Introduction
Inflammatory bowel disease (IBD), including
ulcerative colitis and Crohn's disease (CD), is characterized by
long-term relapsing inflammatory conditions in the gut (1). CD is associated with an increased risk
of general cancer, such as colorectal cancer (2). Recent evidence suggests that the
inflammation in CD, as a comorbidity, may contribute to the
initiation of breast cancer (2). It
has been agreed that CD is an important risk factor for the
development of breast cancer (3).
However, research on the impact of CD on the development of breast
cancer is limited, particularly regarding the underlying molecular
mechanisms.
A current study suggests that certain common genetic
factors may exist that account for the association between Crohn's
disease and breast cancer (4).
Hence, the molecular mechanisms shared between CD and breast cancer
should be investigated. To do this, the present study compared the
differentially expressed genes (DEGs) in the peripheral blood cells
of different cohorts, namely patients with CD vs. healthy controls,
and patients with breast cancer vs. healthy controls. The
overlapping genes and pathways were further studied by using
bioinformatics tools, such as the Kyoto Encyclopedia of Genes and
Genomes (KEGG) database. The present study aimed to analyze gene
expression profiles and related pathways in the peripheral blood
cells of patients with CD and breast cancer. Understanding the
aberrant expression of the genes involved in these pathways may
contribute to developing treatment for breast cancer in patients
with CD.
Materials and methods
Datasets selection and
preprocessing
To search for gene signatures shared by CD and
breast cancer in the peripheral blood of women, the terms ‘PBMC and
Crohn's disease’ or ‘breast cancer’ were used to search for
datasets in Gene Expression Omnibus (5) and Array Express (https://www.ebi.ac.uk/arrayexpress). The dataset
selection criteria were as follows: i) Preference for microarray
datasets; ii) samples using peripheral blood mononuclear cells
(PBMCs); iii) the number of samples was >15 and iv) processed
data were eligible. According to these selection criteria, only two
datasets (GSE3365 and GSE27562) (6,7) were
identified. GSE3365 contained samples from 42 healthy controls and
59 patients with CD. The GSE27562 blood samples were collected from
57 women with breast cancer and 31 healthy women. Normalized
datasets were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds). Datasets
GSE3365 (Crohn's disease) and GSE27562 (breast cancer) were
annotated according to the Affymetrix Human Genome U133A
(https://www.ncbi.nlm.nih.gov/
geo/query/acc.cgi?acc=GPL96) and U133 Plus 2.0 Array platforms
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL570) respectively. Probes were transformed to gene
symbols. If the same gene symbol was mapped by multiple probes,
their maximal value was regarded as the gene expression value. For
the GSE3365 dataset, log conversion and quantile normalization were
applied. Normalized datasets were subjected to principal component
analysis (PCA), an orthogonal linear transformation can convert the
related variables into irrelative variables (8). Samples that were clustered and/or not
separated were discarded.
DEGs analysis
The limma package (version 3.31.22) supplied by
Bioconductor was applied to analyze DEGs in CD or breast cancer
PBMCs compared with normal samples. The list of the DEGs (in the
form of a table) was obtained when the limma top table function was
used. In this table, DEGs were identified according to the
following criteria: i) ‘|Fold-change (FC)|> mean of logFC of
results table + 2× standard derivation of logFC of results table’
and ii) ‘P<0.05’. To obtain overlapping DEGs between the two
individual comparisons (CD vs. healthy controls and breast cancer
vs. healthy controls), a Venn diagram was constructed to identify
the overlapping DEGs in these groups. Then the overlapping DEGs
were used in the following analyses.
Pathway enrichment analysis
To determine the function of identified overlapping
DEGs, ClusterProfiler (http://bioconductor.org/packages/release/bioc/html/
clusterProfiler.html) with a series of strict cut-off (a
variable value that can distinguish positive from negative results)
was used to perform Gene Ontology (GO) and KEGG pathway analysis
(9). GO analysis includes three
categories: Biological process (BP); molecular function (MF) and
cellular component (CC). For all GO term annotation and KEGG
pathway enrichment analysis, false discovery rate <0.05 was
selected as the level of significant difference. Gene abbreviations
are defined in Table SI.
Gene interaction network (GI)
analysis
The overlapping DEGs were selected after pathway
enrichment analysis. Then these DEGs were submitted to the Search
Tool for the Retrieval of Interacting Genes (STRING) database
(http://www.string-db.org/) to construct a
protein-protein interaction network. With a combined score >0.4,
a chart indicating the interactions of DEGs was exported for
downstream analysis using Cytoscape software (version 3.4.0)
(10). Significant modules were
selected according to the default selection criteria (namely,
Molecular Complex Detection scores >2). Next, the Cytoscape
plugin cytoHubba was used to define hub genes of the DEGs
interaction network with a degree topological analysis.
Pathway analysis
The ‘pathview’ package in Bioconductor was used to
display the results, which included expression profiles of the
corresponding genes (11). This tool
links gene expression, gene regulations and related functions to
pathways based on the KEGG analyses. Gene abbreviations are defined
in Tables SII–IV.
Results
Overlapping DEGs
Using limma tools to analyze the GSE3365 and
GSE27562 datasets, DEGs between CD (vs. healthy controls) and
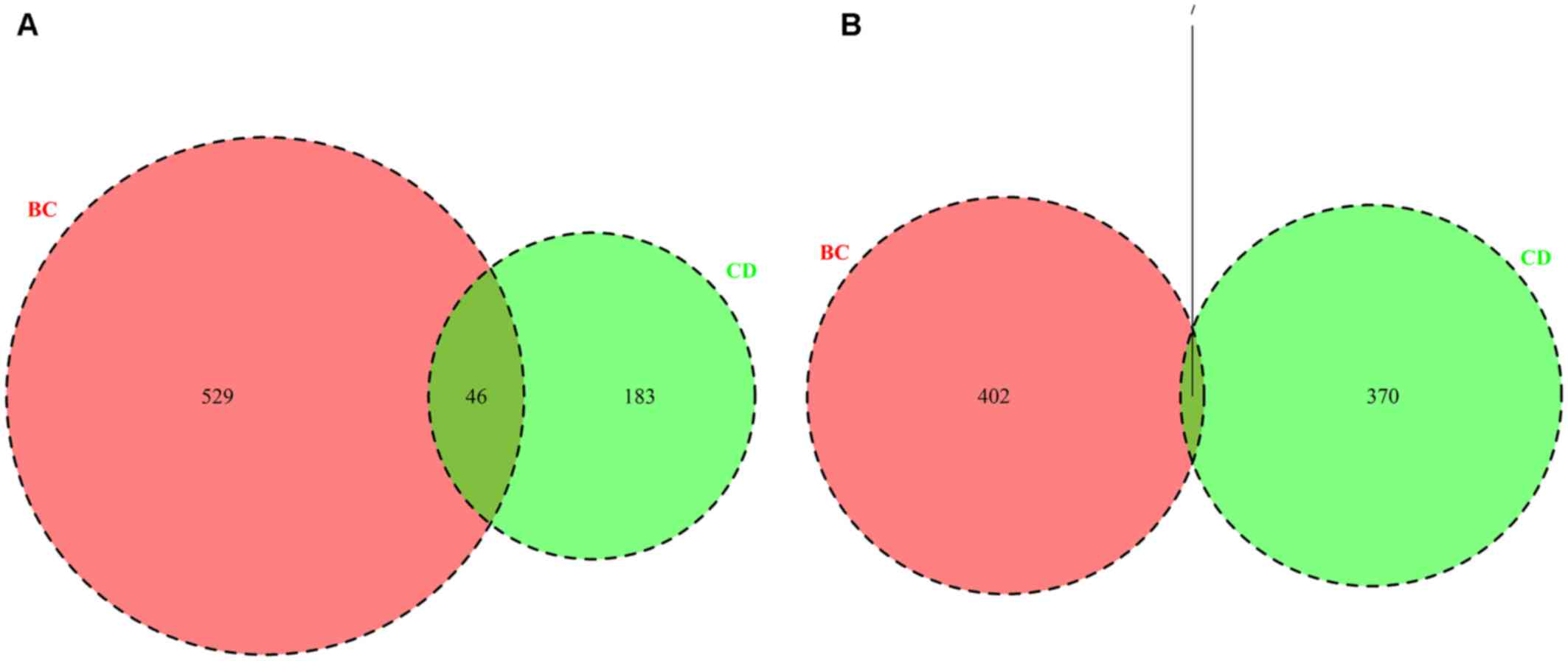
breast cancer (vs. healthy controls) were identified. In total, 606
DEGs, including 229 upregulated DEGs (Fig. 1A) and 377 downregulated DEGs
(Fig. 1B) were identified between
the CD vs. healthy controls (CD group), and 984 DEGs, including 575
upregulated DEGs (Fig. 1A) and 409
downregulated DEGs (Fig. 1B) were
identified in patients with breast cancer vs. healthy controls (BC
group). For the upregulated DEGs, there were 46 overlapping genes
between the BC and CD groups (Fig.
