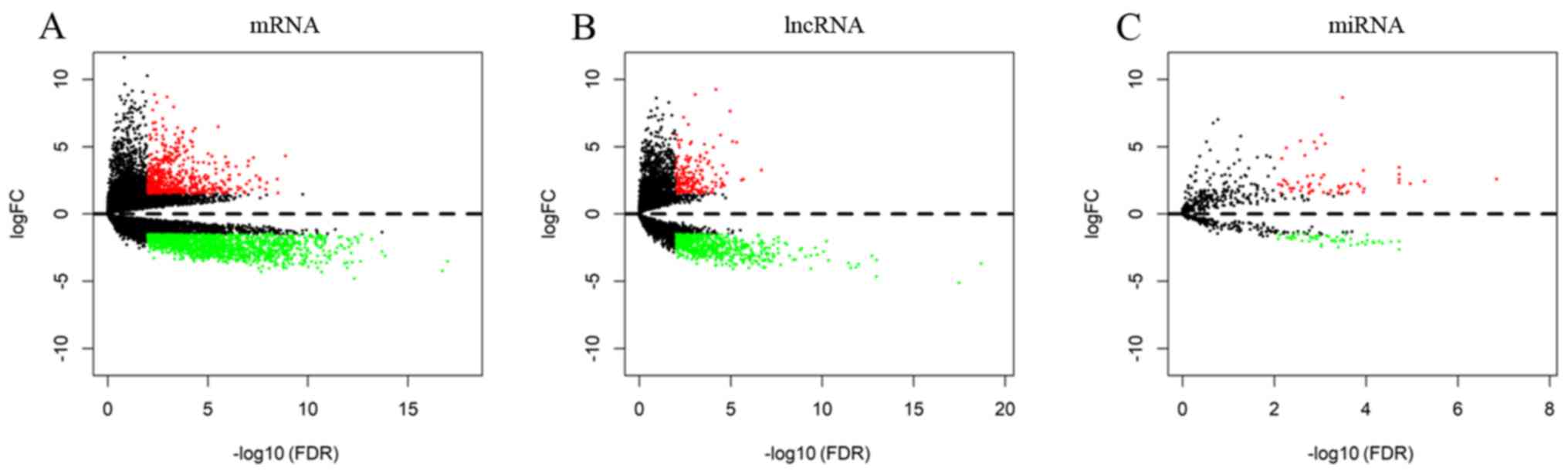
Using the ‘DESeq’ package in R software, significant
DEGs in 598 glioma tissues and 5 normal brain tissues were
identified. A total of 752 DElncRNA (180 upregulated and 572
downregulated), 2,079 DEmRNAs (588 upregulated and 1,491
downregulated) and 113 DEmiRNAs (62 upregulated and 51
downregulated) were identified with thresholds of
|log2FC| >1.5 and adjusted P<0.01. The
distribution of all DEGs on the two dimensions of -log FDR and
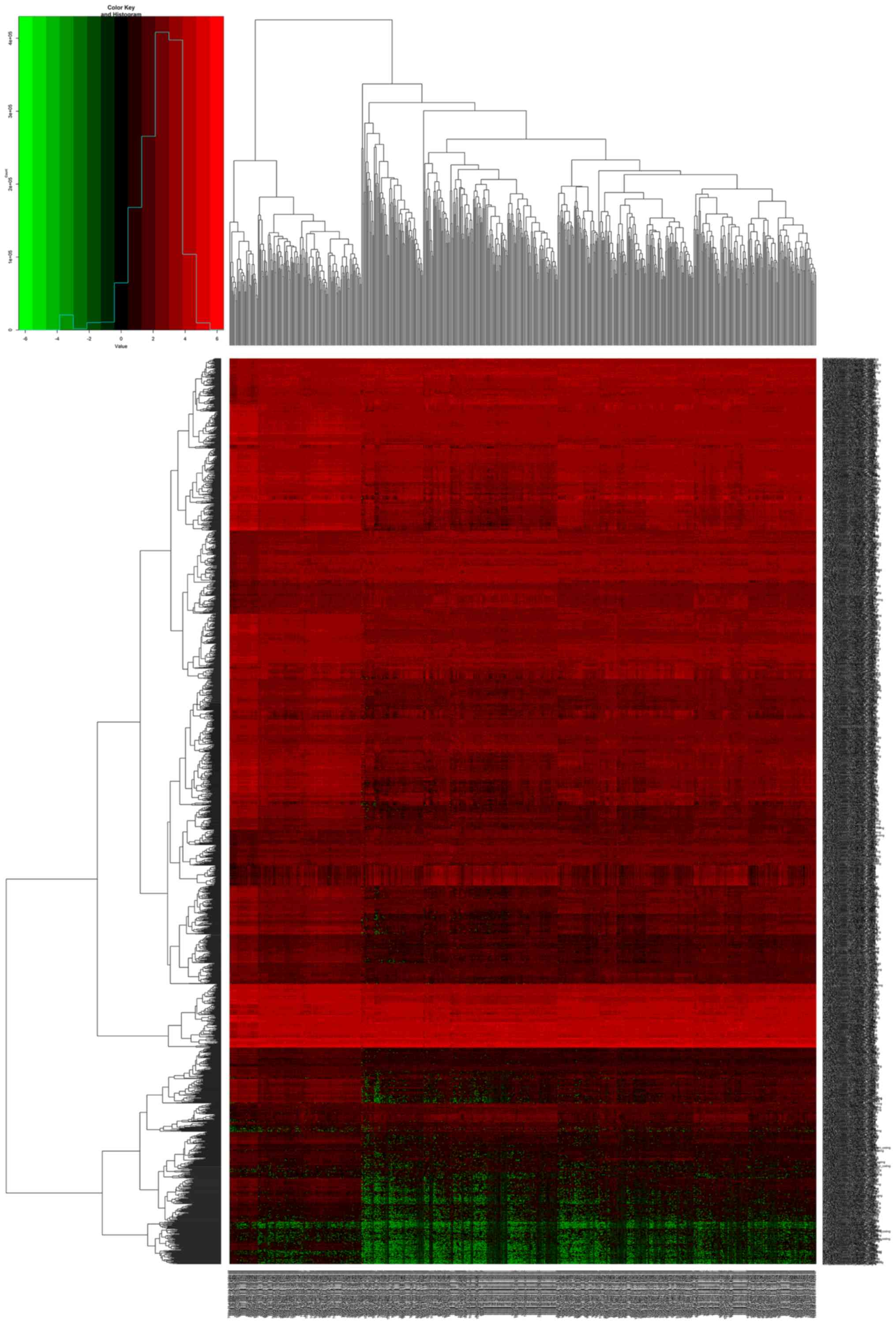
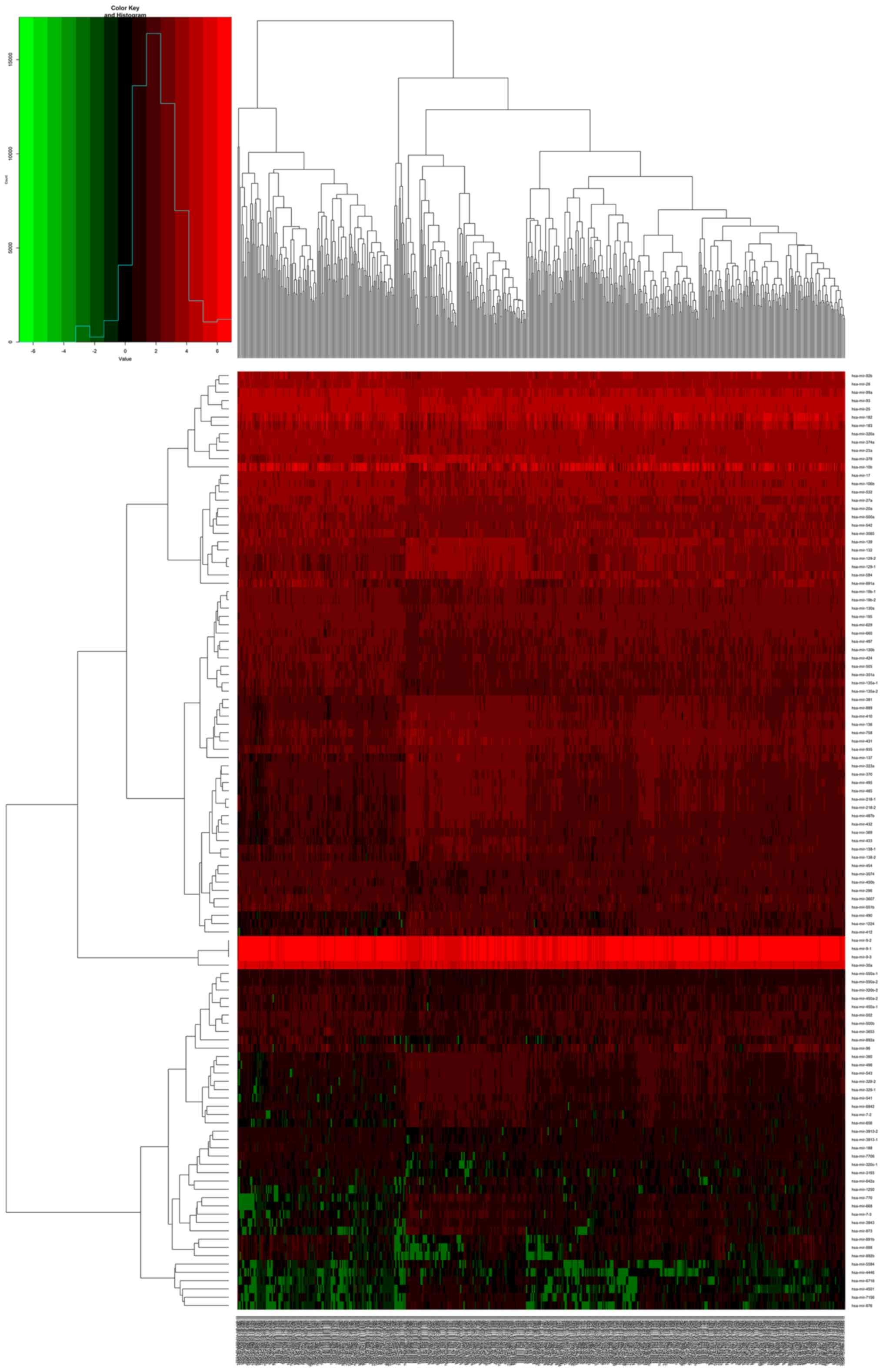
logFC are depicted in the volcano map in Fig. 1. For the heatmaps presented in
Figs. 2–4, the numerical data represent the
expression profiles of DEGs.
