Introduction
Colorectal cancer (CRC) is the third most common
cancer in the world with >1.3 million cases diagnosed every
year, and the incidence of CRC worldwide is predicted to increase
to 2.5 million new cases a year in 2035 (1). Furthermore, CRC accounts for ~10% of
all annually diagnosed cancer types and cancer-related mortalities
worldwide (1,2). Several of the risk factors of CRC, such
as obesity, physical activity, smoking and alcohol use, easily
affect the metabolic environment of the host, leading to
alterations in the intestinal microbial community that may directly
or indirectly cause gut microbiota dysbiosis and trigger the
development of adenoma and CRC (3–6). It has
been reported that ~1014 bacteria live within the human
intestinal tract, which maintain a healthy gastrointestinal system
for regulating processes such as immune regulation, microbial
metabolism and host-derived chemical productions (5,7).
Compared with healthy controls, patients with CRC have an abnormal
gut microbiome structure (8). For
example, patients with CRC can be distinguished from healthy
individuals using specific microbial markers, including
Fusobacterium nucleatum (F. nucleatum), Peptostreptococcus
stomatis, Parvimonas micra (P. micra) and
Solobacterium moorei (2). It
has also been revealed that transplanting fecal bacteria from
patients with CRC into sterile mice results in the formation of
tumors (9). Therefore, these studies
suggest a causal relationship between the presence of specific
microorganisms and the development of cancer.
P. micra is a fastidious, anaerobic,
gram-positive coccus that is found in healthy human oral and
gastrointestinal flora (10).
Previous studies have reported that P. micra is involved in
lung abscesses, iliopsoas abscesses, gastric carcinogenesis and
infections of the periodontal area, soft tissue, bone and joints
(11–14). Currently, based on metagenomic or 16S
RNA sequencing analysis, numerous studies have revealed the
relationship between P. micra and CRC (15–17). By
analyzing the 16S rRNA gene sequence data of 509 fecal samples from
ethnically different cohorts, including those from China and
Austria, Yu et al (2)
observed that the detection rate and abundance of P. micra
were significantly higher in patients with CRC compared with
controls, and these results were further validated using
quantitative PCR (qPCR) in 309 subjects (18). By analyzing the metagenomics
sequencing results from 778 (including 386 samples from patients
with CRC and 392 controls) and 969 (meta-analysis of five publicly
available databases and two new cohorts with validation of the
findings of two additional cohorts) stool samples, two research
groups discovered that CRC-related microbial markers, including
P. micra, could be consistently detected among different
populations, regardless of the detection techniques, diet,
geographical environment, genetics and other factors (19,20).
These results demonstrate that P. micra has an important
relationship with CRC, and may be involved in the development of
CRC.
Most cancer types arise from adenoma, and colorectal
adenoma (CRA) is a critical precursor of CRC (21,22). The
process of CRC development begins with an aberrant crypt, which
evolves into a polyp or adenoma and eventually progresses to CRC
over an estimated 10–15 year period (1). Currently, the microbiota associated
with CRA have not been consistently identified, and the association
between P. micra and CRA remains elusive (23–25).
Therefore, the present study aimed to investigate the association
between P. micra and CRA by measuring the changes in the
relative abundance of P. micra in stool samples obtained
along the adenoma-carcinoma sequence using a qPCR method.
Furthermore, the alteration pattern of the relative abundance of
P. micra were evaluated in patients with CRC or CRA by
analyzing four public 16S rRNA datasets.
Patients and methods
Patient recruitment and sample
collection
An observational case-control study was conducted
between January 2017 and March 2019 at The First Affiliated
Hospital of Soochow University. Stool samples were collected prior
to colonoscopy. All patients with CRC (37 males and 29 females) and
CRA (66 males and 62 females) were first diagnosed via colonoscopy
screening, and the diagnosis was later confirmed by pathology. The
pathological diagnosis was performed by two professionals.
Inclusion criteria were as follows: i) Age ≥18 years old; and ii)
colonoscopy. The exclusion criteria for all participants included
the use of the following medicines: Antibiotics within 1 month of
study participation, non-steroidal anti-inflammatory drugs or
probiotics. Individuals who reported chronic bowel disorders, food
allergies or dietary restrictions were also excluded from the
study. Additional exclusion criteria for patients with CRC included
chemotherapy or radiation treatment prior to surgery. All patients
were categorized according to histopathological features on the
basis of the TNM classification of malignant tumors after surgery
(26). A total of 83 healthy
subjects (43 males and 40 females) were selected as controls by
volunteering during a physical examination, and none of the healthy
subjects had gastrointestinal tract disorders or any antibiotics
treatments in the 3 months before sample collection. The clinical
variables included age, sex and BMI (kg/m2). All 277
participants (age range, 26–88 years) had been local residents of
Suzhou city for >5 years prior to the study. In total, one fecal
sample was self-collected prior to bowel preparation the day before
colonoscopy from each patient or healthy subject. Samples were
transported to the laboratory within 24 h after collection.
All individuals provided written informed consent
prior to participating in the study. All procedures were performed
in accordance with, and were approved by, the ethical standards of
the institutional and/or the national research committee [the
Ethics Committee of The First Affiliated Hospital of Soochow
University; approval no. + 056 (2016)], and with the 1964 Helsinki
declaration and its later amendments or comparable ethical
standards.
