Ovarian cancer (OC) is a life-threatening malignancy
that represents 3.6% female malignancies worldwide (1). It currently ranks as the seventh most
common type of gynecological cancer and 20th as the most common
type of cancer worldwide. OC has the highest mortality of all
gynecologic malignancies (1,2). According to a recent report, it was
estimated that there were 295,414 new cases of OC diagnosed in 2018
and 184,799 cases of mortality resulting from this disease
worldwide (3). Tumor debulking
surgery followed by platinum and paclitaxel chemotherapy is
currently the standard clinical treatment of OC (4). However, the survival rate of patients
with advanced OC remains at ~30%, with the primary reasons being
late discovery and chemoresistance. In particular, chemoresistance
is mediated by both the tumor microenvironment and inherent
resistance of OC cells to chemotherapy (5). Therefore, enhancement of responses to
current treatment and the development of novel therapeutic
strategies are urgently required to improve the survival rate.
Autophagy is a protective, catabolic process that
operates to maintain intracellular homeostasis by recycling
organelles and macromolecules (6).
During this process, defective or aged organelles and other
cytoplasmic components are enclosed by double-membrane vesicles to
form autophagosomes. This then fuses with a lysosome where the
vesicular contents are degraded into amino acids, lipids and
carbohydrates by the lysosomal enzymes. The degradation products
are in turn recycled to make new proteins and organelles (7). A basal level of autophagy is in
operation under physiological conditions (8). However, downregulation or upregulation
of autophagy induced by stress factors, including alterations in
the levels of growth factors, hypoxia and cytotoxic damage, can
result in cell death or cell adaptation in response (9). Autophagy also appears to serve
contradictory roles in the development of cancer. Evidence exists
reporting that inhibition of genes associated with autophagy can
promote tumor development, whereas the expression of proteins
associated with autophagy has also been demonstrated to result in
inhibitory effects in several types of cancers (10–14).
Therefore, autophagy can exert antitumor effects, but in contrast
cancer cells may survive cellular stress in adverse
microenvironments by utilizing autophagy, thereby promoting the
development of tumors (15). In
addition, autophagy is somewhat considered to be a double-edged
sword in the clinical field of cancer. Promotion of autophagy can
induce cell death, in a manner that is similar to apoptosis, whilst
protective cellular autophagy has also been reported to be the
major underlying cause of therapy resistance among cancer cells.
Therefore, increasing the sensitivity of cancer cells to anticancer
therapy by inhibiting autophagy remains a viable option (16).
Currently, there are a number of studies on
autophagy and Sirt3 in cardiovascular diseases, neuronal diseases
and hepatotoxicity (30–32). However, almost no article has
conducted research on the relationship between autophagy, OC and
Sirt3. Therefore, the present review primarily discussed the
potential relationship between Sirt3 and autophagy in OC, with the
aim to provide a possible novel direction for OC research and
therapeutic strategies.
OC poses a significant threat to the health of women
worldwide, and is a disease in which Sirt3 has been reported serve
a regulatory role. Of note, this disease is gradually becoming the
leading cause of mortality associated with gynecological cancer
worldwide in both developing and developed countries (1). Several reports have suggested that OC
is regulated by Sirt3 using a multitude of mechanisms, which is
summarized in this section.
In a previous study, it was found that muscle
tissues after exercise exhibit elevated expression levels of the
Sirt3 protein, which gave rise to the hypothesis that the
expression of Sirt3 is regulated by energy metabolism (33). Energy metabolism is also associated
with the regulation of tumor growth and metastasis. Sirt3 is
regarded as a tumor suppressor, due to a previous finding that its
expression is reduced in tumors (34,35). The
activation of cellular autophagy and apoptosis was demonstrated to
be controlled by Sirt3 via the regulation of several signaling
pathways during the development of OC. A previous study found that
expression of the Sirt3 protein was significantly downregulated in
OC tissues and in highly metastatic HO-8910PM cell lines (35,36). In
addition, Xiang et al (37)
demonstrated that the activation of Sirt3 exerted a proapoptotic
function in SKOV3 cells. These findings suggested that
overexpression of Sirt3 can induce OC cell death. In terms of
mitochondrial dynamics, a previous study revealed that
stabilization of Sirt3 can increase mitochondrial biogenesis and
cristae remodeling in OC tissues (38). Additionally, stabilization of optic
atrophy protein 1, which increased resistance to apoptosis, was
demonstrated to be regulated by increasing the expression of Sirt3
and prohibitin 2 (38). Activation
of Sirt3 has also been found to enhance the sensitivity of OC cells
to cisplatin (39), rendering Sirt3
to be a novel therapeutic target. In addition, Sirt3 was reported
to be a favorable independent prognostic factor for overall
survival for patients with serious OC in a previous study (40). In conclusion, Sirt3 serves an
important role in the development of OC, with therapeutic and
prognostic implications.
Autophagy is a catabolic process that serves to
maintain intracellular homeostasis by recycling damaged cellular
organelles (41), which has been
studied since the 1960s (42). Over
the past decades, the molecular mechanisms underlying this process
have been revealed gradually. It has been suggested that autophagy
is a common phenomenon that occurs during both physiological and
pathological conditions. According to the sizes of the substrates
involved and degradation rate, autophagy can be divided into three
sub-categories: i) Macro-autophagy; ii) microautophagy; and iii)
chaperone-mediated autophagy (43).
Although different types of autophagy utilize distinct mechanisms
to degrade lysosomal proteins, common underlying characteristics
remain (44). The autophagy pathway
consists of the following six steps: i) Initiation of autophagy;
ii) biogenesis of the phagophore; iii) expansion of the phagophore;
iv) formation of the autophagosome; v) fusion with the lysosome;
and vi) reformation of the lysosome (45). Autophagy is constitutively active at
low levels in all cell types under physiological conditions, but
can be potentiated by nutrient deprivation, hypoxia, endoplasmic
reticulum stress, pathogenic toxicity and immune injury, to
maintain intracellular homeostasis (46). Previous studies have demonstrated
that autophagy serves an important role in the pathological
processes of various diseases, including neurodegenerative and
cardiovascular disease, cancer, infectious diseases and immune
deficiency (47–50). Autophagy has been described as a
double-edged sword, since it can exert both tumor suppression and
growth promotion (51). In this
section, the mechanism of autophagy in OC is discussed in
detail.