1A and Table I); whereas there
were seven overlapping downregulated genes (Fig. 1B and Table
I). The results indicated that there existed overlapping DEGs
between the BC and CD groups.
 | Table I.Genes commonly differentially
expressed between Crohn's disease and breast cancer. |
Table I.
Genes commonly differentially
expressed between Crohn's disease and breast cancer.
| Gene symbol | Gene name | Upregulated or
downregulated |
|---|
| AREG | Amphiregulin | Up |
| BAG4 | BCL2 associated
athanogene 4 | Up |
| BCL3 | BCL3 transcription
coactivator | Up |
| C5AR1 | Complement C5a
receptor 1 | Up |
| CCR1 | C-C motif chemokine
receptor 1 | Up |
| CCRL2 | C-C motif chemokine
receptor like 2 | Up |
| CD83 | Cluster of
differentiation 83 molecule | Up |
| CD9 | Cluster of
differentiation 9 molecule | Up |
| CXCL2 | C-X-C motif
chemokine ligand 2 | Up |
| CXCL8 | C-X-C motif
chemokine ligand 8 | Up |
| DNAJC3 | DnaJ heat shock
protein family (Hsp40) member C3 | Up |
| DSC2 | Desmocollin 2 | Up |
| DUSP5 | Dual specificity
phosphatase 5 | Up |
| EGR1 | Early growth
response 1 | Up |
| EGR2 | Early growth
response 2 | Up |
| EGR3 | Early growth
response 3 | Up |
| EPB41L3 | Erythrocyte
membrane protein band 4.1 like 3 | Up |
| FOSB | FosB
proto-oncogene, AP-1 transcription factor subunit | Up |
| FOSL2 | FOS like 2, AP-1
transcription factor subunit | Up |
| G0S2 | G0/G1 switch 2 | Up |
| GAB2 | GRB2 associated
binding protein 2 | Up |
| GABARAPL1 | GABA type A
receptor associated protein like 1 | Up |
| HBEGF | Heparin binding EGF
like growth factor | Up |
| IER3 | Immediate early
response 3 | Up |
| IL1B | Interleukin 1
beta | Up |
| MAFB | MAF bZIP
transcription factor B | Up |
| MARCKS | Myristoylated
alanine rich protein kinase C substrate | Up |
| NAMPT | Nicotinamide
phosphoribosyltransferase | Up |
| NFIL3 | Nuclear factor,
interleukin 3 regulated | Up |
| NR4A2 | Nuclear receptor
subfamily 4 group A member 2 | Up |
| OSM | Oncostatin M | Up |
| PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase
