Introduction
Esophageal cancer (EC) is one of the most common
types of cancer worldwide and is the sixth most common type of
malignancy (1). The incidence of EC
has markedly increased in recent decades. For example, ~572,034 new
case diagnoses and 508,585 mortalities were reported in 2018 alone
(1). EC is histologically
categorized into two subtypes, esophageal squamous cell carcinoma
(ESCC) and esophageal adenocarcinoma (EA), and the incidence varies
by histological type. In addition, the geographical location also
appears to influence the incidence rate of EC. For example,
previous studies have suggested that regions, including China, the
Middle East and Southern Africa exhibit high rates of ESCC
(2–4). As there are initially no detectable
symptoms, the majority of patients are diagnosed at the advanced
stages of EC, resulting in a poor overall prognosis. At present,
available treatments for EC include surgery, chemotherapy and
radiotherapy. Unlike other malignancies, including lung and breast
cancer, there are few molecular markers available for ESCC which
provide guidance for treatment options and facilitate prognostic
predictions (5). Therefore, the
identification of novel biomarkers for ESCC is of considerable
importance.
As a frequent characteristic of human cancer, the
dysregulation of mRNA translation may result in tumor growth,
metastasis, angiogenesis and escape from immune eradication
(6,7). The initiation phase is the
rate-limiting and most tightly regulated step in translational
control. Existing studies have established an associated between
various types of eukaryotic initiation factors (EIFs) and the
genesis and prognosis of different cancer types (8–13). In
eukaryotes, eukaryotic initiation factor 4A (eIF4A) is essential
for translation initiation. It is a canonical DEAD-box helicase,
responsible for unwinding the extensive secondary structures of the
5′ untranslated regions of mRNAs during 40S ribosomal subunit
scanning (14). There are three
eIF4A isoforms in mammals, i.e. eIF4A1-3 (15). As a nuclear protein and component of
the exon junction complex (EJC), eIF4A3 is distinct from the other
isoforms, and is essential in anchoring the EJC to the RNA molecule
(16). Secondly, eIF4A1 and eIF4A2
share 91% amino acid sequence homology and are considered to be
functionally indistinguishable (17). However, recent studies have reported
that unlike eIF4A1, eIF4A2 interacts with the CCR4-NOT
transcription complex and is involved in the inhibition of
miRNA-mediated translation (18). As
such, the upregulation of eIF4A1 is reportedly associated with a
diverse range of malignancies, including non-small cell lung cancer
(NSCLC), endometrial carcinoma, cervical cancer, breast cancer,
malignant peripheral nerve sheath tumors and pancreatic ductal
adenocarcinoma (PDA) (19–24). Several relevant studies have also
demonstrated that the expression of eIF4A2 is positively correlated
with the prognosis of NSCLC and breast cancer (25,26).
However, a recent study revealed that high eIF4A2 expression in
colorectal cancer (CRC) was associated with a poor survival rate.
Additionally, the results of cellular and animal experiments have
confirmed the functions of eIF4A2 in promoting CRC metastasis and
oxaliplatin resistance (27). Such
discrepancies may be attributed to the cancer type or the diversity
of oncogene-stimulated signaling networks. As a result, the
association between eIF4A2 expression and any specific type of
cancer demands independent research and cautious investigation.
In the present study, 253 ESCC patient samples were
collected and the expression of eIF4A2 was detected by
immunohistochemical (IHC) staining. The function of eIF4A2 in the
prognosis of ESCC was then ascertained.
Materials and methods
Patient and tissue specimens
The present study was approved by the Medical Ethics
Committee of Sun Yat-Sen University Cancer Center (Guangzhou,
China), and all enrolled patients provided written informed
consent. A total of 253 patient specimens from 186 males and 67
females; the median patient age was 58 years (age range, 32–80
years) were collected from post-operative patients with ESCC, the
validity of which were confirmed by pathological review following
IHC staining. All 253 patients were observed at Sun Yat-Sen
University Cancer Center (Guangzhou, China) between January 2000
and December 2007, during which they were clinically and
histologically diagnosed with ESCC. The histological grade and
clinical stage of the tumors were recorded according to the 8th
edition of the Tumor-Node-Metastasis (TNM) classification of the
International Union Against Cancer (2018) (28). A patient was selected as a
qualifiable subject if the following five requirements were met: i)
The patient was diagnosed with histologically confirmed primary
ESCC, but had not received any previous treatment elsewhere; ii)
the patient had no family history of cancer; iii) the patient had
undergone radical surgery with lymphadenectomy (limited or
extended); iv) the patient had not received neoadjuvant or adjuvant
treatments; and v) the clinical information and follow-up data of
the patient were documented. As a result, 253 patients constituted
the complete set of specimens, and their clinical data were
obtained from hospital records. The patients were followed up in
2019 to ascertain the latest disease status.
A tissue microarray was constructed according to
previously specified methods (29).
Samples from the 253 patient specimens were fixed in formalin for
24 hours at room temperature and then embedded in paraffin. The
paraffin blocks were sliced into 3-µm sections. The sections were
then reviewed by a senior pathologist and representative tumor
regions were defined by hematoxylin and eosin staining. Next, two
targeted core samples from each tissue specimen were obtained using
a tissue array instrument (MiniCore® instruments;
ALPHELYS). Tissue cylinders with a diameter of 10 mm were punched
and arrayed in a recipient paraffin block. Sections of the tissue
array were then cut and arranged on glass slides.
