Introduction
Hepatocellular carcinoma (HCC) is the fourth most
common cause of cancer-associated death worldwide (1,2). Due to
the limited improvements in the diagnosis and treatment of HCC over
the past two decades, the prognosis and survival rate of patients
with HCC remain poor (3,4). The high mortality rate of HCC, with
~782,000 deaths annually, is largely caused by its metastases and
recurrence (5). Therefore, an
understanding of the mechanisms underlying the invasion and
metastasis of HCC is key to improving the treatment of this
disease.
Previous studies have demonstrated that aberrant
activation of the Hedgehog (Hh) signaling pathway serves a critical
role in the tumorigenesis and progression of various types of
cancer, including pancreatic, breast, lung, ovarian,
gastrointestinal cancer and HCC (6–8). As a
key component of the Hh signaling pathway, glioma-associated
oncogene homolog 1 (Gli1) is a member of the family of zinc finger
transcription factors that functions as a downstream protein in the
Hh signaling pathway. Gli1 is a reliable activation marker of the
Hh signaling pathway (9). Emerging
evidence has demonstrated that Gli1 is overexpressed in a number of
types of cancer, including ovarian (10), prostate (11), gastric (12), esophageal squamous cell (13), pancreatic (14) and liver cancer (15). A recent study concluded that
hyperactivation of Gli1 is sufficient to trigger the uncontrolled
progression of cancer features such as proliferation, migration and
invasion (16). However, the
underlying mechanisms of Gli1 in the invasion and metastasis of HCC
remain to be elucidated.
The focal adhesion kinase-phosphoinositide
3-kinase-AKT (FAK/PI3K/AKT) axis is a critical pathway that
regulates the invasion and metastasis of HCC (17). Briefly, FAK, a non-receptor protein
tyrosine kinase, is an essential factor that regulates the PI3K/AKT
signaling pathway, which is associated with a poor prognosis in
different types of cancer (18).
Activation of FAK can phosphorylate PI3K and subsequently lead to
the phosphorylation of AKT (19).
Phosphorylation of FAK is important for invasion and metastasis as
it regulates the expression of matrix metalloproteinase (MMP)-2 and
MMP-9 in HCC (20). Overexpression
of FAK has been detected in various malignant tumors, including
breast, prostate, colorectal and ovarian cancer (21–24).
Ectopic expression of FAK and phosphorylated (p-)FAK are associated
with the invasion and metastasis of HCC (20,25).
Furthermore, phosphorylation of FAK can activate the PI3K/AKT
signaling pathway, leading to regulation downstream of the pathway
that transduces a β1 integrin viability signal in collagen matrices
(26).
The present study evaluated the roles of Gli1 in
liver cancer cells. Gaining an understanding of the mechanism
underlying the function of Gli1 in the invasion and metastasis of
HCC will provide a potential target to control the invasion and
metastasis of HCC.
Materials and methods
Cell lines and culture
Human liver cancer cell lines (HepG2 and SK-Hep1)
were purchased from the Cell Bank of the Chinese Academy of
Sciences. Cells were cultured in RPMI-1640 (Biological Industries)
supplemented with 10% fetal bovine serum (FBS; Biochrom, Ltd.) and
1% penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) in a humidified 5% CO2 atmosphere at 37°C.
RNA interference
Small interfering (si)RNA specific for Gli1
(si-Gli1) sequences and scrambled negative control (NC) siRNA with
Gli1 analogue nonsense sequences were obtained from Sangon Biotech
Co., Ltd. and were as follows: Gli1 (sense:
5′-CCAGUGUCCUCGACUUGAAdTdT-3′; antisense:
5′-UUCAAGUCGAGGACACUGGTdTd-3′) and NC (sense:
5′-UUCUCCGAACGUGUCACGUTT-3′; antisense:
5′-ACGUGACACGUUCGGAGAATT-3′). Cells were seeded into 6-well plates
and cultured to 80% confluence and then transfected with 50 nmol/l
NC siRNA, and 50 or 100 nmol/l si-Gli1 for 24 h at 37°C using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Briefly, cells were divided into four groups: A blank control
(CTRL) group (only treated with serum-free medium), a NC group
(transfected with NC siRNA), a 50 nmol/l si-Gli1 group and a 100
nmol/l si-Gli1 group. All subsequent experiments were performed 24
h post-transfection.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from liver cancer cells
(HepG2 and SK-Hep1) using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and 5 µg RNA was reverse
transcribed to complementary DNA (cDNA) using a RevertAid First
Strand cDNA synthesis kit (Invitrogen; Thermo Fisher Scientific,
Inc.). Subsequently, qPCR was performed using FastStart Universal
SYBR-Green Master (cat. no. 31168620; Roche Diagnostics GmbH),
according to the manufacturer's protocol, and performed on an ABI
7500 Real-time PCR system (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: 95°C Denaturation for 15 min and
then 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The relative
mRNA expression levels of target genes were normalized to β-actin
and calculated by use of the 2−ΔΔCq method (27). The specific sequences of primers were
as follows: Gli1 forward: 5′-AGGGCTGCAGTAAAGCCTTCA-3′, Gli1
reverse: 5′-CCTGACATGTTTTCGCAGCG-3′; MMP-2 forward:
5′-GACAACGCCCCCATACCAG-3′, MMP-2 reverse:
5′-CACTCGCCCCGTGTGTTAGT-3′; MMP-9 forward:
5′-ACGCAGACATCGTCATCCAGT-3′, MMP-9 reverse:
5′-GGACCACAACTCGTCATCGTC-3′; β-actin forward:
5′-GTGGACATCCGCAAAGAC-3′, β-actin reverse:
5′-GAAAGGGTGTAACGCAACT-3′. All experiments were performed in
triplicate.
Western blotting
Briefly, protein was extracted from cells
(5×106/well) using 6X sample buffer (1 M Tris-HCl, pH
6.8, 10% SDS, 20% glycerol, 0.5% Bromphenol Blue and
2-mercaptoethanol) for 10 min at 100°C and then centrifuged at
10,000 × g for 15 min on ice. The total protein concentrations were
measured using a BCA protein assay kit (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The protein
samples (10 µg/lane) were separated by 10% SDS-PAGE and transferred
to polyvinylidene fluoride membranes (EMD Millipore). Subsequently,
the membranes were incubated with blocking buffer (5% non-fat dry
milk and 0.05% Tween-20 in PBS) for 1 h at 37°C and the following
primary antibodies overnight at 4°C: Rabbit anti-Gli1 (Cell
Signaling Technology, Inc.; cat. no. 3538S; 1:1,000 dilution),
rabbit anti-MMP-2 (Cell Signaling Technology, Inc.; cat. no.