3 | Up |
| PLAUR | Plasminogen
activator, urokinase receptor | Up |
| PPP1R15A | Protein phosphatase
1 regulatory subunit 15A | Up |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2 | Up |
| PTX3 | Pentraxin 3 | Up |
| PVALB | Parvalbumin | Up |
| RAB20 | RAB20, member RAS
oncogene family | Up |
| RGS1 | Regulator of G
protein signaling 1 | Up |
| SAMSN1 | SAM domain, SH3
domain and nuclear localization signals 1 | Up |
| SGK1 |
Serum/glucocorticoid regulated kinase
1 | Up |
| STX11 | Syntaxin 11 | Up |
| TNFRSF21 | TNF receptor
superfamily member 21 | Up |
| TRIB1 | Tribbles
pseudokinase 1 | Up |
| HIST2H2BE | H2B clustered
histone 21? | Up |
| LOC100129518 | SOD2 overlapping
transcript 1, SOD2 | Up |
| ABHD17A | Abhydrolase domain
containing 17A | Down |
| IFT74 | Intraflagellar
transport 74 | Down |
| MPHOSPH8 | M-phase
phosphoprotein 8 | Down |
| RBM41 | RNA binding motif
protein 41 | Down |
| TIPRL | TOR signaling
pathway regulator | Down |
| NOTCH2NL | Notch 2 N-terminal
like A | Down |
| RP11-395B7.7 | Pre-mRNA processing
factor 31 | Down |
GO term annotation and KEGG pathway
enrichment analysis
Functional annotation and pathway analyses of
overlapping genes between the DEGs from the BC and CD groups were
performed using the ClusterProfiler tool. The upregulated
overlapping DEGs were primarily annotated with the GO terms ‘cell
chemotaxis’, ‘positive regulation of response to external stimulus’
and ‘response to lipopolysaccharide’ in terms of BP, and ‘cytokine
activity’, ‘receptor ligand activity’ and ‘receptor regulator
activity’ in terms of MF (Table
II). The downregulated overlapping DEGs were primarily
annotated with the GO terms under the category of MF, such as
‘palmitoyl-(protein) hydrolase activity’ (Table II). The upregulated overlapping DEGs
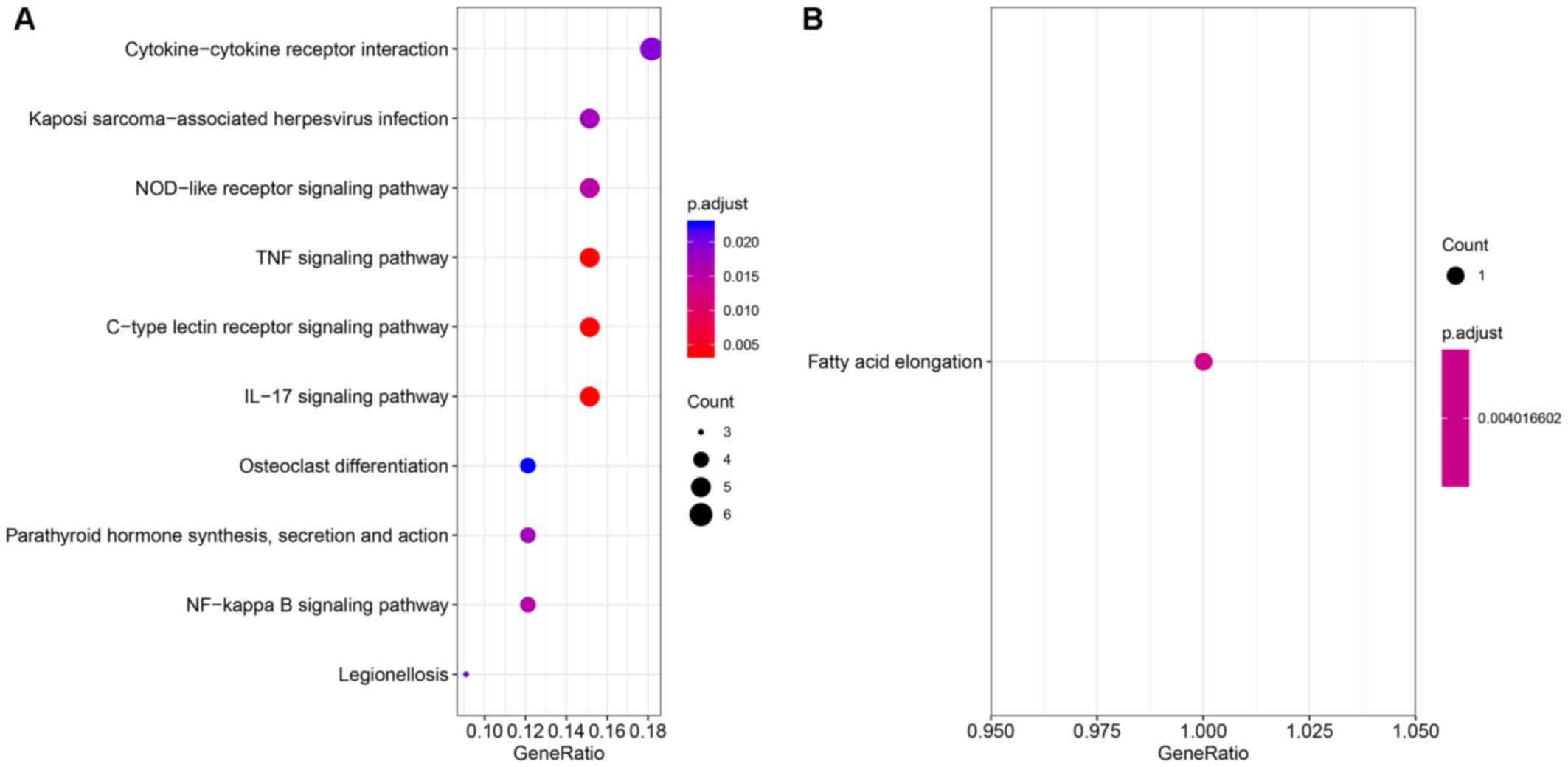
were primarily enriched in the following KEGG signaling pathways:
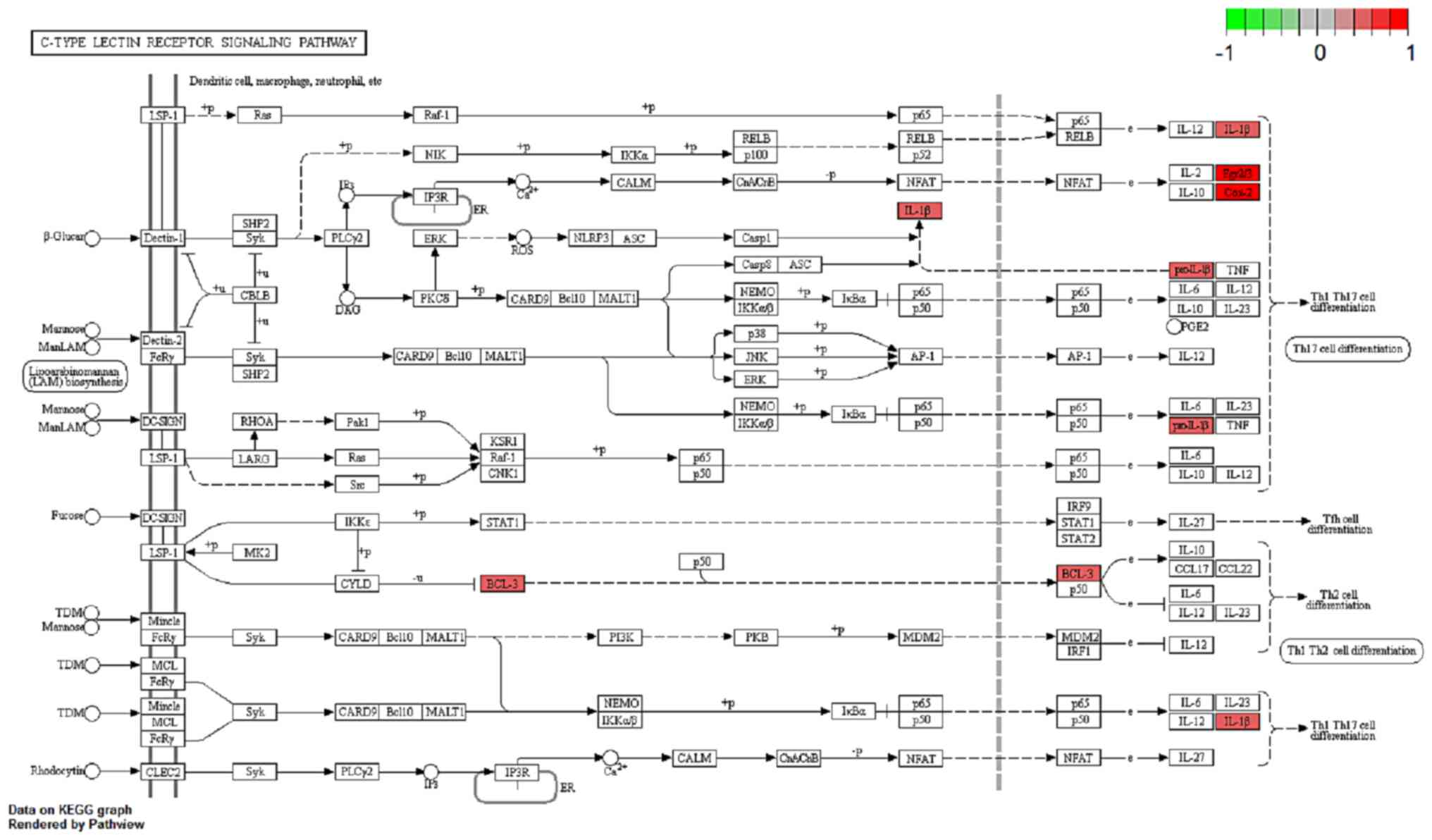
‘IL-17, ‘C-type lectin receptor, ‘tumor necrosis factor (TNF)α’,
‘NF-κB’ and ‘NOD-like receptor’ (Fig.