IHC staining and assessment
Following paraffin embedding, the tissue sections
were dried in incubator at 60°C for 2 h, then deparaffinized in
xylene and rehydrated in a descending graded alcohol series (100,
95, 90, 85 and 75%), and then incubated in 0.3% hydrogen peroxide
for 15 min to block endogenous peroxidase activity. Antigen
retrieval was conducted by heating in EDTA buffer (pH 8.0) for 3
min in a pressure cooker. The sections were blocked for 10 min with
10% normal goat serum (cat. no. ab7481; Abcam) at room temperature,
and then incubated with a polyclonal antibody against eIF4A2
(dilution, 1:1,000; cat. no. ab31218; Abcam) for 12 h in a moist
chamber (4°C). Control samples were incubated in blocking solutions
without the primary antibody. The slides were then incubated with
horseradish peroxidase for 30 min and visualized using
3,30-diaminobenzidine solution. Mayer's hematoxylin was applied as
a counterstain at room temperature for 1 min. Negative control
staining was performed by replacing the primary antibody with
normal murine immunoglobulin G (1:200; cat. no. A7028; Beyotime)
for 12 h in a moist chamber at 4°C. eIF4A2-positive slides were
used as positive controls, and the internal negative controls
consisted of normal squamous esophageal mucosal samples from
cancer-free patients.
Next, two independent pathologists (Lyu and Yan)
blinded to the clinicopathological data evaluated eIF4A2 expression
in the tissue specimens with a light microscope (magnifications,
×100 and ×200). The evaluation approach followed two scoring
criteria: i) The positive cell proportion score was assigned
according to the percentage of positive tumor cells: 0 (0%), 1
(1–10%), 2 (11–50%), 3 (51–70%) and 4 (71–100%); and ii) the
staining intensity score was graded in accordance with the signal
intensity: 0 (no signal), 1 (weak), 2 (medium) and 3 (strong). The
immunoreactivity score (IRS) for eIF4A2 expression was calculated
by multiplying the positive cell proportion and the staining
intensity scores (range, 0–12). The specimens were re-evaluated if
the scores submitted by the two pathologists displayed discrepancy.
In addition, eIF4A2 expression data were downloaded from UALCAN
(http://ualcan.path.uab.edu/index.html), an interactive
web portal for the in-depth analysis of The Cancer Genome Atlas
(TCGA) gene expression data.
Selection of cut-off values
The survival receiver operating characteristic (ROC)
package in R (R software version 3.0.1 and the survival ROC
package; R Foundation for Statistical Computing; www.R-project.org) was used to determine the IRS
cut-off values. The sensitivity and specificity for each outcome
were plotted and ROC curves were generated. The score was selected
as the cut-off value which was closest to the point with both
maximum sensitivity and specificity. For eIF4A2 expression, the
cut-off value for the staining intensity score (0–3), proportion
score (0–4) and IRS (0–12) were assessed separately.
Clinicopathological features, including age (68 years old or >50
years old), sex (male or female), histological grade (1–2 or 3),
tumor status (T1, T2, T3 or T4), nodal status (N0 or N1) and TNM
stage (1–2 or 3) were also evaluated.
Statistical analysis
Survival analysis was performed using SPSS version
25.0 (IBM Corp.). Pearson's χ2 test was used to
determine the association between eIF4A2 expression and patient
clinicopathological features. Disease-free survival (DFS) was
defined as the time between surgery and regional relapse or the
development of distant metastasis. Overall survival (OS) was
defined as the time between surgery and death. The Kaplan-Meier
estimator was used to assess DFS and OS, the results of which were
further compared using the log-rank test. Multivariate survival
analysis was performed using the Cox regression model for all
variables that were identified as significant during univariate
analysis. For all analyses, P<0.05 (two-sided) was considered to
indicate a statistically significant difference.
Results
Demographics
Of the study population, 59 patients were diagnosed
with well-differentiated squamous cell carcinoma (SCC), 163
patients presented with moderately-differentiated SCC and 31 with
poorly-differentiated SCC. The number of patients confirmed to have
T1, T2, T3 and T4 disease was 6, 61, 183 and 3, respectively, and
118 of the 253 patients exhibited nodal metastases. With regards to
TNM staging, 156 patients were diagnosed with stage 1 and 2
disease, and 97 patients presented with stage 3 disease. The
mortality rate across all patients was 52.6% (133 out of 253), and
the median OS and DFS times were 40 (4–115) and 38 (1–115) months,
respectively.