OC ranks as the most lethal gynecological
malignancy, with high morbidity and mortality. Autophagy serves an
important role in OC through the expression of autophagy-associated
proteins, including beclin-1, microtubule-associated proteins 1A/1B
light chain 3B (LC3) and p53. Beclin-1 is a tumor suppressor, which
is an important checkpoint protein that is involved in autophagy
and tumor cell apoptosis (52,53). It
has been reported to mediate various functions in tumors, where its
expression level varies depending on the type of malignancy. A
previous study revealed that the expression level of beclin-1 was
higher in ovarian epithelial cancers, which can be used as an
independent risk factor for the prognosis of patients with this
disease (54). In addition, other
proteins linked to autophagy can mediate functions in OC, which are
in turn associated with a number of signaling pathways, including
the PI3K/AKT/mTOR and p53 signaling pathway. These are summarized
in this section.
The autophagic process in OC is regulated by a
number of factors. The PI3K/AKT/mTOR pathway has frequently been
associated with the majority of human malignancies, studies have
demonstrated that other signaling pathways related to oncogenesis
are also caused by dysregulations in this signaling pathway
(55–57). The reason for this dysregulation is
manifold, including mutations in PI3K, AKT overexpression and the
sustained activation of tyrosine kinase growth factor receptors
(58). The PI3K/AKT/mTOR signaling
pathway has been documented to regulate cell survival,
proliferation, growth, transcription, angiogenesis and metabolism
(55–57). In ~70% cases of OC, the PI3K/AKT/mTOR
pathway has been revealed to be constitutively activated, which has
been considered to be a therapeutic target (59). To verify if the PI3K/AKT pathway is
involved in OC cell proliferation, Hu et al (60) treated OC cell lines with the specific
PI3K inhibitor LY294002 and established a mouse model of OC.
Proliferation of OC cells can be significantly inhibited by
LY294002 treatment in vitro (61). In addition, other studies have found
that AKT inhibitors can prevent the function of mTORC1/2 and AKT
itself to inhibit the PI3K signaling cascade (61–63). In
another study, Ichikawa et al (64) found that the cytotoxic effects of
chemotherapeutic agents can be effectively enhanced by co-treatment
with the selective non-competitive AKT inhibitor TAS-117 in
vivo OC models. In OC, the PI3K/AKT/mTOR signaling pathway is
frequently activated, which indicates that the inhibition of this
signaling pathway could prove to be a potential avenue of treatment
strategies, either as a monotherapy or in combination with other
chemotherapeutic agents.
p53 is a key tumor suppressor that serves an
important regulatory role in autophagy in mammalian cells (65). Expression of the p53 gene is
activated by various intracellular events, including DNA damage,
hypoxia and oncogene activation, to prevent cell damage and
maintain cellular integrity. Numerous types of modifications,
including acetylation, methylation, phosphorylation and
ubiquitination, are involved in regulating the activation of p53 on
a molecular level (66,67). p53 target genes negatively regulate
mTOR activity, which in turn induces autophagy in the nucleus. p53
can promote autophagy by inhibiting mTOR via the 5′AMP-activated
protein kinase (AMPK) pathway (68).
In addition, p53 can also induce autophagy by activating
damage-regulated autophagy modulator (69). Several studies have revealed that
autophagy may be triggered by the inactivation of cytoplasmic p53,
such that extranuclear p53 is an effective inhibitor of autophagy
(70,71). A clinical study previously
demonstrated the upregulated expression of p53 in OC, where it was
revealed that at later tumor stages, the rate of p53-positive
expression was higher (72). These
results suggested that p53 serves an important role in the
development of OC. Additionally, another study previously found
that silencing the p53 signaling pathway may suppress proliferation
whilst facilitating apoptosis and cisplatin chemosensitivity in OC
cells (73). Therefore, these
aforementioned findings suggested that the efficacy of
chemotherapeutic treatments for OC can be improved by inhibiting
the p53-induced autophagic process.
p62 is the selective cargo receptor for autophagy
that is indispensable for the degeneration of misfolded proteins.
Reductions in p62 expression have been previously reported to
activate autophagy (78). In
addition, p62 is considered to be a marker of autophagic flux due
to its differential expression profiles in association with other
proteins linked to autophagy (79).
As for Sirt3, a number of studies have indicated that there is a
close relationship between Sirt3 and the p62-autophagy pathway. A
previous study demonstrated that treatment with ANXA1sp, an
annexin-A1 bioactive peptide, reduced the expression of p62 and
concomitantly upregulated the expression of the mitochondrial
protein deacetylase Sirt3 (80).
This suggested that Sirt3 is involved in autophagy by
downregulating p62. Tong et al (81) also reported a similar finding, where
liraglutide treatment downregulated the expression of p62 to
promote autophagy via the Sirt1/Sirt3-forkhead box O3-p62 pathway
in mice that were fed a normal-fat diet or high-fat diet.
Additionally, a previous investigation focusing on the effects of
melatonin on diabetic cardiomyopathy revealed that melatonin can
upregulate autophagy by increasing the expression of LC3-II whilst
downregulating that of p62 (82),
with the macrophage stimulating 1/Sirt3 signaling pathway being the
likely associated mechanism. These aforementioned pharmacological
studies suggested that Sirt3 is a potent activator of autophagy.
Supporting this, Xiang et al (83) found that small interfering
(si)RNA-mediated silencing of Sirt3 gene expression inhibited the
process of autophagy and p62 degradation in human umbilical vein
endothelial cells.