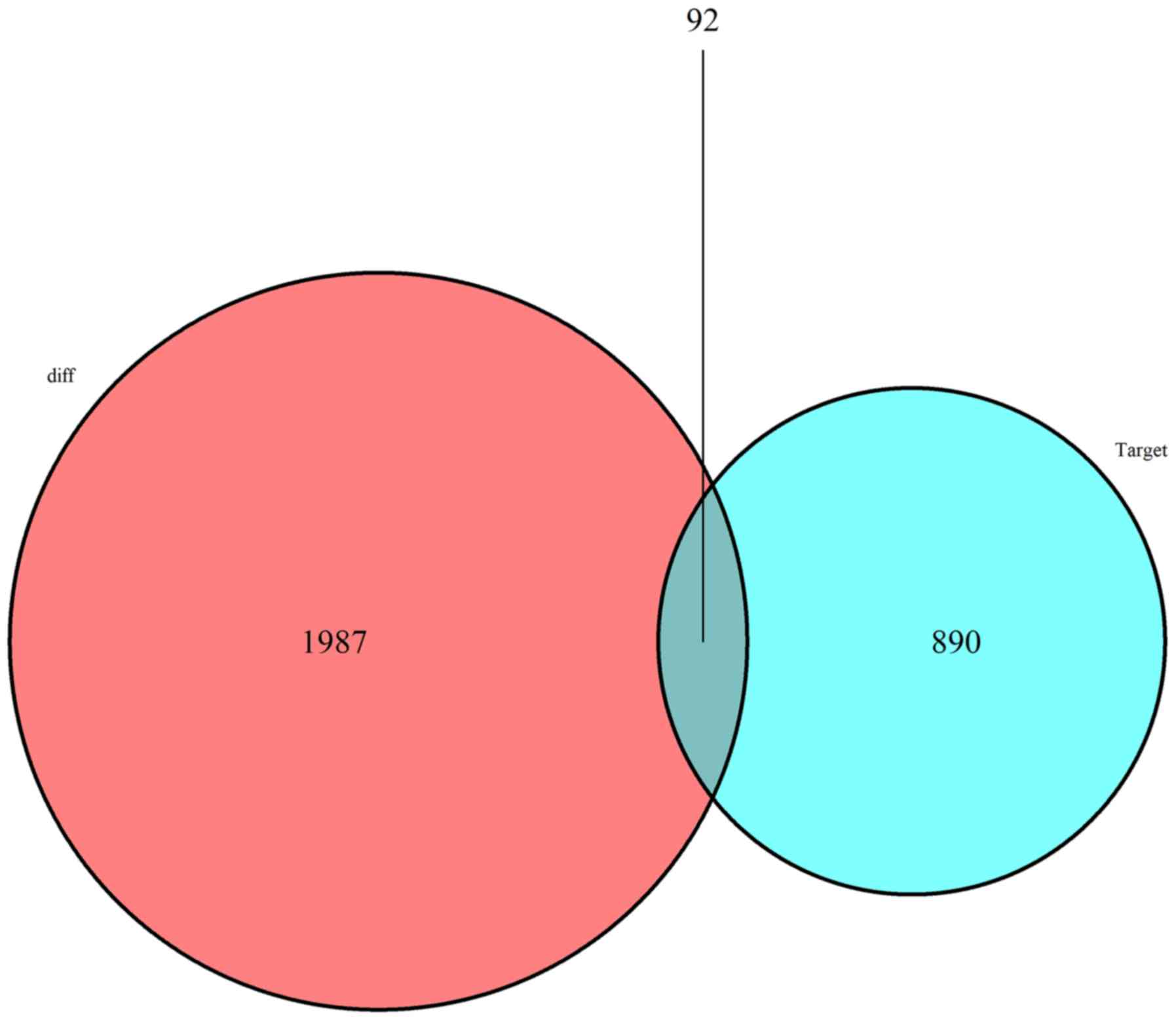
Construction of the ceRNA network
To further examine how lncRNAs interact with miRNAs
to regulate mRNA in glioma, a lncRNA-miRNA-mRNA (ceRNA) network was
constructed based on the aforementioned data and visualized using
Cytoscape v3.6.1. Using the 752 DElncRNAs retrieved from the
miRcode database, the Perl program was applied to identify 211
pairs of interacting lncRNAs and miRNAs. Targeted mRNAs were
screened based on the 12 miRNAs using the miRTarBase, miRDB and
TargetScan database. The final DEmiRNA targeted genes were
selected, which were included in all 3 datasets (miRTarBase, miRDB
and TargetScan). miRNA-targeted mRNAs not included in DEmRNAs were
discarded. Finally, 92 DEmRNAs were included in the ceRNA network
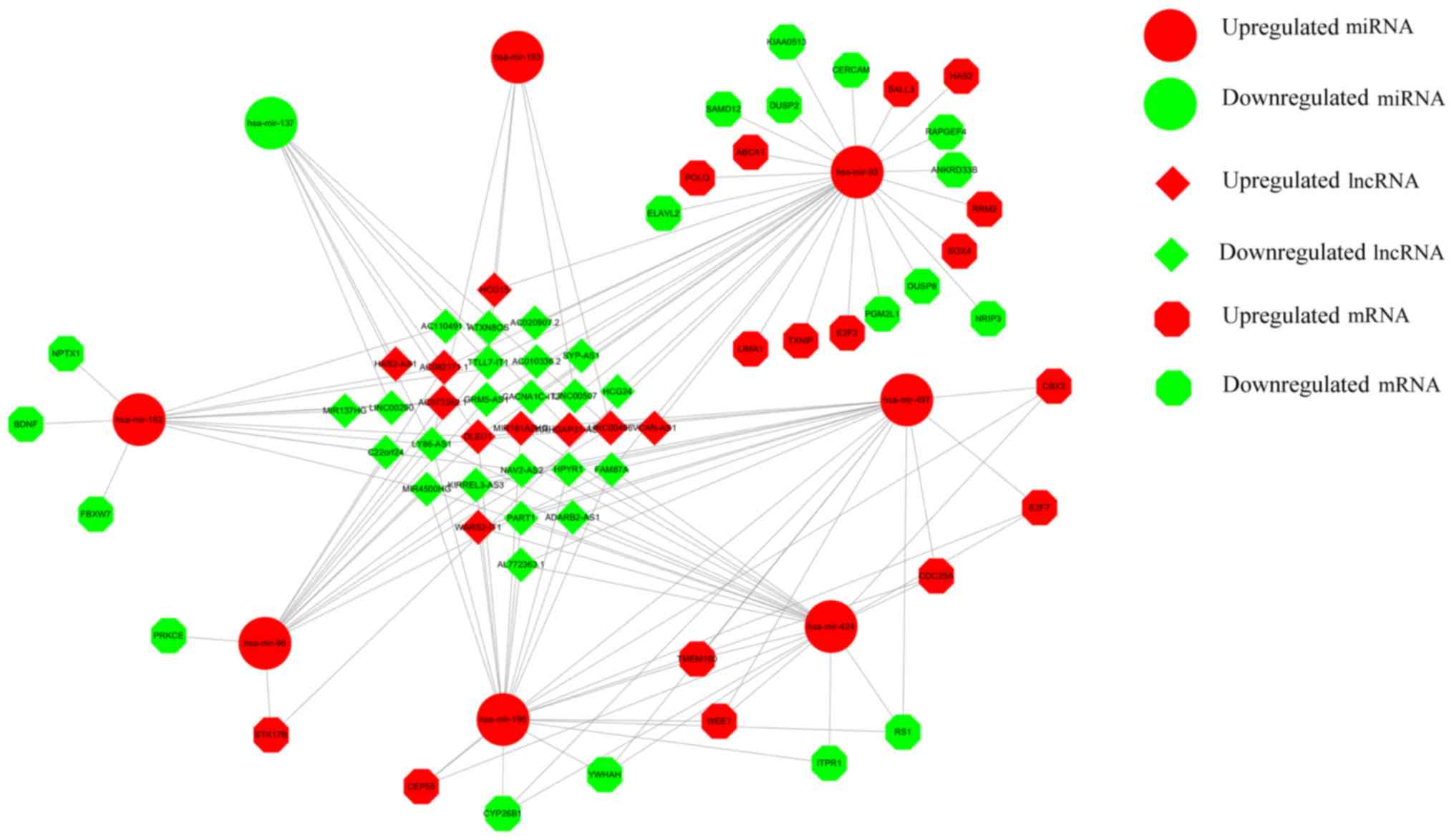
(Fig. 5). Thus, a total of 61 lncRNA
nodes, 12 miRNA nodes and 92 mRNA nodes as differentially expressed
profiles were presented in the ceRNA network (Fig. 6).