Nucleic acid extraction and
storage
Stool samples were immediately frozen in liquid
nitrogen and stored at −80°C. DNA was extracted using a TIANamp
Stool DNA kit (Tiangen Biotech Co., Ltd.) according to the
manufacturer's protocols (27). The
integrity of DNA was measured via 2% (w/v) agarose gel
electrophoresis. Purified nucleic acids were quantified using a
Qubit 3.0 instrument (Thermo Fisher Scientific, Inc.), and stored
at −80°C. Nucleic acids were extracted from all stool samples in a
single batch by one operator to avoid inter-batch variation.
qPCR
All reactions were performed in a 96-well optical
PCR plate. Each reaction contained 40 ng extracted fecal DNA, 250
nM primers and 2X ChamQ Universal SYBR qPCR Master Mix (Vazyme
Biotech Co., Ltd.) in 20 µl reaction volume. Amplification and
detection of DNA was performed with the Applied Biosystems 7500
Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with the following reaction conditions: Initial
denaturation at 50°C for 2 min and 95°C for 2 min, followed by 40
cycles at 95°C for 15 sec and 60°C for 1 min. The primers sequences
(V3-V4) were as follows: P. micra forward,
5′-GTCACTACGGAAGAATTTGTC-3′ and reverse,
5′-GGCTTGAGCGATAATAACTTC-3′; and total bacterial DNA forward,
5′-GTGSTGCAYGGYTGTCGTCA-3′ and reverse, 5′-ACGTCRTCCMCACCTTCCTC-3′.
Each sample was assayed three times. Results were analyzed using
2−ΔΔCq method (28).
Meta-analysis of datasets from
publications
A systematic PubMed search with the terms 16S,
colorectal cancer or adenoma and gut microbiome was performed to
identify studies involving 16S rRNA sequencing of stool samples
from patients with CRC or CRA, and healthy controls. Available data
was only found in four studies: Zeller et al (29) (accession no. ERP005534); Zackular
et al (8) (http://www.mothur.org/MicrobiomeBiomarkerCRC); Baxter
et al (15) (accession no.
SRP062005); and Mori et al (30). All four datasets were obtained from
samples from patients with CRA or CRC, and healthy subjects as
controls (Table I).
 | Table I.Characteristics of the datasets
included in this study. |
Table I.
Characteristics of the datasets
included in this study.
| Named | Author | Country | Healthy | CRA | CRC | Region of 16S
rRNA | Seq platform |
|---|
| crc2 | Zeller et al
(29) | France | 50 | 38 | 41 | V4 | Illumina MiSeq |
| crc4 | Zackular et
al (8) | USA | 30 | 30 | 30 | V4 | Illumina MiSeq |
| crc45 | Baxter et al
(15) | USA+Canada | 87 | 147 | 79 | V4 | Illumina MiSeq |
| crc49 | Mori et al
(30) | Italy | 18 | 38 | 8 | V4 | Illumina MiSeq |
Bioinformatics and sequence
analysis
During data processing, short overlapping forward
and reverse reads from the same fragment were joined together using
PANDAseq (v0.21.1) to form overlapping sequences of the V3-V4 16S
region (31). After joining, the
resulting fragments were trimmed using Trimmomatic (v0.30)
(32). The average probability of a
base being called in error was <0.01, and the minimal length of
a fragment was 100 bp. Next, the chimeric sequences were removed
using Vsearch (v1.9.6) (33). The
samples were uniformly subsampled at a rarefaction level of 30,000
sequences per sample, to mitigate bias of the analyses due to
differences in sampling depth. Samples with <30,000 reads were
removed, and a collection of sequences suitable for further
Quantitative Insights Into Microbial Ecology (QIIME v1.9) analysis
was thus obtained (34). The
sequences were then clustered into operational taxonomic units
(OTUs) using a 99% similarity cutoff, and the relative abundances
were calculated for the OTUs in each sample. The OTUs were
classified using the assign_taxonomy.py script in QIIME using
UCLUST (v1.2.22) (35) as an
assignment method, and the Silva 99% OTU database, which was a
modified version of Silva v132 (36), with the removal of uncultured or
unclassified entries and the addition of extra entries from CORE
database (37).
Statistical analysis
All statistical analyses were conducted in R
software (version 2.15.3; R Foundation for Statistical Computing).
For the qPCR method, the abundance (A) of P. micra in a
sample was calculated as the ΔCq relative to the total bacterial
DNA in the sample, and the relative abundance was calculated as ln
(A × 109+1). For 16S rRNA sequencing data, the A of an
OTU in a sample was calculated as the ratio of the sequence count
of the OTU relative to the total number of sequences in the sample,
and the relative abundance of the OTU was determined as ln (A ×
106 +1) (38).
Differentially abundant OTUs were selected with the Wilcoxon
rank-sum test for the comparison of healthy subjects with the CRC
or CRA group (39). The
Benjamini-Hochberg procedure was used to calculate the false
discovery rate according to the adjustment of the P-values obtained
from the Wilcoxon rank-sum test.
