Introduction
Colorectal cancer (CRC) is the most common
gastrointestinal malignancy, as well as the third most lethal and
fourth most diagnosed type of cancer in the world, despite recent
advancements in the treatment of CRC (1). CRC is caused by genetic and epigenetic
alterations, including histone and chromatin structural
modification, DNA methylation and microRNA (miRNA) aberrations
(2). DNA methylation usually leads
to the hypermethylation of gene promoters and the inactivation of
tumor suppressor genes (TSGs), which plays an important role in the
initiation, development and recurrence of CRC (3). Thus, identification of TSGs that
undergo CpG island hypermethylation and assessment of their roles
and associated molecular mechanisms in tumor progression will help
to develop more effective diagnosis and individualized therapeutic
strategies for patients with CRC (3,4).
The zinc finger proteins (ZNFs) are classified into
eight-fold groups according to the secondary structure around the
zinc-binding site and main chain conformation (5). ZNFs have several functions, including
apoptosis regulation, transcriptional activation, protein folding
and integration, RNA packaging, DNA recognition and lipid binding
(6). ZNFs have been reported to play
important roles in different types of human cancer. Some ZNFs act
as oncogenes, for example, ZNF306 expression is upregulated in CRC,
whereas low ZNF306 expression suppresses tumor development
(7). Furthermore, ZNF307 inhibits
the activity of tumor suppressor genes, p21 and p53 by increasing
the transcription of EP300 and MDM2 (8). Conversely, some ZNFs act as tumor
suppressors, for example, ZNF23 inhibits the proliferation of
SK-OV-3 cells by enhancing p27/kip-1 expression (9). Furthermore, ZNF668 is considered a
tumor suppressor in breast cancer, which stabilizes p53 by
preventing MDM2-mediated ubiquitination and degradation of p53
(10).
ZNF365 contains the N-terminus-C2H2 zinc finger
motif, and was first discovered in 1998 from the human brain cDNA
library (11,12). It has been reported that
polymorphisms in the ZNF365 gene or its locus is associated with
different types of disease. For example, polymorphisms in the
Ala62Throf ZNF365 gene are associated with susceptibility to uric
acid nephrolithiasis (12,13). Furthermore, variants of ZNF365 are
associated with Crohn's disease (14). A total of five single nucleotide
polymorphisms (SNPs) in ADO-ZNF365-EGR2 have been demonstrated to
be associated with Vogt-Koyanagi-Harada (VKH) syndrome in patients
with VKH, in Thailand (15). It has
been reported that genetic variations in ZNF365 affected the risk
of breast cancer by influencing the proportion of dense tissue in
the breast (16). Additionally,
variants at the ZNF365 loci are associated with estrogen receptor
subtypes of breast cancer risk in BRCA1 and BRCA2 mutation carriers
(17).
Although ZNF365 is known to play important roles in
different types of human cancer, its function in CRC remains
unknown. Thus, the present study aimed to investigate the
association between ZNF365 expression and tumor progression of CRC,
and determine its underlying molecular mechanism.
Materials and methods
Cell lines and culture conditions
The CRC cell lines (Colo320, SW620, SW480, HCT116,
SW48, LOVO, HCT8, DLD1, HT29 and RKO) were purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
Cells were maintained in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) or RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium
supplemented with 10% fetal bovine serum (cat. no. VS500T;
Ausbian), in a humidified 5% CO2 incubator at 37°C.
Tissue specimens
For immunohistochemistry (IHC) analysis, 120
patients with CRC who underwent surgery at the Sir Run Run Shaw
Hospital between February 2004 and June 2006 were recruited in the
present study, and 10 normal colonic mucosa biopsy samples were
selected as the normal controls (4 males and 6 females; mean age,
51.12 years; age range, 32–65 years). The present study was
approved by the Ethics Committee of Sir Run Run Shaw Hospital
(Hangzhou, China) and all patients provided written informed
consent prior to the study start (approval no. 2019ZNF365-1).
Patients who had received preoperative chemotherapy, radiotherapy
or immunotherapy prior to surgery were excluded. A total of 79 men
and 41 women, with a mean age of 63.6 years (age range, 28–89
years) were included in the present study.
Following surgical resection, the tissue samples
were fixed at room temperature in 10% formalin for 24 h, embedded
in paraffin and sectioned into 4-um-thick slices. The intensity of
ZNF365 immunostaining was scored by two experienced pathologists
from Sir Run Run Shaw Hospital (Hangzhou, China) who were unaware
of the clinicopathological outcomes of the patients, using the
World Health Organization classification guidelines (18). A typical section for each case was
selected for IHC analysis.
Differentiation status was divided into three
subtypes: i) Well differentiated, including papillary
adenocarcinoma and high differentiated tubular adenocarcinoma; ii)
moderately differentiated, including highly to moderately
differentiated tubular adenocarcinoma and iii) poorly
differentiated, including poorly differentiated adenocarcinoma,
signet-ring cell carcinoma, mucinous adenocarcinoma and
undifferentiated carcinoma. According to these criteria, there were
82 well differentiated, 25 well/moderately differentiated and 13
poorly differentiated types of cancer of the 120 cases. Lymph node
metastasis and depth of invasion were graded based on the 7th
edition of the International Union Against Cancer
tumor-node-metastasis (TNM) system (19). All patients were followed up via
telephone for 36 months.
ZNF365 methylation was assessed in normal colorectal
(NC) tissues and paired CRC tissues, which were obtained from 30
patients in July 2009. The patients included 11 males and 19
females with a mean age of 56.2 years (age range, 41–68 years). The
corresponding NC tissues were removed from the margin of the
resection with a distance >10 cm away from the tumor.
Semi-quantitative reverse
transcription (RT)-PCR
Semi-quantitative RT-PCR was performed as previously
described (20). Total RNA was
extracted from fresh cells (Colo320, SW620, SW480, HCT116, SW48,
LOVO, HCT8, DLD1, HT29 and RKO) using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, RNA was
converted to cDNA using RevertAid First Strand cDNA Synthesis kit
(cat. no. K1622; Thermo Fisher Scientific, Inc.). GAPDH mRNA was
used as a control. The primers used in the present study are listed
in Table SI.
Methylation analysis of ZNF365
To determine the molecular mechanism underlying
aberrant ZNF365 expression in colorectal cancer, the association
between ZNF365 mRNA expression and DNA methylation was assessed
using the cBioPortal online database (www.cbioportal.org).
GeneMANIA analysis
GeneMANIA is a commonly used website for
constructing protein-protein interaction networks and predicting
the function of favorite genes (21). The GeneMANIA database (http://genemania.org/) was used to assess the
association between ZNF365 and p53 expression levels.
5-Aza-2-deoxycytidine and trichostatin
A treatment
RKO cells that do not express ZNF365 were seeded
into a 10-cm dish at a density of 1×106 cells/ml and
cultured overnight in a humidified incubator at 37°C with 5%
CO2. Cells were subsequently treated with demethylating
agent 5-aza (Sigma-Aldrich; Merck KGaA) at a final concentration of
10 mM for 3 days and further treated with the histone deacetylase
inhibitor TSA (Sigma-Aldrich; Merck KGaA) at a final concentration
of 300 nmol/l for an additional 24 h at 37°C. Cells were collected
for DNA and RNA extraction.
Bisulfite treatment and promoter
methylation analysis
Methylation-specific PCR (MSP), bisulfite
modification of DNA and bisulfite genomic sequencing (BGS) were
performed as previously described (22). The primer sequences used for MSP and
BGS are listed in Table SI.