2A). The downregulated overlapping DEGs were primarily enriched
in the pathway of ‘fatty acid elongation’ (Fig. 2B). The gene functional enrichment
analysis of theses DEGs suggested that they were mainly involved in
inflammatory pathways.
 | Table II.Gene Ontology analysis terms for the
shared differentially expressed genes between the Crohn's disease
and breast cancer datasets. |
Table II.
Gene Ontology analysis terms for the
shared differentially expressed genes between the Crohn's disease
and breast cancer datasets.
| A, Biological
process |
|---|
|
|---|
| Term | Gene ID | Count | P-value |
|---|
| GO:0060326-cell
chemotaxis |
CXCL2/HBEGF/CXCL8/IL1B/EGR3/C5AR1/CCR1 | 7 |
5.53×10−06 |
| GO:0032103-positive
regulation of response to external stimulus |
CXCL2/CXCL8/IL1B/PTGS2/C5AR1/CCR1/OSM | 7 |
7.19×10−06 |
| GO:0032496-response
to lipopolysaccharide |
CXCL2/CXCL8/TRIB1/IL1B/TNFRSF21/PTGS2/C5AR1 | 7 |
1.54×10−05 |
|
|---|
| B, Molecular
function |
|
| Term | Gene ID | Count | P-value |
|
|---|
| GO:0005125-cytokine
activity |
CXCL2/CXCL8/IL1B/NAMPT/OSM/AREG | 6 | 2.09×10
−05 |
| GO:0048018-receptor
ligand activity |
CXCL2/HBEGF/CXCL8/IL1B/NAMPT/OSM/AREG | 7 |
1.58×10−4 |
| GO:0030545-receptor
regulator activity |
CXCL2/HBEGF/CXCL8/IL1B/NAMPT/OSM/AREG | 7 |
2.31×10−4 |
|
GO:0008474-palmitoyl-(protein) hydrolase
activity | ABHD17A | 1 |
3.453×10−3 |
|
GO:0098599-palmitoyl hydrolase
activity | ABHD17A | 1 |
3.453×10−3 |
|
GO:0016790-thiolester hydrolase
activity | ABHD17A | 1 |
1.2042×10−2 |
Hub genes and significant modules
identified from the GI network
Functional overlapping DEGs were submitted to the
STRING database. For overlapping DEGs between the CD vs. BC group,
network states showed 30 nodes and 67 edges. Based on this network,
cytoscape software was applied to construct the GI network
(Fig. 3A). Among the overlapping
DEGs, ten hub DEGs according to the node degree were identified
(Fig. 3B; Table III). Furthermore, significant
modules that met the default selection criteria were obtained
(Fig. 3C). For the intersecting DEGs
between the CD vs. BC groups, the CXCL8, IL1β and
prostaglandin G/H synthase 2 (PTGS2) genes were identified
in the module (Table III). Based
on the analysis of the hub gene and significant module, the
interactions between the DEGs were identified.
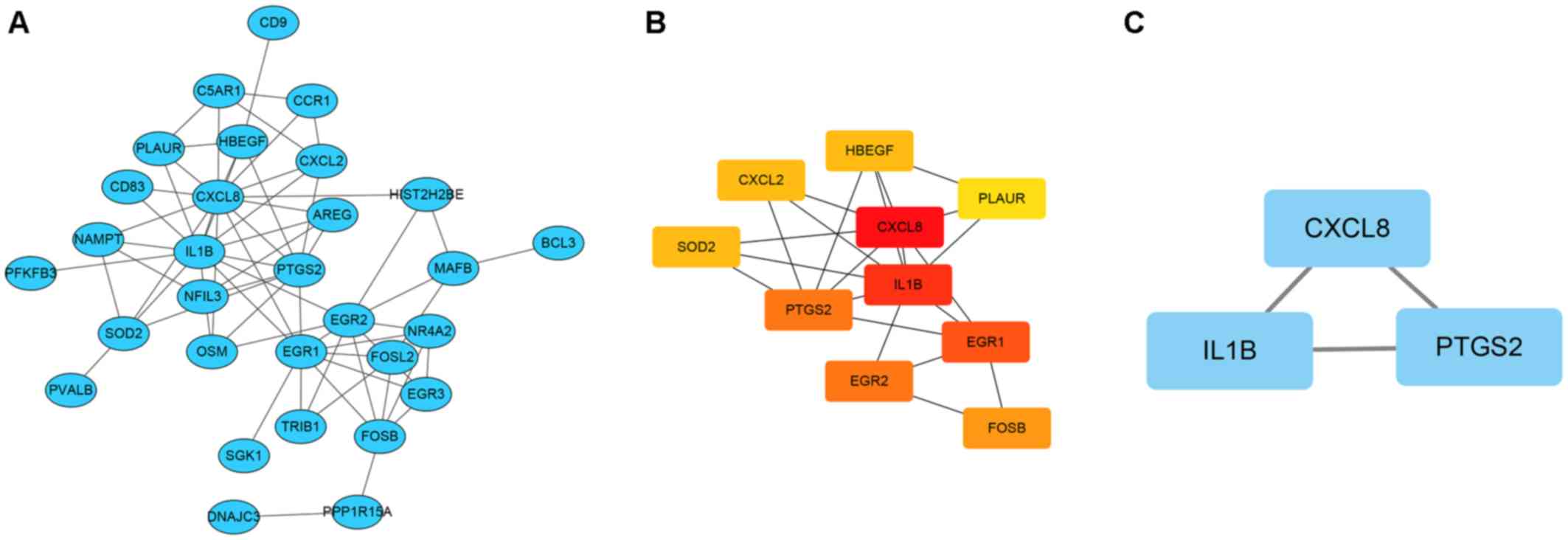 | Figure 3.Gene interaction network and the
significant module in Crohn's disease vs. breast cancer. (A) Gene
interaction network of the DEGs. (B) Hub gene network in the gene
interaction network. (C) The significant module of the gene
interaction network. All nodes represent upregulated genes. DNAJC3,
DnaJ heat shock protein family (Hsp40) member C3; CXCL1, C-X-C
motif chemokine ligand 1; CXCL2, C-X-C motif chemokine ligand 2;
PLAUR, plasminogen activator, urokinase receptor; SGK1,
serum/glucocorticoid regulated kinase 1; FOSL2, FOS-like 2; HBEGF,
heparin-binding EGF-like growth factor; NR4A2, nuclear receptor
subfamily 4 group A member 2; MAFB, MAF bZIP transcription factor
B; CXCL8, C-X-C motif chemokine ligand 8; PFKFB3,
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3; TRIB1,
tribbles pseudokinase 1; IL1B, interleukin 1 β; NFIL3, nuclear
factor, interleukin 3-regulated; EGR3, early growth response 3;
EGR1, early growth response 1; PTGS2, prostaglandin-endoperoxide
synthase 2; EGR2, early growth response 2; BCL3, BCL3 transcription
coactivator; HIST2H2BE, histone cluster 2 H2B family member e;
PPP1R15A, protein phosphatase 1 regulatory subunit 15A;
C5AR1,complement C5a receptor 1; CCR1, C-C motif chemokine receptor
1; CD9, cluster of differentiation 9; FOSB, FosB proto-oncogene,
AP-1 transcription factor subunit; NAMPT, nicotinamide
phosphoribosyltransferase; PVALB, parvalbumin; OSM, oncostatin M;
CD83, cluster of differentiation 83; AREG, amphiregulin; SOD2,
superoxide dismutase 2. |
 | Table III.Ten hub differentially expressed
genes shared between the Crohn's disease and breast cancer
datasets. |
Table III.
