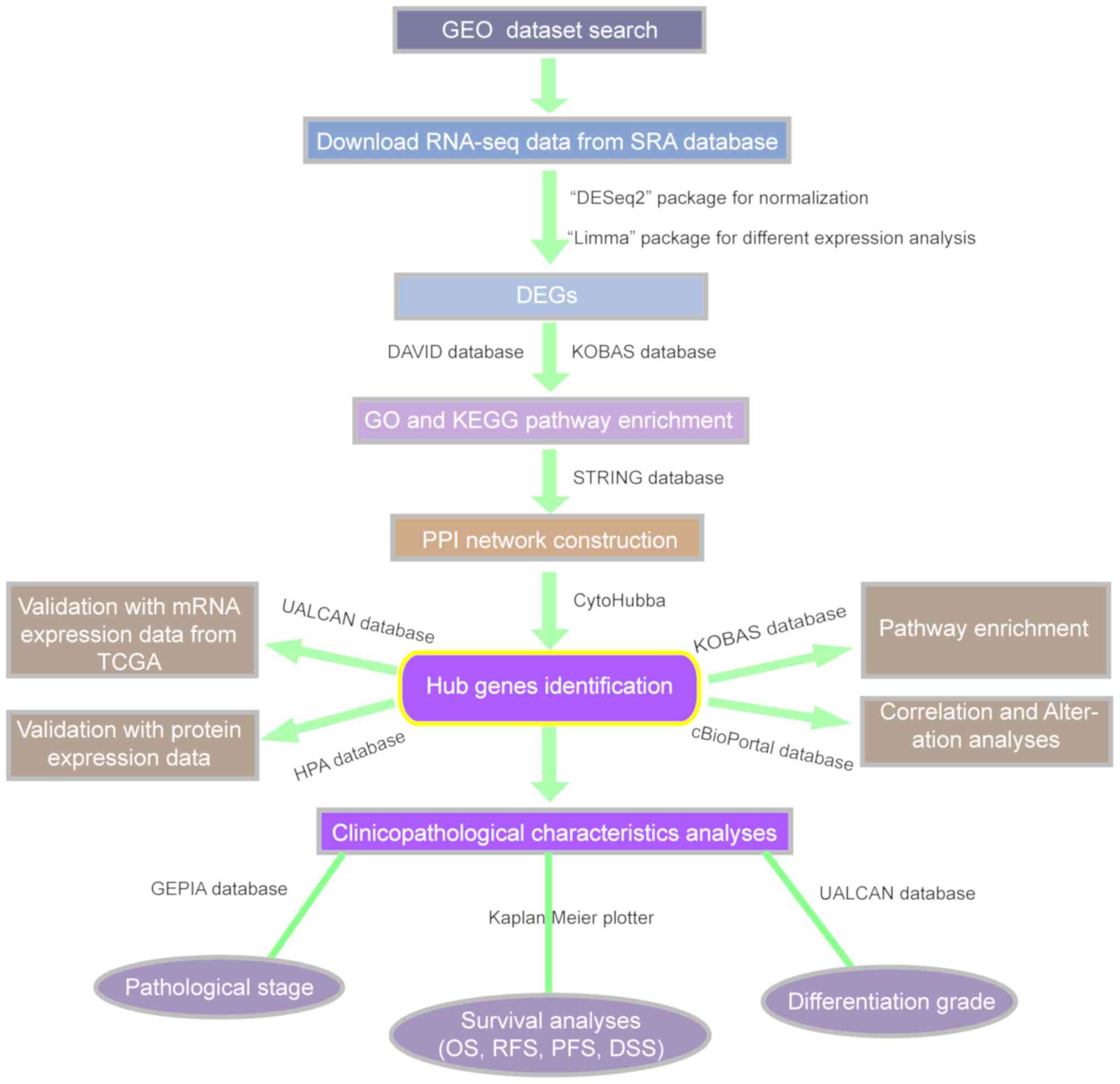
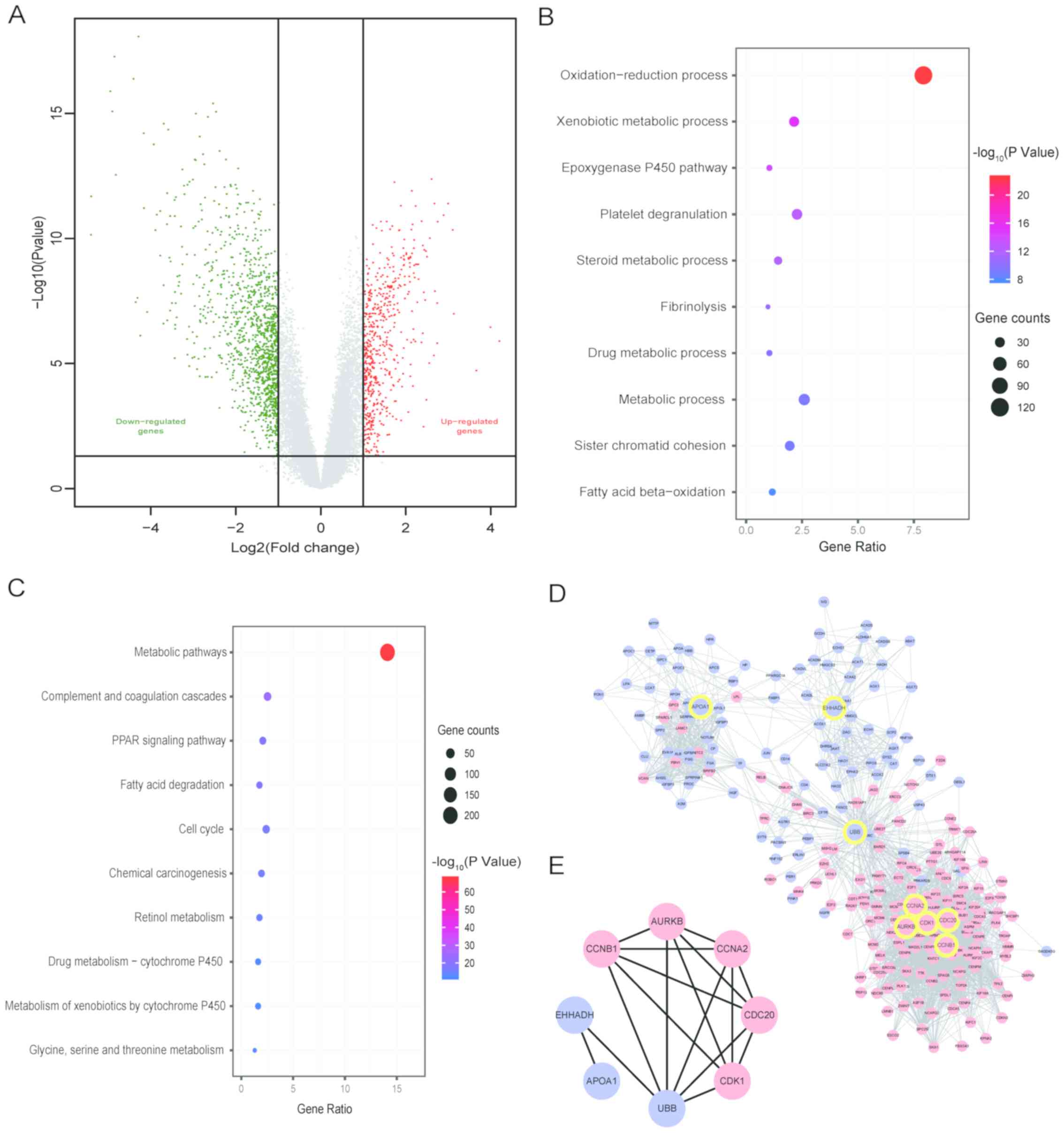
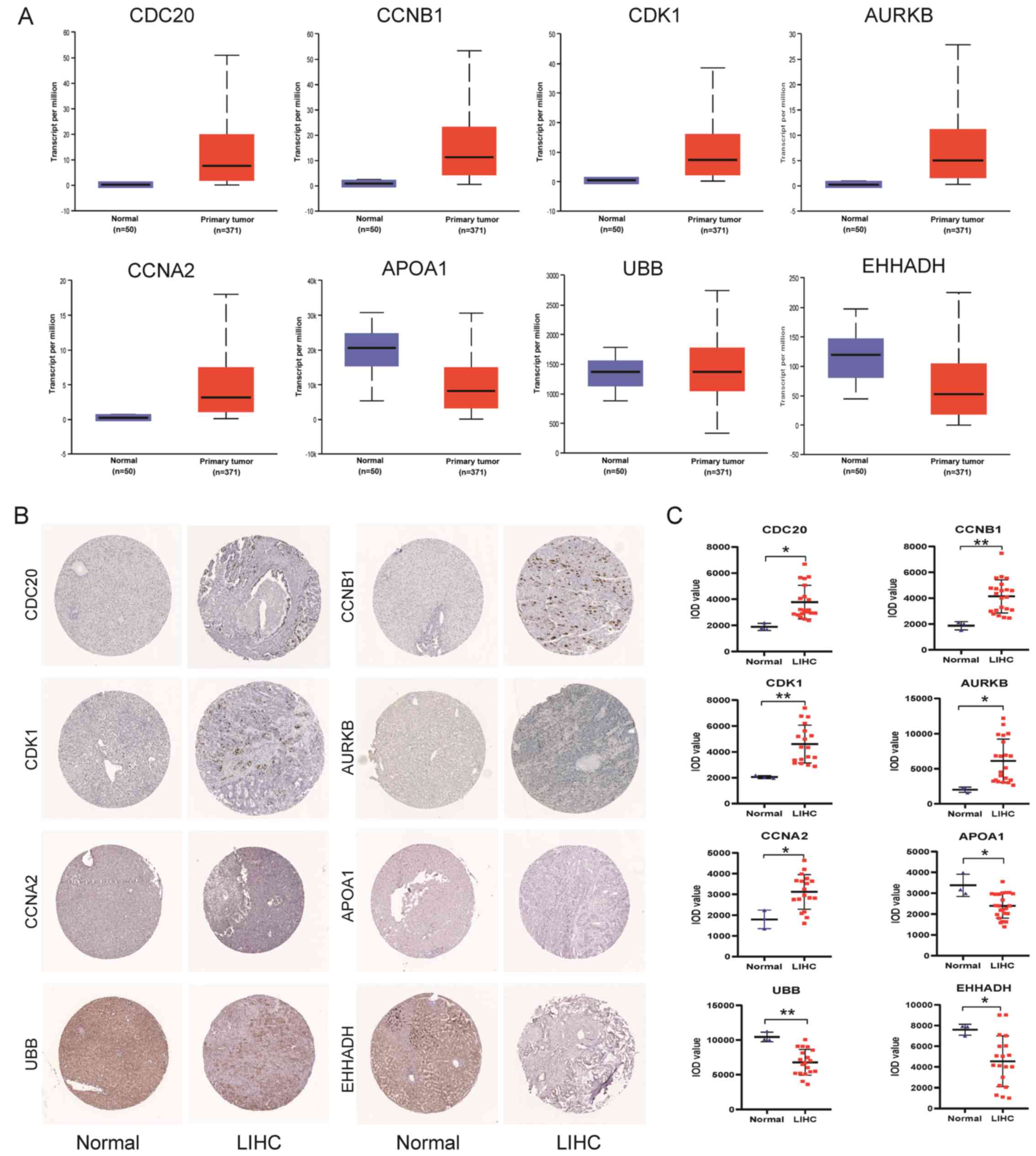
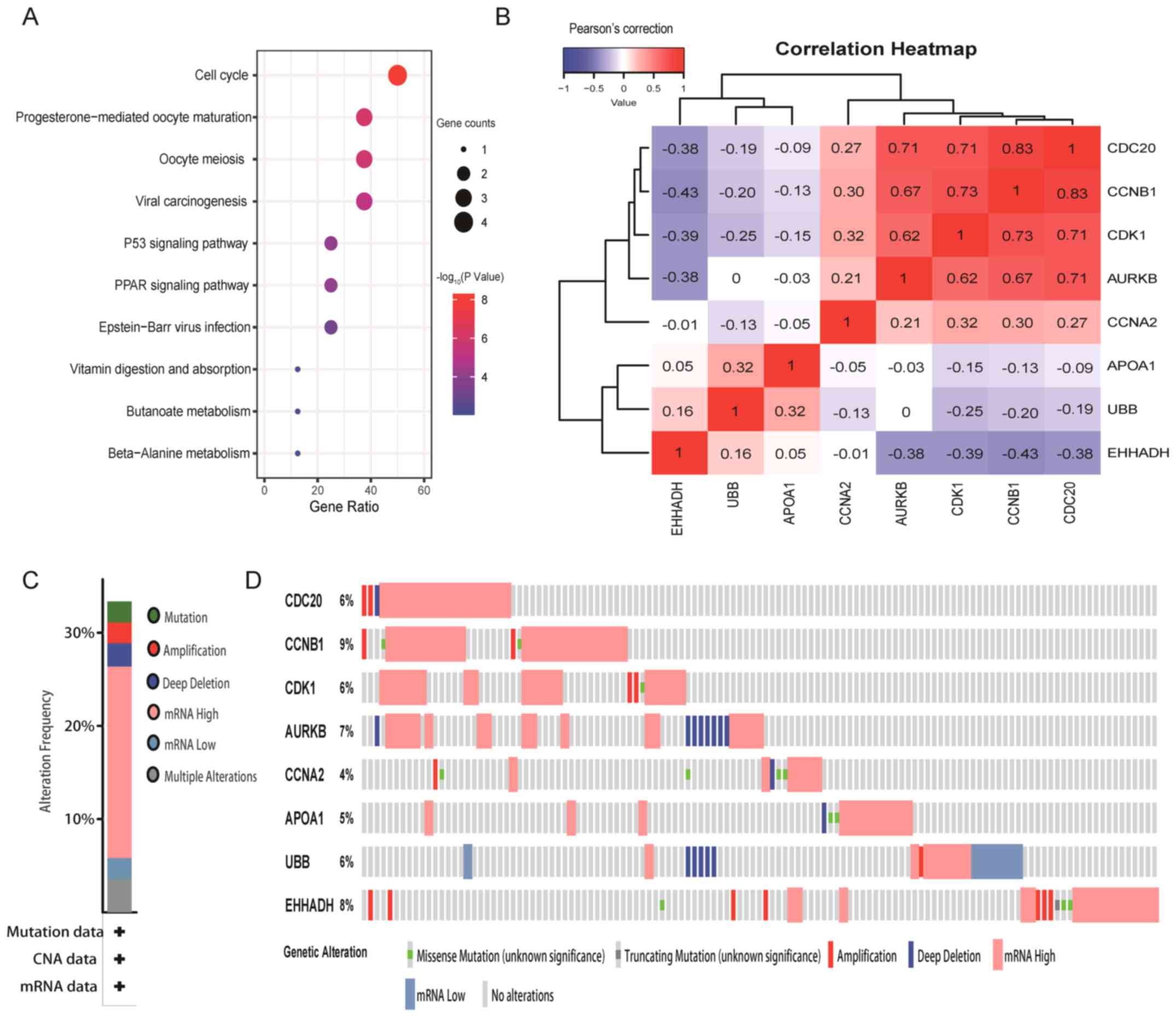
|
1
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bruix J and Sherman M; Practice Guidelines
Committee, American Association for the Study of Liver Diseases, :
Management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vogel A, Cervantes A, Chau I, Daniele B,
Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, et al:
Hepatocellular carcinoma: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 29 (Suppl
4):iv238–iv255. 2018. View Article : Google Scholar
|
|
7
|