40994S; 1:1,000 dilution), rabbit anti-MMP-9 (Abcam; cat. no.
ab38898; 1:1,000 dilution), rabbit anti-FAK (Abcam; cat. no.
ab40794; 1:1,000 dilution), rabbit anti-p-FAK (Abcam; cat. no.
ab39967; 1:1,000 dilution), rabbit anti-PI3K (Cell Signaling
Technology, Inc.; cat. no. 4257S; 1:1,000 dilution), rabbit
anti-p-PI3K (Cell Signaling Technology, Inc.; cat. no. 4228S;
1:1,000 dilution), mouse anti-AKT (Cell Signaling Technology, Inc.;
cat. no. 2920S; 1:1,000 dilution), rabbit anti-p-AKT (Abcam; cat.
no. ab81283; 1:1,000 dilution) and mouse anti-β-Actin (Abcam; cat.
no. ab8226; 1:2,000 dilution). Finally, the membranes were
incubated with goat anti-rabbit or goat anti-mouse horseradish
peroxidase-conjugated secondary antibodies (Sigma-Aldrich; Merck
KGaA; cat. nos. A8919 and AP186P; 1:10,000 dilution) for 1 h at
room temperature. The immunoblot bands were visualized using an
enhanced chemiluminescence kit (cat. no. E1328; Beijing Lilye
Science & Technology Co., Ltd., http://www.micro-helix.com) and exposed on the gel
imaging analysis system (Amersham Imager 600; Cytiva). The
intensity of bands was quantified using ImageJ software (version
1.48; National Institutes of Health).
Immunofluorescence staining
The cells transfected with or without si-Gli1 were
fixed in 4% paraformaldehyde/PBS for 15 min at −20°C and then
washed three times using PBS containing 0.1% Triton X-100 (PBST)
for 15 min at room temperature. Following blocking in PBS
containing 5% BSA for 30 min at 37°C, cells were incubated with the
primary antibodies rabbit anti-FAK (Abcam; cat. no. ab40794;
1:1,000 dilution), rabbit anti-p-FAK (Abcam; cat. no. ab39967;
1:1,000 dilution) and rabbit anti-p-AKT (Abcam; cat. no. ab81283;
1:1,000 dilution) for 1 h at room temperature. The labeled cells
were washed three times with PBST and then incubated with Alexa
Fluor 594 (goat anti-rabbit IgG; Abcam, cat. no. ab150088; 1:1,000
dilution) for 1 h at room temperature in the dark. After washing
three times with PBST, the cell nuclei were stained with DAPI (1
µg/ml, Abcam, cat. no. ab228549) for 10 min at room temperature in
the dark and detected with a confocal laser scanning microscope
(LSM 880; Zeiss AG) at ×63 magnification. Cell clusters were
counted in five randomly selected fields using a fluorescence
microscope and the mean intensity was calculated using ImageJ
software.
Cell viability assay
Cell viability was determined by Cell Counting Kit
(CCK)-8 assay (Beijing Solarbio Science & Technology Co., Ltd.;
cat. no. CA-1210-500) following the manufacturer's instructions.
Briefly, liver cancer cells with or without transfection of si-Gli1
were incubated in 96-plates for 24 h at 37°C. Then, cells were
added to 10 µl CCK-8 staining reagent and then incubated for
another 4 h at 37°C. Absorbance at 450 nm was measured using a
microplate reader (Varioskan Lux; Thermo Fisher Scientific,
Inc.).
Adhesion assay
Cell adhesive abilities of SK-Hep1 cells in
different groups were determined using an adhesion assay. In brief,
50 µl Matrigel (BD Biosciences; cat. no. 8036008) was added to each
well in a 96-well plate for 30 min at 37°C and then