Comparisons between groups were performed with
unpaired Student's t-tests and χ2 tests for quantitative
and categorical variables, respectively. Variables that followed a
Gaussian distribution were compared with one-way ANOVA. The
correlation between the quantity of the P. micra and the
characteristics of patients such as age, BMI, sex and tumor
progression were calculated using Kendall, Pearson or Mann-Whitney
analysis. As the majority of the datasets did not meet the
assumptions of a normal distribution, non-parametric Dunn's tests
with Kruskal-Wallis tests or the Mann-Whitney U test were used,
where applicable. P<0.05 was considered to indicate a
statistically significant difference. Meta-analysis was performed
using the meta for package (v2.4-0) (40). The DESeq2 package (v1.28.1) was used
to conduct the difference analysis on the OTUs of crc2, crc4, crc45
and crc49, and the log2 fold change of each OTU in each sample was
obtained (41). Receiver operating
characteristic (ROC) curves were drawn using the pROC package
(v1.16.2) (42). Other diagrams were
generated using the ggplot2 (v3.3.2) and ggpubr packages (v0.4.0)
(43,44).
Results
Evaluation of P. micra using qPCR
To investigate the associations between P.
micra and CRA and CRC, qPCR was performed to detect the
relative abundance of P. micra in the fecal samples of 277
subjects (including 83 healthy controls, 128 patients with CRA and
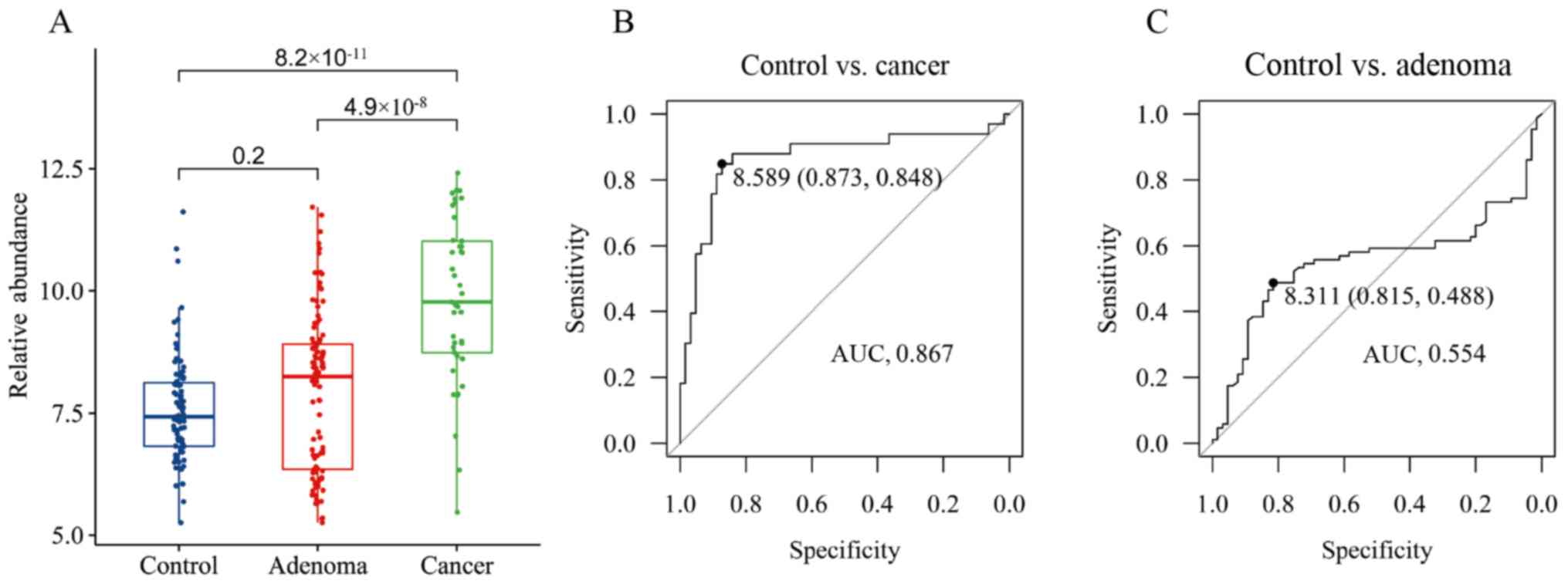
66 patients with CRC) recruited from Suzhou (Table II). The results demonstrated that
the relative abundance of P. micra in patients with CRC was
significantly higher compared with the healthy controls and CRA
(CRC vs. control, P=8.2×10−11; CRC vs. CRA,
P=4.9×10−8; Fig. 1A),
while the relative abundance in patients with CRA was not different
from the healthy controls (P=0.2; Fig.
1A). Then, 80% of the samples were used as the training set and
the rest as the test set to establish a prediction model, and it
was found that CRC samples could be distinguished from healthy
control samples with an area under the curve (AUC) of 0.867 and a
cutoff of 8.589 (Fig. 1B). The model
performed well for the test set with an FPR (false positive rate)
of 0.053 and an FNR (false negative rate) of 0.3 (Table III). However, the CRA samples were
poorly distinguished from the healthy control samples (AUC, 0.554
at a cutoff of 8.311) with an FPR of 0.105 and an FNR of 0.615
(Fig. 1C; Table III). These results suggested that
P. micra may serve as a diagnostic marker for CRC, but not
for CRA.
 | Table II.Demography of patients. |
Table II.