Construction of the cytohubba
network
Based on the cytohubba plugin, the top-10 network
was obtained (Fig. 7). The network
consisted of 6 miRNAs, including hsa-mir-93, hsa-mir-424,
hsa-mir-497, hsa-mir-195, hsa-mir-182 and hsa-mir-96, and 4
lncRNAs, AC092171.1, family with sequence similarity 87 member A
(FAM87A), deleted in lymphocytic leukemia 1 (DLEU1) and lymphocyte
antigen 86 antisense RNA 1 (LY86-AS1).
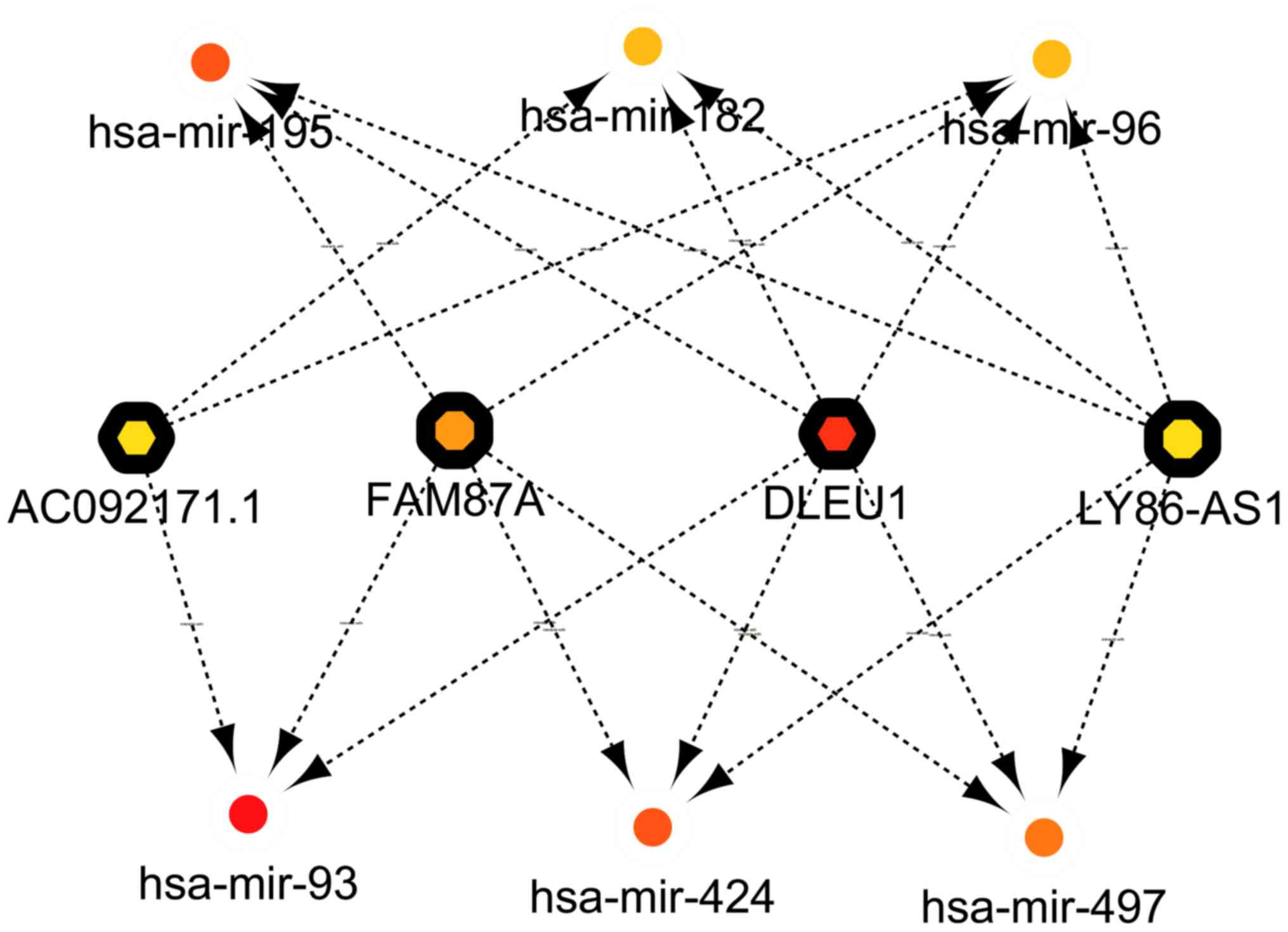 | Figure 7.Network of top-10 based on closeness
in the competing endogenous RNA network. The network consisted of 6
miRNAs, including hsa-mir-93, hsa-mir-424, hsa-mir-497,
hsa-mir-195, hsa-mir-182 and hsa-mir-96 and 4 lncRNAs, FAM87A,
family with sequence similarity 87 member A; DLEU1, deleted in
lymphocytic leukemia 1; LY86-AS1, lymphocyte antigen 86 antisense
RNA 1; AC092171.1, non-protein coding RNA. Boxes with rounded
edges, miRNAs; boxes with hexagon edges, lncRNAs. |
Functional analysis of DEmRNAs in the
ceRNA network