Ten hub differentially expressed
genes shared between the Crohn's disease and breast cancer
datasets.
| Gene | Gene name | Degree |
|---|
| CXCL8 | C-X-C motif
chemokine ligand 8 | 14 |
| IL1B | Interleukin 1
β | 13 |
| EGR1 | Early growth
response 1 | 10 |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2 | 9 |
| EGR2 | Early growth
response 2 | 9 |
| FOSB | FosB
proto-oncogene, AP-1 transcription factor subunit | 6 |
|
LOC100129518 | SOD2 overlapping
transcript 1, SOD2 | 5 |
| HBEGF | Heparin binding EGF
like growth factor | 5 |
| CXCL2 | C-X-C motif
chemokine ligand 2 | 5 |
| PLAUR | Plasminogen
activator, urokinase receptor | 4 |
Regulation of overlapping DEGs in
enriched pathways
Several studies have shown that IL-17, C-type lectin
receptor and NF-κB signaling pathways serve an important role in
both CD and breast cancer pathology (12–16).
Thus, the regulation and potential function of the overlapping DEGs
between the BC and CD groups were analyzed in these enriched
pathways in the BC group (Figs.
4–6). Indeed, similar pathway
patterns were also identified in the CD group (data not shown). In
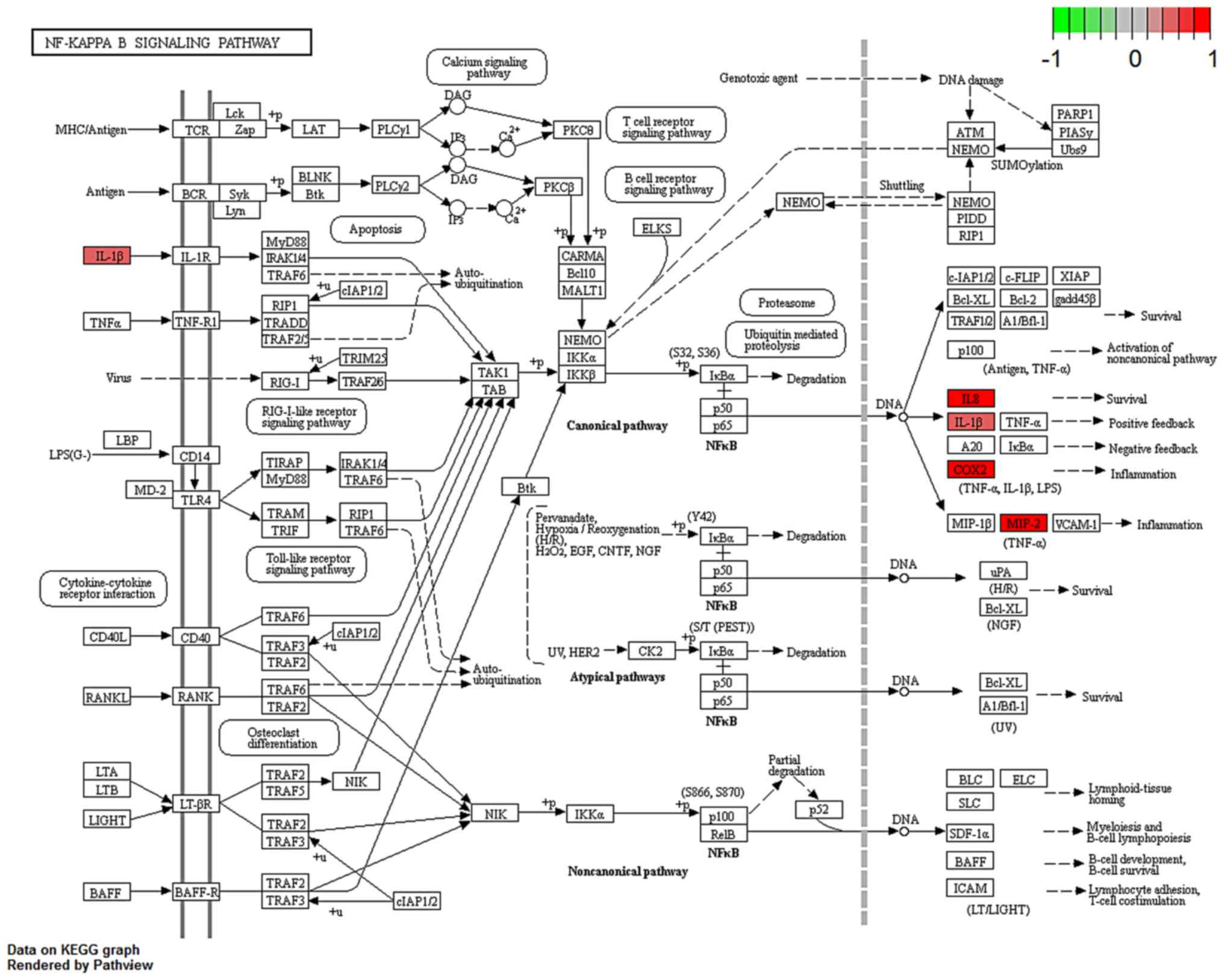
Fig. 4, a number of upstream genes
induced the degradation of inhibitor of NF-κB (IκB), thereby
allowing NF-κB translocation into the nucleus to activate
inflammatory gene expression in the NF-κB signaling pathway. The
inflammatory genes CXCL8 (encoding the IL-8 protein), IL1β and
PTGS2 were mainly involved in cell survival, positive/negative
feedback loops of inflammation processes in different types of
disease, such as colorectal cancer (17) and rheumatoid arthritis (18). In Fig.