IHC staining
IHC staining was performed using the ChemMate™
EnVision™ detection kit (Dako; Agilent Technologies, Inc.)
according to the manufacturer's instructions. Briefly, the CRC
sections and the normal colonic mucosa biopsy samples were dewaxed
and hydrated with 100% dimethylbenzene (cat. no. 1330-20-7;
Shanghai Macklin Biochemical Co., Ltd.), and rehydrated in a
descending ethanol series. Tissue sections were washed with
deionized water and phosphate buffered saline (PBS). The antigen
retrieval process was performed with 0.01 M citrate buffer (pH 6.0;
cat. no. C1013; Beijing Solarbio Science & Technology Co.,
Ltd.). After cooling to room temperature, the tissue sections were
blocked with 3% hydrogen peroxidase-methanol solution for 30 min to
inhibit endogenous peroxidase activity, and subsequently incubated
in preimmunized goat serum (cat. no. C0265; Beyotime Institute of
Biotechnology) for 30 min, both at room temperature. Tissue
sections were incubated with rabbit polyclonal IgG primary antibody
directed against ZNF365 (1:50; cat. no. HPA052446; Atlas
Antibodies) overnight at 4°C. After warming to room temperature,
the tissue sections were washed five times with PBS and
subsequently incubated with ChemMateEnVision/HRP, Rabbit/Mouse
reagent (Dako; Agilent Technologies, Inc.) at room temperature for
30 min. The sections were stained with ChemMate DAB+chromogen
(Dako; Agilent Technologies, Inc.) and counterstained with
hematoxylin at room temperature for 1 min each. Tissue sections
were dehydrated in an ascending ethanol series and dimethylbenzene,
and were observed using a light microscope (magnification,
×200).
Evaluation of staining
A total of two independent pathologists from Sir Run
Run Shaw Hospital (Hangzhou, China) blindly assessed the slides
three times to determine the percentage of positive cells, staining
intensity and subcellular localization. ZNF365 expression was
scored using the World Health Organization classification system.
The percentage of positive cells was scored as follows: 0, 0–10; 1,
11–25; 2, 26–50; and 3, 51–100%. The intensity of staining was
scored as follows: 0, negative; 1, weak; 2, moderate and 3, strong.
The final immunoreactivity score (IRS) was equal to the sum of both
scores, which ranged from 0–9. The specimens were divided into two
groups according to the IRS value, high expression group (IRS 6–9)
and low expression group (IRS 0–5) in order to assess the
association between ZNF365 expression and clinicopathological
characteristics.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.). The Kaplan-Meier method was used to assess
the survival curve and statistical differences were determined
using the log-rank test. Pearson's χ2 test and Fisher's
exact test were used to assess the association between ZNF365
expression with clinicopathological characteristics. Cox's
proportional hazards regression model was used to perform
univariate (depth of invasion, tumor location, distant metastasis,
sex, age, differentiation, lymph node metastasis, TNM stage and
ZNF365 expression) and multivariate analyses. Relative risk of
mortality is presented as adjusted hazard ratios (HRs) and
corresponding 95% confidence intervals (CI). P<0.05 was
considered to indicate a statistically significant difference.
Results
ZNF365 expression in CRC tissues
ZNF365 expression was assessed in 120 cases of CRC
and 10 normal colonic mucosa biopsy samples were used as the normal
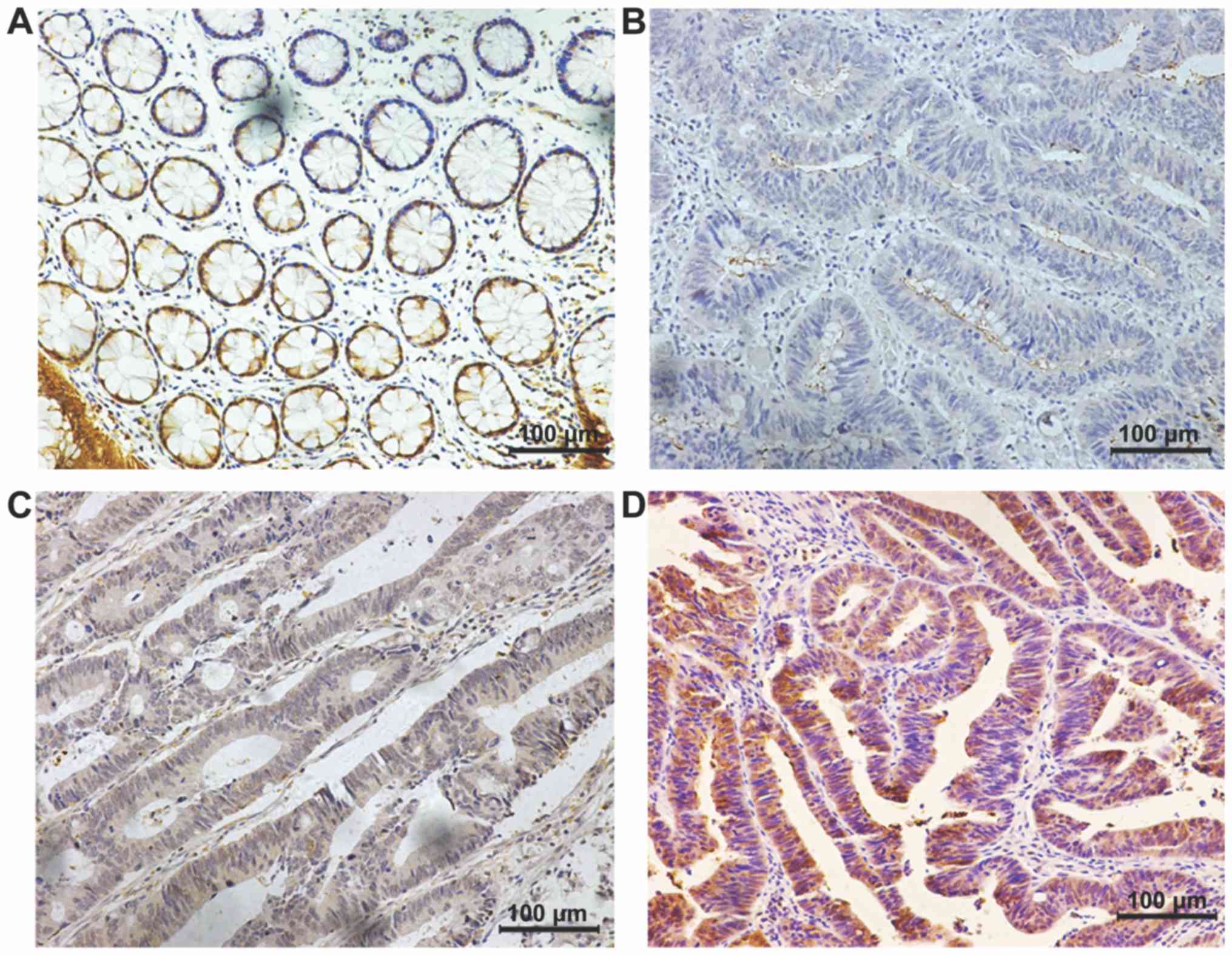
controls. Representative immunostaining images of ZNF365 in CRC
tissues are presented in Fig. 1. The
results demonstrated that ZNF365 was expressed in the cytoplasm and
nucleus of cancer tissues. Patients were divided into two groups
according to the IRS value, high expression group (IRS 6–9) and low
expression group (IRS 0–5). A total of 54 cases of CRC tissues
(45%) were classified into the low ZNF365 expression group, while
the remaining 66 cases (55%) were classified into the high ZNF365
expression group (Table I).
 | Table I.Association between ZNF365 expression
and clinicopathological characteristics of patients with colorectal
cancer (n=120). |
Table I.