IHC detection of eIF4A2 expression in
ESCC
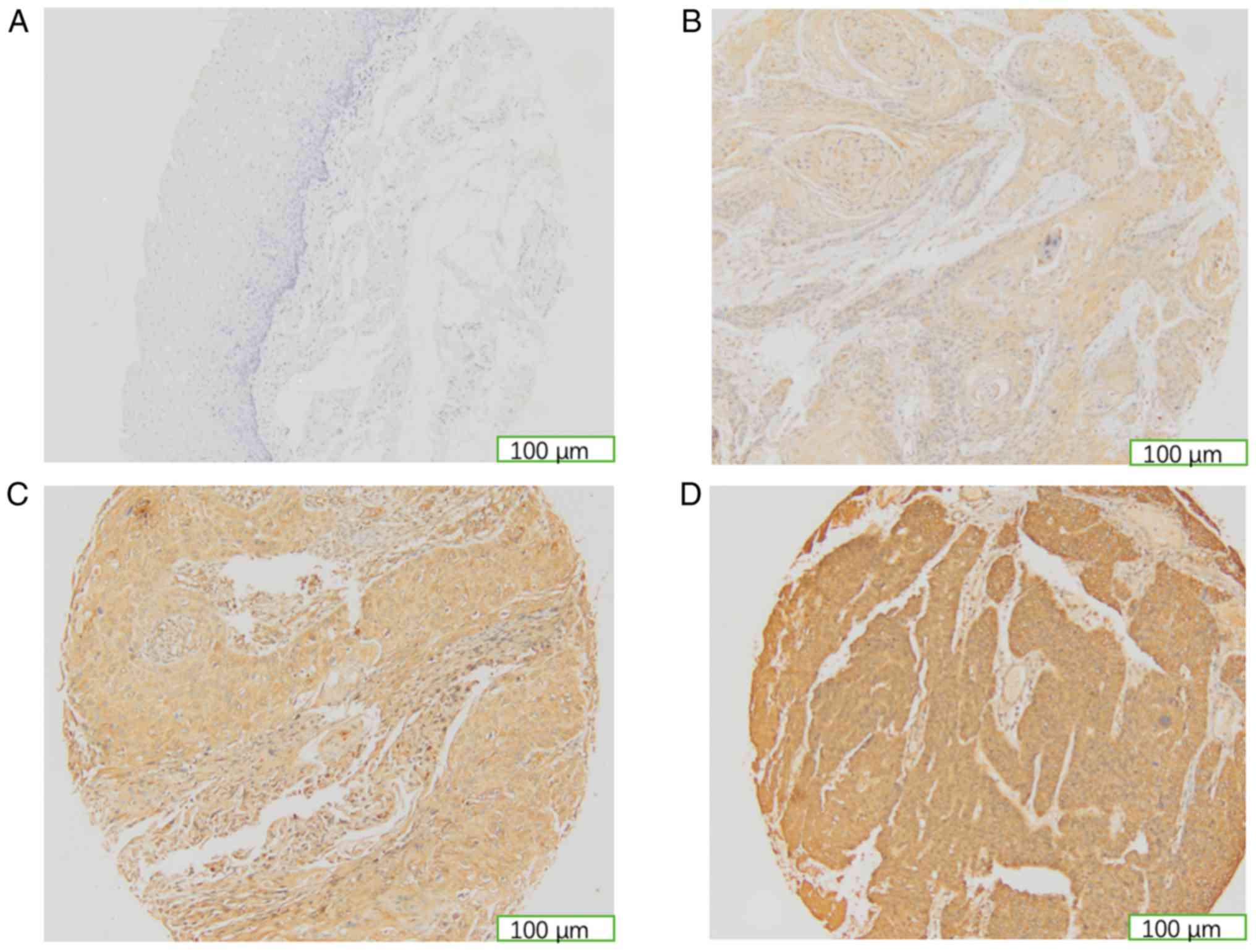
The results of the present study demonstrated that
eIF4A2 expression is localized to the cytoplasm of ESCC cells.
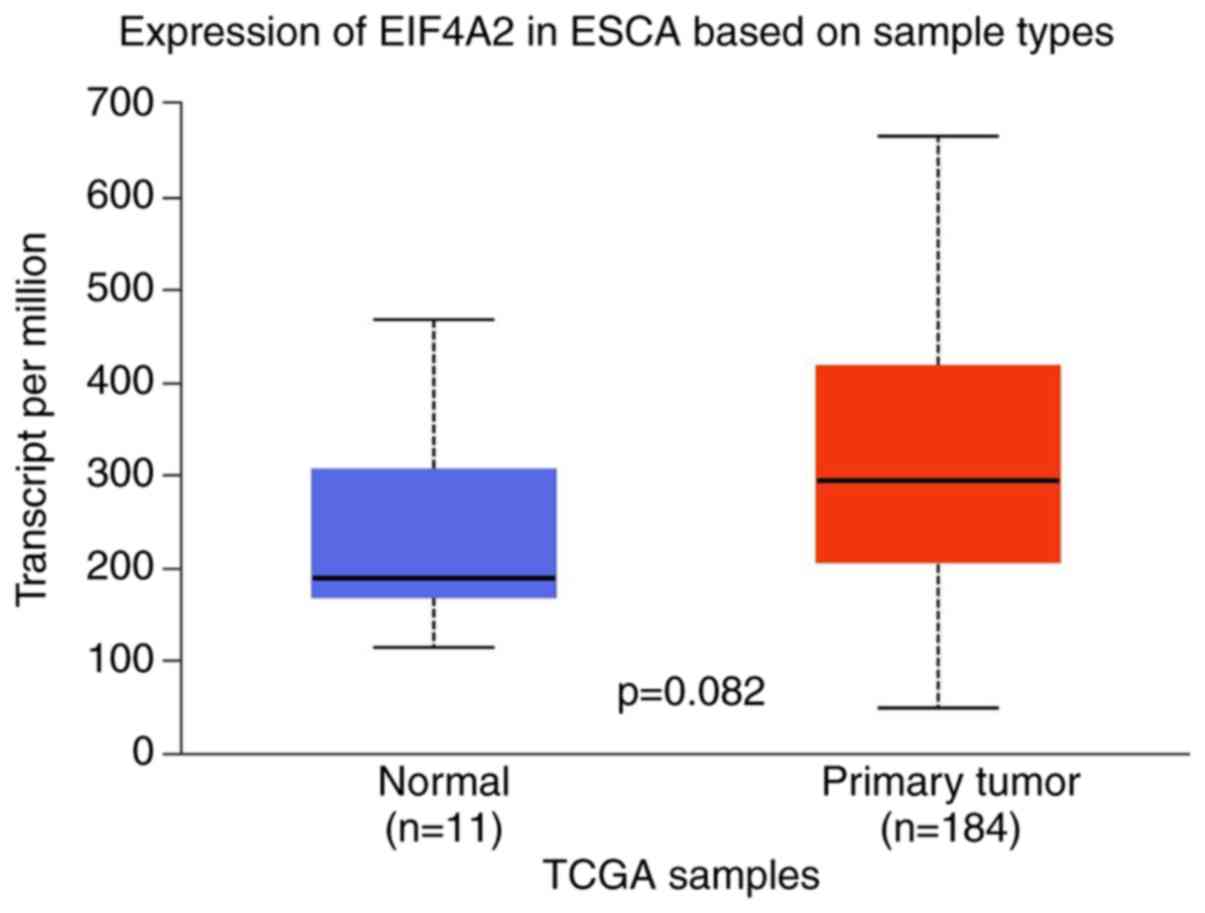
eIF4A2 was also revealed to be more highly expressed in esophageal
carcinoma tissues than in the normal control samples, displaying a
similar expression trend to that determined by TCGA database
analysis, though not statistically significant (Figs. 1 and 2). In the present study, samples with a
signal intensity score of 3 (strong) were considered to exhibit
high eIF4A2 expression, while those with a score of 1 (weak) or 2
(medium) were considered to exhibit low expression intensity. High
eIF4A2 expression was detected in 53 patients, while the remaining
200 samples presented with low eIF4A2 expression. None of the
samples scored 0 (Fig. 2). The
cut-off value for the proportion score (0–4) was 1, indicating that
samples with a score ≤1 were considered to exhibit a low proportion
of eIF4A2 expression, and those with a score of >1 were regarded
to express a high proportion of eIF4A2. There were 10 patients with
a low, and 243 patients with a high proportion of eIF4A2
expression. For the IRS (0–12), ESCC samples with a score >9
were defined as highly eIF4A2-immunoreactive. Therefore, in the
present study, 214 patients possessed high eIF4A2 immunoreactivity
and the remaining 39 patients presented with low immunoreactivity.
However, Kaplan-Meier analysis revealed that only the intensity
score was significantly associated with DFS and OS, hence a high
eIF4A2 intensity was defined as high eIF4A2 expression, and low