Demography of patients.
| Group | Healthy | CRA | CRC | P-value |
|---|
| Sample | Stool | Stool | Stool |
|
| Sex |
|
|
| 0.882 |
|
Male | 43 | 66 | 37 |
|
|
Female | 40 | 62 | 29 |
|
| Age, years | 55.2±3.7 | 56.8±10.9 | 59.2±10.4 | 0.196 |
| Height, cm | 165.6±7.8 | 163.8±8 | 164.2±8.7 | 0.443 |
| Weight, kg | 63.9±16.3 | 74.2±28 | 65.4±16.7 | 0.026 |
| BMI,
kg/m2 | 22.7±3.1 | 24.1±3.8 | 23.6±3 | 0.069 |
 | Table III.Diagnostic performance of
Parvimonas micra. |
Table III.
Diagnostic performance of
Parvimonas micra.
| Value | Group | CRC vs.
healthy | CRA vs.
healthy |
|---|
| Actual value |
|
| Healthy | 19 | 19 |
|
| CRC/CRA | 10 | 26 |
| Predicted
value | Healthy | 18 | 17 |
|
| CRC/CRA | 7 | 10 |
| False positive
rate |
| 0.3 | 0.615 |
| False negative
rate |
| 0.053 | 0.105 |
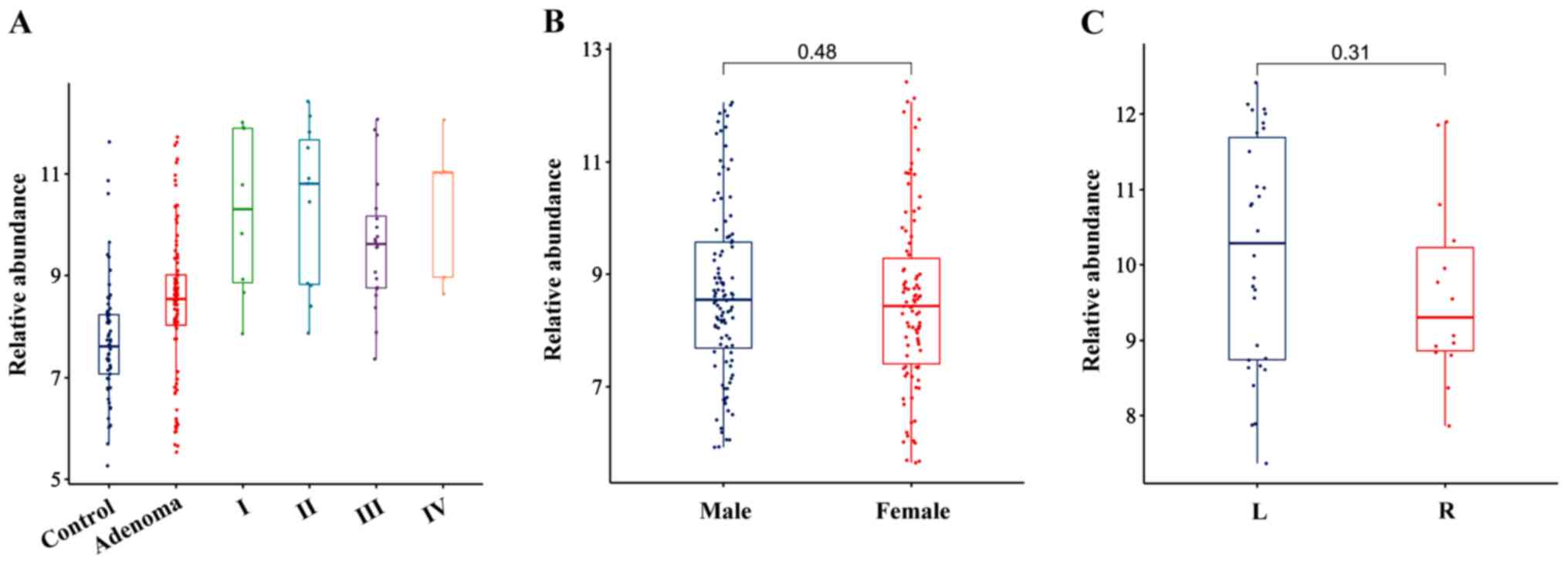
It was also identified that P. micra was
predominantly enriched in stages I/II and III/IV of CRC compared
with the healthy controls (Fig. 2A),
and the relative abundance of P. micra was not affected by
the sex, BMI or the site of cancer origin (right and left) of the
patients but was affected by age when a Pearson correlation was
used (Fig. 2B and C; Table IV).
 | Table IV.A correlation between the relative
abundance of Parvimonas micra and the characteristics of
patients. |
Table IV.