Bruix J and Sherman M; American
Association for the Study of Liver Diseases, : Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Varela M, Reig M, de la Mata M, Matilla A,
Bustamante J, Pascual S, Turnes J, Aracil C, Del Val A, Pascasio
JM, et al: Treatment approach of hepatocellular carcinoma in Spain.
Analysis of 705 patients from 62 centers. Med Clin (Barc).
134:569–576. 2010.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bargellini I, Sacco R, Bozzi E, Bertini M,
Ginanni B, Romano A, Cicorelli A, Tumino E, Federici G, Cioni R, et
al: Transarterial chemoembolization in very early and early-stage
hepatocellular carcinoma patients excluded from curative treatment:
A prospective cohort study. Eur J Radiol. 81:1173–1178. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song YG, Shin SW, Cho SK, Choi D, Rhim H,
Lee MW, Kim YS, Park KB, Park HS, Choo SW, et al: Transarterial
chemoembolization as first-line therapy for hepatocellular
carcinomas infeasible for ultrasound-guided radiofrequency
ablation: A retrospective cohort study of 116 patients. Acta
Radiol. 56:70–77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim JW, Kim JH, Sung KB, Ko HK, Shin JH,
Kim PN, Choi HK, Ko GY, Yoon HK, Chun SY and Gwon DI: Transarterial
chemoembolization vs. radiofrequency ablation for the treatment of
single hepatocellular carcinoma 2 cm or smaller. Am J
Gastroenterol. 109:1234–1240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gopal P, Yopp AC, Waljee AK, Chiang J,
Nehra M, Kandunoori P and Singal AG: Factors that affect accuracy
of α-fetoprotein test in detection of hepatocellular carcinoma in
patients with cirrhosis. Clin Gastroenterol Hepatol. 12:870–877.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang TS, Wu YC, Tung SY, Wei KL, Hsieh
YY, Huang HC, Chen WM, Shen CH, Lu CH, Wu CS, et al:
Alpha-fetoprotein measurement benefits hepatocellular carcinoma
surveillance in patients with cirrhosis. Am J Gastroenterol.