intensity samples were defined by low eIF4A2 expression (Table I).
 | Table I.Univariate Cox regression analysis of
DFS and OS in patients with esophageal squamous cell carcinoma. |
Table I.
Univariate Cox regression analysis of
DFS and OS in patients with esophageal squamous cell carcinoma.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Ages, years |
|
>68 | 2.053 | 0.002 | 1.929 | 0.004 |
| ≤68 | (1.297–3.250) |
| (1.218–3.054) |
|
| Sex |
| Male | 1.202 | 0.359 | 1.240 | 0.284 |
|
Female | (0.811–1.782) |
| (0.837–1.839) |
|
| Histological
grade |
| 1-2 | 1.791 | 0.012 | 1.699 | 0.022 |
| 3 | (1.132–2.883) |
| (1.074–2.689) |
|
| Tumor status |
| T1 | 1.456 | 0.047 | 1.523 | 0.026 |
| T2 | (1.004–2.114) |
| (1.050–2.211) |
|
| T3 |
| T4 |
| Nodal status |
| N0 | 3.306 | <0.001 | 3.452 | <0.001 |
| N1 | (2.299–4.754) |
| (2.399–4.976) |
|
| TNM stage |
|
1-2 | 3.494 | <0.001 | 3.751 | <0.001 |
| 3 | (2.456–4.972) |
| (2.631–5.349) |
|
| eIF4A2
expression |
|
Low | 1.948 | <0.001 | 1.956 | <0.001 |
|
High | (1.347–2.818) |
| (1.352–2.829) |
|
| Proportion of
eIF4A2 expression |
|
Low | 0.520 | 0.086 | 0.576 | 0.151 |
|
High | (0.243–1.113) |
| (0.269–1.234) |
|
| IRS for eIF4A2
expression |
|
Low | 1.441 | 0.089 | 1.473 |
|
|
High | (0.944–2.189) |
| (0.965–2.248) | 0.071 |
Association between eIF4A2 expression
and the clinicopathological characteristics of patients with
ESCC
There were no statistically significant associations
between the general clinicopathological characteristics of the 253
patients and eIF4A2 expression (Table
II).
 | Table II.Association between specific
clinicopathological characteristics and eIF4A2 expression in
patients with esophageal squamous cell carcinoma. |
Table II.