A correlation between the relative
abundance of Parvimonas micra and the characteristics of
patients.
| Factors | r | P-value | Method |
|---|
| Age, years | 0.2812 | 0.00006 | Pearson |
| BMI,
kg/m2 | −0.0319 | 0.6759 | Pearson |
| Sex |
| 0.4845 | Mann-Whitney |
| Tumor stage, I, II,
III, IV | −0.0720 | 0.5383 | Kendall |
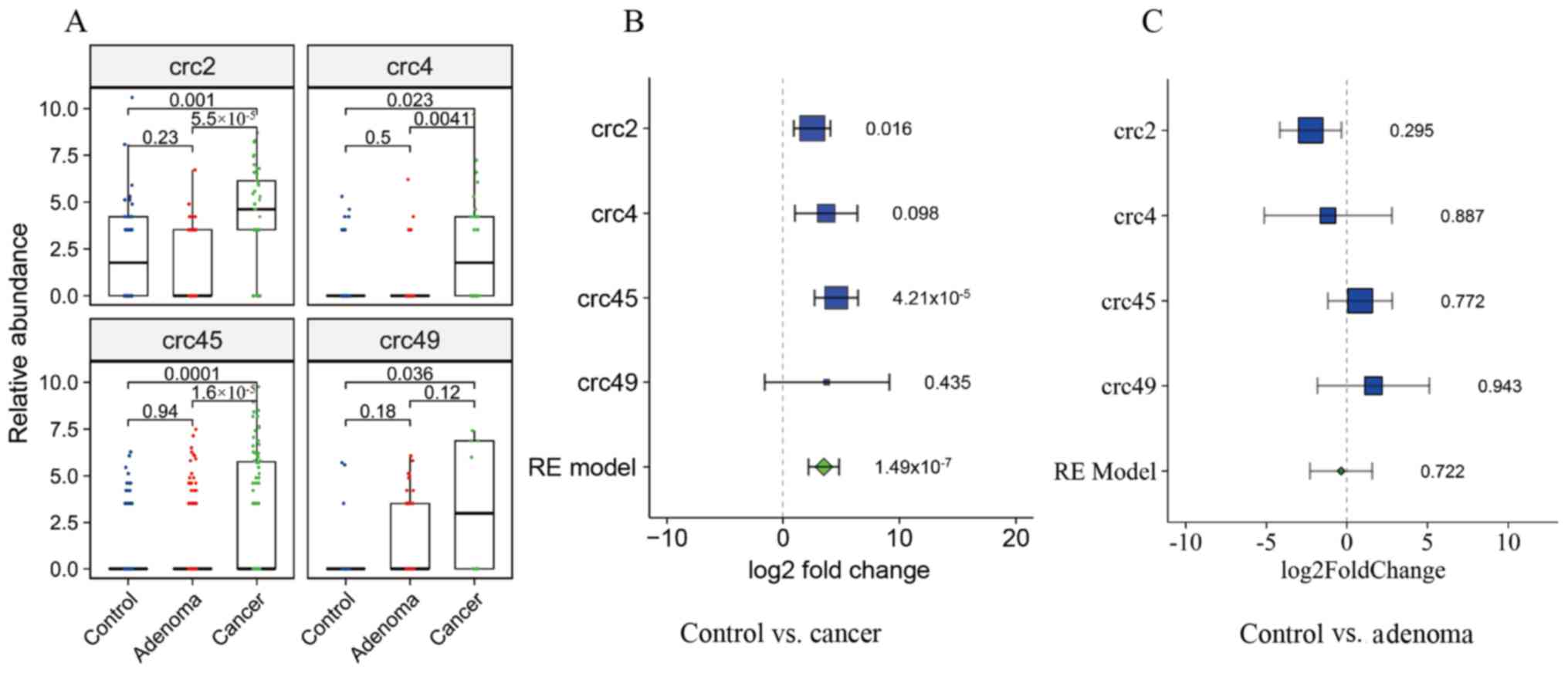
Meta-analysis of 16S rRNA sequencing
datasets
The association between P. micra and CRC was
analyzed in four public datasets. A total of 596 samples, including
158 CRC, 253 CRA and 185 healthy control samples, were included in
the analysis after quality filtering. Compared with the healthy
controls, the relative abundance of P. micra in patients
with CRC was significantly higher in all four datasets (crc2,
P=0.001; crc4, P=0.023; crc45, P=0.0001; crc49, P=0.036) but was
not different in patients with CRA (P=0.18-0.94) (Fig. 3A). Furthermore, there were
significant increases in the fold changes in the relative abundance
of P. micra in the CRC group compared with the healthy
control group in all four datasets, while there were few changes
between the healthy control and CRA groups (Fig. 3B and C).
Discussion
The present study used qPCR to measure the relative
abundance of P. micra in healthy individuals, patients with
CRA and patients with CRC, and demonstrated that the relative
abundance of P. micra was similar in the healthy and CRA
groups, but significantly increased in the CRC group. The fecal
level of P. micra could effectively distinguish patients
with CRC from healthy controls (AUC, 0.867) but could only poorly
distinguish patients with CRA from healthy controls (AUC, 0.554).
The same alteration pattern in fecal P. micra abundance,
which was low in healthy controls and patients with CRA, but
elevated in patients with CRC, was identified in all four public
16S RNA sequencing datasets. These results suggested that P.
micra was closely associated with, and may serve as a
diagnostic marker of, CRC but not CRA. To the best of our
knowledge, the present study was the first to demonstrate the
association of P. micra with CRC but not CRA in a large
number of cases and using two different methods. The present study
had some limitations, such as the mechanism of P. micra in
CRC initiation and development was not clear and whether changes in
its abundance are influenced by a number of host extrinsic factors,
including diet medications and other lifestyle components, such as
exercise, smoking, and sleep cycles was not assessed. The
aforementioned points should be explored in future studies.