5×104 cells were seeded in the coated plates for
incubation for 4 h at 37°C. Finally, cells were washed with PBS to
wash away the unattached cells. The attached cells were analyzed
using the CCK-8 assay as described above.
Wound healing assay
Cell migratory abilities were detected using wound
healing assay. The prepared cells treated with or without si-Gli1
were cultured in 6-well plates to ~90% confluence using serum-free
medium. The confluent monolayer cells were scratched gently with a
100 µl pipette tip and images captured using an inverted microscope
(DMi8; Leica Microsystems GmbH; magnification, ×100) at 0 and 24 h.
The wound area was quantified using ImageJ software.
Transwell invasion assay
Cell invasion assay was performed using 24-well
Transwell plates with 8-µm pore filters (Corning Life Sciences;
cat. no. 3422) precoated with Matrigel Basement Membrane Matrix (BD
Biosciences) at 37°C for 1–4 h according to the manufacturer's
protocols. Briefly, 1×105 cells were suspended in 100 µl
of serum-free medium and added to the upper chamber and then 600 µl
RPMI-1640 supplemented with 20% FBS was loaded into the lower
chamber. After 24 h of incubation, the non-invaded cells in the top
chamber were removed with a cotton swab and the cells fixed with
100% methanol for 20 min at room temperature and stained with 0.1%
crystal violet (Beijing Solarbio Science & Technology Co.,
Ltd.; cat. no. G1063) for 30 min at room temperature. The stained
cells were imaged under an inverted light microscope (DMi8; Leica
Microsystems GmbH; magnification, ×100) and analyzed using ImageJ
software.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (version 7.0; GraphPad Software, Inc.). All
experiments were performed in triplicate and data are presented as
the mean ± standard deviation. Unpaired Student's t-test was used
to compare differences between two groups, whilst one-way analysis
of variance followed by Dunnett's post-hoc test were performed to
compare differences between multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
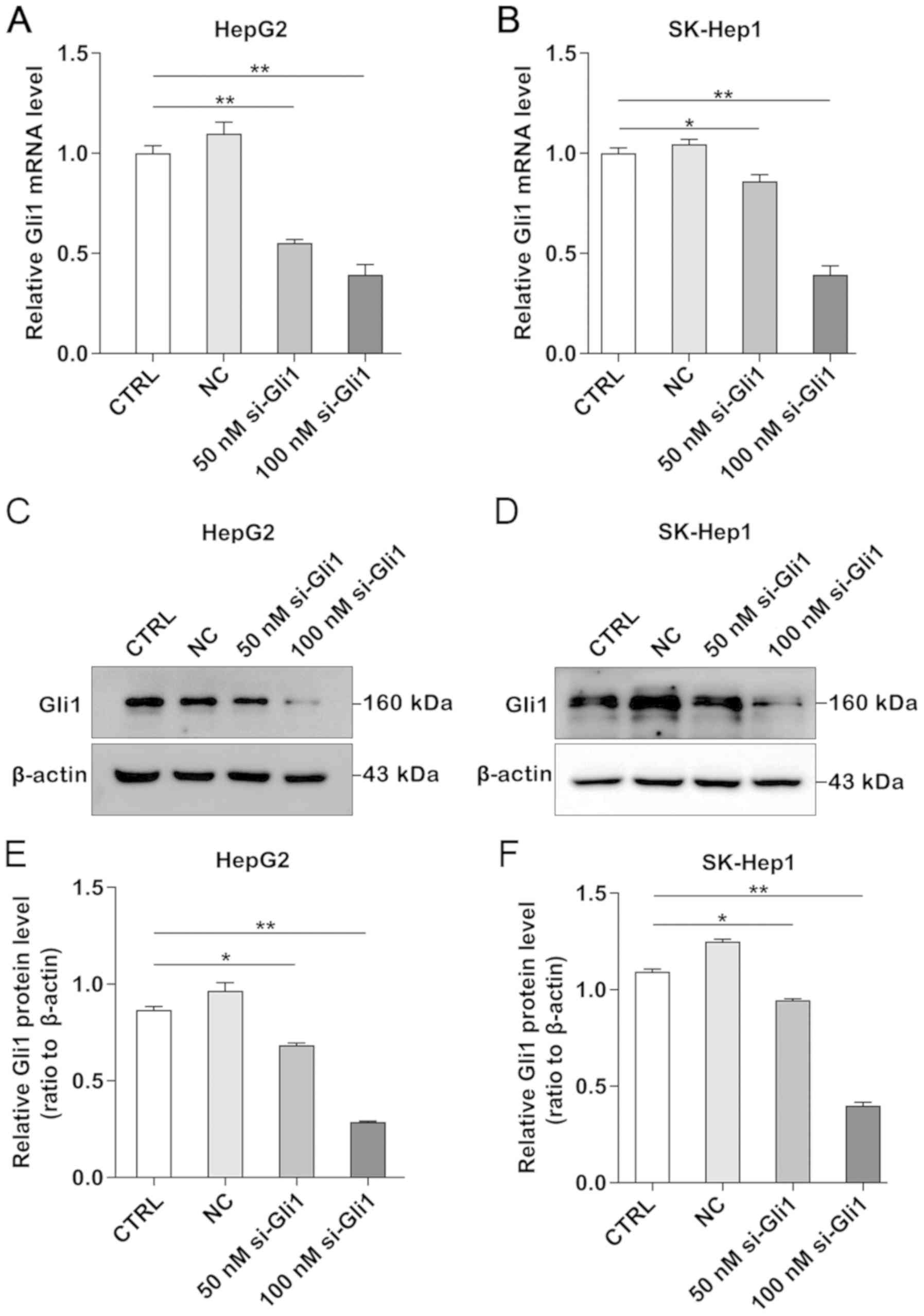
Knockdown of Gli1 inhibits Gli1
expression in HepG2 and SK-Hep1 cells
To understand the potential function of Gli1 in
liver cancer cells, Gli1 expression was downregulated by RNA
interference. The knockdown efficiency of Gli1 in liver cancer
cells was determined using RT-qPCR and western blotting. The
results demonstrated that the mRNA and protein expression levels
were significantly decreased in a dose-dependent manner and the
most efficient concentration for Gli1 silencing was 100 nmol/l in
both HepG2 and SK-Hep1 cells (P<0.05 and P<0.01; Fig. 1).
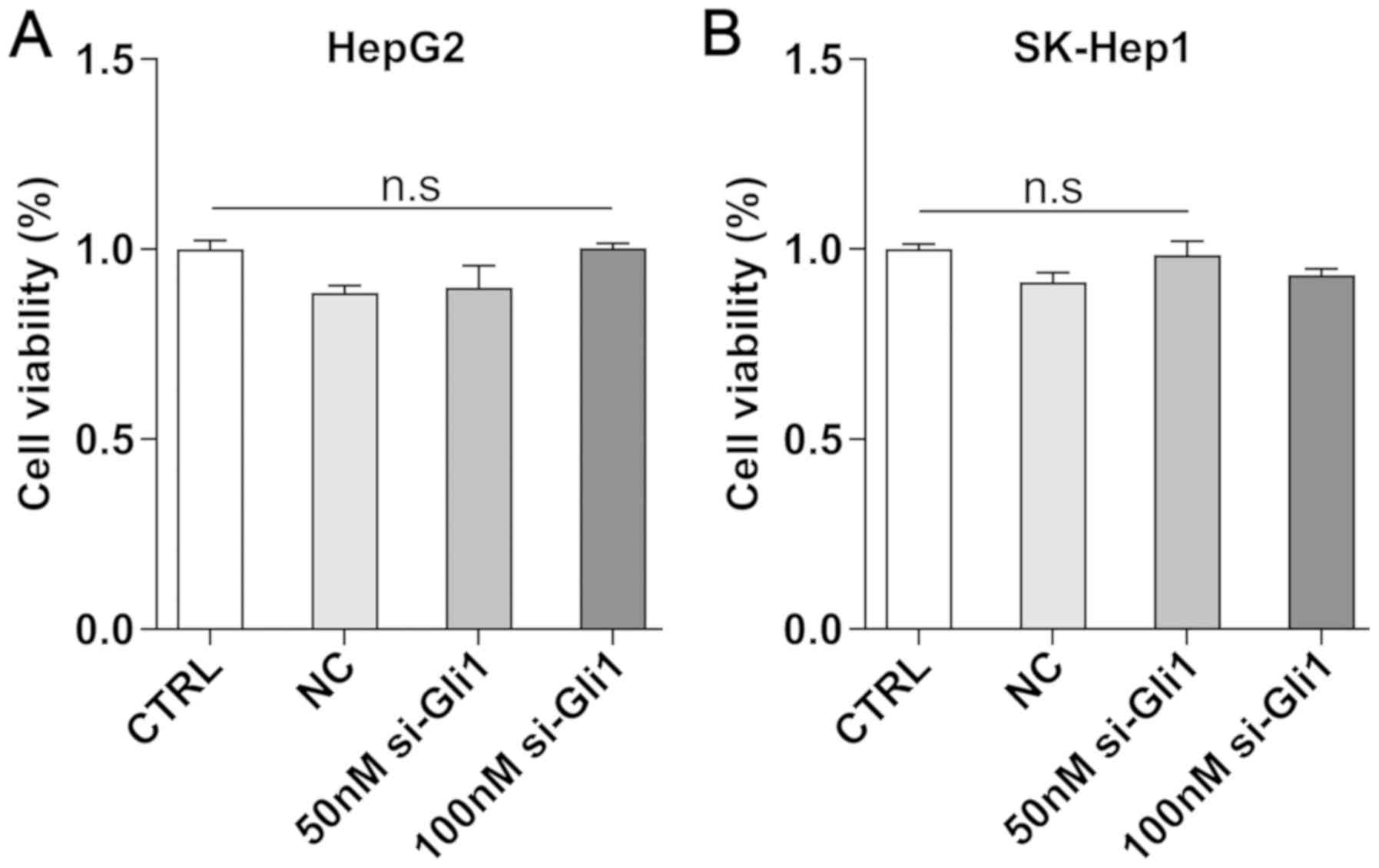
To investigate whether the efficiency of Gli1
transfected with siRNA was dependent on its cellular cytotoxicity,
CCK-8 assay was performed to examine the viability of HepG2 and
SK-Hep1 cells. The results demonstrated that the 50- and 100-nmol/l
si-Gli1 groups were not significantly different compared with the
CTRL group (P>0.05; Fig. 2),
which indicated that the effect of Gli1 transfection and the
following experiments using liver cancer cells was independent of
its cellular cytotoxicity.