Association between specific
clinicopathological characteristics and eIF4A2 expression in
patients with esophageal squamous cell carcinoma.
|
|
| eIF4A2
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Cases, n | Low (n=200) | High (n=53) | P-value |
|---|
| Ages, years |
|
|
| 0.353 |
|
>68 | 29 | 21 | 8 |
|
|
≤68 | 224 | 179 | 45 |
|
| Sex |
|
|
| 0.301 |
|
Male | 186 | 150 | 36 |
|
|
Female | 67 | 50 | 17 |
|
| Histological
grade |
|
|
| 0.071 |
| 1 | 59 | 51 | 8 |
|
| 2 | 163 | 127 | 36 |
|
| 3 | 31 | 22 | 9 |
|
| Tumor status |
|
|
| 0.279 |
| T1 | 6 | 4 | 2 |
|
| T2 | 61 | 53 | 8 |
|
| T3 | 183 | 141 | 42 |
|
| T4 | 3 | 2 | 1 |
|
| Nodal status |
|
|
| 0.482 |
| N0 | 135 | 109 | 26 |
|
| N1 | 118 | 91 | 27 |
|
| TNM stage |
|
|
| 0.116 |
| 1 | 9 | 8 | 1 |
|
| 2 | 147 | 120 | 27 |
|
| 3 | 97 | 72 | 25 |
|
Association between eIF4A2 expression
and the prognosis of patients with ESCC
According to the results of Kaplan-Meier analysis,
specific clinicopathological characteristics, including age,
histological grade, tumor, nodal and TNM staging were revealed to
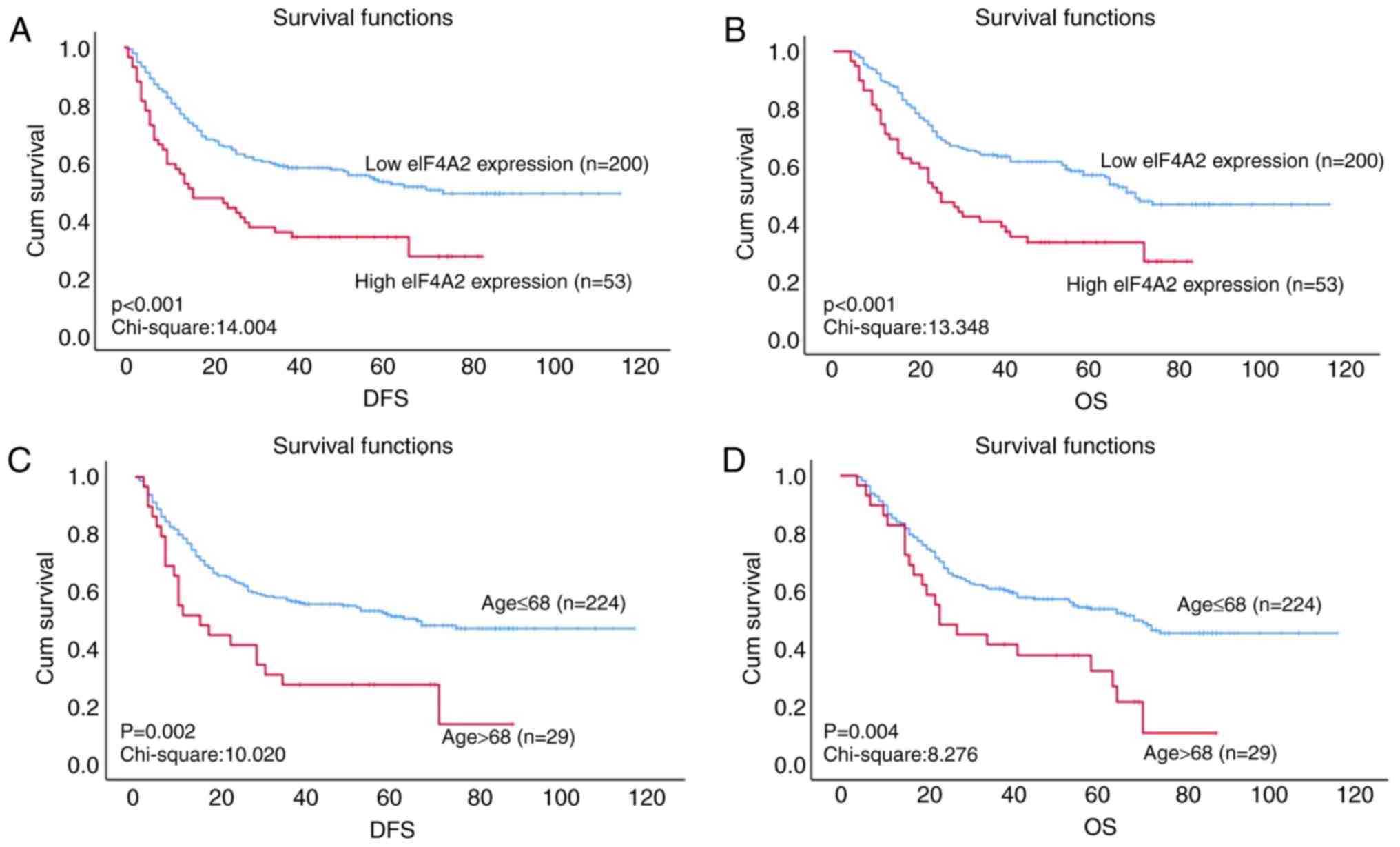
be significantly associated with DFS and OS time(P=0.002; Fig. 3C and D); the cut-off value for age
was set at 68 years. eIF4A2 expression was also revealed to be
associated with the prognosis of ESCC. The median DFS time was 40
months for patients with low eIF4A2 expression and 16 months for
those with high eIF4A2 expression (P<0.001; Fig. 3A). In addition, the median OS time
was 48 months for patient with low eIF4A2 expression, but 25 months
for those with high eIF4A2 expression (P<0.001; Fig. 3B). When patient specimens were
stratified into N stage subgroups, the association between eIF4A2
expression and DFS or OS became more significant. In the N0
subgroup, the median DFS and OS times of patients with low eIF4A2
expression were 62 and 65 months, respectively, while the median
DFS and OS times of those with high eIF4A2 expression were both 40
months (both P=0.002). In the N1 subgroup, the median DFS and OS
times of patients with low eIF4A2 expression were 19 and 27 months,
respectively, while the median DFS and OS times of those with high
eIF4A2 expression were 10 and 15 months, respectively (both
P=0.002). These data indicate that high expression levels of eIF4A2
are associated with a poor prognosis in patients with ESCC
(Fig. 3).