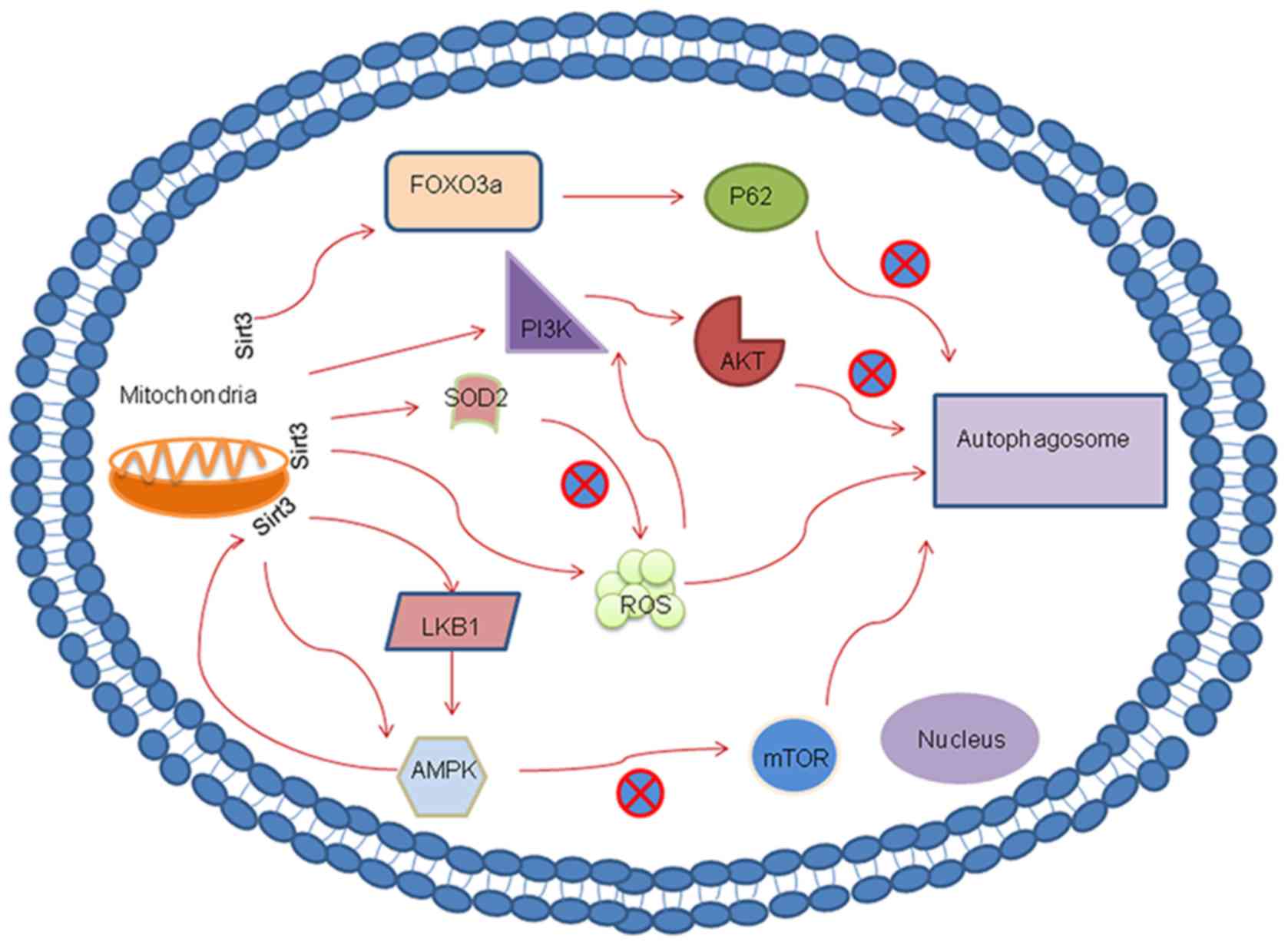
The PI3K/AKT pathway serves an important inhibitory
role in autophagy, and is also involved in a variety of
physiological and pathological processes (84,85). By
regulating the PI3K/AKT pathway, Sirt3 can function as an autophagy
suppressor. A previous report that investigated hepatocellular
carcinoma found that the expression levels of Sirt3 was higher in
adjacent non-cancerous tissues compared with those in
hepatocellular carcinoma tissues (86). Furthermore, using Sirt3 knockdown,
this previous study also demonstrated that Sirt3 may serve as a
suppressor of autophagy in hepatocellular carcinoma by targeting
the PI3K/AKT pathway. Another study revealed a consistent finding,
where Sirt3 functioned as a growth suppressor in prostate cancer by
inhibiting the activation of PI3K/AKT both in vitro and
in vivo (87). This previous
study also showed that the progression of prostate cancer may be
downregulated via the Sirt3/AKT/c-Myc signaling axis (87). Wang et al (88) demonstrated that glioblastoma
multiforme cell growth was inhibited through the Sirt3/p53-mediated
PI3K/AKT/ERK and mitochondrial signaling pathway. In addition,
Sirt3 was reported to indirectly regulate AKT hyperactivation by
regulating mitochondrial ROS production upstream of the
ROS-mediated Ras-PI3K-AKT activation (89). In summary, Sirt3 can regulate
autophagy by either directly or indirectly regulating the PI3K/AKT
pathway.
AMPK is a conserved a intracellular energy sensor
that mediates energy homeostasis by regulating lipid and glucose
metabolism (90). Dysregulation of
AMPK has previously been associated with accelerated aging
(91). In previous years, a number
of studies have attempted to unravel the mechanism underlying the
regulation of autophagy. There is increasing consensus that AMPK
and mTOR are regarded as the main regulators of autophagic
degradation (92,93). A close mechanistic relationship has
been reported to exist between Sirt3 and the AMPK-mTOR-autophagy
pathway. Zhao et al (94)
previously found that induction of autophagy was directly
controlled by Sirt3 via the AMPK-mTOR pathway during acute kidney
injury. In addition, another study reported that Sirt3-indcued
autophagy protected against oxygen and glucose deprivation by
regulating the AMPK-mTOR pathway (30).
Previous studies have found that AMPK can negatively
regulate mTORC1 activity via two different mechanisms. AMPK can
phosphorylate Thr1227 and Ser1345 residues to activate tuberous
sclerosis complex (TSC) 2, thereby promoting the formation of the
TSC1/TSC2 heterodimer to inhibit mTORC1 activity (95). By contrast, AMPK can also
phosphorylate regulatory-associated protein of mTOR on its Ser722
and Ser792 residues to inhibit mTORC1 (96). Liver kinase B1 (LKB1) is a tumor
suppressor and an upstream regulator of AMPK that has an essential
role in the control of redox homeostasis (97). Incidentally, LKB1 can also be
activated by Sirt3. A previous investigation into a
rotenone-induced SH-SY5Y cell injury model found that the
overexpression of Sirt3 promoted LKB1 phosphorylation, which
activated AMPK and reduced the phosphorylation of mTOR. This
observation suggested that the LKB1-AMPK-mTOR pathway can be
regulated by Sirt3 (98). In
addition, another previous study documented that activation of the
LKB1-AMPK-mTOR-mediated autophagy signaling pathway can be induced
by Sirt3 (99). Notably, AMPK can
function upstream of Sirt3 during the regulation of insulin
sensitivity in skeletal muscle via the AMPK-peroxisome
proliferator-activated receptor γ coactivator 1α-Sirt3 autophagy
signaling pathway in Sirt3-/-mice (100). Collectively, this indicated that
Sirt3 and autophagy have a complex mutual regulatory
relationship.
ROS consists of a group of highly reactive chemical
entities, including oxygen radicals, hydroxyl, peroxyl, alkoxyl,
non-radicals, singlet oxygen and hydrogen peroxide. These molecules
are primarily produced from redox transactions as part of the
oxidative phosphorylation system in mitochondria (100). A number of studies have
demonstrated that ROS can act as a signal to trigger autophagy
through autophagy-regulating protease 4, which serves as part of
the autophagy process (101,102).