The biological functions of the 92 DEmRNAs were
further explored using GO and KEGG analysis, which demonstrated
that these DEmRNAs were enriched in 42 GO biological process
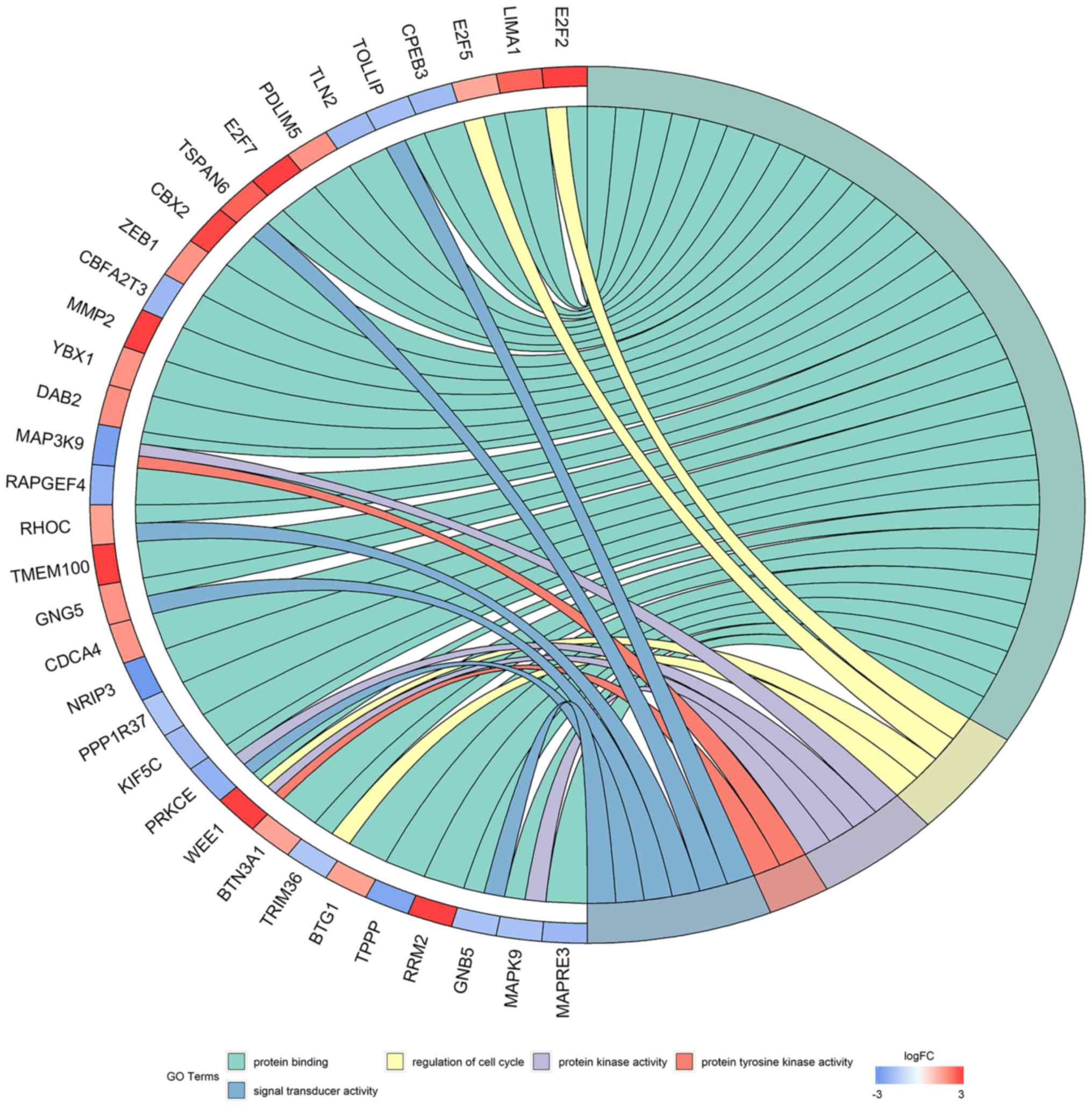
categories (P<0.05). In the GO analysis, a total of 5
significantly enriched pathways were obtained (Table I; Fig.
8). The most enriched GO term was ‘protein binding’. In the
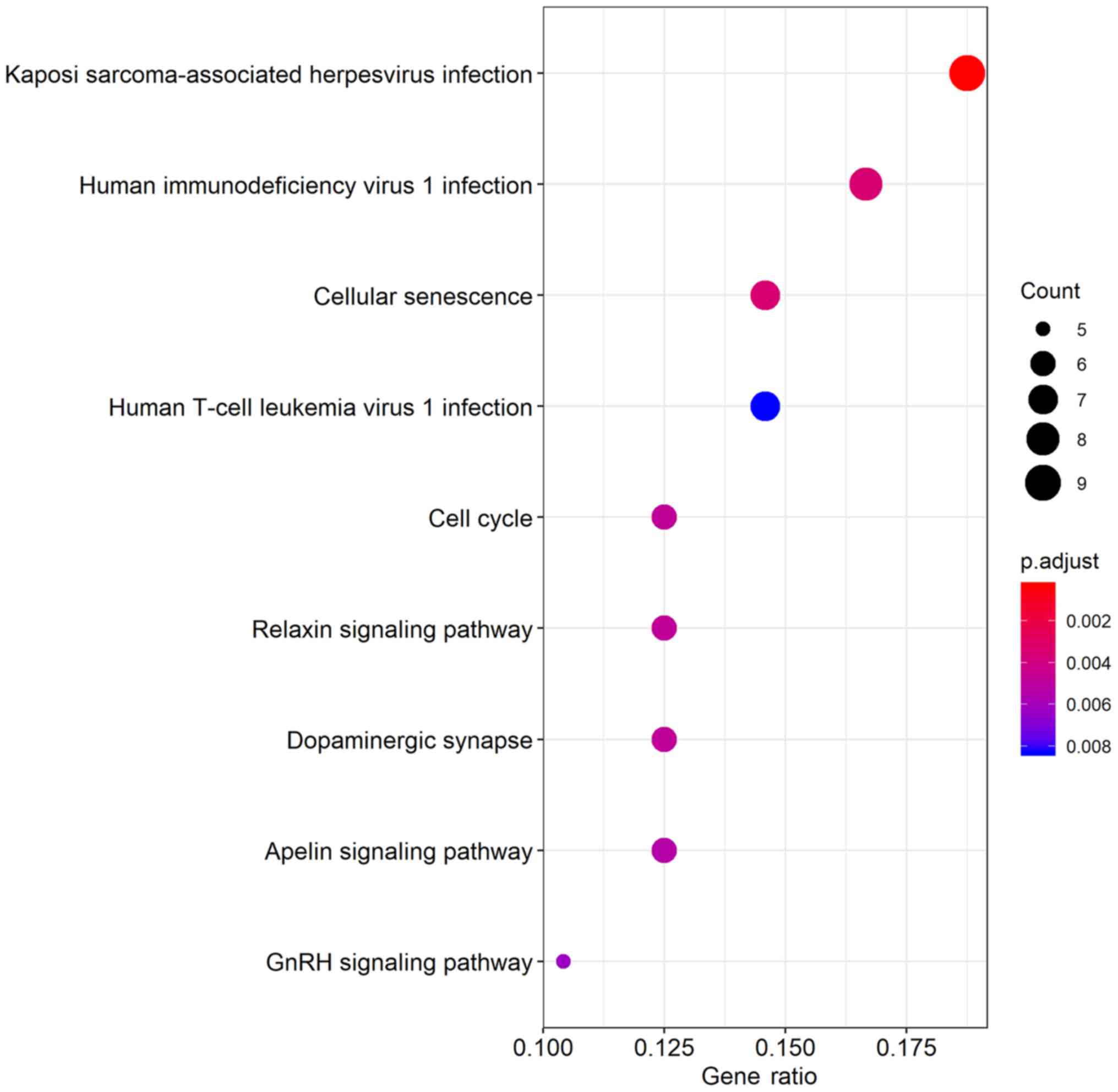
KEGG pathway analysis, a total of 9 significantly enriched pathways
were obtained (Table II; Fig. 9); Among the 9 pathways, ‘cell cycle’,
‘dopaminergic synapse’ and ‘Kaposi sarcoma-associated herpesvirus
infection’ were linked with the progression of glioma.
Additionally, other pathways such as ‘relaxin signaling pathway’,
‘cellular senescence’ and ‘human T-cell leukemia virus 1 infection’
were also tumor-related pathways.
 | Table I.GO pathways enriched with
differentially expressed mRNA involved in the competing endogenous
RNA network. |
Table I.