Early screening is essential for the prevention of
CRC and the survival of patients with CRC, as the 5-year survival
rate >90% if CRC is detected at an early stage but decreases to
10% if it is discovered at an advanced metastatic stage (24). Currently, the methods for CRC
screening are the fecal occult blood test (FOBT), fecal DNA test,
detection of tumor markers and colonoscopy. However, these methods
suffer from high costs, invasiveness and/or low sensitivity
(8,25). The FOBT is currently the standard
non-invasive screening test, which has limited sensitivity and
specificity for CRC and does not reliably detect precancerous
lesions (29). A previous study
indicated that the accuracy of fecal microbiota detection was
similar to that of the standard FOBT, and when both approaches were
combined, the sensitivity can be ≤45% while maintaining the
specificity of FOBT (29). In
addition, combining a fecal immunochemical test (FIT) with the
detection of diagnostic markers, such as F. nucleatum,
Peptostreptococcus anaerobius and P. micra, can
significantly increase the detection rate for CRC with a
sensitivity of 92.3% and a specificity of 93.0% (18). The combined test identifies >75%
of the CRC samples missed by the stand-alone FIT (18). Similarly, the present results
suggested that the fecal level of P. micra can effectively
distinguish patients with CRC, indicating that P. micra can
be used as a diagnostic marker for CRC screening.
To evaluate the role of the gut microbiota in CRC
initiation and development, researchers have proposed a number of
models (45–47), including the ‘driver-passenger’
model, first suggested by Tjalsma et al (48). In the ‘driver-passenger’ model, the
‘drivers’ are defined as microbial species that increase in
abundance in the early stage of CRC, such as adenoma, while the
‘passengers’ are defined as those species that increase in
abundance in the late stage of CRC (46). Drivers are the primary pathogens that
cause the initiation of tumors, and passengers are more suited to
survive in the gut microenvironment resulting from tumorigenesis
(46). An example of a passenger is
F. nucleatum, which is enriched in CRC but not in CRA cases
(49). The present study identified
a significant elevation of P. micra in CRC but not in CRA
cases. Consistent with these findings, it has been shown that P.
micra is predominantly enriched in stages I/II and III/IV, and
its abundance is decreased after tumor resection, indicating that
P. micra is not the cause of carcinogenesis but is adapted
to the CRC microenvironment (50,51).
Therefore, P. micra may be a passenger in the
driver-passenger model.
P. micra is a component of the healthy
commensal flora of the gastrointestinal tract, and an opportunistic
pathogen (10). As types of
periodontal bacteria, P. micra and F. nucleatum have
synergistic effects on biofilm formation, which is important for
the colonization by these two species of apical periodontitis
lesions (52). P. micra
significantly enhances the activity of gingipains, which are
virulence factors in Porphyromonas gingiva that are
important in periodontal disease (53). P. micra may also promote
cancer development, although the exact mechanism it yet to be fully
elucidated. Moreover, P. micra may contribute to the
pathogenesis of periodontitis by stimulating Toll-like receptor 4,
nucleotide binding oligomerization domain containing (NOD)1 and
NOD2 (54). It has also been
reported that P. micra may be involved in gut bacterial
translocation and the upregulation of interleukins in the tumor
microenvironment (55). A previous
study demonstrated that APC Min/+ mice gavaged
with P. micra exhibited a significantly higher tumor burden
and tumor load, and cell proliferation was significantly higher in
the colon tissues of P. micra gavaged germ-free mice
compared with control mice (56).
Furthermore, the tumor promoting effect of P. micra has been
reported to be associated with altered immune responses and
increased inflammation in the gut (50,56).
These findings indicate that P. micra is primarily adapted
to the CRC microenvironment and could contribute to a pro-tumoral
inflammatory environment in patients susceptible to developing
CRC.
In conclusion, the present study identified that
P. micra was associated with CRC and may serve as a
diagnostic marker for CRC. In addition, P. micra was not
enriched in patients with CRA, suggesting that it serves a limited
role in the tumorigenesis of CRA.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81672372).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX, MY and SY analyzed and interpreted the patient
data from patients with CRC. DW, YZ and WC wrote the manuscript.
JX, MY and DW participated in the experimental study and data
analysis. JX, MY and SZ participated in data collection and
statistical analysis. JX, YZ and WC conceived the idea of, and
designed the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All individuals provided written informed consent
prior to participating in the study. All procedures were performed
in accordance with and were approved by the ethical standards of
the institutional and/or the national research committee [the
Ethics Committee of The First Affiliated Hospital of Soochow
University; approval no. + 056 (2016)], and with the 1964 Helsinki
declaration and its later amendments or comparable ethical
standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu J, Feng Q, Wong SH, Zhang D, Liang QY,
Qin Y, Tang L, Zhao H, Stenvang J, Li Y, et al: Metagenomic
analysis of faecal microbiome as a tool towards targeted
non-invasive biomarkers for colorectal cancer. Gut. 66:70–78. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis P, Hold GL and Flint HJ: The gut
microbiota, bacterial metabolites and colorectal cancer. Nat Rev
Microbiol. 12:661–672. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niederreiter L, Adolph TE and Tilg H:
Food, microbiome and colorectal cancer. Dig Liver Dis. 50:647–652.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saus E, Iraola-Guzmán S, Willis JR,
Brunet-Vega A and Gabaldón T: Microbiome and colorectal cancer:
Roles in carcinogenesis and clinical potential. Mol Aspects Med.