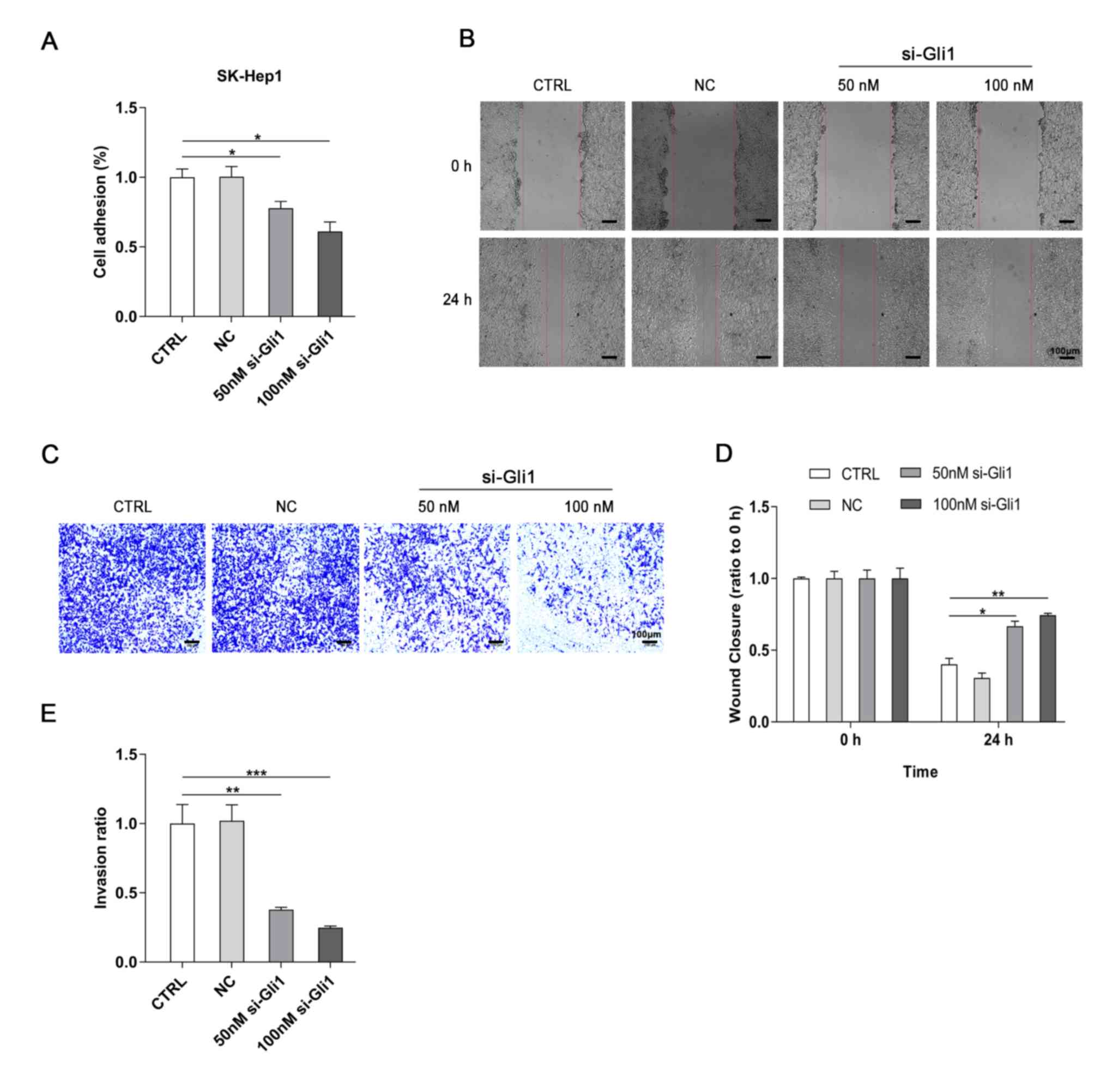
Reduced Gli1 reduces the adhesion,
migration and invasion of SK-Hep1 cells
Our previous findings demonstrated that the aberrant
activation of Gli1 is correlated with invasion and metastasis in
HCC tissues (28). Therefore, the
effects of Gli1-knockdown on the adhesive, migratory and invasive
abilities of hepatoma cells in vitro were determined. First,
a cell adhesion assay was performed to test the adhesive ability of
the SK-Hep1 cells. The results demonstrated that the adhesive
ability was decreased in both 50- and 100 nmol/l si-Gli1 groups
compared with the CTRL group (P<0.05; Fig. 3A). The wound healing assay
demonstrated that the wound healing rates were markedly inhibited
when Gli1 was knocked down in hepatoma cells compared with the CTRL
group (P<0.05 and P<0.01; Fig. 3B
and D), suggesting that downregulation of Gli1 expression may
suppress the motility of SK-Hep1 cells. Finally, the results of the
invasion assay revealed that the invasive capability of the cells
was also significantly inhibited after Gli1 was knocked down
(P<0.01 and P<0.001; Fig. 3C and
E). In addition, NC siRNA had no significant effects on the
cell adhesion, migration and invasion of SK-Hep1 cells.