GO pathways enriched with
differentially expressed mRNA involved in the competing endogenous
RNA network.
| GO ID | Term | Genes | P-value |
|---|
| 0005515 | Protein
binding | E2F2, LIMA1, E2F5,
CPEB3, TOLLIP, TLN2, PDLIM5, E2F7, TSPAN6, CBX2, ZEB1, CBFA2T3,
MMP2, YBX1, DAB2, MAP3K9, RAPGEF4, RHOC, TMEM100, GNG5, CDCA4,
NRIP3, PPP1R37, KIF5C, PRKCE, WEE1, BTN3A1, TRIM36, BTG1, TPPP,
RRM2, GNB5, MAPK9, MAPRE3, PPP3R1, SOX4, BCL2L2, CHEK1, CEP55,
ABCA1, ATP6V1B2, HPRT1, FBXW7, MOAP1, BCL11B, ETV1, SSX2IP, POLQ,
TXNIP, GABARAPL1, ADARB1, MAP2K1, MAP2K4, ELAVL2, MAFK, CDC25A,
ITPR1, EPHA4, EPHA7, YWHAH, DUSP2, SLC16A9, FAM126B, PLEKHA1,
TP53INP1 | <0.001 |
| 0051726 | Regulation of cell
cycle | E2F2, ADARB1,
TRIM36, E2F5, CDC25A, WEE1 | <0.001 |
| 0004672 | Protein kinase
activity | EPHA4, MAP2K1,
MAP3K9, MAP2K4, STK17B, MAPK9, CHEK1, PRKCE, WEE1 | <0.001 |
| 0004713 | Protein tyrosine
kinase activity | EPHA4, EPHA7,
MAP2K1, MAP3K9, MAP2K4, WEE1 | <0.001 |
| 0004871 | Signal transducer
activity | GNAL, TOLLIP,
TSPAN6, GNB5, RHOC, PRKCE, GNG5 | <0.001 |
 | Table II.Kyoto Encyclopedia of Genes and
Genomes pathways enriched with differentially expressed mRNA
involved in the competing endogenous RNA network. |
Table II.
Kyoto Encyclopedia of Genes and
Genomes pathways enriched with differentially expressed mRNA
involved in the competing endogenous RNA network.
| Pathway ID | Description | Genes | Count | P-value |
|---|
| hsa05167 | Kaposi
sarcoma-associated herpesvirus infection | MAP2K1, MAPK9,
PPP3R1, MAP2K4, GABARAPL1, GNB5, ITPR1, E2F2, GNG5 | 9 | <0.001 |
| hsa05170 | Human
immunodeficiency virus 1 infection | MAP2K1, MAPK9,
PPP3R1, GNB5, ITPR1, WEE1, GNG5, CHEK1 | 8 | <0.001 |
| hsa04218 | Cellular
senescence | MAP2K1, PPP3R1,
ITPR1, E2F5, E2F2, CDC25A, CHEK1 | 7 | <0.001 |
| hsa04110 | Cell cycle | YWHAH, E2F5, E2F2,
WEE1, CDC25A, CHEK1 | 6 | <0.001 |
| hsa04926 | Relaxin signaling
pathway | MAP2K1, MAPK9,
MAP2K4, GNB5, MMP2, GNG5 | 6 | <0.001 |
| hsa04728 | Dopaminergic
synapse | MAPK9, GNB5, ITPR1,
KIF5C, GNG5, GNAL | 6 | <0.001 |
| hsa04371 | Apelin signaling
pathway | MAP2K1,
GABARAPL1/GNB5, PRKCE, ITPR1, GNG5 | 6 | <0.001 |
| hsa04912 | GnRH signaling
pathway | MAP2K1, MAPK9,
MAP2K4, ITPR1, MMP2 | 5 | <0.001 |
| hsa05166 | Human T-cell
leukemia virus 1 infection | MAP2K1, MAPK9,
PPP3R1, MAP2K4, TLN2, E2F2, CHEK1 | 7 | <0.001 |
Survival associated lncRNAs in the
ceRNA network
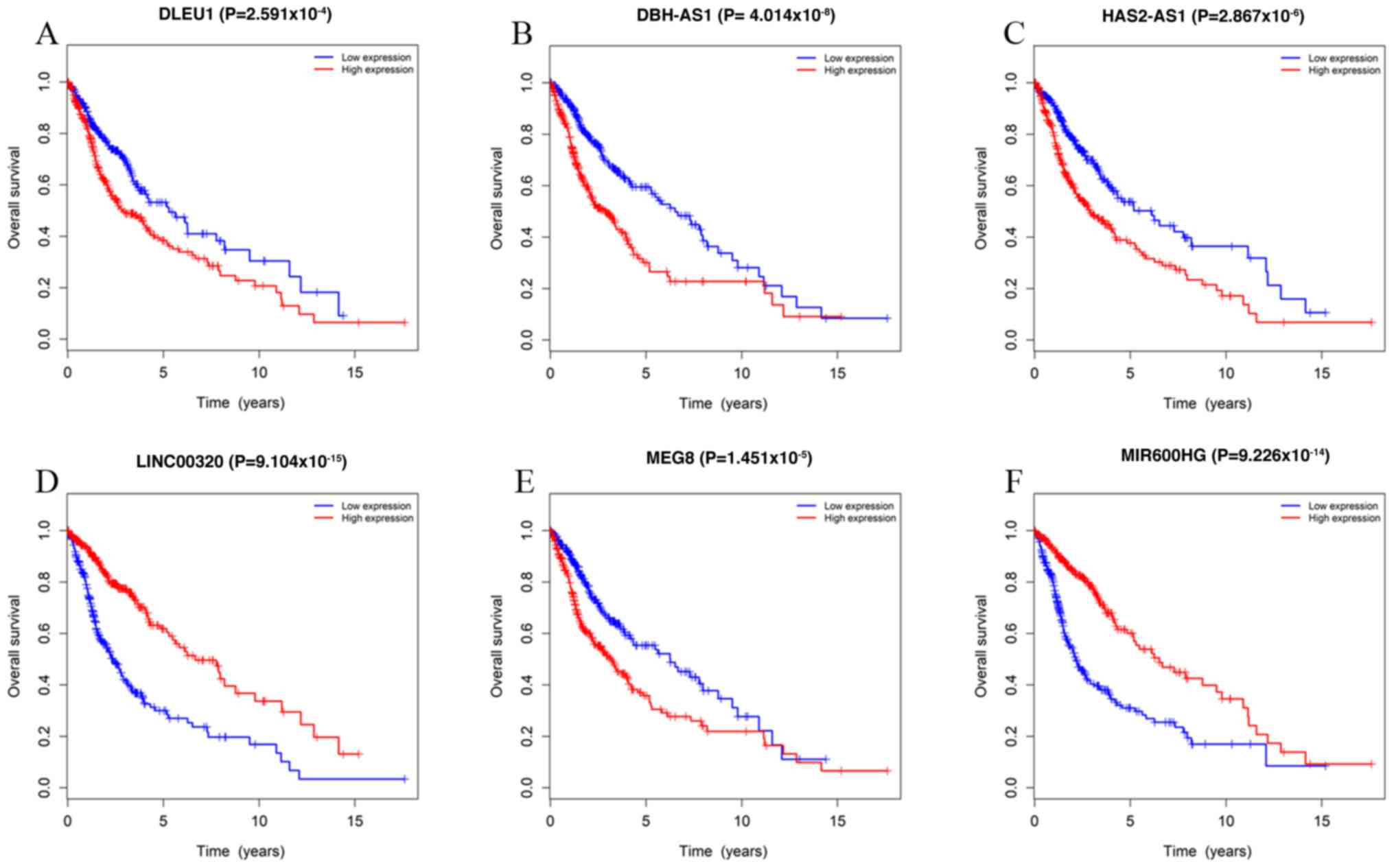
To identify the association between DElncRNAs in the
ceRNA network and the prognosis of patients with gliomas, a
Kaplan-Meier survival analysis was conducted. A total of 36 out of
752 DElncRNAs were significantly associated with OS. Among the 36
significant DElncRNAs, 13 lncRNAs, including DLEU1, dopamine
β-hydroxylase antisense RNA1 (DBH-AS1), hyaluronan synthase 2
antisense RNA1 (HAS2-AS1), LINC02875, AL117190.1, chromosome 9 open
reading frame 147, cytochrome P450 family 1 subfamily B member 1
antisense RNA 1, human leukocyte antigen group (HCG) 15, HCG23,
LINC00466, tryptophanyl tRNA synthetase 2, maternally expressed
(MEG) 3 and MEG8, were negatively associated with OS. The remaining
23 lncRNAs [CCD26 lncRNA, long intergenic non-protein coding RNA
(LINC00320), AC011374.1, AC020907.2, AC022400.1, AC092171.1,
AC110491.1, ArfGAP with coiled-coil, ankyrin repeat and PH domains
2 intronic transcript 1, adenosine deaminase RNA specific B2
antisense RNA 1, AL359541.1, AL772363.1, Rho GTPase activating
protein 31 antisense RNA 1, ZNF22 antisense RNA 1, calcium
voltage-gated channel subunit α 1 C intronic transcript, glutamate
metabotropic receptor 5 antisense RNA 1, LINC00501, MIR4500HG,
versican antisense RNA 1, helicobacter pylori responsive 1,
LINC00461, myocardial infarction associated transcript (MIAT),
small nucleolar (sno) RNA host gene 1 and MIR600HG)] were
positively associated with OS. The top 6 most significant
survival-associated DElncRNAs are presented in Fig. 10.