Autophagy may be suppressed by Sirt3 via the regulation of
mitochondrial ROS (mROS) production. Recently, a study
investigating the potential antineoplastic properties of metformin
combined with nelfinavir revealed that this drug combination can
increase the expression level of Sirt3. This increment in
Sirt3-mediated mROS production served a vital role in the
autophagic mechanism within human cervical cancer cells (103). Upstream of ROS, SOD2 activity can
moderately reduce cellular ROS levels. Sirt3-mediated deacetylation
can significantly potentiate SOD2 activity to ultimately break down
intracellular ROS (104). A
previous study demonstrated that Sirt3 protein expression and
activity was downregulated by cadmium (Cd), which can also
concurrently promote the acetylation of SOD2 to suppress its
activity, thereby increasing mROS production (31). In summary, mitochondrial-derived
ROS-dependent autophagic cell death can be induced by Cd.
Consistent with this notion, Cd-induced hepatotoxicity has been
reported to be alleviated by the protective properties of melatonin
(31). In this particular study, the
activity, but not the expression of Sirt3, was revealed to be
enhanced by melatonin treatment (31). Additionally, melatonin was also
demonstrated to inhibit mitochondria-derived O2
production, reduced the acetylation of SOD2 and suppressed
autophagy (31). This finding
suggested that melatonin exerts hepatoprotective effects by
regulating mitochondria-derived O2-stimulated autophagic
cell death via the Sirt3-SOD2 pathway. According to the
aforementioned findings, SOD2 activity and intracellular mROS
homeostasis may underlie the Sirt3 downregulation of autophagy
(31).
Autophagy is a fundamental catabolic process that
has been reported to be involved in the progression of a variety of
diseases. It can serve a protective role in OC cells from cell
death since it may enhance resistance to cisplatin (105). A previous investigation revealed
that cisplatin treatment activated autophagy, whereas
Bcl-2-associated athanogene 3 attenuated cisplatin resistance by
inhibiting autophagy (106).
However, recent studies have also suggested that autophagy can
inhibit the growth of OC, since it has been found to promote
OVCAR-3 cell death (107). By
contrast, findings from another previous study suggested that
inhibition of autophagy promoted the proliferation and invasion of
OC cells via the PI3K/AKT/mTOR pathway (108). These findings demonstrated that
autophagy is a double-edged sword in the regulation of OC
physiology. Sirt3 serves an important role in the maintenance of
intracellular homeostasis in OC. Previous studies have indicated
that there is a close mutual regulatory relationship between Sirt3
and autophagy, which are linked by the aforementioned signaling
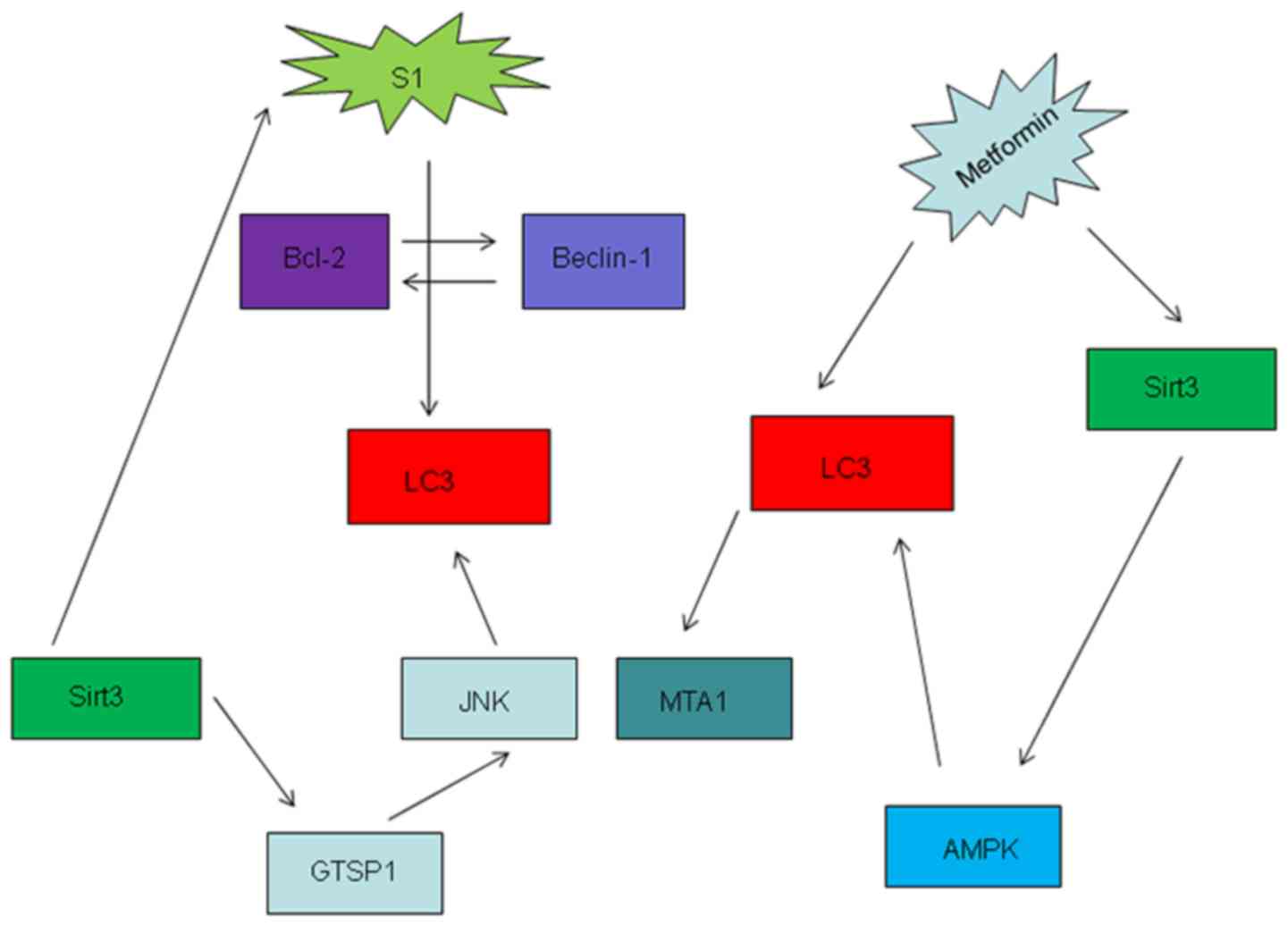
pathways in the present review. Metformin-induced overexpression of
Sirt3 promoted apoptosis and mitochondrial dysfunction whilst
increasing the activation of AMPK in OC cell lines (109). In addition, metformin has been
documented to promote autophagy in OC (110). S1, a novel Bcl-2 inhibitor, has
also been reported to exert autophagic effects in OC by
interrupting the interaction between Bcl-2 and beclin1 in OC to
promote apoptosis (111); however,
high doses and longer exposure of S1 can overpower the protective
function of autophagy and induce apoptosis (112,113).