Association between ZNF365 expression
and clinicopathological characteristics of patients with colorectal
cancer (n=120).
|
|
| ZNF365
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Number of patients,
n (%) | Low, n (%) | High, n (%) | χ2 | P-value |
|---|
| Total | 120 | 54 (45.000) | 66 (55.000) |
|
|
| Sex |
|
|
|
|
|
|
Male | 79 (65.833) | 33 (41.772) | 46 (58.228) | 0.973 | 0.324 |
|
Female | 41 (34.167) | 21 (51.220) | 20 (48.780) |
|
|
| Age, years |
|
|
|
|
|
|
≥63 | 62 (51.667) | 28 (45.162) | 34 (54.838) | 0.001 | 0.971 |
|
<63 | 58 (48.333) | 26 (44.828) | 32 (55.172) |
|
|
| Tumor location |
|
|
|
|
|
|
Rectum | 77 (64.167) | 32 (41.558) | 45 (58.442) | 1.028 | 0.311 |
|
Colon | 43 (35.833) | 22 (51.163) | 21 (48.837) |
|
|
| Histopathological
grading |
|
|
|
|
|
| G1
(Well) | 82 (68.333) | 35 (42.683) | 47 (57.317) | 6.349 | 0.042a |
| G2
(Moderate) | 25 (20.833) | 16 (64.000) | 9
(36.000) |
|
|
| G3
(Poor) | 13 (10.833) | 3 (23.077) | 10 (76.923) |
|
|
| Depth of
invasion |
|
|
|
|
|
|
pT1/T2 | 34 (28.333) | 10 (29.412) | 24 (70.588) | 4.658 | 0.031a |
|
pT3/T4 | 86 (71.667) | 44 (51.163) | 42 (48.837) |
|
|
| Lymph node
status |
|
|
|
|
|
| N0 | 68 (56.667) | 24 (35.294) | 44 (64.706) | 5.973 | 0.015a |
|
N1/2 | 52 (43.333) | 30 (57.692) | 22 (42.308) |
|
|
| Distant
metastasis |
|
|
|
|
|
| M0 | 101 (84.167) | 43 (42.574) | 58 (57.426) | 1.517 | 0.218 |
| M1 | 19 (15.833) | 11 (57.895) | 8
(42.105) |
|
|
| TNM stage |
|
|
|
|
|
|
I/II | 63 (52.500) | 24 (38.095) | 39 (61.905) | 2.555 | 0.110 |
|
III/IV | 57 (47.500) | 30 (52.632) | 27 (47.368) |
|
|
Association between ZNF365 expression
and clinicopathological characteristics of patients with CRC
The association between ZNF365 expression and
clinicopathological characteristics of patients with CRC are
presented in Table I. The results
demonstrated that ZNF365 was significantly associated with lymph
node metastasis (P=0.015), depth of invasion (P=0.031) and
histopathological grading (P=0.042). However, there were no
significant associations between ZNF365 expression and age
(P=0.971), sex (P=0.324), distant metastasis (P=0.218), tumor
location (P=0.311) or TNM stages (P=0.11).
Downregulation of ZNF365 is associated
with poor survival of patients with CRC
All patients were followed up for overall survival
following surgery, to further determine the role ZNF365 plays in
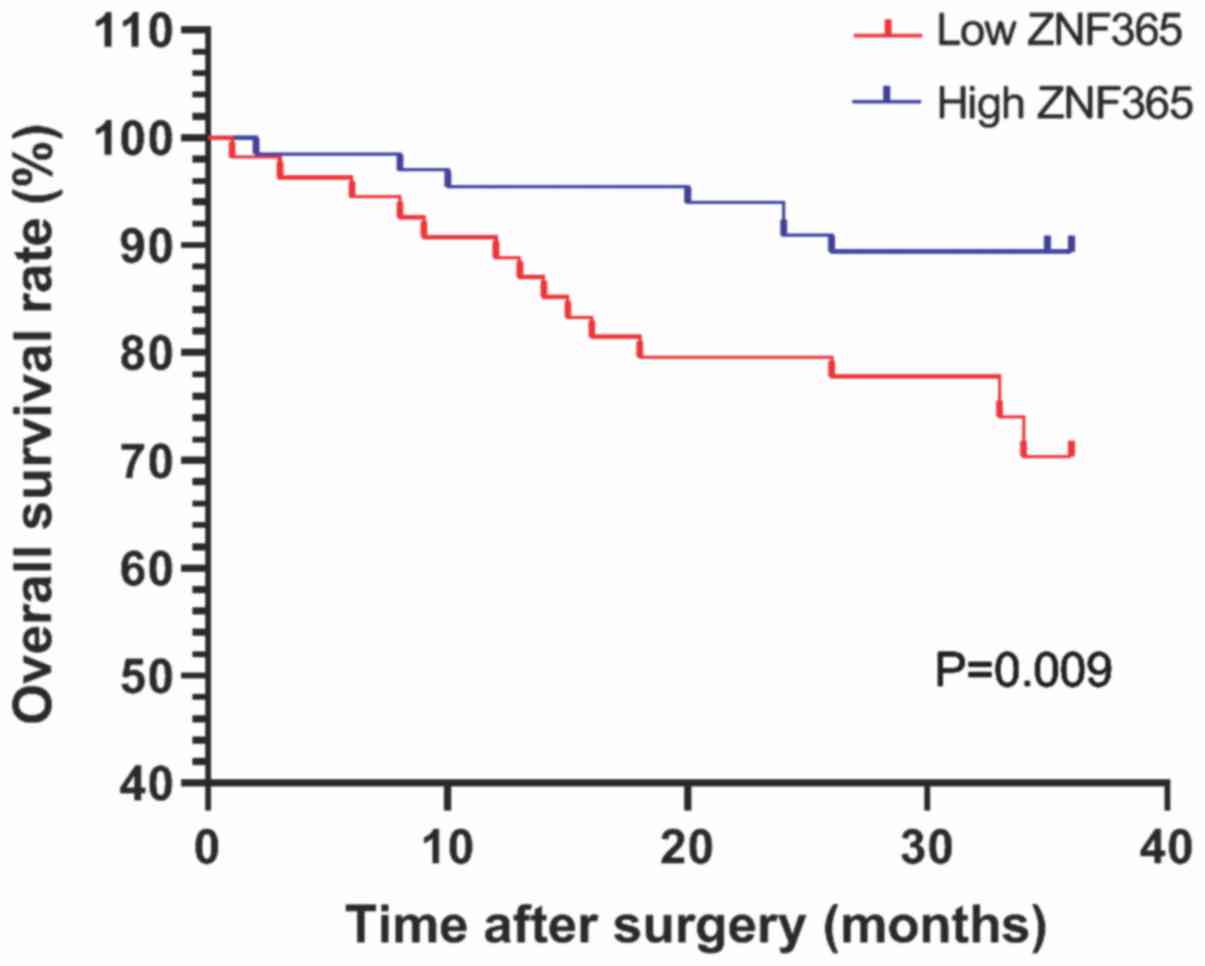
the progression of CRC. The prognosis of patients with high and low
ZNF365 expression was assessed (Fig.
2). Kaplan-Meier survival analysis demonstrated that the 3-year
survival rate was higher in patients with high ZNF365 expression
than those with low ZNF365 expression (Fig. 2; P=0.009). Univariate analysis
demonstrated that in addition to ZNF365 expression (P=0.013), lymph
node metastasis (P<0.001), tumor histopathological grading
(P=0.011), distant metastasis (P<0.001), depth of invasion
(P=0.044) and TNM stages (P<0.001) were also significantly
associated with 3-year overall survival rates (Table II).
 | Table II.Univariate survival analysis of
prognostic factors in colorectal cancer. |
Table II.
Univariate survival analysis of
prognostic factors in colorectal cancer.
|
Characteristics | HR | 95% CI | P-value |
|---|
| Sex |
|
| 0.419 |
| Male
vs. Female | 1.468 | 0.579-3.723 |
|
| Age, years |
|
| 0.561 |
| ≥63 vs.