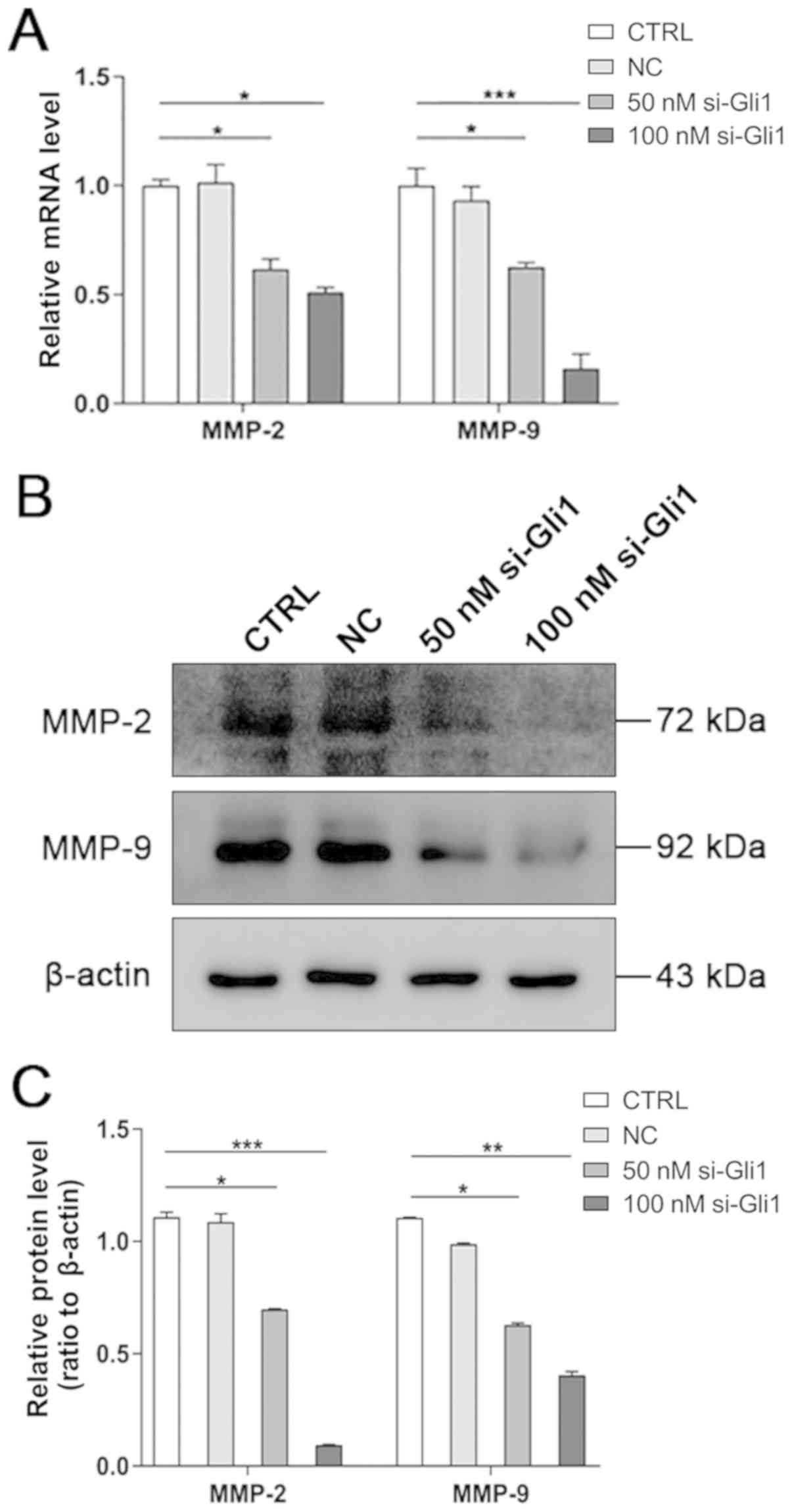
Knockdown of Gli1 decreases the
expression levels of MMP-2 and MMP-9
Accumulating evidence has elucidated that MMPs serve
a critical role in the invasion and metastasis of HCC by degrading
the extracellular matrix (ECM) components (29,30).
Therefore, to determine if Gli1 silencing-mediated inhibition of
cell migration and invasion was associated with MMP-2 and MMP-9,
RT-qPCR and western blot analyses was performed to detect their
expression. The results revealed that the mRNA and protein
expression levels of MMP-2 and MMP-9 were significantly decreased
in both si-Gli1 groups compared with the CTRL group (P<0.05,
P<0.01 and P<0.001; Fig. 4),
indicating that inhibition of the migration and invasion of
hepatoma cells by Gli1 depletion may be associated with the
downregulation of MMP-2 and MMP-9.
Silencing Gli1 inhibits FAK/AKT
signaling
To explore the crosstalk between Gli1 and the
FAK/AKT pathway, FAK, p-FAK, PI3K, p-PI3K, AKT and p-AKT protein
expression was detected in SK-Hep1 cells following Gli1-knockdown.
The western blotting results demonstrated that the protein levels
of p-FAK, p-PI3K and p-AKT were markedly decreased in both si-Gli1
groups compared with the CTRL group, whereas those of FAK, PI3K and
AKT were not significantly different from the CTRL group
(P<0.05, P<0.01 and P<0.001; Fig. 5A and B).
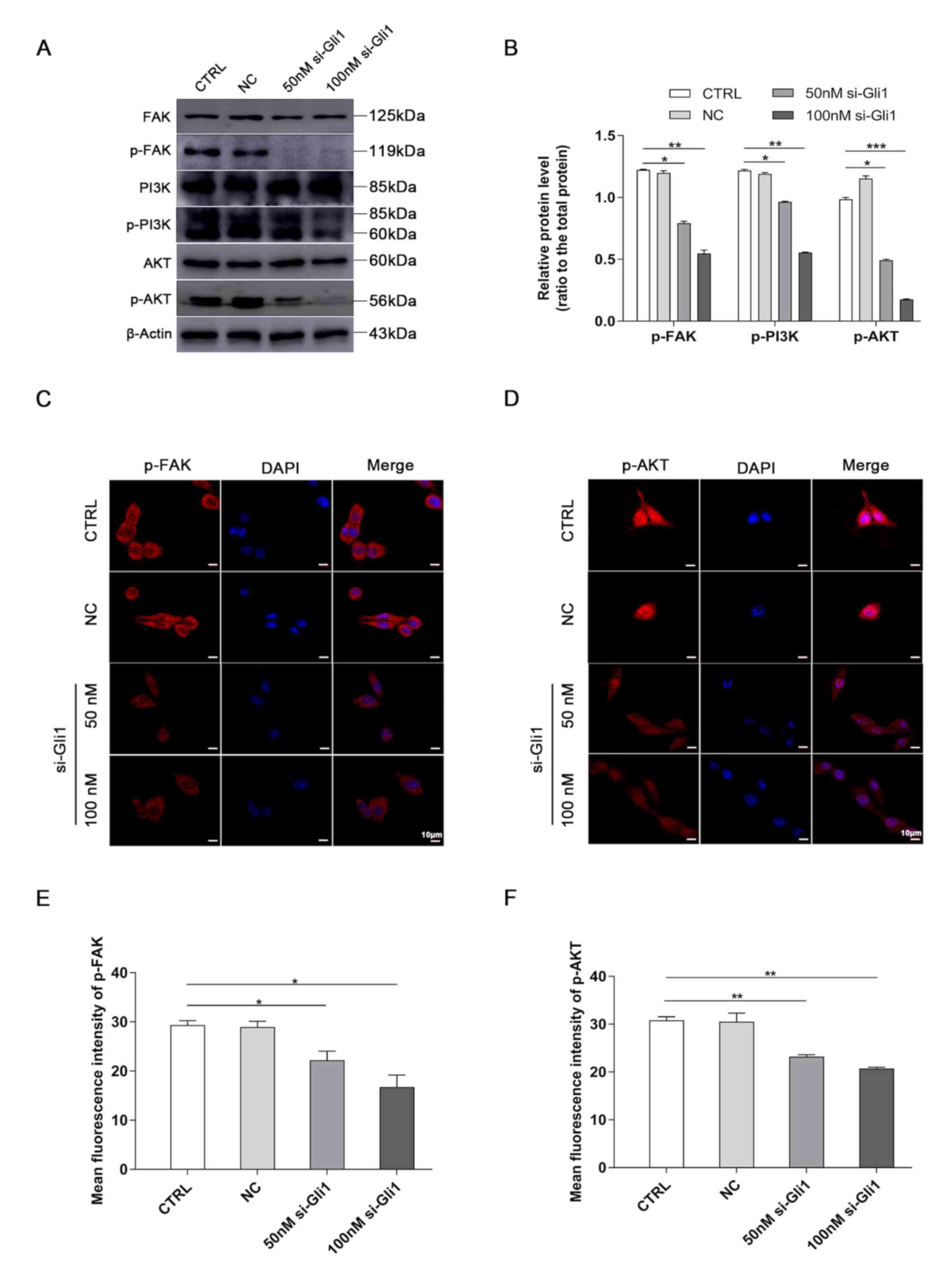 | Figure 5.Downregulation of Gli1 blocks the
FAK/AKT signaling pathway. (A) The protein expression levels of
FAK, p-FAK, PI3K, p-PI3K, AKT, and p-AKT in SK-Hep1 cells were
determined by western blotting. (B) Quantification analysis of
representative western blot images using ImageJ software. The
phosphorylated epitopes were normalized to the total protein
expression, respectively. β-actin served as a loading control.