5, the expression of the inflammatory genes IL1β, PTGS2 and E3
SUMO-protein ligase EGR2/3 were associated with T helper (Th)17
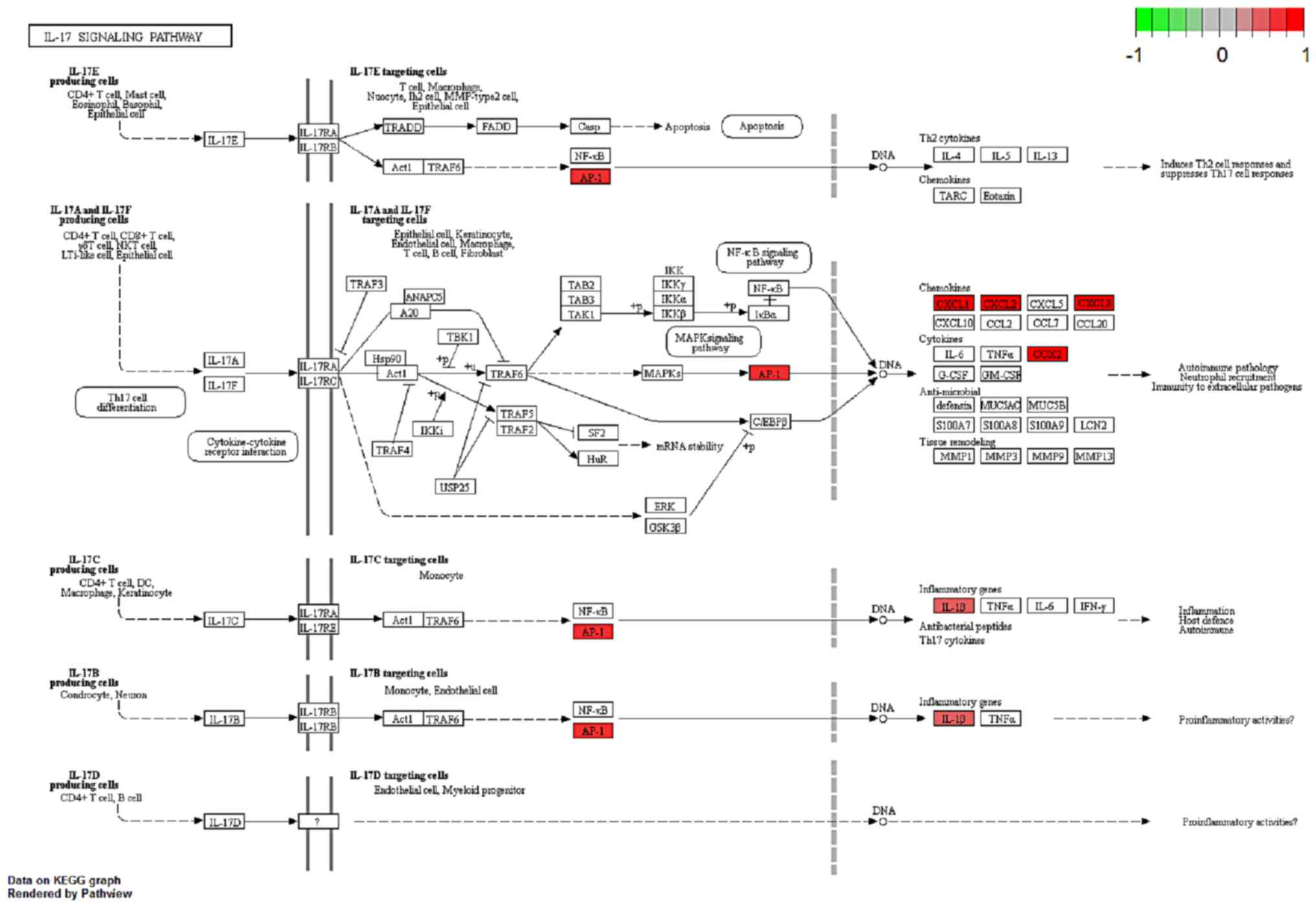
cell differentiation function. In Fig.
6, IL-17B/IL-17RB activated IL1β through AP1 complex
activation, which leads to inflammation. A comprehensive pathway
and network analysis of hub genes were identified.
Discussion
Patients with IBD have a greater risk of developing
cancer compared with patients without IBD (2). A previous study showed that patients
with CD have a high risk of developing colorectal cancer (19). Patients with CD also have a higher
risk of developing breast cancer than those without CD (2). Based on these observations, new
treatment strategies need to be developed to treat patients with
IBD and breast cancer. The molecular mechanisms underlying the
association between CD and breast cancer may provide important
information to inform such therapy. The present study identified 53
overlapping DEGs between the CD and BC group. A gene interaction
network based on the overlapping genes was constructed, which
demonstrated how the molecular mechanisms underpinning CD are
associated with breast cancer.
GO term annotation and KEGG pathway enrichment
analyses were then performed. The results showed that the
upregulated overlapping DEGs are annotated with ‘cell chemotaxis’.
This suggested that they may be mainly involved in inflammatory
pathways. Moreover, the KEGG analysis showed that the upregulated
overlapping DEGs in the CD vs. BC groups were enriched in the NF-κB
signaling pathway. These findings are consistent with the results
of previous studies. According to a previous study, the NF-κB
signaling pathway demonstrates unregulated activation and plays a
key role in the pathogenesis of Crohn's disease (20). Interestingly, in the peripheral blood
of patients with breast cancer, it was reported that certain genes
were enriched in the GO term ‘NF-κB’ and some upregulated genes
were involved in the canonical mitogen-activated protein kinase
signaling pathway, which is associated with NF-κB pathway signaling
(7). This consistency implies that
similar molecular mechanisms are shared between CD and breast
cancer pathogenesis.
The present study presented the transcriptional
regulation associations of the hub DEGs in the NF-κB and IL-17
signaling pathways in patients with CD and breast cancer. In the
colon of patients with CD, the predominance of Th1/Th17 cells lead
to the upregulation of multiple cytokines, including TNF-α, IFN-γ,
IL-1, IL-12 and IL-17. In mononuclear and epithelial cells of the
inflamed colon, inflammatory cytokines, such as TNF-α, may activate
the NF-κB transcription factor to induce expression of the general
inflammatory gene (21,22). Numerous blood Th17 cells infiltrate
the inflamed mucosa of patients with Crohn's Disease (23). When compared with controls, the
peripheral CD4+ T cells of patients with CD show increased
bacterial responses due to overexpression of Th17-associated genes,
including IL-17A, IL17F, nuclear receptor ROR-γ, C-C
chemokine receptor type 6 and C-C motif chemokine 20 (24). Moreover, the peripheral blood
monocytes of patients with CD express higher levels of IL1β, CCL2
and CCL5 (25). It is well known
that IL1β is a factor that promotes the expansion of Th17 cells
(26). Furthermore, when the
circulating Th17 cells of patients with CD reach breast tumor
tissue, Th17 cell activation may be exacerbated (26). Breast tumor cells can interact with
activated T cells via CD40-CD40L, by which TGFβ production and the
differentiation of Th17 cells increases (27). In the end, inflammatory blood Th17
cells, together with NF-κB signaling, may serve a role in linking
inflammation, immunity and breast oncogenesis (28). In fact, Th17 cells infiltrate breast
cancer tissues, including the inflammatory tumor microenvironment.
For example, IL-17 can upregulate the expression of CXCL1 on breast
cancer cells, which can activate the AKT/NF-κB signaling pathway to
enhance breast cancer growth and metastasis (29). Th17-associated inflammation may also
lead to the upregulation of IL17B, another member of the IL-17
cytokine family, and its receptor (IL17RB) in breast epithelial
malignant cells, which can induce the activation of the ERK1/2,
NF-κB and Bcl-2 pathways and enhance the inflammation and
development of breast cancer (30).