<63 | 1.277 | 0.560-2.912 |
|
| Tumor location |
|
| 0.067 |
| Rectum
vs. Colon | 2.148 | 0.948-4.871 |
|
| Histopathological
grading |
|
| 0.011a |
| Well
vs. Moderate vs. Poor | 1.914 | 1.163-3.15 |
|
| Depth of
invasion |
|
| 0.044a |
| T1+T2
vs. T3+T4 | 2.629 | 1.025-6.744 |
|
| Lymph node
metastasis |
|
|
<0.001b |
| N0 vs.
N1/2 | 3.111 | 1.84-5.258 |
|
| Distant
metastasis |
|
|
<0.001b |
| M0 vs.
M1 | 6.326 | 2.792-14.496 |
|
| TNM stage |
|
|
<0.001b |
| I/II
vs. III/IV | 2.863 | 1.723-4.756 |
|
| ZNF365
expression |
|
| 0.013a |
| Low vs.
High | 0.324 | 0.133-0.787 |
|
Multivariate analysis demonstrated that tumor
location (HR, 2.818; 95% CI, 1.173–6.770; P=0.021),
histopathological grading (HR, 1.907; 95% CI, 1.07–3.389; P=0.028),
TNM stage (HR, 4.801; 95% CI, 1.912–12.053; P=0.001) and ZNF365
expression (HR, 0.386; 95% CI, 0.152–0.980; P=0.045) were all
statistically significant prognostic factors in CRC (Table III). Collectively, these results
indicate that ZNF365 may be a valuable prognostic factor in
CRC.
 | Table III.Multivariate survival analysis of
prognostic factors in colorectal cancer. |
Table III.
Multivariate survival analysis of
prognostic factors in colorectal cancer.
| Characteristic | HR | 95% CI | P-value |
|---|
| Sex |
|
| 0.784 |
| Male
vs. Female | 1.149 | 0.424-3.115 |
|
| Age, years |
|
| 0.616 |
| ≥63 vs.
<63 | 1.249 | 0.524-2.973 |
|
| Tumor location |
|
| 0.021a |
| Rectum
vs. Colon | 2.818 | 1.173-6.770 |
|
| Histopathological
grading |
|
| 0.028a |
| Well
vs. Moderate vs. Poor | 1.907 | 1.073-3.389 |
|
| Depth of
invasion |
|
| 0.336 |
| T1 vs.
T2+T3+T4 | 0.582 | 0.193-1.753 |
|
| TNM stage |
|
| 0.001b |
| I+II
vs. III+IV | 4.801 | 1.912-12.053 |
|
| ZNF365
expression |
|
| 0.045a |
| Low vs.
High | 0.386 | 0.152-0.980 |
|
Correlation between ZNF365 expression
and p53 expression in CRC patients
p53 plays a significant role in the development of
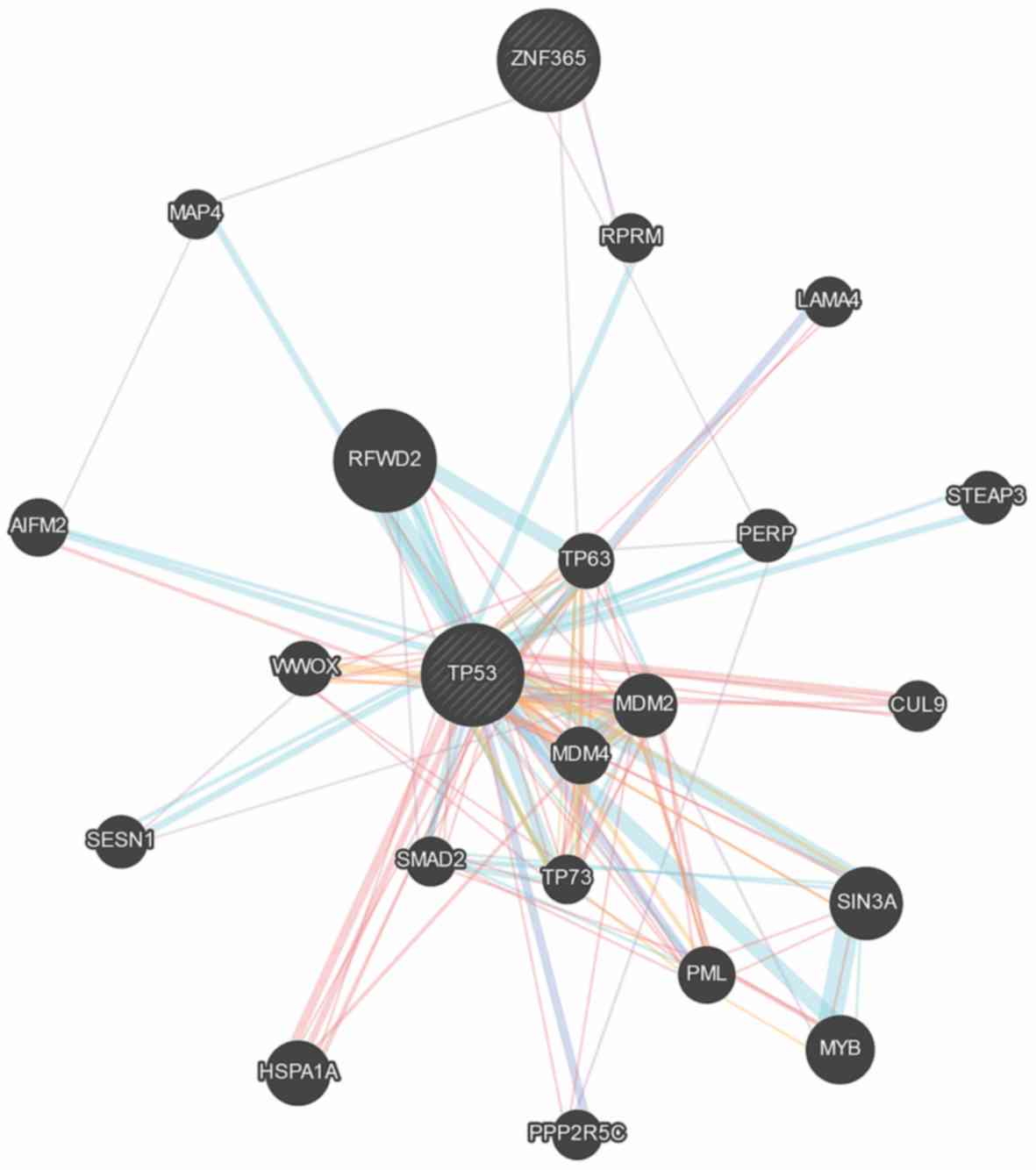
tumors (23). The GeneMANIA database
was used to assess the association between ZNF365 and p53
expression levels. The results demonstrated that ZNF365 can
interact with p53 via RPRM and MAP4 (Fig. 3).