Representative images of (C) p-FAK and (D) p-AKT immunofluorescence
staining in hepatoma SK-Hep1 cells. Nuclei were stained with DAPI.
Scale bar, 10 µm. Semi-quantitative analysis of (E) p-FAK and (F)
p-AKT fluorescence intensity using ImageJ software. *P<0.05,
**P<0.01 and ***P<0.001 vs. CTRL group. Gil1,
glioma-associated oncogene homolog 1; p-, phosphorylated; si, small
interference; CTRL, blank control group; NC, negative control siRNA
group. |
The results of immunofluorescence staining were in
agreement and demonstrated that the fluorescence intensity of p-FAK
localized in the cell membrane was significantly weaker compared
with the CTRL group (P<0.05; Fig. 5C
and E) and that of p-AKT located in the nucleus was also
significantly weaker in both si-Gli1 groups compared with the CTRL
group (P<0.01; Fig. 5D and F).
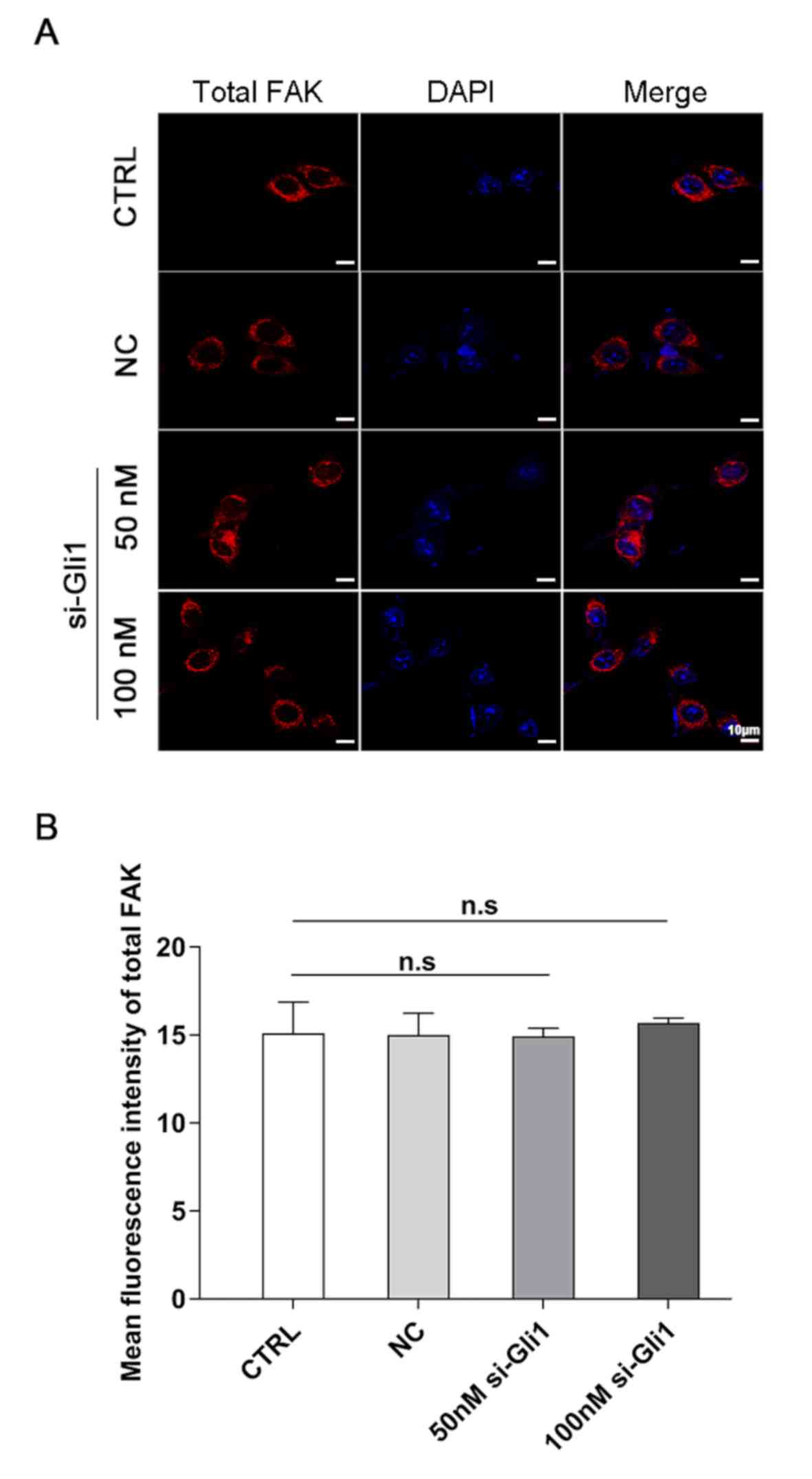
Notably, the intensity of total FAK localized in the cell membrane
did not reveal any significant change (P>0.05; Fig. 6), which was consistent with the
results of western blotting above, suggesting that the observed
decrease in p-FAK protein level was not due to a decrease in the
amount of total FAK protein.
Discussion
The results of the present study demonstrated that
the efficiency of Gli1 transfected with siRNA was feasible and was
independent of its cellular cytotoxicity. Loss of function
experiments were performed to elucidate the effects of Gli1 on the
biological functions of hepatoma cells. By performing cell
adhesion, wound healing and invasion assays after knockdown of
Gli1, the data revealed that knockdown significantly reduced the
motility and invasion of hepatoma cells. These results indicated
that Gli1 serves a crucial role in cell migration and invasion, and
provides a potential therapeutic target for HCC.