In addition, tumor-associated monocytes and macrophages can create
a cancer stem cell (CSC) niche through juxtacrine signaling,
including NF-κB (31). The present
hub DEGs in the gene interaction networks may participate in this
process. In the CSC niche, the NF-κB may translocate into the
nucleus and subsequently activate the expression of inflammatory
genes CXCL8, IL1β and PTGS2 (32,33).
Hub genes in significant modules overexpressed by
peripheral blood cells that are involved in the mentioned pathways
may contribute to development of breast cancer in CD. These
inflammatory mediators are CXCL8, IL1β and PTGS2. CXCL8, also known
as IL-8, may serve an important role in cancer progression by
regulating CSC proliferation and self-renewal (34). In addition, CXCL8 also has a role in
neovascularization, which contributes to tumor growth and
metastasis in the tumor microenvironment (35). IL-1β is another proinflammatory
mediator that can enhance tumor-promoting inflammation in breast
cancer (36). IL-1β can activate the
β-catenin signaling pathway to induce the epithelial-mesenchymal
transition (EMT) in breast cancer cells (37). IL-1β induces the migration of breast
cancer cells through the induction of hypoxia-inducible factor
(HIF)-1α (38). However, inhibition
of HIF-1α cannot prevent the breast cancer cell migration activated
by IL-1β under hypoxic conditions, in which NFκB/CXCL8 signaling
may play a compensatory role (39).
PTGS2, formerly known as cyclooxygenase 2 (COX-2), catalyzes the
production of inflammatory prostaglandins that exacerbate
inflammation (40). PTGS2-induced
PGE2 production promotes the migration and EMT of human breast
cancer cells (41). The inhibition
of the PTGS2 signaling pathway can inhibit the growth of
inflammatory breast cancer in vivo (42). PTGS2 plays a role in HIF1 activation
and promotes tumor angiogenesis in breast carcinoma (43). PTGS2 can also interact with other
crucial genes, such as IL8, IL1β and CXCL2 in tumor-associated
inflammation. Expression of IL8/CXCL8 increases in PTGS2
overexpressed NSCLC cell lines (44). Taken together, the results of the
present study demonstrated that a series of inflammatory proteins
(such as CXCL8, IL1β and PTGS2) overexpressed by peripheral blood
cells may increase systemic inflammation in female patients with CD
(45). Systemic inflammation can
affect tumorigenesis and progression in breast cancer by
establishing a tumor immune microenvironment (TIME) (46).
The limitations of the present study include the
existence of batch effect, which refers to non-biological variation
between measurements of different groups of samples, and the lack
of samples from patients with breast cancer with comorbid CD, thus
indicating that present data may be insufficient to highlight the
molecular basis underlying the genetic interaction between these
two diseases. To overcome these limitations, further studies should
use microarray datasets in which batch effects are removed, and
focus on investigating individuals who have both CD and breast
cancer.
In summary, a total of 53 overlapping DEGs were
identified between Crohn's disease and breast cancer. Functional
analyses of the upregulated overlapping DEGs showed that the two
diseases are associated with the IL-17 and NF-κB signaling
pathways. Gene interaction network and module analysis demonstrated
that the major hub genes were CXCL8, IL1β and PTGS2. The genes and
pathways identified in the present study may help inform our
understanding of the possible mechanisms underlying why Crohn's
disease can be a risk factor for developing breast cancer.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (NSFC; grant nos. 81970457,
91842302, 31470876 and 91629102).
Availability of data and materials
All datasets for this study (GSE3365 and GSE27562)
are included in the GEO program (https://www.ncbi.nlm.nih.gov/geo).
Authors' contributions
RY and JZ designed the study. JZ acquired, analyzed
and interpreted the data. RY and JZ wrote the paper. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Weimers P and Munkholm P: The natural
history of IBD: Lessons learned. Curr Treat Options Gastroenterol.
16:101–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hovde Ø, Høivik ML, Henriksen M, Solberg
IC, Småstuen MC and Moum BA: Malignancies in patients with
inflammatory bowel disease: Results from 20 Years of follow-up in
the IBSEN Study. J Crohns Colitis. 11:571–577. 2017.PubMed/NCBI
|
|
3
|
Pellino G, Sciaudone G, Patturelli M,
Candilio G, De Fatico GS, Landino I, Facchiano A, Vastarella A,
Canonico S, Riegler G and Selvaggi F: Relatives of Crohn's disease
patients and breast cancer: An overlooked condition. Int J Surg. 12
(Suppl 1):S156–S158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Riegler G, Caserta L, Castiglione F,
Esposito I, Valpiani D, Annese V, Zoli G, Gionchetti P, Viscido A,
Sturniolo GC, et al: Increased risk of breast cancer in
first-degree relatives of Crohn's disease patients. An IG-IBD
study. Dig Liver Dis. 38:18–23. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database Issue)):
D991–D995. 2013.PubMed/NCBI
|
|
6
|
Burczynski ME, Peterson RL, Twine NC,
Zuberek KA, Brodeur BJ, Casciotti L, Maganti V, Reddy PS, Strahs A,
Immermann F, et al: Molecular classification of Crohn's disease and
ulcerative colitis patients using transcriptional profiles in
peripheral blood mononuclear cells. J Mol Diagn. 8:51–61. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Labreche HG, Nevins JR and Huang E:
Integrating factor analysis and a transgenic mouse model to reveal
a peripheral blood predictor of breast tumors. BMC Med Genomics.
4:612011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lenz M, Müller FJ, Zenke M and Schuppert
A: Principal components analysis and the reported low intrinsic
dimensionality of gene expression microarray data. Sci Rep.
6:256962016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo W and Brouwer C: Pathview: An
R/Bioconductor package for pathway-based data integration and
visualization. Bioinformatics. 29:1830–1831. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Welte T and Zhang XHF: Interleukin-17
could promote breast cancer progression at several stages of the
disease. Mediators Inflamm. 2015:8043472015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wolfkamp SC, Verstege MI, Vogels EW,
Meisner S, Verseijden C, Stokkers PC and te Velde AA: Single
nucleotide polymorphisms in C-type lectin genes, clustered in the
IBD2 and IBD6 susceptibility loci, may play a role in the
pathogenesis of inflammatory bowel diseases. Eur J Gastroenterol
Hepatol. 24:965–970. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hohenberger M, Cardwell LA, Oussedik E and
Feldman SR: Interleukin-17 inhibition: Role in psoriasis and
inflammatory bowel disease. J Dermatolog Treat. 29:13–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tambuwala MM: Natural nuclear factor kappa
beta inhibitors: Safe therapeutic options for inflammatory bowel
disease. Inflamm Bowel Dis. 22:719–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang W, Nag SA and Zhang R: Targeting the
NFκB signaling pathways for breast cancer prevention and therapy.