110:836–845. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou J, Sun HC, Wang Z, Cong WM, Wang JH,
Zeng MS, Yang JM, Bie P, Liu LX, Wen TF, et al: Guidelines for
diagnosis and treatment of primary liver cancer in China (2017
edition). Liver Cancer. 7:235–260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takikawa Y and Suzuki K: Is AFP a new
reliable marker of liver regeneration in acute hepatic failure? J
Gastroenterol. 37:681–682. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Minguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Sun A, Zhao Y, Ying W, Sun H,
Yang X, Xing B, Sun W, Ren L, Hu B, et al: Proteomics identifies
new therapeutic targets of early-stage hepatocellular carcinoma.
Nature. 567:257–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anders S, Pyl PT and Huber W: HTSeq-a
python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bringmann LF, Elmer T, Epskamp S, Krause
RW, Schoch D, Wichers M, Wigman JTW and Snippe E: What do
centrality measures measure in psychological networks? J Abnorm
Psychol. 128:892–903. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Subramanian A, Narayan R, Corsello SM,
Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK,
et al: A next generation connectivity map: L1000 platform and the
first 1,000,000 profiles. Cell. 171:1437–1452 e17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu SC, Huang YW, Wang TC, Hu JT, Chen DS
and Yang SS: Increased risk of hepatocellular carcinoma in chronic
hepatitis B patients with new onset diabetes: A nationwide cohort
study. Aliment Pharmacol Ther. 41:1200–1209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang YW, Wang TC, Yang SS, Lin SY, Fu SC,
Hu JT, Liu CJ, Kao JH and Chen DS: Increased risk of hepatocellular
carcinoma in chronic hepatitis C patients with new onset diabetes:
A nation-wide cohort study. Aliment Pharmacol Ther. 42:902–911.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xia H, Chen J, Sekar K, Shi M, Xie T and
Hui KM: Clinical and metabolomics analysis of hepatocellular
carcinoma patients with diabetes mellitus. Metabolomics.
15:1562019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou QC, Shi B, Jiao LF, Jin M, Sun P,
Ding LY and Yuan Y: Hepatopancreas and ovarian transcriptome
response to different dietary soybean lecithin levels in portunus
trituberculatus. Comp Biochem Physiol Part D Genomics Proteomics.
31:1006002019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamashita T, Honda M, Takatori H, Nishino
R, Minato H, Takamura H, Ohta T and Kaneko S: Activation of
lipogenic pathway correlates with cell proliferation and poor
prognosis in hepatocellular carcinoma. J Hepatol. 50:100–110. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li N, Li L and Chen Y: The identification
of core gene expression signature in hepatocellular carcinoma. Oxid
Med Cell Longev. 2018:34783052018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong H, Xiao M, Zarkovic K, Zhu M, Sa R,
Lu J, Tao Y, Chen Q, Xia L, Cheng S, et al: Mitochondrial control
of apoptosis through modulation of cardiolipin oxidation in
hepatocellular carcinoma: A novel link between oxidative stress and
cancer. Free Radic Biol Med. 102:67–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J, Zhang C and Feng Z: Tumor
suppressor p53 and its gain-of-function mutants in cancer. Acta
Biochim Biophys Sin (Shanghai). 46:170–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rebouissou S and Nault JC: Advances in
molecular classification and precision oncology in hepatocellular
carcinoma. J Hepatol. 72:215–229. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shao Y, Song X, Jiang W, Chen Y, Ning Z,
Gu W and Jiang J: MicroRNA-621 acts as a tumor radiosensitizer by
directly targeting SETDB1 in hepatocellular carcinoma. Mol Ther.
27:355–364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pan YH, Yang M, Liu LP, Wu DC, Li MY and
Su SG: UBE2S enhances the ubiquitination of p53 and exerts
oncogenic activities in hepatocellular carcinoma. Biochem Biophys
Res Commun. 503:895–902. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun G, Sui X, Han D, Gao J, Liu Y and Zhou
L: TRIM59 promotes cell proliferation, migration and invasion in
human hepatocellular carcinoma cells. Pharmazie. 72:674–679.