Univariate analysis was performed using the Cox
proportional hazards model to assess the importance of multiple
factors on the survival times of patients with ESCC. The results
indicated that age, histological grade, tumor and nodal status, TNM
stage and eIF4A2 expression are significantly associated with DFS
and OS time (Table I). Multivariate
analysis was then applied to investigate these identified
parameters. The results verified that only eIF4A2 expression and
age were independent prognostic factors for DFS and OS time in ESCC
(Table III).
 | Table III.Multivariate Cox regression analysis
of DFS and OS in patients with esophageal squamous cell
carcinoma. |
Table III.
Multivariate Cox regression analysis
of DFS and OS in patients with esophageal squamous cell
carcinoma.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Ages, years |
|
>68 | 2.0210 | 0.003 | 1.985 | 0.004 |
|
≤68 | (1.269–3.219) |
| (1.246–3.162) |
|
| Histological
grade |
|
1-2 | 1.577 | 0.052 | 1.537 | 0.068 |
| 3 | (0.995–2.500) |
| (0.969–2.437) |
|
| Tumor status |
| T1 | 1.030 | 0.905 | 1.088 | 0.733 |
| T2 | (0.636–1.667) |
| (0.670–1.768) |
|
| T3 |
| T4 |
| Nodal status |
| N0 | 2.063 | 0.06 | 2.128 | 0.054 |
| N1 | (0.969–4.391) |
| (0.989–4.581) |
|
| TNM stage |
|
1-2 | 1.772 | 0.175 | 1.828 | 0.16 |
| 3 | (0.775–4.050) |
| (0.789–4.239) |
|
| eIF4A2
expression |
|
Low | 1.755 | 0.003 | 1.804 | 0.002 |
|
High | (1.210–2.545) |
| (1.244–2.618) |
|
Discussion
In the present study, the expression of eIF4A2 in
ESCC samples was investigated by IHC staining. TCGA database
analysis revealed that eIF4A2 is more highly expressed in tumor
tissues than in normal control samples, and that the highest
amplification frequency of eIF4A2 is detected in ESCC (Figs. 1 and 2). Biologically identified as an
ATP-dependent RNA helicase, eIF4A2 belongs to the eukaryotic
initiation factor (eIF) family, which is essential for translation
initiation (30). Existing findings
have established various associations between the eIFs and
carcinogenesis, as well as patient prognosis (11,13,24). For
example, the upregulation of eIF4E functions as an effective
indicator for ~30% of all cancer cases tested, and the
phosphorylation of eIF4E promotes cellular proliferation and
inhibits apoptosis (12). eIF4A2 was
the focus of the present study, and is a subunit of eIF4A that is
associated with the oncogenic translation of PDA (24). eIF4A is a subunit of eIF4F that also
comprises eIF4E (a cap binding protein) and eIF4G (a regulatory
scaffold protein). The eIF4F family serves an important role in the
translation process. eIF4A comprises three subunits, eIF4A1, eIF4A2
and eIF4A3. Within the eIF4A family, the amino acid sequences of
eIF4A1 and eIF4A2 are homologues and are highly conserved (8,17), and
dysregulation of either of these molecules is associated with a
diverse range of malignancies (20,22,25,27). In
the present study, the expression rate of eIF4A2 in the ESCC cohort
was 100% (253 out of 253). Additionally, 20.9% (53 out of 253) of
patient specimens exhibited high eIF4A2 expression. However, no
statistically significant correlations were detected between eIF4A2
expression and general clinicopathological characteristics. An
existing study demonstrated that eIF4A2 is more highly expressed in
lung SCC than in adenocarcinoma, and that eIF4A2 downregulation
indicates a poor prognosis in patients with NSCLC (25). The present study had a distinct focus
and established that high eIF4A2 expression is associated with a
poor patient prognosis in ESCC.
Another aim of the present study was to investigate
the association between abnormal eIF4A2 expression and the
prognosis of patients with ESCC. High eIF4A2 expression was
revealed to be significantly associated with a poor outcome in
ESCC, as determined by Kaplan-Meier and log-rank analysis. However,
this analytical approach tends to produce false positive results.