Curr Med Chem. 22:264–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abdulamir AS, Hafidh RR and Bakar FA:
Molecular detection, quantification, and isolation of Streptococcus
gallolyticus bacteria colonizing colorectal tumors:
Inflammation-driven potential of carcinogenesis via IL-1, COX-2,
and IL-8. Mol Cancer. 9:2492010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee YA, Choi HM, Lee SH, Yang HI, Yoo MC,
Hong SJ and Kim KS: Synergy between adiponectin and interleukin-1β
on the expression of interleukin-6, interleukin-8, and
cyclooxygenase-2 in fibroblast-like synoviocytes. Exp Mol Med.
44:440–447. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kameyama H, Nagahashi M, Shimada Y, Tajima
Y, Ichikawa H, Nakano M, Sakata J, Kobayashi T, Narayanan S, Takabe
K and Wakai T: Genomic characterization of colitis-associated
colorectal cancer. World J Surg Oncol. 16:1212018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han YM, Koh J, Kim JW, Lee C, Koh SJ, Kim
B, Lee KL, Im JP and Kim JS: NF-kappa B activation correlates with
disease phenotype in Crohn's disease. PLoS One. 12:e01820712017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kmieć Z, Cyman M and Ślebioda TJ: Cells of
the innate and adaptive immunity and their interactions in
inflammatory bowel disease. Adv Med Sci. 62:1–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schreiber S, Nikolaus S and Hampe J:
Activation of nuclear factor κB in inflammatory bowel disease. Gut.
42:477–484. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gálvez J: Role of Th17 cells in the
pathogenesis of human IBD. ISRN Inflamm. 2014:9284612014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bassolas-Molina H, Raymond E, Labadia M,
Labadia M, Wahle J, Ferrer-Picón E, Panzenbeck M, Zheng J, Harcken
C, Hughes R, et al: An RORγ oral inhibitor modulates IL-17
responses in peripheral blood and intestinal Mucosa of Crohn's
disease patients. Front Immunol. 9:23072018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schwarzmaier D, Foell D, Weinhage T, Varga
G and Dabritz J: Peripheral monocyte functions and activation in
patients with quiescent Crohn's disease. PLoS One. 8:e627612013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brand S: Crohn's disease: Th1, Th17 or
Both? The change of a paradigm: New immunological and genetic
insights implicate Th17 cells in the pathogenesis of Crohn's
disease. Gut. 58:1152–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim H, Kim Y, Bae S, Kong JM, Choi J, Jang
M, Choi J, Hong JM, Hwang YI, Kang JS and Lee WJ: Direct
interaction of CD40 on tumor cells with CD40L on T cells increases
the proliferation of tumor cells by enhancing TGF-β production and
Th17 differentiation. PLoS One. 10:e01257422015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang J, Cai D, Ma B, Wu G and Wu J:
Skewing the balance of regulatory T-cells and T-helper 17 cells in
breast cancer patients. J Int Med Res. 39:691–701. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma K, Yang L, Shen R, Kong B, Chen W,
Liang J, Tang G and Zhang B: Th17 cells regulate the production of
CXCL1 in breast cancer. Int Immunopharmacol. 56:320–329. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alinejad V, Dolati S, Motallebnezhad M and
Yousefi M: The role of IL17B-IL17RB signaling pathway in breast
cancer. Biomed Pharmacother. 88:795–803. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu H, Clauser KR, Tam WL, Fröse J, Ye X,
Eaton EN, Reinhardt F, Donnenberg VS, Bhargava R, Carr SA and
Weinberg RA: A breast cancer stem cell niche supported by
juxtacrine signalling from monocytes and macrophages. Nat Cell
Biol. 16:1105–1117. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li B, Lu Y, Yu L, Han X, Wang H, Mao J,
Shen J, Wang B, Tang J, Li C and Song B: miR-221/222 promote cancer
stem-like cell properties and tumor growth of breast cancer via
targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chem Biol
Interact. 277:33–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nomura A, Gupta VK, Dauer P, Sharma NS,
Dudeja V, Merchant N, Saluja AK and Banerjee S: NFκB-mediated
invasiveness in CD133+ pancreatic TICs is regulated by
autocrine and paracrine activation of IL1 signaling. Mol Cancer
Res. 16:162–172. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ha H, Debnath B and Neamati N: Role of the
CXCL8-CXCR1/2 axis in cancer and inflammatory diseases.
Theranostics. 7:1543–1588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dinarello CA: An interleukin-1 signature
in breast cancer treated with interleukin-1 receptor blockade:
Implications for treating cytokine release syndrome of checkpoint
inhibitors. Cancer Res. 78:5200–5202. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Perez-Yepez EA, Ayala-Sumuano JT, Lezama R
and Meza I: A novel β-catenin signaling pathway activated by IL-1β
leads to the onset of epithelial-mesenchymal transition in breast
cancer cells. Cancer Lett. 354:164–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naldini A, Filippi I, Miglietta D,
Moschetta M, Giavazzi R and Carraro F: Interleukin-1β regulates the
migratory potential of MDAMB231 breast cancer cells through the
hypoxia-inducible factor-1α. Eur J Cancer. 46:3400–3408. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Filippi I, Carraro F and Naldini A:
Interleukin-1β affects MDAMB231 breast cancer cell migration under
hypoxia: Role of HIF-1α and NFκB transcription factors. Mediators
Inflamm. 2015:7894142015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
O'Connor PM, Lapointe TK, Beck PL and
Buret AG: Mechanisms by which inflammation may increase intestinal
cancer risk in inflammatory bowel disease. Inflamm Bowel Dis.
16:1411–1420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Reader J, Holt D and Fulton A:
Prostaglandin E2 EP receptors as therapeutic targets in breast
cancer. Cancer Metastasis Rev. 30:449–463. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang X, Reyes ME, Zhang D, Funakoshi Y,
Trape AP, Gong Y, Kogawa T, Eckhardt BL, Masuda H, Pirman DA Jr, et
al: EGFR signaling promotes inflammation and cancer stem-like
activity in inflammatory breast cancer. Oncotarget. 8:67904–67917.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Maroni P, Bendinelli P, Matteucci E and
Desiderio MA: The therapeutic effect of miR-125b is enhanced by the
prostaglandin endoperoxide synthase 2/cyclooxygenase 2 blockade and
hampers ETS1 in the context of the microenvironment of bone
metastasis. Cell Death Dis. 9:4722018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Põld M, Zhu LX, Sharma S, Burdick MD, Lin
Y, Lee PP, Põld A, Luo J, Krysan K, Dohadwala M, et al:
Cyclooxygenase-2-dependent expression of angiogenic CXC chemokines
ENA-78/CXC Ligand (CXCL) 5 and interleukin-8/CXCL8 in human
non-small cell lung cancer. Cancer Res. 64:1853–1860. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gross V, Andus T, Leser HG, Roth M and
Schölmerich J: Inflammatory mediators in chronic inflammatory bowel
diseases. Klin Wochenschr. 69:981–987. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Deshmukh SK, Srivastava SK, Poosarla T,
Dyess DL, Holliday NP, Singh AP and Singh S: Inflammation,
immunosuppressive microenvironment and breast cancer: Opportunities
for cancer prevention and therapy. Ann Transl Med. 7:5932019.
View Article : Google Scholar : PubMed/NCBI
|