2017.PubMed/NCBI
|
|
44
|
Trachootham D, Lu W, Ogasawara MA, Nilsa
RD and Huang P: Redox regulation of cell survival. Antioxid Redox
Signal. 10:1343–1374. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Oztopcu-Vatan P, Sayitoglu M, Gunindi M
and Inan E: Cytotoxic and apoptotic effects of menadione on rat
hepatocellular carcinoma cells. Cytotechnology. 67:1003–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu D and Xu Y: p53, oxidative stress, and
aging. Antioxid Redox Signal. 15:1669–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
D'Souza LC, Mishra S, Chakraborty A,
Shekher A, Sharma A and Gupta SC: Oxidative stress and cancer
development: Are noncoding RNAs the missing links? Antioxid Redox
Signal. Jan 24–2020.(Epub ahead of print). doi:
10.1089/ars.2019.7987. View Article : Google Scholar
|
|
48
|
Maurya AK and Vinayak M: Anticarcinogenic
action of quercetin by downregulation of phosphatidylinositol
3-kinase (PI3K) and protein kinase C (PKC) via induction of p53 in
hepatocellular carcinoma (HepG2) cell line. Mol Biol Rep.
42:1419–1429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu S, Yang TB, Nan YL, Li AH, Pan DX, Xu
Y, Li S, Li T, Zeng XY and Qiu XQ: Genetic variants of cell cycle
pathway genes predict disease-free survival of hepatocellular
carcinoma. Cancer Med. 6:1512–1522. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Maddika S, Ande SR, Panigrahi S,
Paranjothy T, Weglarczyk K, Zuse A, Eshraghi M, Manda KD, Wiechec E
and Los M: Cell survival, cell death and cell cycle pathways are
interconnected: Implications for cancer therapy. Drug Resist Updat.
10:13–29. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Giono LE and Manfredi JJ: The p53 tumor
suppressor participates in multiple cell cycle checkpoints. J Cell
Physiol. 209:13–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen J: The cell-cycle arrest and
apoptotic functions of p53 in tumor initiation and progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ito Y, Takeda T, Sakon M, Monden M,
Tsujimoto M and Matsuura N: Expression and prognostic role of
cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma.
Oncology. 59:68–74. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
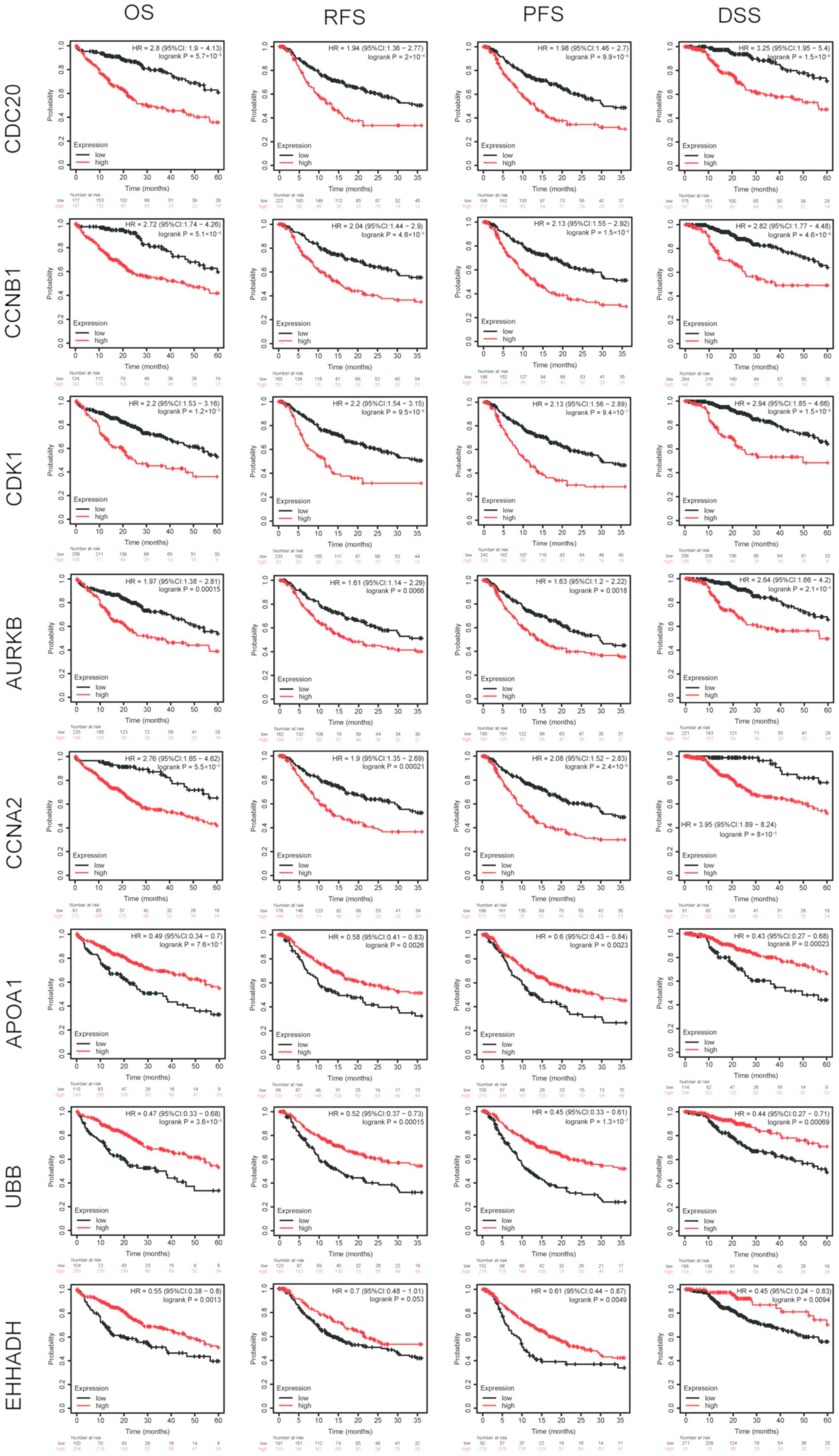
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gu J, Liu X, Li J and He Y: MicroRNA-144
inhibits cell proliferation, migration and invasion in human
hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int.
19:152019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bayard Q, Meunier L, Peneau C, Renault V,
Shinde J, Nault JC, Mami I, Couchy G, Amaddeo G, Tubacher E, et al:
Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass