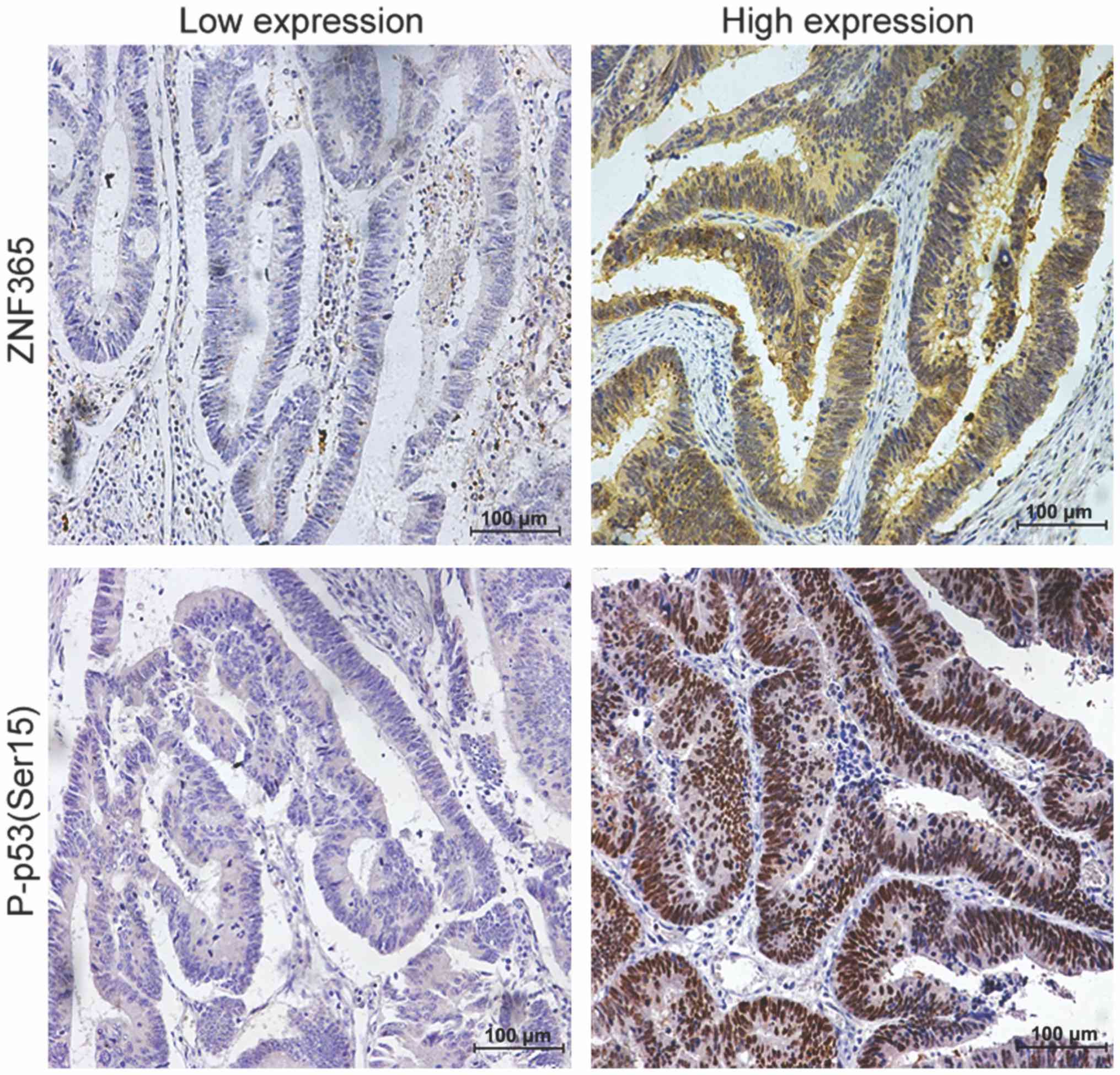
IHC analysis was performed to confirm the
correlation between ZNF365 protein expression and total p53 and
P-p53 (Ser15) protein expression in 120 cases of CRC. No
significant correlation was observed between ZNF365 expression and
total p53 expression, while ZNF365 expression was positively
correlated with P-p53 (Ser15) protein expression, with a
correlation coefficient of 0.189 (P=0.038; Table IV). Representative IHC staining
images of ZNF365 and P-p53 (Ser15) in CRC tissues are presented in
Fig. 4.
 | Table IV.Correlation between ZNF365 expression
and p53 expression in patients with colorectal cancer (n=120). |
Table IV.
Correlation between ZNF365 expression
and p53 expression in patients with colorectal cancer (n=120).
|
|
| ZNF365
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| p53 expression | Number of patients,
n (%) | Low, n (%) | High, n (%) | r value | P-value |
|---|
| Total p53 |
|
|
|
|
|
|
Low | 40 (33.333) | 21 (52.500) | 19 (47.500) | 0.107 | 0.247 |
|
High | 80 (66.667) | 33 (41.250) | 47 (58.750) |
|
|
| Phospho-p5
(Ser15) |
|
|
|
|
|
|
Low | 52 (43.333) | 29 (55.769) | 23 (44.231) | 0.189 | 0.038 |
|
High | 68 (56.667) | 25 (36.765) | 43 (63.235) |
|
|
ZNF365 is downregulated by methylation
in most CRC cell lines and tissues
In order to determine the molecular mechanism by
which ZNF365 expression is decreased in CRC, the cBioPortal
database was used to assess the association between ZNF365
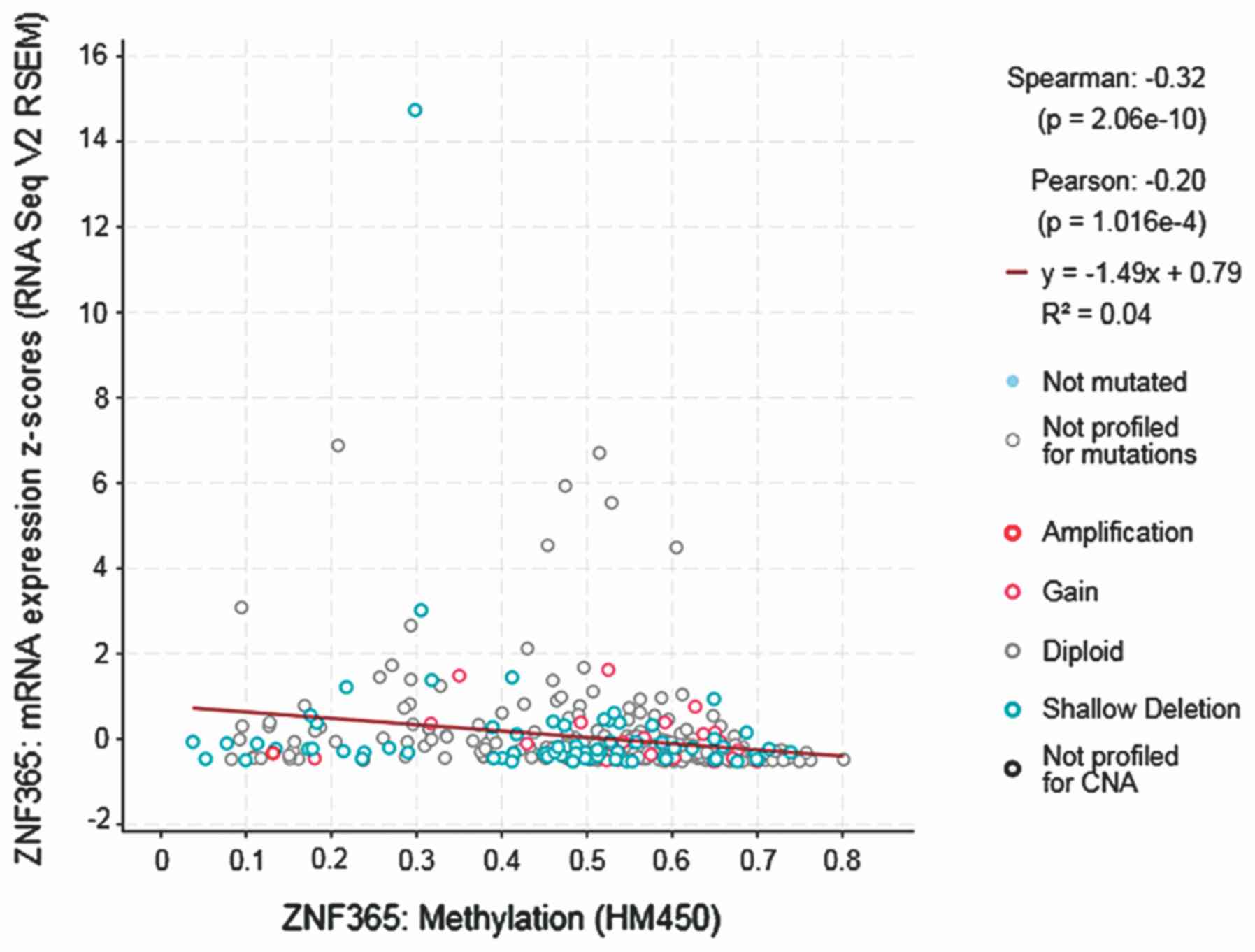
expression and DNA methylation. The results demonstrated a
statistically significant negative correlation between ZNF365 gene
expression and DNA methylation (Spearman, −0.32;
P=1.016×10−4; Fig. 5).