Yang et al (114) previously
found that S1 promoted JNK3 expression, thus increasing cell
sensitivity to apoptosis. Sirt3 can also regulate autophagy via the
glutathione S-transferase P/JNK/autophagy pathway, such that Sirt3
knockdown has been demonstrated to alleviate S1-induced apoptosis
(Fig. 2) (37).
Autophagy serves an important role in recycling
damaged organelles and maintaining intracellular homeostasis. Sirt3
is a potential therapeutic target, since it has been previously
reported to activate autophagy. In the present review, although the
potential relationship between Sirt3 and autophagy in OC was
explored, there remains an insufficient number of studies on this
topic. The associated underlying mechanism in the Sirt3-induced
autophagic process in OC remains unclear. With further study, novel
insights into the molecular relationship between Sirt3 and
autophagy may contribute to the development of novel therapeutic
interventions for OC.
Not applicable.
The present project was mainly supported by the
National Natural Science Foundation of China (grant no.
81802586).
Not applicable.
YS and RH contributed to the conception of this
manuscript and wrote the draft. YY contributed to the literature
collection and preparation. YH contributed to the revision of this
manuscript. LZ and BW provided funding and proofed the manuscript.
All authors read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hackshaw A, Gershenson D and Ledermann J:
Mucinous ovarian carcinoma. N Engl J Med. 381:e32019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang L, Wang Q, Xu Y and Han L: Advances
in the treatment of ovarian cancer using PARP inhibitors and the
underlying mechanism of resistance. Curr Drug Targets. 21:167–178.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hansen JM, Coleman RL and Sood AK:
Targeting the tumour microenvironment in ovarian cancer. Eur J
Cancer. 56:131–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Ren Y, Hou Y, Zhang C, Wang B, Li
X, Sun R and Liu J: Dihydroartemisinin induces endothelial cell
autophagy through suppression of the Akt/mTOR pathway. J Cancer.
10:6057–6064. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ravikumar B, Sarkar S, Davies JE, Futter
M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M,
Korolchuk VI, Lichtenberg M, Luo S, et al: Regulation of mammalian
autophagy in physiology and pathophysiology. Physiol Rev.
90:1383–1435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Melendez A and Neufeld TP: The cell
biology of autophagy in metazoans: A developing story. Development.
135:2347–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boya P, González-Polo RA, Casares N,
Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D,
Souquere S, Yoshimori T, et al: Inhibition of macroautophagy
triggers apoptosis. Mol Cell Biol. 25:1025–1040. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coppola D, Khalil F, Eschrich SA, Boulware
D, Yeatman T and Wang HG: Down-regulation of Bax-interacting
factor-1 in colorectal adenocarcinoma. Cancer. 113:2665–2670. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boteon YL, Laing R, Mergental H, Reynolds
GM, Mirza DF, Afford SC and Bhogal RH: Mechanisms of autophagy
activation in endothelial cell and their targeting during
normothermic machine liver perfusion. World J Gastroenterol.
23:8443–8451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nussenzweig SC, Verma S and Finkel T: The
role of autophagy in vascular biology. Circ Res. 116:480–488. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang C, Feng P, Ku B, Dotan I, Canaani D,
Oh BH and Jung JU: Autophagic and tumour suppressor activity of a
novel Beclin1-binding protein UVRAG. Nat Cell Biol. 8:688–699.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bishop E and Bradshaw TD: Autophagy
modulation: A prudent approach in cancer treatment? Cancer
Chemother Pharmacol. 82:913–922. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Haigis MC, Deng CX, Finley LW, Kim HS and
Gius D: SIRT3 is a mitochondrial tumor suppressor: A scientific
tale that connects aberrant cellular ROS, the Warburg effect, and
carcinogenesis. Cancer Res. 72:2468–2472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sack MN and Finkel T: Mitochondrial
metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol.
4:a0131022012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Verdin E, Hirschey MD, Finley LW and
Haigis MC: Sirtuin regulation of mitochondria: Energy production,
apoptosis, and signaling. Trends Biochem Sci. 35:669–675. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Anderson KA and Hirschey MD: Mitochondrial
protein acetylation regulates metabolism. Essays Biochem. 52:23–35.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alhazzazi TY, Kamarajan P, Joo N, Huang
JY, Verdin E, D'Silva NJ and Kapila YL: Sirtuin-3 (SIRT3), a novel
potential therapeutic target for oral cancer. Cancer.
117:1670–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi T, Wang F, Stieren E and Tong Q:
SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial
function and thermogenesis in brown adipocytes. J Biol Chemy.
280:13560–13567. 2005. View Article : Google Scholar
|
|
24
|
Kong X, Wang R, Xue Y, Liu X, Zhang H,
Chen Y, Fang F and Chang Y: Sirtuin 3, a new target of PGC-1alpha,
plays an important role in the suppression of ROS and mitochondrial
biogenesis. PLoS One. 5:e117072010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guarente L: Introduction: Sirtuins in
aging and diseases. Methods Mol Biol. 1077:3–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Resendis-Antonio O, Checa A and
Encarnacion S: Modeling core metabolism in cancer cells: Surveying
the topology underlying the Warburg effect. PLoS One. 5:e123832010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Madhok BM, Yeluri S, Perry SL, Hughes TA
and Jayne DG: Targeting glucose metabolism: An emerging concept for
anticancer therapy. Am J Clin Oncol. 34:628–635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hirschey MD, Shimazu T, Goetzman E, Jing
E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S and
Ilkayeva OR: SIRT3 regulates mitochondrial fatty-acid oxidation by
reversible enzyme deacetylation. Nature. 464:121–125. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bagul PK, Katare PB, Bugga P, Dinda AK and
Banerjee SK: SIRT-3 modulation by resveratrol improves
mitochondrial oxidative phosphorylation in diabetic heart through
deacetylation of TFAM. Cells. 7:2352018. View Article : Google Scholar
|
|
30
|
Dai SH, Chen T, Li X, Yue KY, Luo P, Yang
LK, Zhu J, Wang YH, Fei Z and Jiang XF: Sirt3 confers protection
against neuronal ischemia by inducing autophagy: Involvement of the
AMPK-mTOR pathway. Free Radic Biol Med. 108:345–353. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pi H, Xu S, Reiter RJ, Guo P, Zhang L, Li
Y, Li M, Cao Z, Tian L, Xie J, et al: SIRT3-SOD2-mROS-dependent
autophagy in cadmium-induced hepatotoxicity and salvage by
melatonin. Autophagy. 11:1037–1051. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Ma Y, Song L, Yu L, Zhang L, Zhang
Y, Xing Y, Yin Y and Ma H: SIRT3 deficiency exacerbates
p53/Parkin-mediated mitophagy inhibition and promotes mitochondrial
dysfunction: Implication for aged hearts. Int J Mol Med.