with a rearrangement signature of replication stress. Nat Commun.
9:52352018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shi Y, Li Y, Huang C, Ying L, Xue J, Wu H,
Chen Z and Yang Z: Resveratrol enhances HBV replication through
activating Sirt1-PGC-1α-PPARα pathway. Sci Rep. 6:247442016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang JY, Chou SF, Lee JW, Chen HL, Chen
CM, Tao MH and Shih C: MicroRNA-130a can inhibit hepatitis B virus
replication via targeting PGC1α and PPARγ. RNA. 21:385–400. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kanakkanthara A, Jeganathan KB, Limzerwala
JF, Baker DJ, Hamada M, Nam HJ, van Deursen WH, Hamada N, Naylor
RM, Becker NA, et al: Cyclin A2 is an RNA binding protein that
controls Mre11 mRNA translation. Science. 353:1549–1552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yan H, Li Z, Shen Q, Wang Q, Tian J, Jiang
Q and Gao L: Aberrant expression of cell cycle and material
metabolism related genes contributes to hepatocellular carcinoma
occurrence. Pathol Res Pract. 213:316–321. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW,
Lai PL, Lee PH, Cheng AL and Hsu HC: Significance of Aurora B
overexpression in hepatocellular carcinoma. Aurora B overexpression
in HCC. BMC Cancer. 10:4612010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yasen M, Mizushima H, Mogushi K, Obulhasim
G, Miyaguchi K, Inoue K, Nakahara I, Ohta T, Aihara A, Tanaka S, et
al: Expression of Aurora B and alternative variant forms in
hepatocellular carcinoma and adjacent tissue. Cancer Sci.
100:472–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tanaka S, Arii S, Yasen M, Mogushi K, Su
NT, Zhao C, Imoto I, Eishi Y, Inazawa J, Miki Y and Tanaka H:
Aurora kinase B is a predictive factor for the aggressive
recurrence of hepatocellular carcinoma after curative hepatectomy.
Br J Surg. 95:611–619. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY,
Chan ACY, Ma KW, Xia W and Cheung TT: Blocking CDK1/PDK1/β-Catenin
signaling by CDK1 inhibitor RO3306 increased the efficacy of
sorafenib treatment by targeting cancer stem cells in a preclinical
model of hepatocellular carcinoma. Theranostics. 8:3737–3750. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang W, Zhang L, Guo Q, Wang H, Ma M, Sun
J and Chen C: Identification of the pathogenic biomarkers for
hepatocellular carcinoma based on RNA-seq analyses. Pathol Oncol
Res. 25:1207–1213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ganapathy E, Su F, Meriwether D, Devarajan
A, Grijalva V, Gao F, Chattopadhyay A, Anantharamaiah GM, Navab M,
Fogelman AM, et al: D-4F, an apoA-I mimetic peptide, inhibits
proliferation and tumorigenicity of epithelial ovarian cancer cells
by upregulating the antioxidant enzyme MnSOD. Int J Cancer.