69:93–106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Almeida CV, de Camargo MR, Russo E and
Amedei A: Role of diet and gut microbiota on colorectal cancer
immunomodulation. World J Gastroenterol. 25:151–162. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gagnière J, Raisch J, Veziant J, Barnich
N, Bonnet R, Buc E, Bringer MA, Pezet D and Bonnet M: Gut
microbiota imbalance and colorectal cancer. World J Gastroenterol.
22:501–518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zackular JP, Rogers MA, Ruffin MT IV and
Schloss PD: The human gut microbiome as a screening tool for
colorectal cancer. Cancer Prev Res (Phila). 7:1112–1121. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wong SH, Zhao L, Zhang X, Nakatsu G, Han
J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wu WKK, et al: Gavage of fecal
samples from patients with colorectal cancer promotes intestinal
carcinogenesis in germ-free and conventional mice.
Gastroenterology. 153:1621–1633.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baghban A and Gupta S: Parvimonas
micra: A rare cause of native joint septic arthritis. Anaerobe.
39:26–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khan MS, Ishaq M, Hinson M, Potugari B and
Rehman AU: Parvimonas micra bacteremia in a patient with
colonic carcinoma. Caspian J Intern Med. 10:472–475.
2019.PubMed/NCBI
|
|
12
|
Yun SS, Cho HS, Heo M, Jeong JH, Lee HR,
Ju S, Kim JY, You JW, Cho YJ, Jeong YY, et al: Lung abscess by
actinomyces odontolyticus and Parvimonas micra co-infection
presenting as acute respiratory failure: A case report. Medicine
(Baltimore). 98:e169112019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Coker OO, Dai Z, Nie Y, Zhao G, Cao L,
Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY and Yu J: Mucosal
microbiome dysbiosis in gastric carcinogenesis. Gut. 67:1024–1032.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sawai T, Koga S, Ide S, Yoshioka S, Matsuo
N and Mukae H: An iliopsoas abscess caused by Parvimonas
micra: A case report. J Med Case Rep. 13:472019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baxter NT, Koumpouras CC, Rogers MA,
Ruffin MT IV and Schloss PD: DNA from fecal immunochemical test can
replace stool for detection of colonic lesions using a
microbiota-based model. Microbiome. 4:592016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Yu X, Yu E, Wang N, Cai Q, Shuai
Q, Yan F, Jiang L, Wang H, Liu J, et al: Changes in gut microbiota
and plasma inflammatory factors across the stages of colorectal
tumorigenesis: A case-control study. BMC Microbiol. 18:922018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baxter NT, Ruffin MT IV, Rogers MA and
Schloss PD: Microbiota-based model improves the sensitivity of
fecal immunochemical test for detecting colonic lesions. Genome
Med. 8:372016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai
RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, et al:
Quantitation of faecal Fusobacterium improves faecal
immunochemical test in detecting advanced colorectal neoplasia.
Gut. 66:1441–1448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wirbel J, Pyl PT, Kartal E, Zych K,
Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R,
et al: Meta-analysis of fecal metagenomes reveals global microbial
signatures that are specific for colorectal cancer. Nat Med.
25:679–689. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thomas AM, Manghi P, Asnicar F, Pasolli E,
Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, et
al: Metagenomic analysis of colorectal cancer datasets identifies
cross-cohort microbial diagnostic signatures and a link with
choline degradation. Nat Med. 25:667–678. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Castellarin M, Warren RL, Freeman JD,
Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P,
Allen-Vercoe E, Moore RA and Holt RA: Fusobacterium
nucleatum infection is prevalent in human colorectal carcinoma.
Genome Res. 22:299–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valcz G, Sipos F, Krenács T, Molnár J,
Patai AV, Leiszter K, Tóth K, Solymosi N, Galamb O, Molnár B and
Tulassay Z: Elevated osteopontin expression and
proliferative/apoptotic ratio in the colorectal
adenoma-dysplasia-carcinoma sequence. Pathol Oncol Res. 16:541–545.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sze MA and Schloss PD: Leveraging existing
16S rRNA gene surveys to identify reproducible biomarkers in
individuals with colorectal tumors. mBio. 9:e00630–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang B, Xu S, Xu W, Chen Q, Chen Z, Yan
C, Fan Y, Zhang H, Liu Q, Yang J, et al: Leveraging fecal bacterial
survey data to predict colorectal tumors. Front Genet. 10:4472019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shah MS, DeSantis TZ, Weinmaier T,
McMurdie PJ, Cope JL, Altrichter A, Yamal JM and Hollister EB:
Leveraging sequence-based faecal microbial community survey data to
identify a composite biomarker for colorectal cancer. Gut.