41:3517–3526. 2018.PubMed/NCBI
|
|
33
|
Edgett BA, Hughes MC, Matusiak JB, Perry
CG, Simpson CA and Gurd BJ: SIRT3 gene expression but not SIRT3
subcellular localization is altered in response to fasting and
exercise in human skeletal muscle. Exp Physiol. 101:1101–1113.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y, Fu LL, Wen X, Wang XY, Liu J,
Cheng Y and Huang J: Sirtuin-3 (SIRT3), a therapeutic target with
oncogenic and tumor-suppressive function in cancer. Cell Death Dis.
5:e10472014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dong XC, Jing LM, Wang WX and Gao YX:
Down-regulation of SIRT3 promotes ovarian carcinoma metastasis.
Biochem Biophys Res Commun. 475:245–250. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang Y, Cao Y, Chen L, Liu F, Qi Z, Cheng
X and Wang Z: Cryptotanshinone suppresses cell proliferation and
glucose metabolism via STAT3/SIRT3 signaling pathway in ovarian
cancer cells. Cancer Med. 7:4610–4618. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiang XY, Kang JS, Yang XC, Su J, Wu Y,
Yan XY, Xue YN, Xu Y, Liu YH, Yu CY, et al: SIRT3 participates in
glucose metabolism interruption and apoptosis induced by BH3
mimetic S1 in ovarian cancer cells. Int J Oncol. 49:773–784. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Signorile A, De Rasmo D, Cormio A, Musicco
C, Rossi R, Fortarezza F, Palese LL, Loizzi V, Resta L, Scillitani
G, et al: Human ovarian cancer tissue exhibits increase of
mitochondrial biogenesis and cristae remodeling. Cancers (Basel).
11:13502019. View Article : Google Scholar
|
|
39
|
Hou L, Wang R, Wei H, Li S, Liu L, Lu X,
Yu H and Liu Z: ABT737 enhances ovarian cancer cells sensitivity to
cisplatin through regulation of mitochondrial fission via Sirt3
activation. Life Sci. 232:1165612019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Yue H, Yu H, Lu X and Xue X:
Development and validation of SIRT3-related nomogram predictive of
overall survival in patients with serous ovarian cancer. J Ovarian
Res. 12:472019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Y and Klionsky DJ: Autophagy and
disease: Unanswered questions. Cell Death Differ. 27:858–871. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wilde L, Tanson K, Curry J and
Martinez-Outschoorn U: Autophagy in cancer: A complex relationship.
Biochem J. 475:1939–1954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Glick D, Barth S and Macleod KF:
Autophagy: Cellular and molecular mechanisms. J Pathol. 221:3–12.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bednarczyk M, Zmarzly N, Grabarek B,
Mazurek U and Muc-Wierzgon M: Genes involved in the regulation of
different types of autophagy and their participation in cancer
pathogenesis. Oncotarget. 9:34413–34428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhi X, Feng W, Rong Y and Liu R: Anatomy
of autophagy: From the beginning to the end. Cell Mol Life SciS.
75:815–831. 2018. View Article : Google Scholar
|
|
46
|
Burman C and Ktistakis NT: Autophagosome
formation in mammalian cells. Semin Immunopathol. 32:397–413. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schaeffer V, Lavenir I, Ozcelik S, Tolnay
M, Winkler DT and Goedert M: Stimulation of autophagy reduces
neurodegeneration in a mouse model of human tauopathy. Brain.
135:2169–2177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liao X, Sluimer JC, Wang Y, Subramanian M,
Brown K, Pattison JS, Robbins J, Martinez J and Tabas I: Macrophage
autophagy plays a protective role in advanced atherosclerosis. Cell
Metab. 15:545–553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dikic I, Johansen T and Kirkin V:
Selective autophagy in cancer development and therapy. Cancer Res.
70:3431–3434. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim JJ, Lee HM, Shin DM, Kim W, Yuk JM,
Jin HS, Lee SH, Cha GH, Kim JM, Lee ZW, et al: Host cell autophagy
activated by antibiotics is required for their effective
antimycobacterial drug action. Cell Host Microbe. 11:457–468. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pu Z, Wu L, Guo Y, Li G, Xiang M, Liu L,
Zhan H, Zhou X and Tan H: LncRNA MEG3 contributes to
adenosine-induced cytotoxicity in hepatoma HepG2 cells by
downregulated ILF3 and autophagy inhibition via regulation
PI3K-AKT-mTOR and beclin-1 signaling pathway. J Cell Biochem.
120:18172–18185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li X, Lou X, Xu S, Wang Q, Shen M and Miao
J: Knockdown of miR-372 Inhibits nerve cell apoptosis induced by
spinal cord ischemia/reperfusion injury via enhancing autophagy by
Up-regulating Beclin-1. J Mol Neurosci. 66:437–444. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cai M, Hu Z, Liu J, Gao J, Liu C, Liu D,
Tan M, Zhang D and Lin B: Beclin 1 expression in ovarian tissues
and its effects on ovarian cancer prognosis. Int J Mol Sci.
15:5292–5303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and Gonzalez-Baron M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/AKT signalling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Arico S, Petiot A, Bauvy C, Dubbelhuis PF,
Meijer AJ, Codogno P and Ogier-Denis E: The tumor suppressor PTEN
positively regulates macroautophagy by inhibiting the
phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol
Chem. 276:35243–35246. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li H, Zeng J and Shen K: PI3K/AKT/mTOR
signaling pathway as a therapeutic target for ovarian cancer. Arch
Gynecol Obstet. 290:1067–1078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hu L, Zaloudek C, Mills GB, Gray J and
Jaffe RB: In vivo and in vitro ovarian carcinoma growth inhibition
by a phosphatidylinositol 3-kinase inhibitor (LY294002). Clin
Cancer Res. 6:880–886. 2000.PubMed/NCBI
|
|
61
|
Engel JB, Schönhals T, Häusler S,
Krockenberger M, Schmidt M, Horn E, Köster F, Dietl J, Wischhusen J
and Honig A: Induction of programmed cell death by inhibition of
AKT with the alkylphosphocholine perifosine in in vitro models of
platinum sensitive and resistant ovarian cancers. Arch Gynecol
Obstet. 283:603–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun H, Yu T and Li J: Co-administration of
perifosine with paclitaxel synergistically induces apoptosis in
ovarian cancer cells: More than just AKT inhibition. Cancer Lett.
310:118–128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yu Y, Hall T, Eathiraj S, Wick MJ,
Schwartz B and Abbadessa G: In-vitro and in-vivo combined effect of
ARQ 092, an AKT inhibitor, with ARQ 087, a FGFR inhibitor.