Discussion
Non-coding RNAs include several types of RNAs, and
can be classified into lncRNAs and short non-coding RNAs. The
latter can be further categorized into transfer RNAs, ribosomal
RNAs, miRNAs, small interfering RNAs and snoRNAs (24). Previous studies demonstrated that
lncRNAs played a key role in gene transcriptional control,
epigenetic regulation and post-transcriptional modification
(25,26). The present study analyzed RNA
sequencing and clinical data from patients with gliomas acquired
from TCGA in order to investigate the functions of lncRNAs in the
ceRNA network. DElncRNAs, DEmRNAs and DEmiRNAs were identified
using R packages and were used to construct a ceRNA network. The
biological functions of the DEmRNAs were determined by GO and KEGG
analyses. Further survival analysis was performed to investigate
the association between DElncRNAs and OS.
miRNAs are involved in the regulation of a number of
biological processes, including cell differentiation, proliferation
and apoptosis, by binding with target mRNAs at the transcriptional
or post-transcriptional levels (27). A previous study demonstrated that
aberrantly expressed miRNAs were closely associated with several
types of cancer (28). Moreover,
growing evidence suggests that several miRNAs are implicated in the
progression of glioma (29–31). hsa-mir-183 is frequently methylated
and was correlated with poor outcome in hepatocellular carcinoma
(32). Moreover, hsa-miR-183-5p was
involved in cell cycle regulation via the MAPK and glioma signaling
pathways (33). In addition,
multivariative analysis revealed that low serum hsa-mir-137 levels
were closely associated with high clinical grades and poor survival
in patients with GBM (34).
Moreover, hsa-miR-93-5p upregulation was correlated with poor
prognosis in non-small cell lung cancer (35), while hsa-miR-93-3p was recognized as
a diagnostic biomarker in triple negative breast cancer (36). Overexpression of hsa-mir-497 promoted
the proliferation of glioma cells (37) and has been demonstrated to be an
unfavorable prognostic biomarker in human glioma (38). In the present study, 6 DEmiRNAs,
which may serve as promising prognostic biomarkers, were involved
in the ceRNA network.
In the present study, 92 DEmRNAs were identified in
the ceRNA network. The biological processes and pathways of the
DEmRNAs were assessed by GO and KEGG pathway analyses. The most
enriched GO biological processes included ‘protein binding’,
‘signal transducer activity’, ‘protein kinase activity’ and
‘regulation of cell cycle’. The pathway analysis revealed that
several genes were involved in cancer-related pathways, such as
‘cell cycle’, ‘cellular senescence’, ‘Kaposi sarcoma-associated
herpesvirus infection’, ‘relaxin signaling pathway’ and ‘human
T-cell leukemia virus 1 infection’. Among the enriched pathways,
‘Kaposi sarcoma-associated herpesvirus infection’ has been reported
to modulate the proliferation of glioma stem-like cells (39). Kaposi's sarcoma-associated
herpesvirus exhibits neurotropism and its mRNA was detected in the
plasma of patients with glioma in previous studies (39–41).
Another previous study suggested that viral infection was a risk
factor for glioma (42). The
potential association between Kaposi's sarcoma-associated
herpesvirus infection and glioma progression warrants further
investigation. Among the DEmRNAs in the ceRNA network, tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
(YWHA)-H, E2F transcription factor (E2F) 5, E2F2, WEE1
G2 checkpoint kinase (WEE1), cell division cycle 25A
(CDC25A) and checkpoint kinase 1 (CHEK1) were enriched in cell
cycle-associated pathways. The present study demonstrated that
YWHAH was significantly downregulated and associated with
hsa-mir-424, hsa-mir-497 and hsa-mir-195 in the ceRNA network. A
previous study demonstrated that YWHAH was involved in several
cellular processes, including G1/S and G2/M
cell cycle transition (43).
Additionally, YWHAE, another YWHA isoform involved in cell cycle
control and signal transduction (44,45), was
associated with copy number aberrations in astrocytoma pathogenesis
(46). E2F5 is a key transcription
factor involved in cell cycle progression. In a previous study,
E2F5 silencing inhibited the proliferation of GBM cells and induced
cell cycle arrest (47). miR-218
inhibited the growth and metabolism of glioma cells by targeting
E2F2 (48). WEE1, a regulator of the
G2 checkpoint in GBM cells, was associated with
malignancy and poor outcomes of patients with GBM (49). CDC25A and CHEK1 are significant
regulators of the cell cycle and apoptosis in glioma cells
(50).
The top 10 RNAs in the ceRNA network were identified
using the cytohubba plugin. The network consisted of 6 miRNAs,
including hsa-mir-93, hsa-mir-424, hsa-mir-497, hsa-mir-195,
hsa-mir-182 and hsa-mir-96, and 4 lncRNAs, including AC092171.1,
FAM87A, DLEU1 and LY86-AS1. LY86-AS1 and FAM87A were demonstrated
to have a high degree of closeness with hsa-mir-424, hsa-mir-497
and hsa-mir-195. A previous study revealed that hsa-mir-497
demonstrated high sensitivity and specificity for the prediction of
malignant astrocytomas (51).
Moreover, overexpression of hsa-mir-497 promoted the proliferation
of U87 glioma cells by targeting neuregulin receptor degradation
protein 1 (37). Previous studies
held conflicting views on the role of aberrantly expressed
hsa-mir-195 in gliomas. For instance, hsa-mir-195 was demonstrated
to inhibit the proliferation of human glioma cells by directly
targeting cyclin D1 and cyclin E1 mRNA (52), and was positively correlated with the
OS of patients with GBM (53).
Additionally, Zhang et al (54) revealed that overexpression of
hsa-mir-195 resulted in cell cycle arrest and significantly
decreased invasion of GBM cell lines. By contrast, hsa-mir-195
promoted the proliferation and invasion of the human glioma cell
line U87 via the transforming growth factor-β (TGF-β) signaling
pathway (55). The present results
are in agreement with those reported by Zhang et al
(54) and suggested that hsa-mir-195
was upregulated in glioma tissues. The mechanisms underlying the
interactions between hsa-mir-195, YWHAH and other potential mRNAs
involved in cell cycle regulation require further
investigation.
Protein-coding genes have long been recognized as
conventional cancer biomarkers. Nevertheless, lncRNAs, which play a
major role in the regulation of gene expression, have become the
focus of predictive biomarker investigation due to their ability to
better reflect tumor prognosis (11). Accumulating evidence suggests that
dysregulation of lncRNA expression is associated with
carcinogenesis and progression of glioma. Therefore, lncRNAs may
serve as potential biomarkers for prognosis (56–58). For
instance, colorectal neoplasia differentially expressed (CRNDE),
the most upregulated lncRNA in glioma (59), facilitated cancer cell proliferation,
migration and invasion by downregulating miR-384 expression. CRNDE
knockdown combined with miR-384 overexpression significantly
attenuated tumor progression in vivo (57). lncRNA activated by TGF-β (lncRNA-ATB)
served as a ceRNA and a sponge for miR-200a, promoting cell
proliferation and invasion in glioma. Upregulation of lncRNA-ATB
was associated with worse OS in patients with glioma (60).