The log-rank test can more sensitively predict long-term
differences, while the Breslow test is more appropriately used to
predict short-term differences in ending events. Therefore, when
analyzing survival curves with similar data values over a given
time frame, the log-rank test is more likely to generate a
significant result than the Breslow test. By contrast, for survival
curves that differ considerably at the initial time points, but
draw closer over time, significant results are more likely to be
obtained using the Breslow method. The results of the present study
were verified using the Breslow test (P<0.001), and univariate
analysis was performed using the Cox proportional hazards model to
correct confounding factors (P=0.003). Therefore, the final results
were concluded to be reliable.
The association between eIF4A2 and
clinicopathological characteristics became statistically
significant when the patients were classified into different N
stage subgroups, which was also consistent with the survival curve
trend shown in TCGA database. eIF4A1 and eIF4A2 share a 91%
homologous amino acid sequence and are considered to be
functionally indistinguishable (17). Increased expression of eIF4A1 has
been associated with NSCLC metastasis and the poor outcomes of
patients with cervical and breast cancer. Knocking down eIF4A2 has
also been reported to inhibit sphere formation and experimental
metastasis in CRC, and eIF4A2 expression was revealed to be
positively correlated with the prognosis of patients with NSCLC and
breast cancer (20,22,27).
Furthermore, TCGA analysis revealed that high eIF4A2 expression is
negatively correlated with the prognosis of those with liver
cancer, head and neck cancer, melanoma and prostate cancer. The
Bushell lab demonstrated that eIF4A2 contributes toward the
repression of miRNA translation initiation via an interaction
between DDX6 and the CCR4-NOT complex, and that eIF4A2 inhibits the
deadenylation activity of CNOT7 subunit, which further results in
translational repression (31).
eIF4A2 has also been hypothesized to function through the Myc
pathway, though further investigation is required to confirmation
this (27). In the present study,
the Cox proportional hazards model identified eIF4A2 expression as
an independent prognostic factor for ESCC. However, the molecular
mechanisms by which eIF4A2 dysregulation contributes toward the
prognosis of ESCC remain unclear. The discrepancy in the
correlation between eIF4A2 expression and cancer may be attributed
to the nature of the different cancer types, or the diversity of
oncogene-stimulated signaling networks.
In the present study, age was also identified as an
independent prognostic factor, and was significantly correlated
with DFS and OS time. The cut-off value for age was set at 68
years, indicating that patients >68 years old with ESCC had
poorer outcomes. Previous studies have also identified sex, T
staging and N staging as independent prognostic indicators of ESCC
(4,32). Due to the limited sample size of the
present study, univariate analysis revealed an association between
eIF4A2 expression and patient clinicopathological characteristics,
including histological grade, tumor, nodal and TNM staging, and
survival without characterizing these factors as independent
indicators.
In conclusion, there are two major limitations to
the present study: i) The molecular mechanisms by which eIF4A2
dysregulation contributes toward the prognosis of ESCC remain
unclear, and require further investigation; and ii) the sample size
of the study was not sufficient to establish the correlation
between eIF4A2 expression and general clinicopathological
characteristics in survival analysis. However, these findings
demonstrate the association between high eIF4A2 expression and the
poor prognosis of patients, as well as the potential of eIF4A2 as
an effective prognostic indicator in ESCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SY and SX are responsible for the study design. SL
and JL performed the experiments and draft the manuscript. WC, WH
and HH participated in the data analysis and interpretation. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Sun Yat-Sen University Cancer Center (approval no.
GZR2018-220) (Guangzhou, China) and all patients provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gholipour C, Shalchi RA and Abbasi M: A
histopathological study of esophageal cancer on the western side of
the Caspian littoral from 1994 to 2003. Dis Esophagus. 21:322–327.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pickens A and Orringer MB: Geographical
distribution and racial disparity in esophageal cancer. Ann Thorac
Surg. 76:S1367–S1369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen MF, Yang YH, Lai CH, Chen PC and Chen
WC: Outcome of patients with esophageal cancer: A nationwide
analysis. Ann Surg Oncol. 20:3023–3030. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smyth EC, Lagergren J, Fitzgerald RC,
Lordick F, Shah MA, Lagergren P and Cunningham D: Oesophageal
cancer. Nat Rev Dis Primers. 3:170482017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhat M, Robichaud N, Hulea L, Sonenberg N,
Pelletier J and Topisirovic I: Targeting the translation machinery
in cancer. Nat Rev Drug Discov. 14:261–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spilka R, Ernst C, Mehta AK and Haybaeck
J: Eukaryotic translation initiation factors in cancer development
and progression. Cancer Lett. 340:9–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aitken CE and Lorsch JR: A mechanistic
overview of translation initiation in eukaryotes. Nat Struct Mol
Biol. 19:568–576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hinnebusch AG and Lorsch JR: The mechanism
of eukaryotic translation initiation: New insights and challenges.
Cold Spring Harb Perspect Biol. 4:a0115442012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Donzé O, Jagus R, Koromilas AE, Hershey JW
and Sonenberg N: Abrogation of translation initiation factor eIF-2
phosphorylation causes malignant transformation of NIH 3T3 cells.