Anticancer Drugs. 28:503–513. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ichikawa K, Abe T, Nagase H, Saito H,
Fujita R, Okada M, Yonekura K, Shimomura T and Utsugi T: Abstract
C177: TAS-117, a highly selective non-ATP competitive inhibitor of
AKT demonstrated antitumor activity in combination with
chemotherapeutic agents and molecular targeted drugs. Mol Cancer
Ther. 12 (Suppl 11):C1772013.
|
|
65
|
Son Y, An Y, Jung J, Shin S, Park I, Gwak
J, Ju BG, Chung YH, Na M and Oh S: Protopine isolated from Nandina
domestica induces apoptosis and autophagy in colon cancer cells by
stabilizing p53. Phytother Res. 33:1689–1696. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Brooks CL and Gu W: Ubiquitination,
phosphorylation and acetylation: The molecular basis for p53
regulation. Curr Opin Cell Biol. 15:164–171. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Brooks CL and Gu W: The impact of
acetylation and deacetylation on the p53 pathway. Protein Cell.
2:456–462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Feng Z, Hu W, de Stanchina E, Teresky AK,
Jin S, Lowe S and Levine AJ: The regulation of AMPK beta1, TSC2,
and PTEN expression by p53: Stress, cell and tissue specificity,
and the role of these gene products in modulating the
IGF-1-AKT-mTOR pathways. Cancer Res. 67:3043–3053. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Crighton D, Wilkinson S, O'Prey J, Syed N,
Smith P, Harrison PR, Gasco M, Garrone O, Crook T and Ryan KM:
DRAM, a p53-induced modulator of autophagy, is critical for
apoptosis. Cell. 126:121–134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tasdemir E, Maiuri MC, Galluzzi L, Vitale
I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C,
Harper F, et al: Regulation of autophagy by cytoplasmic p53. Nat
Cell Biol. 10:676–687. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen N and Karantza-Wadsworth V: Role and
regulation of autophagy in cancer. Biochim Biophys Acta.
1793:1516–1523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liang M and Zhao J: Protein expressions of
AIB1, p53 and Bcl-2 in epithelial ovarian cancer and their
correlations with the clinical pathological features and prognosis.
Eur Rev Med Pharmacol Sci. 22:5134–5139. 2018.PubMed/NCBI
|
|
73
|
Chen YN, Ren CC, Yang L, Nai MM, Xu YM,
Zhang F and Liu Y: MicroRNA let7d5p rescues ovarian cancer cell
apoptosis and restores chemosensitivity by regulating the p53
signaling pathway via HMGA1. Int J Oncol. 54:1771–1784.
2019.PubMed/NCBI
|
|
74
|
Cho CS, Lombard DB and Lee JH: SIRT3 as a
regulator of hepatic autophagy. Hepatology. 66:700–702. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gao J, Feng Z, Wang X, Zeng M, Liu J, Han
S, Xu J, Chen L, Cao K, Long J, et al: SIRT3/SOD2 maintains
osteoblast differentiation and bone formation by regulating
mitochondrial stress. Cell Death Differ. 25:229–240. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liu H, Li S, Liu X, Chen Y and Deng H:
SIRT3 overexpression inhibits growth of kidney tumor cells and
enhances mitochondrial biogenesis. J Proteome Res. 17:3143–3152.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Shi H, Deng HX, Gius D, Schumacker PT,
Surmeier DJ and Ma YC: Sirt3 protects dopaminergic neurons from
mitochondrial oxidative stress. Hum Mol Genet. 26:1915–1926. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Duran A, Amanchy R, Linares JF, Joshi J,
Abu-Baker S, Porollo A, Hansen M, Moscat J and Diaz-Meco MT: p62 is
a key regulator of nutrient sensing in the mTORC1 pathway. Mol
Cell. 44:134–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang ZG, Li H, Huang Y, Li R, Wang XF, Yu
LX, Guang XQ, Li L, Zhang HY, Zhao YZ, et al: Nerve growth
factor-induced Akt/mTOR activation protects the ischemic heart via
restoring autophagic flux and attenuating ubiquitinated protein
accumulation. Oncotarget. 8:5400–5413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ma Q, Zhang Z, Shim JK, Venkatraman TN,
Lascola CD, Quinones QJ, Mathew JP, Terrando N and Podgoreanu MV:
Annexin A1 bioactive peptide promotes resolution of
neuroinflammation in a rat model of exsanguinating cardiac arrest
treated by emergency preservation and resuscitation. Front
Neurosci. 13:6082019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tong W, Ju L, Qiu M, Xie Q, Chen Y, Shen
W, Sun W, Wang W and Tian J: Liraglutide ameliorates non-alcoholic
fatty liver disease by enhancing mitochondrial architecture and
promoting autophagy through the SIRT1/SIRT3-FOXO3a pathway. Hepatol
Res. 46:933–943. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang M, Lin J, Wang S, Cheng Z, Hu J,
Wang T, Man W, Yin T, Guo W, Gao E, et al: Melatonin protects
against diabetic cardiomyopathy through Mst1/Sirt3 signaling. J
Pineal Res. May 8–2017.(Epub ahead of print). doi:
10.1111/jpi.12418. View Article : Google Scholar
|
|
83
|
Xiang X, Huang J, Song S, Wang Y, Zeng Y,
Wu S and Ruan Y: 17β-estradiol inhibits
H2O2-induced senescence in HUVEC cells
through upregulating SIRT3 expression and promoting autophagy.