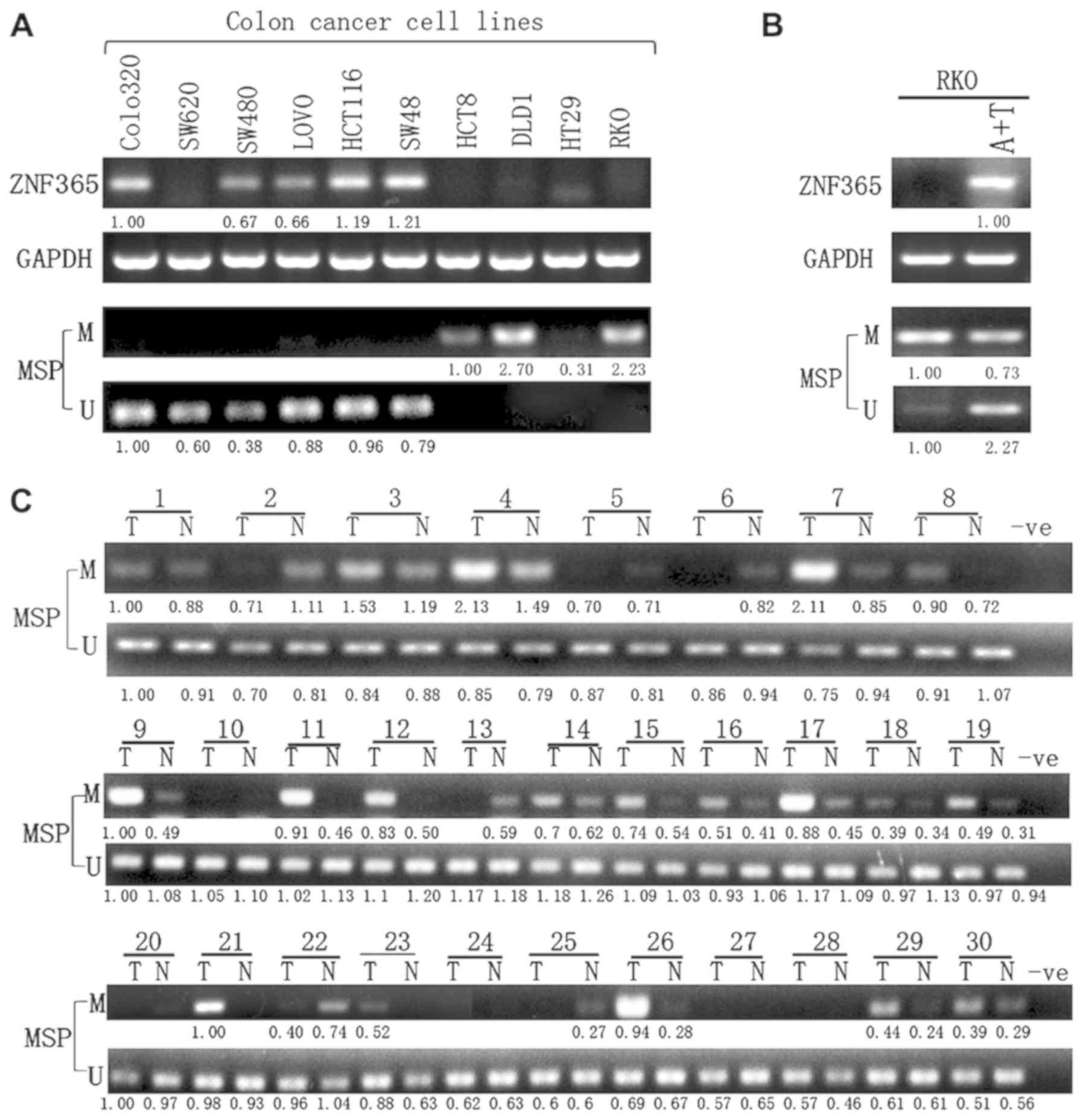
Semi-quantitative RT-PCR analysis was subsequently performed to
assess ZNF365 expression in CRC cell lines (Colo320, SW620, SW480,
LOVO, HCT116, SW48, HCT8, DLD1, HT29 and RKO). The results
demonstrated that ZNF365 expression was downregulated or even
silenced in most cell lines (Fig.
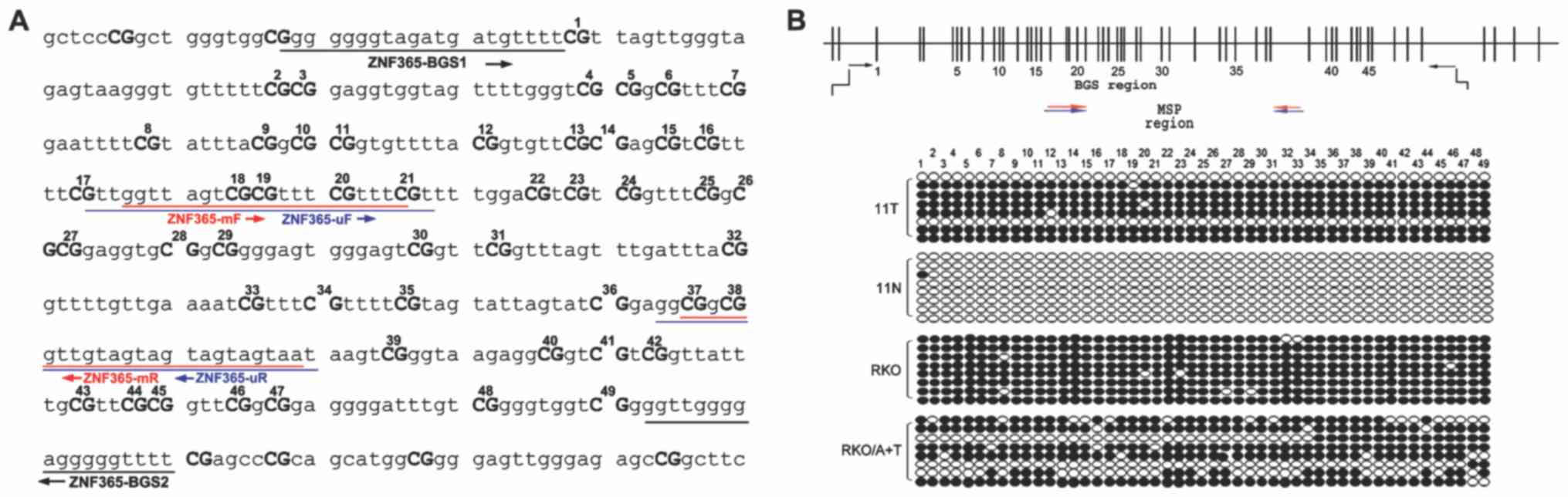
6A). In addition, MSP primers of ZNF365 were designed to
determine its methylation status according to the ZNF365 CpG island
(CGI) sequence (Fig. 7A). As
expected, the ZNF365 promoter was methylated in the cell lines with
decreased ZNF365 expression or ZNF365 silenced (Fig. 6A).