Cancer invasion and metastasis are complicated
multi-step processes in which tumor cells separate from the
original site, intravasate into blood vessels, extravasate to a
systemic location and colonize at a secondary site (31). Degradation of ECM and proteolysis of
the basement membrane are early events in cancer invasion and
metastasis (32,33). It has been found that MMPs contain
three domains: A catalytic domain, pro-peptide domain and
hemagglutinin-like C-terminal domain (34). Tumor cells undergo invasion and
metastasis during the early stages of cancer based on their ability
to degrade ECM components (35).
Among MMPs, MMP-2 and MMP-9 are the two most important factors that
degrade type IV collagen, which is the main component of the
basement membrane, and serve an essential role in tumor invasion
and metastasis (36). Chen et
al (15) found that
overexpression of MMP-2 and MMP-9 is observed in HCC tissues and is
associated with the invasion and metastasis of malignant hepatoma.
The present study found that silencing of Gli1 markedly inhibited
the mRNA and protein expression of MMP-2 and MMP-9, suggesting that
low expression of Gli1 may suppress HCC migration and invasion by
regulating the expression of MMP-2 and MMP-9.
A previous study demonstrated that blockade of the
FAK/AKT signaling pathway inhibits head and neck cancer metastasis
(37). Another study demonstrated
that activating the FAK/AKT pathway promotes the
epithelial-mesenchymal transition and invasion of HCC (38). Consequently, identifying the
crosstalk between Gli1 and FAK/AKT pathway may provide a further
understanding of the molecular mechanism of HCC invasion and
metastasis. In the present study, Gli1 interference decreased the
expression of p-FAK, p-PI3K and p-AKT as known as the active
ingredients of the FAK/AKT signaling, while total FAK, PI3K and AKT
protein expression was unaltered in the HCC cells. The data from
the present study indicated that siRNA-mediated depletion of Gli1
may significantly inhibit the FAK/AKT signaling pathway in
vitro. In addition, a previous study demonstrated that
phosphorylation of FAK promotes the invasion and metastasis of HCC
by upregulating the expression of MMP-2 and MMP-9 (20). Abnormal activation of the PI3K/AKT
signaling pathway can promote the proliferation and invasion of
tumor cells by regulating the expression of MMP-2 and MMP-9
(39–41). Thus, it was hypothesized that the
downregulation of Gli1 suppressed the invasion and metastasis of
HCC cells, which may be achieved by blocking the expression of
MMP-2 and MMP-9 mediated through the FAK/AKT signaling pathway.
The present study is not without limitations. First,
the present study only assessed the effects of Gli1 knockdown on
the biological functions of hepatoma cells. Prospective studies
will upregulate the Gli1 gene to determine the effects of Gli1 on
the migratory and invasive abilities of liver cancer cells.
Secondly, the molecular mechanism in the present study is not fully
elucidated. Whether downregulation of Gli1 blocks the FAK/AKT
signaling pathway, directly or indirectly, requires further
investigation via co-immunoprecipitation. Furthermore,
understanding protein interactions between the Gli1 and FAK/AKT
pathway components may help determine the underlying molecular
mechanism presented here.
In conclusion, the results of the present study
demonstrated that abnormal Gli1 expression resulted in the
significant repression of hepatoma cell migration and invasion via
downregulating MMP-2 and MMP-9 by blocking the FAK/AKT signaling
pathway. These observations revealed novel biological insights into
the mechanisms underlying HCC invasion and metastasis, and
highlight Gli1 as a potential therapeutic target for HCC.
Acknowledgements
Not applicable.
Funding
The present study was funded by grants from the
National Natural Science Foundation of China (grant no. 81760160),
the Natural Science Foundation of Jiangxi Province (grant no.
20151BAB205044), the Science and Technology Research Project of
Jiangxi Provincial Education Department (grant no. 180797), the
Provincial Innovation Training Project of Jiangxi Provincial
Undergraduates (grant no. S202010413009) and the Science and
Technology Innovation Project for Undergraduates of Gannan Medical
University (grant no. BKSZR201902).
Availability of data and materials
All data generated or analyzed during the present
study are available from the corresponding authors upon reasonable
request.
Authors' contributions
JNZ and BC conceived and designed the study. ZH, FX,
AH and MX performed the experiments and collected the data. YL and
JKZ analyzed the data and ZH wrote the manuscript. JX and YS
replicated the experiments and revised the figures. JNZ and BC
critically revised the manuscript for important intellectual
content. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mikhail S, Cosgrove D and Zeidan A:
Hepatocellular carcinoma: Systemic therapies and future
perspectives. Expert Rev Anticancer Ther. 14:1205–1218. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baek KK, Kim JH, Uhm JE, Park SH, Lee J,
Park JO, Park YS, Kang WK and Lim HY: Prognostic factors in
patients with advanced hepatocellular carcinoma treated with
sorafenib: A retrospective comparison with previously known
prognostic models. Oncology. 80:167–174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ikeda M, Morizane C, Ueno M, Okusaka T,
Ishii H and Furuse J: Chemotherapy for hepatocellular carcinoma:
Current status and future perspectives. Jpn J Clin Oncol.
48:103–114. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Skoda AM, Simovic D, Karin V, Kardum V,
Vranic S and Serman L: The role of the Hedgehog signaling pathway
in cancer: A comprehensive review. Bosn J Basic Med Sci. 18:8–20.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Han C, Lu L, Magliato S and Wu T:
Hedgehog signaling pathway regulates autophagy in human
hepatocellular carcinoma cells. Hepatology. 58:995–1010. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma Y, Yu W, Shrivastava A, Alemi F,
Lankachandra K, Srivastava RK and Shankar S: Sanguinarine inhibits
pancreatic cancer stem cell characteristics by inducing oxidative
stress and suppressing sonic hedgehog-Gli-Nanog pathway.
Carcinogenesis. 38:1047–1056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Magistri P, Leonard SY, Tang CM, Chan JC,
Lee TE and Sicklick JK: The glypican 3 hepatocellular carcinoma
marker regulates human hepatic stellate cells via Hedgehog
signaling. J Surg Res. 187:377–385. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke Z, Caiping S, Qing Z and Xiaojing W:
Sonic hedgehog-Gli1 signals promote epithelial-mesenchymal
transition in ovarian cancer by mediating PI3K/AKT pathway. Med
Oncol. 32:3682015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanchez P, Hernández AM, Stecca B, Kahler
AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S and Ruiz i
Altaba A: Inhibition of prostate cancer proliferation by
interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci
USA. 101:12561–12566. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jian-Hui C, Er-Tao Z, Si-Le C, Hui W,
Kai-Ming W, Xin-Hua Z, Chuang-Qi C, Shi-Rong C and Yu-Long H: CD44,
sonic hedgehog, and Gli1 expression are prognostic biomarkers in
gastric cancer patients after radical resection. Gastroenterol Res
Pract. 2016:10130452016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Min S, Xiaoyan X, Fanghui P, Yamei W,
Xiaoli Y and Feng W: The glioma-associated oncogene homolog 1
promotes epithelial-mesenchymal transition in human esophageal
squamous cell cancer by inhibiting E-cadherin via Snail. Cancer
Gene Ther. 20:379–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sheng W, Dong M, Zhou J, Li X, Liu Q, Dong
Q and Li F: The clinicopathological significance and relationship
of Gli1, MDM2 and p53 expression in resectable pancreatic cancer.