Biogerontology. Mar 14–2020.(Epub ahead of print). doi:
10.1007/s10522-020-09868-w. View Article : Google Scholar
|
|
84
|
Zhang Y, Kwok-Shing Ng P, Kucherlapati M,
Chen F, Liu Y, Tsang YH, de Velasco G, Jeong KJ, Akbani R,
Hadjipanayis A, et al: A pan-cancer proteogenomic atlas of
PI3K/AKT/mTOR pathway alterations. Cancer Cell. 31:820–832.e3.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun Y, Huang YH, Huang FY, Mei WL, Liu Q,
Wang CC, Lin YY, Huang C, Li YN, Dai HF and Tan GH:
3′-epi-12β-hydroxyfroside, a new cardenolide, induces
cytoprotective autophagy via blocking the Hsp90/Akt/mTOR axis in
lung cancer cells. Theranostics. 8:2044–2060. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang
L, Ma J, Li X, Zeng Y, Yang Z, et al: A circular RNA promotes
tumorigenesis by inducing c-myc nuclear translocation. Cell Death
Differ. 24:1609–1620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Quan Y, Wang N, Chen Q, Xu J, Cheng W, Di
M, Xia W and Gao WQ: SIRT3 inhibits prostate cancer by
destabilizing oncoprotein c-MYC through regulation of the PI3K/Akt
pathway. Oncotarget. 6:26494–26507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang G, Wang JJ, Wang YZ, Feng S, Jing G
and Fu XL: Myricetin nanoliposomes induced SIRT3-mediated
glycolytic metabolism leading to glioblastoma cell death. Artif
Cells Nanomed Biotechnol. 46 (Suppl 3):S180–S191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Diotte NM, Xiong Y, Gao J, Chua BH and Ho
YS: Attenuation of doxorubicin-induced cardiac injury by
mitochondrial glutaredoxin 2. Biochim Biophys Acta. 1793:427–438.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhao M and Klionsky DJ: AMPK-dependent
phosphorylation of ULK1 induces autophagy. Cell Metab. 13:119–120.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Garcia D and Shaw RJ: AMPK: Mechanisms of
cellular energy sensing and restoration of metabolic balance. Mol
Cell. 66:789–800. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Inoki K, Kim J and Guan KL: AMPK and mTOR
in cellular energy homeostasis and drug targets. Annu Rev Pharmacol
Toxicol. 52:381–400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kim J, Kundu M, Viollet B and Guan KL:
AMPK and mTOR regulate autophagy through direct phosphorylation of
Ulk1. Nat Cell Biol. 13:132–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhao W, Zhang L, Chen R, Lu H, Sui M, Zhu
Y and Zeng L: SIRT3 protects against acute kidney injury via
AMPK/mTOR-regulated autophagy. Front Physiol. 9:15262018.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Inoki K, Zhu T and Guan KL: TSC2 mediates
cellular energy response to control cell growth and survival. Cell.
115:577–590. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gwinn DM, Shackelford DB, Egan DF,
Mihaylova MM, Mery A, Vasquez DS, Turk BE and Shaw RJ: AMPK
phosphorylation of raptor mediates a metabolic checkpoint. Mol
Cell. 30:214–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shackelford DB and Shaw RJ: The LKB1-AMPK
pathway: Metabolism and growth control in tumour suppression. Nat
Rev Cancer. 9:563–575. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang M, Deng YN, Zhang JY, Liu J, Li YB,
Su H and Qu QM: SIRT3 protects rotenone-induced injury in SH-SY5Y
cells by promoting autophagy through the LKB1-AMPK-mTOR pathway.
Aging Dis. 9:273–286. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Bazhin AV, Philippov PP and Karakhanova S:
Reactive oxygen species in cancer biology and anticancer therapy.
Oxid Med Cell Longev. 2016:41978152016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shi L, Zhang T, Zhou Y, Zeng X, Ran L,
Zhang Q, Zhu J and Mi M: Dihydromyricetin improves skeletal muscle
insulin sensitivity by inducing autophagy via the
AMPK-PGC-1alpha-Sirt3 signaling pathway. Endocrine. 50:378–389.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Lv W, Sui L, Yan X, Xie H, Jiang L, Geng
C, Li Q, Yao X, Kong Y and Cao J: ROS-dependent Atg4 upregulation
mediated autophagy plays an important role in Cd-induced
proliferation and invasion in A549 cells. Chem Biol Interact.
279:136–144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li ZY, Yang Y, Ming M and Liu B:
Mitochondrial ROS generation for regulation of autophagic pathways
in cancer. Biochem Biophys Res Commun. 414:5–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Xia C, He Z, Liang S, Chen R, Xu W, Yang
J, Xiao G and Jiang S: Metformin combined with nelfinavir induces
SIRT3/mROS-dependent autophagy in human cervical cancer cells and
xenograft in nude mice. Eur J Pharmacol. 848:62–69. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Qiu X, Brown K, Hirschey MD, Verdin E and
Chen D: Calorie restriction reduces oxidative stress by
SIRT3-mediated SOD2 activation. Cell Metab. 12:662–667. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Tan WX, Xu TM, Zhou ZL, Lv XJ, Liu J,
Zhang WJ and Cui MH: TRP14 promotes resistance to cisplatin by
inducing autophagy in ovarian cancer. Oncol Rep. 42:1343–1354.
2019.
|
|
106
|
Qiu S, Sun L, Zhang Y and Han S:
Downregulation of BAG3 attenuates cisplatin resistance by
inhibiting autophagy in human epithelial ovarian cancer cells.
Oncol Lett. 18:1969–1978. 2019.PubMed/NCBI
|
|
107
|
Xie Z, Guo Z, Wang Y, Lei J and Yu J:
Protocatechuic acid inhibits the growth of ovarian cancer cells by
inducing apoptosis and autophagy. Phytother Res. 32:2256–2263.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhu H, Diao S, Lim V, Hu L and Hu J:
FAM83D inhibits autophagy and promotes proliferation and invasion
of ovarian cancer cells via PI3K/AKT/mTOR pathway. Acta Biochim
Biophys Sin (Shanghai). 51:509–516. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wu Y, Gao WN, Xue YN, Zhang LC, Zhang JJ,
Lu SY, Yan XY, Yu HM, Su J and Sun LK: SIRT3 aggravates
metformin-induced energy stress and apoptosis in ovarian cancer
cells. Exp Cell Res. 367:137–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zou G, Bai J, Li D and Chen Y: Effect of
metformin on the proliferation, apoptosis, invasion and autophagy
of ovarian cancer cells. Exp Ther Med. 18:2086–2094.
2019.PubMed/NCBI
|
|
111
|
Li X, Su J, Xia M, Li H, Xu Y, Ma C, Ma L,
Kang J, Yu H, Zhang Z and Sun L: Caspase-mediated cleavage of
Beclin1 inhibits autophagy and promotes apoptosis induced by S1 in
human ovarian cancer SKOV3 cells. Apoptosis. 21:225–238. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Liu N, Xu Y, Sun JT, Su J, Xiang XY, Yi
HW, Zhang ZC and Sun LK: The BH3 mimetic S1 induces endoplasmic
reticulum stress-associated apoptosis in cisplatin-resistant human
ovarian cancer cells although it activates autophagy. Oncol Rep.
30:2677–2684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Yang X, Xiang X, Xia M, Su J, Wu Y, Shen
L, Xu Y and Sun L: Inhibition of JNK3 promotes apoptosis induced by
BH3 mimetic S1 in chemoresistant human ovarian cancer cells. Anat
Rec (Hoboken). 298:386–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yang Y, Li N, Chen T, Zhang C, Li J, Liu
L, Qi Y, Zheng X, Zhang C and Bu P: Sirt3 promotes sensitivity to
sunitinib-induced cardiotoxicity via inhibition of
GTSP1/JNK/autophagy pathway in vivo and in vitro. Arch Toxicol.
93:3249–3260. 2019. View Article : Google Scholar : PubMed/NCBI
|