EMBO J. 14:3828–2834. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shahbazian D, Parsyan A, Petroulakis E,
Topisirovic I, Martineau Y, Gibbs BF, Svitkin Y and Sonenberg N:
Control of cell survival and proliferation by mammalian eukaryotic
initiation factor 4B. Mol Cell Biol. 30:1478–1485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bitterman PB and Polunovsky VA:
eIF4E-mediated translational control of cancer incidence. Biochim
Biophys Acta. 1849:774–780. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bao Y, Lu Y, Wang X, Feng W, Sun X, Guo H,
Tang C, Zhang X, Shi Q and Yu H: Eukaryotic translation initiation
factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer
through epithelial mesenchymal transition. Cancer Cell Int.
15:1092015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wolfe AL, Singh K, Zhong Y, Drewe P,
Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van
der Meulen J, et al: RNA G-quadruplexes cause eIF4A-dependent
oncogene translation in cancer. Nature. 513:65–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meijer HA, Kong YW, Lu WT, Wilczynska A,
Spriggs RV, Robinson SW, Godfrey JD, Willis AE and Bushell M:
Translational repression and eIF4A2 activity are critical for
microRNA-mediated gene regulation. Science. 340:82–85. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chan CC, Dostie J, Diem MD, Feng W, Mann
M, Rappsilber J and Dreyfuss G: eIF4A3 is a novel component of the
exon junction complex. RNA. 10:200–209. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Q, Imataka H, Morino S, Rogers GW Jr,
Richter-Cook NJ, Merrick WC and Sonenberg N: Eukaryotic translation
initiation factor 4AIII (eIF4AIII) is functionally distinct from
eIF4AI and eIF4AII. Mol Cell Biol. 19:7336–7346. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilczynska A, Gillen SL, Schmidt T, Meijer
HA, Jukes-Jones R, Langlais C, Kopra K, Lu WT, Godfrey JD, Hawley
BR, et al: eIF4A2 drives repression of translation at initiation by
Ccr4-Not through purine-rich motifs in the 5′UTR. Genome Biol.
20:2622019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang S, Zhou Y, Chen Y, Ke G, Wen H and
Wu X: Decreased expression of EIF4A1 after preoperative
brachytherapy predicts better tumor-specific survival in cervical
cancer. Int J Gynecol Cancer. 24:908–915. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lomnytska MI, Becker S, Gemoll T, Lundgren
C, Habermann J, Olsson A, Bodin I, Engström U, Hellman U, Hellman
K, et al: Impact of genomic stability on protein expression in
endometrioid endometrial cancer. Br J Cancer. 106:1297–1305. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Modelska A, Turro E, Russell R, Beaton J,
Sbarrato T, Spriggs K, Miller J, Gräf S, Provenzano E, Blows F, et
al: The malignant phenotype in breast cancer is driven by
eIF4A1-mediated changes in the translational landscape. Cell Death
Dis. 6:e16032015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oblinger JL, Burns SS, Akhmametyeva EM,
Huang J, Pan L, Ren Y, Shen R, Miles-Markley B, Moberly AC,
Kinghorn AD, et al: Components of the eIF4F complex are potential
therapeutic targets for malignant peripheral nerve sheath tumors
and vestibular schwannomas. Neuro Oncol. 18:1265–1277. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chan K, Robert F, Oertlin C,
Kapeller-Libermann D, Avizonis D, Gutierrez J, Handly-Santana A,
Doubrovin M, Park J, Schoepfer C, et al: eIF4A supports an
oncogenic translation program in pancreatic ductal adenocarcinoma.
Nat Commun. 10:51512019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shaoyan X, Juanjuan Y, Yalan T, Ping H,
Jianzhong L and Qinian W: Downregulation of EIF4A2 in
non-small-cell lung cancer associates with poor prognosis. Clin
Lung Cancer. 14:658–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ,
Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, et al: Knockdown of miR-21 in
human breast cancer cell lines inhibits proliferation, in vitro
migration and in vivo tumor growth. 13(R2)2011.
|
|
27
|
Chen ZH, Qi JJ, Wu QN, Lu JH, Liu ZX, Wang
Y, Hu PS, Li T, Lin JF, Wu XY, et al: Eukaryotic initiation factor
4A2 promotes experimental metastasis and oxaliplatin resistance in
colorectal cancer. J Exp Clin Cancer Res. 38:1962019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rice TW, Kelsen DP and Blackstone EH:
Esophagus and Esophagogastric Junction. AJCC Cancer Staging Manual.
8th. Amin MB, Edge SB and Greene FL: New York, NY: Springer; pp.
185–202. 2017, View Article : Google Scholar
|
|
29
|
Dancau AM, Simon R, Mirlacher M and Sauter
G: Tissue microarrays. Methods Mol Biol. 576:49–60. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hinnebusch AG: The scanning mechanism of
eukaryotic translation initiation. Annu Rev Biochem. 83:779–812.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meijer HA, Schmidt T, Gillen SL, Langlais
C, Jukes-Jones R, de Moor CH, Cain K, Wilczynska A and Bushell M:
DEAD-box helicase eIF4A2 inhibits CNOT7 deadenylation activity.
Nucleic Acids Res. 47:8224–8238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng YF, Chen HS, Wu SC, Chen HC, Hung
WH, Lin CH and Wang BY: Esophageal squamous cell carcinoma and
prognosis in Taiwan. Cancer Med. 7:4193–4201. 2018. View Article : Google Scholar : PubMed/NCBI
|