In the present study, 92 DElncRNAs were involved in
the ceRNA network. Kaplan-Meier curve analysis indicated that 36
DElncRNAs were associated with OS. Therefore, these lncRNAs may
serve as potential prognostic biomarkers in patients with glioma.
Tian et al demonstrated that LINC00320, a tumor-suppressive
lncRNA, was downregulated in glioma tissues and inhibited the
proliferation of glioma cells by repressing the Wnt/β-catenin
signaling pathway in vitro and in vivo (26). Moreover, Mills et al (61) demonstrated that full-length LINC00320
was only expressed in human brain tissue, suggesting that it may be
a highly specific biomarker for glioma. Zhu et al (62) have demonstrated that the expression
level of HAS2-AS1 was closely associated with lymph node metastasis
and hypoxic tumor status in patients with oral squamous cell
carcinoma. Notably, the expression level of HAS2-AS1 was negatively
associated with OS in the present study. DLEU1 was identified to
act as a sponge for miR-490 and contributed to the development of
endometrial cancer (63).
Furthermore, DLEU1 has been widely accepted as an oncogenic lncRNA
and its aberrant upregulation was associated with the outcome of
various types of cancer, including gastric (64), pancreatic (65) and colorectal cancer (66). Terashima et al (67) demonstrated that MEG8 was required for
the epithelial-mesenchymal transition in lung and pancreatic cancer
cells. DBH-AS1 was demonstrated to promote cell proliferation and
survival through the MAPK signaling pathway in hepatocellular
carcinoma cells (68). MIR600HG
independently correlated with the outcome of patients with
pancreatic cancer (69). In the
present study, low MEG8 and DBH-AS1 expression, as well as high
MIR600HG expression, were associated with an improved outcome.
Therefore, further investigation is required to validate whether
these lncRNAs may serve as potential predictive biomarkers for
glioma.
However, there were some limitations in the present
study. Indeed, the present study was designed as a bioinformatics
analysis based on a small sample size, which also lacked
experimental verification. Further experiment validation in larger
samples is required to verify the present findings.
In conclusion, the present study constructed a
specific ceRNA network consisting of lncRNAs, miRNAs and mRNAs, and
the mechanisms of ceRNA regulation were further analyzed. Survival
analysis indicated that lncRNAs may serve as potential biomarkers
for predicting the outcomes of patients with glioma. To the best of
the authors' knowledge, the present study was the first to attempt
to construct a specific ceRNA network and to investigate the
interactions between miRNAs and lncRNAs in glioma based on TCGA
data.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Science and
Education for Health Foundation of Suzhou for Youth (grant nos.
kjxw2018030 and kjxw2018032), The Science and Technology Project
Foundation of Suzhou (grant no. SS201651), The Medical Key
Discipline Foundation of Jiangsu Province (grant no. ZDXKC2016007)
and The Education Research Project Foundation of Nanjing Medical
University (grant no. FZS-ZD-201701).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZLD and WJW contributed to the conception and design
of the study. JZ and YQH performed data analysis and curation. HW
performed the statistical analysis. WS and YFL carried out the
concept, design, definition of intellectual content, manuscript
preparation and wrote the first draft of the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ostrom QT, Gittleman H, Truitt G, Boscia
A, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical report:
Primary brain and other central nervous system tumors diagnosed in
the United States in 2011–2015. Neuro Oncol. 20 (Suppl 4):iv1–iv86.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vigneswaran K, Neill S and Hadjipanayis
CG: Beyond the World Health Organization grading of infiltrating
gliomas: Advances in the molecular genetics of glioma
classification. Ann Transl Med. 3:952015.PubMed/NCBI
|
|
3
|
National Brain Tumor Association, . Quick
brain tumor facts. 2018.Available at: http:
//braintumor.org/brain-tumor-information/brain-tumor-facts/.
November 27–2017
|
|
4
|
American Brain Tumor Association (ABTA), .
Brain tumor statistics. 2017.Available at:
http://www.abta.org/about-us/news/brain-tumor-statistics/.
22–November. 2017
|
|
5
|
McNeill KA: Epidemiology of brain tumors.
Neurol Clin. 34:981–998. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barone F, Alberio N, Iacopino DG,
Giammalva GR, D'Arrigo C, Tagnese W, Graziano F, Cicero S and
Maugeri R: Brain mapping as helpful tool in brain glioma surgical
treatment-toward the ‘Perfect Surgery’? Brain Sci. 8:1922018.
View Article : Google Scholar
|
|
7
|
Ostrom QT, Gittleman H, Liao P,
Vecchione-Koval T, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and other central nervous
system tumors diagnosed in the United States in 2010–2014. Neuro
Oncol. 19 (Suppl 5):v1–v88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Venur VA, Peereboom DM and Ahluwalia MS:
Current medical treatment of glioblastoma. Cancer Treat Res.
163:103–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kang JH and Adamson C: Novel
chemotherapeutics and other therapies for treating high-grade
glioma. Expert Opin Investig Drugs. 24:1361–1379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sotillo E and Thomas-Tikhonenko A: The
long reach of noncoding RNAs. Nat Genet. 43:616–617. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and
Peterson KJ: A uniform system for the annotation of vertebrate
microRNA genes and the evolution of the human microRNAome. Annu Rev
Genet. 49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43((Database issue)): D146–D152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar
|
|
25
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian S, Liu W, Pan Y and Zhan S: Long
non-coding RNA Linc00320 inhibits glioma cell proliferation through
restraining Wnt/β-catenin signaling. Biochem Biophys Res Commun.
508:458–464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao
HP, Rycaj K, Takata Y, Lin K, Lu Y, et al: MicroRNA-141 suppresses
prostate cancer stem cells and metastasis by targeting a cohort of
pro-metastasis genes. Nat Commun. 8:142702017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi L, Zhang J, Pan T, Zhou J, Gong W, Liu
N, Fu Z and You Y: MiR-125b is critical for the suppression of
human U251 glioma stem cell proliferation. Brain Res. 1312:120–126.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L, Shi ZM, Jiang CF, Liu X, Chen QD,
Qian X, Li DM, Ge X, Wang XF, Liu LZ, et al: MiR-143 acts as a
tumor suppressor by targeting N-RAS and enhances
temozolomide-induced apoptosis in glioma. Oncotarget. 5:5416–5427.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu X, Bao Z, Liu Y, Ji J and Liu N:
MicroRNA-98 attenuates cell migration and invasion in glioma by
directly targeting Pre-B cell leukemia homeobox 3. Cell Mol
Neurobiol. 37:1359–1371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anwar SL, Krech T, Hasemeier B, Schipper
E, Schweitzer N, Vogel A, Kreipe H, Buurman R, Skawran B and
Lehmann U: hsa-mir-183 is frequently methylated and related to poor
survival in human hepatocellular carcinoma. World J Gastroenterol.
23:1568–1575. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang BL, Dong FL, Guo TW, Gu XH, Huang LY
and Gao DS: MiRNAs mediate GDNF-induced proliferation and migration
of glioma cells. Cell Physiol Biochem. 44:1923–1938. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li HY, Li YM, Li Y, Shi XW and Chen H:
Circulating microRNA-137 is a potential biomarker for human
glioblastoma. Eur Rev Med Pharmacol Sci. 20:3599–3604.
2016.PubMed/NCBI
|
|
35
|
Yang W, Bai J, Liu D, Wang S, Zhao N, Che
R and Zhang H: MiR-93-5p up-regulation is involved in non-small
cell lung cancer cells proliferation and migration and poor
prognosis. Gene. 647:13–20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li HY, Liang JL, Kuo YL, Lee HH, Calkins
MJ, Chang HT, Lin FC, Chen YC, Hsu TI, Hsiao M, et al:
miR-105/93-3p promotes chemoresistance and circulating
miR-105/93-3p acts as a diagnostic biomarker for triple negative
breast cancer. Breast Cancer Res. 19:1332017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao Y, Sun L, Wu Z and Chen X:
Over-expression of miR-497 promotes the proliferation of U87 glioma
cells by targeting neuregulin receptor degradation protein 1. Xi
Bao Yu Fen Zi Mian Yi Xue Za Zhi. 33:1051–1055. 2017.(In Chinese).