In order to further determine whether promoter
methylation directly mediated silencing of ZNF365, its expression
was compared before and after treatment in these cell lines, with
5-aza and TSA. The results demonstrated that ZNF365 expression
significantly recovered following demethylation treatment in the
assessed cell lines, (Fig. 6B). BGS
analysis of 49 CpG sites was performed to determine the methylation
profiles of ZNF365 CGI, including those CpG sites analyzed by MSP
(Fig. 7). Densely methylated CpG
sites were detected in the cell lines without ZNF365 expression.
Both BGS and MSP analyses demonstrated that the ZNF365 CGI was
significantly demethylated following TSA and 5-aza treatment,
suggesting a direct association between ZNF365 silencing and CGI
methylation (Figs. 6B and 7B). Furthermore, MSP analysis was performed
to detect ZNF365 methylation in 30 primary colorectal tumors (T)
and paired adjacent normal tissues (N). In 63.3% (19/30) of cases,
ZNF365 methylated bands in tumor tissues were stronger than the
paired adjacent normal tissues (Fig.
6C), which was confirmed following BGS analysis (Fig. 7B).
Discussion
ZNF365 serves as a transcription factor, playing key
roles in transcriptional activation, protein folding, DNA
recognition, lipid binding, RNA packaging and apoptosis regulation
(7). The present study aimed to
investigate the association between ZNF365 protein expression and
the clinicopathological characteristics of patients with CRC.
ZNF365, which is predominantly expressed in the cytoplasm and
nucleus of CRC tissues (24), was
significantly associated with histopathological grading, lymph node
metastasis and depth of invasion. However, no significant
associations were observed between ZNF365 expression and age, sex,
tumor location, distant metastasis or TNM stage. Furthermore, the
survival rate of patients with high ZNF365 expression was
significantly higher than that of patients with low ZNF365
expression. ZNF365 expression was downregulated in tumor tissues,
particularly in poorly differentiated and advanced CRC. The
association between ZNF365 expression and CRC from a histological
level was also assessed. Multivariate survival analysis
demonstrated that ZNF365 protein expression may be used as a
prognostic and diagnostic marker for CRC.
Previous studies have reported that genetic
polymorphisms of ZNF365 are associated with immune-related diseases
in Latin America and Asia (25,26). It
has been demonstrated that ZNF365 is associated with atopic
dermatitis (AD) in Japanese (26)
and overall susceptibility to Crohn's disease in Canadian children
(25). Variants of ZNF365 are also
associated with susceptibility to breast cancer (16). A total of five SNPs in
ADO-ZNF365-EGR2 have been reported to be associated with
Vogt-Koyanagi-Harada (VKH) syndrome in Thai patients with VKH but
not in other Asian patients (15).
In addition, Zhang et al (24) demonstrated that ZNF365 acts as a
transcriptional target of p53, which is a novel factor caused
genomic instability (24). A
mechanistic study demonstrated that ZNF365 can suppress expression
of a group of common fragile sites including telomeres, and thus,
that polymorphisms in the ZNF365 locus are associated with an
increased risk of cancer that may result from telomere dysfunction
(24). Another study revealed that
when induced by DNA double strand break signals, ZNF365 can
participate in the homologous recombination repair pathway and
maintain genomic integrity during DNA replication by interacting
with poly(ADP-ribose) polymerase 1 to tether MRE11 to the DNA end
resection site (27). Telomere
dysfunction triggers inaccurate DNA repair followed by genomic
instability, which is a feature of almost all types of human cancer
(28).
p53, activated as a transcription factor, plays an
important role in preventing tumorigenesis and tumor progression
(29). Following DNA damage, p53
protein rapidly accumulates through post-transcriptional
mechanisms, which induces growth arrest or apoptosis (30,31). In
some cells with defective DNA repair, p53 accumulation is absent or
delayed (32). However, mutant p53,
detected in >50% of cases of solid cancers, loses its ability to
specifically bind to sequences of genes that respond to senescence,
cell cycle arrest and apoptosis, which results in the
transformation of this tumor suppressor into an oncogenic factor
(33–35). The results of the present study
demonstrated that ZNF365 expression did not correlate with total
p53 expression; however, it was positively correlated with
phospho-p53 (Ser15) expression. Upon DNA damage, p53 is
phosphorylated at serine double 15, which results in decreased
interaction of p53 with its negative regulator, the tumor protein
MDM2, both in vitro and in vivo (32). This suggests that ZNF365 acts as a
suppressor in CRC tumorigenesis and progression by downregulating
phospho-p53 (Ser15) expression.
Regarding the expression control mechanisms, the
results of the present study demonstrated that ZNF365 expression
was downregulated or even silenced in most cell lines. ZNF365
expression was also downregulated in tumor tissues, particularly in
advanced cancer and poorly differentiated cancer. Downregulation of
ZNF365 expression may be due to its methylation (36), which was detected in 63.3% of CRC
tumors and only 36.7% of adjacent non-tumor tissues. DNA
methylation is a vital component in multilevel gene control in
eukaryotes (37). Increasing
evidence demonstrates that methylation levels and patterns are
deranged in tumor cells (38,39). DNA
methylation, together with other epigenetic changes such as RNA
editing, affects chromatin structure and thus regulates processes,
including allele-specific expression of imprinted genes,
X-chromosome inactivation, transcription and inactivation of tumor
suppressor genes (40). It was also
demonstrated that in the Colo320 and SW620 cell lines, unmethylated
alleles coexisted with silencing, suggesting that other expression
control mechanisms, such as deletions may also be involved.
However, promoter methylation is the main cause of downregulation
of ZNF365 (36), and the results of
the present study suggest that ZNF365 methylation is a common
cancer-specific event in CRC.
The present study is not without limitations. First,
as this was a single retrospective study, more research is required
to determine the underlying molecular mechanisms between ZNF365 and
P-p53. To the best of our knowledge, the present study was the
first to investigate the association between ZNF365 expression and
CRC. Taken together, the results suggest that downregulation of
ZNF365 by methylation may independently predict poor prognosis in
patients with CRC, by decreasing P-p53 (Ser15) expression.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81672362) and the
Test Programs of Science and Technology Commission Foundation of
Zhejiang Province (grant no. 2018C37063).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XH, CW, SL, YK and XF designed the study, analyzed
data and approved the version to be published. CW performed the
experiments and drafted the manuscript. SL and YK revised it
critically for important intellectual content. XF reviewed the
manuscript and gave final approval of the version to be published.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Sir Run Run Shaw Hospital (Hangzhou, China) and all
patients provided written informed consent prior to the study start
(approval no. 2019ZNF365-1).
Patient consent for publication
Patients agreed to the use of their samples in
scientific research and provided consent for publication of their
information.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Khodavirdipour A, Zarean R and
Safaralizadeh R: Evaluation of the anti-cancer effect of Syzygium
cumini ethanolic extract on HT-29 colorectal cell line. J
Gastrointest Cancer. Jun 6–2020.(Epub ahead of print). View Article : Google Scholar
|
|
2
|
Zoratto F, Rossi L, Verrico M, Papa A,
Basso E, Zullo A, Tomao L, Romiti A, Lo Russo G and Tomao S: Focus
on genetic and epigenetic events of colorectal cancer pathogenesis:
Implications for molecular diagnosis. Tumour Biol. 35:6195–6206.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palii SS and Robertson KD: Epigenetic
control of tumor suppression. Crit Rev Eukaryot Gene Expr.