Histopathology. 64:523–535. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen JS, Huang XH, Wang Q, Huang JQ, Zhang
LJ, Chen XL, Lei J and Cheng ZX: Sonic hedgehog signaling pathway
induces cell migration and invasion through focal adhesion
kinase/AKT signaling-mediated activation of matrix
metalloproteinase (MMP)-2 and MMP-9 in liver cancer.
Carcinogenesis. 34:10–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu CH, Lan CT, Chou JF, Tseng TJ and Liao
WC: CHSY1 promotes aggressive phenotypes of hepatocellular
carcinoma cells via activation of the hedgehog signaling pathway.
Cancer Lett. 403:280–288. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo D, Zhang D, Ren M, Lu G, Zhang X, He S
and Li Y: THBS4 promotes HCC progression by regulating ITGB1 via
FAK/PI3K/AKT pathway. FASEB J. Jun 22–2020.(Epub ahead of print).
View Article : Google Scholar
|
|
18
|
Tokunaga E, Oki E, Egashira A, Sadanaga N,
Morita M, Kakeji Y and Maehara Y: Deregulation of the Akt pathway
in human cancer. Curr Cancer Drug Targets. 8:27–36. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen HC, Appeddu PA, Isoda H and Guan JL:
Phosphorylation of tyrosine 397 in focal adhesion kinase is
required for binding phosphatidylinositol 3-kinase. J Biol Chem.
271:26329–26334. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen JS, Huang XH, Wang Q, Chen XL, Fu XH,
Tan HX, Zhang LJ, Li W and Bi J: FAK is involved in invasion and
metastasis of hepatocellular carcinoma. Clin Exp Metastasis.
27:71–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cance WG, Harris JE, Iacocca MV, Roche E,
Yang X, Chang J, Simkins S and Xu L: Immunohistochemical analyses
of focal adhesion kinase expression in benign and malignant human
breast and colon tissues: Correlation with preinvasive and invasive
phenotypes. Clin Cancer Res. 6:2417–2423. 2000.PubMed/NCBI
|
|
22
|
Owens LV, Xu L, Craven RJ, Dent GA, Weiner
TM, Kornberg L, Liu ET and Cance WG: Overexpression of the focal
adhesion kinase (p125FAK) in invasive human tumors. Cancer Res.
55:2752–2755. 1995.PubMed/NCBI
|
|
23
|
Owens LV, Xu L, Dent GA, Yang X, Sturge
GC, Craven RJ and Cance WG: Focal adhesion kinase as a marker of
invasive potential in differentiated human thyroid cancer. Ann Surg
Oncol. 3:100–105. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tremblay L, Hauck W, Aprikian AG, Begin
LR, Chapdelaine A and Chevalier S: Focal adhesion kinase (pp125FAK)
expression, activation and association with paxillin and p50CSK in
human metastatic prostate carcinoma. Int J Cancer. 68:164–171.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ayaki M, Komatsu K, Mukai M, Murata K,
Kameyama M, Ishiguro S, Miyoshi J, Tatsuta M and Nakamura H:
Reduced expression of focal adhesion kinase in liver metastases
compared with matched primary human colorectal adenocarcinomas.
Clin Cancer Res. 7:3106–3112. 2001.PubMed/NCBI
|
|
26
|
Xia H, Nho RS, Kahm J, Kleidon J and Henke
CA: Focal adhesion kinase is upstream of phosphatidylinositol
3-kinase/Akt in regulating fibroblast survival in response to
contraction of type I collagen matrices via a beta 1 integrin
viability signaling pathway. J Biol Chem. 279:33024–33034. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen B and Hu Z, Li B, Lin X, Luo Z and Hu
Z: The expressions of Hedgehog and PI3K-AKT pathway components
correlate with invasion and metastasis in hepatocellular carcinoma.
Int J Clin Exp Pathol. 12:2381–2388. 2019.PubMed/NCBI
|
|
29
|
Yamamoto H, Itoh F, Adachi Y, Sakamoto H,
Adachi M, Hinoda Y and Imai K: Relation of enhanced secretion of
active matrix metalloproteinases with tumor spread in human
hepatocellular carcinoma. Gastroenterology. 112:1290–1296. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Coussens LM, Fingleton B and Matrisian LM:
Matrix metalloproteinase inhibitors and cancer: Trials and
tribulations. Science. 295:2387–2392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stetler-Stevenson WG: Type IV collagenases
in tumor invasion and metastasis. Cancer Metastasis Rev. 9:289–303.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vihinen P and Kähäri VM: Matrix
metalloproteinases in cancer: Prognostic markers and therapeutic
targets. Int J Cancer. 99:157–166. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hadler-Olsen E, Winberg JO and
Uhlin-Hansen L: Matrix metalloproteinases in cancer: Their value as
diagnostic and prognostic markers and therapeutic targets. Tumour
Biol. 34:2041–2051. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Parnham A and Bousfield C: The influence
of matrix metalloproteases and biofilm on chronic wound healing: A
discussion. Br J Community Nurs. 23 (Suppl 3):S22–S29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim SA, Kwon SM, Kim JA, Kang KW, Yoon JH
and Ahn SG: 5′-Nitro-indirubinoxime, an indirubin derivative,
suppresses metastatic ability of human head and neck cancer cells
through the inhibition of Integrin β1/FAK/Akt signaling. Cancer
Lett. 306:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang R, Yu Z, Chen F, Xu H, Shen S, Chen
W, Chen L, Su Q, Zhang L, Bi J, et al: miR-300 regulates the
epithelial-mesenchymal transition and invasion of hepatocellular
carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed
Pharmacother. 103:1632–1642. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cheng JC, Chou CH, Kuo ML and Hsieh CY:
Radiation-enhanced hepatocellular carcinoma cell invasion with
MMP-9 expression through PI3K/Akt/NF-kappaB signal transduction
pathway. Oncogene. 25:7009–7018. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shukla S, Maclennan GT, Hartman DJ, Fu P,
Resnick MI and Gupta S: Activation of PI3K-Akt signaling pathway
promotes prostate cancer cell invasion. Int J Cancer.
121:1424–1432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li H, Zhang Y, Hai J, Wang J, Zhao B, Du L
and Geng X: Knockdown of TRIM31 suppresses proliferation and
invasion of gallbladder cancer cells by down-regulating MMP2/9
through the PI3K/Akt signaling pathway. Biomed Pharmacother.
103:1272–1278. 2018. View Article : Google Scholar : PubMed/NCBI
|