PubMed/NCBI
|
|
38
|
Feng F, Kuai D, Wang H, Li T, Miao W, Liu
Y and Fan Y: Reduced expression of microRNA-497 is associated with
greater angiogenesis and poor prognosis in human gliomas. Hum
Pathol. 58:47–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jeon H, Kang YH, Yoo SM, Park MJ, Park JB,
Lee SH and Lee MS: Kaposi's sarcoma-associated herpesvirus
infection modulates the proliferation of glioma stem-like cells. J
Microbiol Biotechnol. 28:165–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chan PK, Ng HK, Cheung JL and Cheng AF:
Survey for the presence and distribution of human herpesvirus 8 in
healthy brain. J Clin Microbiol. 38:2772–2773. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Merelli E, Bedin R, Sola P, Barozzi P,
Mancardi GL, Ficarra G and Franchini G: Human herpes virus 6 and
human herpes virus 8 DNA sequences in brains of multiple sclerosis
patients, normal adults and children. J Neurol. 244:450–454. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
McFaline-Figueroa JR and Wen PY: The viral
connection to glioblastoma. Curr Infect Dis Rep. 19:52017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lee CG, Park GY, Han YK, Lee JH, Chun SH,
Park HY, Lim KH, Kim EG, Choi YJ, Yang K and Lee CW: Roles of
14-3-3η in mitotic progression and its potential use as a
therapeutic target for cancers. Oncogene. 32:1560–1569. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jin J, Smith FD, Stark C, Wells CD,
Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor
L, et al: Proteomic, functional, and domain-based analysis of in
vivo 14-3-3 binding proteins involved in cytoskeletal regulation
and cellular organization. Curr Biol. 14:1436–1450. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mackintosh C: Dynamic interactions between
14-3-3 proteins and phosphoproteins regulate diverse cellular
processes. Biochem J. 381:329–342. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pećina-Šlaus N, Kafka A, Gotovac Jerčić K,
Logara M, Bukovac A, Bakarić R and Borovečki F: Comparable genomic
copy number aberrations differ across astrocytoma malignancy
grades. Int J Mol Sci. 20:12512019. View Article : Google Scholar
|
|
47
|
Xu X, Cai N, Zhi T, Bao Z, Wang D, Liu Y,
Jiang K, Fan L, Ji J and Liu N: MicroRNA-1179 inhibits glioblastoma
cell proliferation and cell cycle progression via directly
targeting E2F transcription factor 5. Am J Cancer Res. 7:1680–1692.
2017.PubMed/NCBI
|
|
48
|
Zhang Y, Han D, Wei W, Cao W, Zhang R,
Dong Q, Zhang J, Wang Y and Liu N: MiR-218 inhibited growth and
metabolism of human glioblastoma cells by directly targeting E2F2.
Cell Mol Neurobiol. 35:1165–1173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Music D, Dahlrot RH, Hermansen SK,
Hjelmborg J, de Stricker K, Hansen S and Kristensen BW: Expression
and prognostic value of the WEE1 kinase in gliomas. J Neurooncol.
127:381–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Krell A, Wolter M, Stojcheva N, Hertler C,
Liesenberg F, Zapatka M, Weller M, Malzkorn B and Reifenberger G:
MiR-16-5p is frequently down-regulated in astrocytic gliomas and
modulates glioma cell proliferation, apoptosis, and response to
cytotoxic therapy. Neuropathol Appl Neurobiol. 45:441–458.
2019.PubMed/NCBI
|
|
51
|
Yang C, Wang C, Chen X, Chen S, Zhang Y,
Zhi F, Wang J, Li L, Zhou X, Li N, et al: Identification of seven
serum microRNAs from a genome-wide serum microRNA expression
profile as potential noninvasive biomarkers for malignant
astrocytomas. Int J Cancer. 132:116–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hui W, Yuntao L, Lun L, WenSheng L,
ChaoFeng L, HaiYong H and Yueyang B: MicroRNA-195 inhibits the
proliferation of human glioma cells by directly targeting cyclin D1
and cyclin E1. PLoS One. 8:e549322013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lakomy R, Sana J, Hankeova S, Fadrus P,
Kren L, Lzicarova E, Svoboda M, Dolezelova H, Smrcka M, Vyzula R,
et al: MiR-195, miR-196b, miR-181c, miR-21 expression levels and
O-6-methylguanine-DNA methyltransferase methylation status are
associated with clinical outcome in glioblastoma patients. Cancer
Sci. 102:2186–2190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ,
Yao Q, Zhou H and Qu LH: MicroRNA-195 plays a tumor-suppressor role
in human glioblastoma cells by targeting signaling pathways
involved in cellular proliferation and invasion. Neuro Oncol.
14:278–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Duan Y and Chen Q: TGF-β1 regulating
miR-205/miR-195 expression affects the TGF-β signal pathway by
respectively targeting SMAD2/SMAD7. Oncol Rep. 36:1837–1844. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J,
Duan R, Pu P, Kang C and Han L: A novel cell cycle-associated
lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA
transcript and is a biomarker of progression in glioma. Cancer
Lett. 373:251–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C
and Liu Y: CRNDE promotes malignant progression of glioma by
attenuating miR-384/PIWIL4/STAT3. Mol Ther. 24:1199–1215. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brodie S, Lee HK, Jiang W, Cazacu S, Xiang
C, Poisson LM, Datta I, Kalkanis S, Ginsberg D and Brodie C: The
novel long non-coding RNA TALNEC2, regulates tumor cell growth and
the stemness and radiation response of glioma stem cells.
Oncotarget. 8:31785–31801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li DX, Fei XR, Dong YF, Cheng CD, Yang Y,
Deng XF, Huang HL, Niu WX, Zhou CX, Xia CY and Niu CS: The long
non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy
by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2.
Oncotarget. 8:88163–88178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ma CC, Xiong Z, Zhu GN, Wang C, Zong G,
Wang HL, Bian EB and Zhao B: Long non-coding RNA ATB promotes
glioma malignancy by negatively regulating miR-200a. J Exp Clin
Cancer Res. 35:902016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mills JD, Chen J, Kim WS, Waters PD,
Prabowo AS, Aronica E, Halliday GM and Janitz M: Long intervening
non-coding RNA 00320 is human brain-specific and highly expressed
in the cortical white matter. Neurogenetics. 16:201–213. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhu G, Wang S, Chen J, Wang Z, Liang X,
Wang X, Jiang J, Lang J and Li L: Long noncoding RNA HAS2-AS1
mediates hypoxia-induced invasiveness of oral squamous cell
carcinoma. Mol Carcinog. 56:2210–2222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shao W, Li Y, Chen F, Jia H, Jia J and Fu
Y: Long non-coding RNA DLEU1 contributes to the development of
endometrial cancer by sponging miR-490 to regulate SP1 expression.
Pharmazie. 73:379–385. 2018.PubMed/NCBI
|
|
64
|
Li X, Li Z, Liu Z, Xiao J, Yu S and Song
Y: Long non-coding RNA DLEU1 predicts poor prognosis of gastric
cancer and contributes to cell proliferation by epigenetically
suppressing KLF2. Cancer Gene Ther. 25:58–67. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Gao S, Cai Y, Zhang H, Hu F, Hou L and Xu
Q: Long noncoding RNA DLEU1 aggravates pancreatic ductal
adenocarcinoma carcinogenesis via the miR-381/CXCR4 axis. J Cell
Physiol. 234:6746–6757. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu T, Han Z, Li H, Zhu Y, Sun Z and Zhu
A: LncRNA DLEU1 contributes to colorectal cancer progression via
activation of KPNA3. Mol Cancer. 17:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Terashima M, Ishimura A, Wanna-Udom S and
Suzuki T: MEG8 long noncoding RNA contributes to epigenetic
progression of the epithelial-mesenchymal transition of lung and
pancreatic cancer cells. J Biol Chem. 293:18016–18030. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM,
Hu YW, Lin L, Chen J, Zheng L and Wang Q: HBx-related long
non-coding RNA DBH-AS1 promotes cell proliferation and survival by
activating MAPK signaling in hepatocellular carcinoma. Oncotarget.
6:33791–33804. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Song J, Xu Q, Zhang H, Yin X, Zhu C, Zhao
K and Zhu J: Five key lncRNAs considered as prognostic targets for
predicting pancreatic ductal adenocarcinoma. J Cell Biochem.
119:4559–4569. 2018. View Article : Google Scholar : PubMed/NCBI
|