17:295–316. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer-a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Krishna SS, Majumdar I and Grishin NV:
Structural classification of zinc fingers: Survey and summary.
Nucleic Acids Res. 31:532–550. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laity JH, Lee BM and Wright PE: Zinc
finger proteins: New insights into structural and functional
diversity. Curr Opin Struct Biol. 11:39–46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang L, Hamilton SR, Sood A, Kuwai T,
Ellis L, Sanguino A, Lopez-Berestein G and Boyd DD: The previously
undescribed ZKSCAN3 (ZNF306) is a novel ‘driver’ of colorectal
cancer progression. Cancer Res. 68:4321–4330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Wang Y, Fan X, Mo X, Wang Z, Li Y,
Yin Z, Deng Y, Luo N, Zhu C, et al: ZNF307, a novel zinc finger
gene suppresses p53 and p21 pathway. Biochem Biophys Res Commun.
363:895–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang C, Jia Y, Yang S, Chen B, Sun H,
Shen F and Wang Y: Characterization of ZNF23, a KRAB-containing
protein that is downregulated in human cancers and inhibits cell
cycle progression. Exp Cell Res. 313:254–263. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu R, Peng G, Dai H, Breuer EK,
Stemke-Hale K, Li K, Gonzalez-Angulo AM, Mills GB and Lin SY:
ZNF668 functions as a tumor suppressor by regulating p53 stability
and function in breast cancer. Cancer Res. 71:6524–6534. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nagase T, Ishikawa K, Suyama M, Kikuno R,
Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N and Ohara O:
Prediction of the coding sequences of unidentified human genes.
XII. The complete sequences of 100 new cDNA clones from brain which
code for large proteins in vitro. DNA Res. 5:355–364. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gianfrancesco F, Esposito T, Ombra MN,
Forabosco P, Maninchedda G, Fattorini M, Casula S, Vaccargiu S,
Casu G, Cardia F, Deiana I, et al: Identification of a novel gene
and a common variant associated with uric acid nephrolithiasis in a
Sardinian genetic isolate. Am J Hum Genet. 72:1479–1491. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Medina-Escobedo M, González-Herrera L,
Villanueva-Jorge S and Martín-Soberanis G: Metabolic abnormalities
and polymorphisms of the vitamin D receptor (VDR) and ZNF365 genes
in children with urolithiasis. Urolithiasis. 42:395–400. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Haritunians T, Jones MR, McGovern DP, Shih
DQ, Barrett RJ, Derkowski C, Dubinsky MC, Dutridge D, Fleshner PR,
Ippoliti A, et al: Variants in ZNF365 isoform D are associated with
Crohn's disease. Gut. 60:1060–1067. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao S, Chee SP, Yu HG, Sukavatcharin S, Wu
L, Kijlstra A, Hou S and Yang P: Investigation of the association
of Vogt-Koyanagi-Harada syndrome with IL23R-C1orf141 in Han Chinese
Singaporean and ADO-ZNF365-EGR2 in Thai. Br J Ophthalmol.
100:436–442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lindström S, Vachon CM, Li J, Varghese J,
Thompson D, Warren R, Brown J, Leyland J, Audley T, Wareham NJ, et
al: Common variants in ZNF365 are associated with both mammographic
density and breast cancer risk. Nat Genet. 43:185–187. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Couch FJ, Gaudet MM, Antoniou AC, Ramus
SJ, Kuchenbaecker KB, Soucy P, Beesley J, Chen X, Wang X, Kirchhoff
T, et al: Common variants at the 19p13.1 and ZNF365 loci are
associated with ER subtypes of breast cancer and ovarian cancer
risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol
Biomarkers Prev. 21:645–657. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kleihues P and Sobin LH: World Health
Organization classification of tumors. Cancer. 88:28872000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tao Q, Huang H, Geiman TM, Lim CY, Fu L,
Qiu GH and Robertson KD: Defective de novo methylation of viral and
cellular DNA sequences in ICF syndrome cells. Hum Mol Genet.
11:2091–2102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38((Web Server Issue)): W214–W220. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ying J, Li H, Seng TJ, Langford C,
Srivastava G, Tsao SW, Putti T, Murray P, Chan AT and Tao Q:
Functional epigenetics identifies a protocadherin PCDH10 as a
candidate tumor suppressor for nasopharyngeal, esophageal and
multiple other carcinomas with frequent methylation. Oncogene.
25:1070–1080. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Naccarati A, Polakova V, Pardini B,
Vodickova L, Hemminki K, Kumar R and Vodicka P: Mutations and
polymorphisms in TP53 gene-an overview on the role in colorectal
cancer. Mutagenesis. 27:211–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Shin SJ, Liu D, Ivanova E,
Foerster F, Ying H, Zheng H, Xiao Y, Chen Z, Protopopov A, et al:
ZNF365 promotes stability of fragile sites and telomeres. Cancer
Discov. 3:798–811. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amre DK, Mack DR, Morgan K, Israel D,
Deslandres C, Seidman EG, Lambrette P, Costea I, Krupoves A, Fegury
H, et al: Susceptibility loci reported in genome-wide association
studies are associated with Crohn's disease in Canadian children.
Aliment Pharmacol Ther. 31:1186–1191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hirota T, Takahashi A, Kubo M, Tsunoda T,
Tomita K, Sakashita M, Yamada T, Fujieda S, Tanaka S, Doi S, et al:
Genome-wide association study identifies eight new susceptibility
loci for atopic dermatitis in the Japanese population. Nat Genet.
44:1222–1226. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Park E, Kim CS and Paik JH:
ZNF365 promotes stalled replication forks recovery to maintain
genome stability. Cell Cycle. 12:2817–2828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability-an evolving hallmark of cancer. Nat Rev Mol
Cell Biol. 11:220–228. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Zhang J, Tong JHM, Chan AWH, Yu J,
Kang W and To KF: Targeting the oncogenic p53 mutants in colorectal
cancer and other solid tumors. Int J Mol Sci. 20:59992019.
View Article : Google Scholar
|
|
30
|
Lane DP: Cancer. p53, guardian of the
genome. Nature. 358:15–16. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harris SL and Levine AJ: The p53 pathway:
Positive and negative feedback loops. Oncogene. 24:2899–2908. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shieh SY, Ikeda M, Taya Y and Prives C:
DNA damage-induced phosphorylation of p53 alleviates inhibition by
MDM2. Cell. 91:325–334. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soussi T and Beroud C: Assessing TP53
status in human tumours to evaluate clinical outcome. Nat Rev
Cancer. 1:233–240. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sigal A and Rotter V: Oncogenic mutations
of the p53 tumor suppressor: The demons of the guardian of the
genome. Cancer Res. 60:6788–6793. 2000.PubMed/NCBI
|
|
35
|
Liu YY, Patwardhan GA, Bhinge K, Gupta V,
Gu X and Jazwinski SM: Suppression of glucosylceramide synthase
restores p53-dependent apoptosis in mutant p53 cancer cells. Cancer
Res. 71:2276–2285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vedeld HM, Andresen K, Eilertsen IA,
Nesbakken A, Seruca R, Gladhaug IP, Thiis-Evensen E, Rognum TO,
Boberg KM and Lind GE: The novel colorectal cancer biomarkers CDO1,
ZSCAN18 and ZNF331 are frequently methylated across
gastrointestinal cancers. Int J Cancer. 136:844–853. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jones PA: DNA methylation and cancer.
Cancer Res. 46:461–466. 1986.PubMed/NCBI
|
|
38
|
Riggs AD and Jones PA: 5-methylcytosine,
gene regulation, and cancer. Adv Cancer Res. 40:1–30. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2 (Suppl 1):S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|