67:882–891. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Russo E, Bacci G, Chiellini C, Fagorzi C,
Niccolai E, Taddei A, Ricci F, Ringressi MN, Borrelli R, Melli F,
et al: Preliminary comparison of oral and intestinal human
microbiota in patients with colorectal cancer: A pilot study. Front
Microbiol. 8:26992018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sheng Q, Du H, Cheng X, Cheng X, Tang Y,
Pan L, Wang Q and Lin J: Characteristics of fecal gut microbiota in
patients with colorectal cancer at different stages and different
sites. Oncol Lett. 18:4834–4844. 2019.PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeller G, Tap J, Voigt AY, Sunagawa S,
Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, et
al: Potential of fecal microbiota for early-stage detection of
colorectal cancer. Mol Syst Biol. 10:7662014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mori G, Rampelli S, Orena BS, Rengucci C,
De Maio G, Barbieri G, Passardi A, Casadei Gardini A, Frassineti
GL, Gaiarsa S, et al: Shifts of faecal microbiota during sporadic
colorectal carcinogenesis. Sci Rep. 8:103292018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Masella AP, Bartram AK, Truszkowski JM,
Brown DG and Neufeld JD: PANDAseq: Paired-end assembler for
illumina sequences. BMC Bioinformatics. 13:312012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rognes T, Flouri T, Nichols B, Quince C
and Mahé F: VSEARCH: A versatile open source tool for metagenomics.
PeerJ. 4:e25842016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Edgar RC: Search and clustering orders of
magnitude faster than BLAST. Bioinformatics. 26:2460–2461. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Quast C, Pruesse E, Yilmaz P, Gerken J,
Schweer T, Yarza P, Peplies J and Glöckner FO: The SILVA ribosomal
RNA gene database project: Improved data processing and web-based
tools. Nucleic Acids Res. 41((Database Issue)): D590–D596.
2013.PubMed/NCBI
|
|
37
|
Griffen AL, Beall CJ, Firestone ND, Gross
EL, Difranco JM, Hardman JH, Vriesendorp B, Faust RA, Janies DA and
Leys EJ: CORE: A phylogenetically-curated 16S rDNA database of the
core oral microbiome. PLoS One. 6:e190512011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liang JQ, Li T, Nakatsu G, Chen YX, Yau
TO, Chu E, Wong S, Szeto CH, Ng SC, Chan FKL, et al: A novel faecal
lachnoclostridium marker for the non-invasive diagnosis of
colorectal adenoma and cancer. Gut. 69:1248–1257. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Paulson JN, Stine OC, Bravo HC and Pop M:
Differential abundance analysis for microbial marker-gene surveys.
Nat Methods. 10:1200–1202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Viechtbauer W: Conducting meta-analyses in
R with the metafor package. J Stat Softw. 36:1–48. 2010. View Article : Google Scholar
|
|
41
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wickham H: ggplot2: Elegant Graphics for
Data Analysis. Springer-Verlag. (New York, NY). 2016.
|
|
44
|
Alboukadel K: ggpubr: ‘ggplot2’ based
publication ready plots. R package version 0.4.0. 2018.
|
|
45
|
Sears CL and Pardoll DM: Perspective:
Alpha-bugs, their microbial partners, and the link to colon cancer.
J Infect Dis. 203:306–311. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Vogelstein B and Kinzler KW: The multistep
nature of cancer. Trends Genet. 9:138–141. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tjalsma H, Boleij A, Marchesi JR and
Dutilh BE: A bacterial driver-passenger model for colorectal
cancer: Beyond the usual suspects. Nat Rev Microbiol. 10:575–582.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Amitay EL, Werner S, Vital M, Pieper DH,
Höfler D, Gierse IJ, Butt J, Balavarca Y, Cuk K and Brenner H:
Fusobacterium and colorectal cancer: Causal factor or passenger?
Results from a large colorectal cancer screening study.
Carcinogenesis. 38:781–788. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mizutani S, Yamada T and Yachida S:
Significance of the gut microbiome in multistep colorectal
carcinogenesis. Cancer Sci. 111:766–773. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yachida S, Mizutani S, Shiroma H, Shiba S,
Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M,
et al: Metagenomic and metabolomic analyses reveal distinct
stage-specific phenotypes of the gut microbiota in colorectal
cancer. Nat Med. 25:968–976. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Horiuchi A, Kokubu E, Warita T and
Ishihara K: Synergistic biofilm formation by Parvimonas
micra and Fusobacterium nucleatum. Anaerobe.
62:1021002020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Neilands J, Davies JR, Bikker FJ and
Svensäter G: Parvimonas micra stimulates expression of
gingipains from Porphyromonas gingivalis in multi-species
communities. Anaerobe. 55:54–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Marchesan J, Jiao Y, Schaff RA, Hao J,
Morelli T, Kinney JS, Gerow E, Sheridan R, Rodrigues V, Paster BJ,
et al: TLR4, NOD1 and NOD2 mediate immune recognition of putative
newly identified periodontal pathogens. Mol Oral Microbiol.
31:243–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ge W, Hu H, Cai W, Xu J, Hu W, Weng X, Qin
X, Huang Y, Han W, Hu Y, et al: High-risk stage III colon cancer
patients identified by a novel five-gene mutational signature are
characterized by upregulation of IL-23A and gut bacterial
translocation of the tumor microenvironment. Int J Cancer.
146:2027–2035. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yu J, Zhao LY, Zhao RS, Long XH, Coker O
and Sung JJY: The role of Parvimonas micra in intestinal
tumorigenesis in germ-free and conventional APCmin/+
mice. J Clin Oncol. 37 (Suppl 4):S5312019. View Article : Google Scholar
|