130:1071–1081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cedó L, García-León A, Baila-Rueda L,
Santos D, Grijalva V, Martínez-Cignoni MR, Carbó JM, Metso J,
López-Vilaró L, Zorzano A, et al: ApoA-I mimetic administration,
but not increased apoA-I-containing HDL, inhibits tumour growth in
a mouse model of inherited breast cancer. Sci Rep. 6:363872016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gao F, Vasquez SX, Su F, Roberts S, Shah
N, Grijalva V, Imaizumi S, Chattopadhyay A, Ganapathy E, Meriwether
D, et al: L-5F, an apolipoprotein A-I mimetic, inhibits tumor
angiogenesis by suppressing VEGF/basic FGF signaling pathways.
Integr Biol (Camb). 3:479–489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zamanian-Daryoush M, Lindner D, Tallant
TC, Wang Z, Buffa J, Klipfell E, Parker Y, Hatala D,
Parsons-Wingerter P, Rayman P, et al: The cardioprotective protein
apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol
Chem. 288:21237–21252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Oh C, Park S, Lee EK and Yoo YJ:
Downregulation of ubiquitin level via knockdown of polyubiquitin
gene Ubb as potential cancer therapeutic intervention. Sci Rep.
3:26232013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tang Y, Geng Y, Luo J, Shen W, Zhu W, Meng
C, Li M, Zhou X, Zhang S and Cao J: Downregulation of ubiquitin
inhibits the proliferation and radioresistance of non-small cell
lung cancer cells in vitro and in vivo. Sci Rep. 5:94762015.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tian Y, Ding W, Wang Y, Ji T, Sun S, Mo Q,
Chen P, Fang Y, Liu J, Wang B, et al: Ubiquitin B in cervical
cancer: Critical for the maintenance of cancer stem-like cell
characters. PLoS One. 8:e844572013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Valdagni R, Rancati T, Ghilotti M,
Cozzarini C, Vavassori V, Fellin G, Fiorino C, Girelli G, Barra S,
Zaffaroni N, et al: To bleed or not to bleed. A prediction based on
individual gene profiling combined with dose-volume histogram
shapes in prostate cancer patients undergoing three-dimensional
conformal radiation therapy. Int J Radiat Oncol Biol Phys.
74:1431–1440. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Daniele B, De Vivo R, Perrone F, Lastoria
S, Tambaro R, Izzo F, Fiore F, Vallone P and Pignata S: Phase I
clinical trial of liposomal daunorubicin in hepatocellular
carcinoma complicating liver cirrhosis. Anticancer Res.
20:1249–1251. 2000.PubMed/NCBI
|
|
76
|
Brandi G, Biasco G, Mirarchi MG, Golfieri
R, Di Paolo A, Borghi A, Fanello S, Derenzini E, Agostini V,
Giampalma E, et al: A phase I study of continuous hepatic arterial
infusion of Irinotecan in patients with locally advanced
hepatocellular carcinoma. Dig Liver Dis. 43:1015–1021. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Tak WY, Lin SM, Wang Y, Zheng J, Vecchione
A, Park SY, Chen MH, Wong S, Xu R, Peng CY, et al: Phase III HEAT
study adding lyso-thermosensitive liposomal doxorubicin to
radiofrequency ablation in patients with unresectable
hepatocellular carcinoma lesions. Clin Cancer Res. 24:73–83. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lin HC, Chen YF, Hsu WH, Yang CW, Kao CH
and Tsai TF: Resveratrol helps recovery from fatty liver and
protects against hepatocellular carcinoma induced by hepatitis B
virus X protein in a mouse model. Cancer Prev Res (Phila).
5:952–962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bhattacharya S, Mondal L, Mukherjee B,
Dutta L, Ehsan I, Debnath MC, Gaonkar RH, Pal MM and Majumdar S:
Apigenin loaded nanoparticle delayed development of hepatocellular
carcinoma in rats. Nanomedicine. 14:1905–1917. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Balamurugan K and Karthikeyan J:
Evaluation of luteolin in the prevention of
N-nitrosodiethylamine-induced Hepatocellular carcinoma using animal
model system. Indian J Clin Biochem. 27:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen F, Wang H, Zhu J, Zhao R, Xue P,
Zhang Q, Bud Nelson M, Qu W, Feng B and Pi J: Camptothecin
suppresses NRF2-ARE activity and sensitises hepatocellular
carcinoma cells to anticancer drugs. Br J Cancer. 117:1495–1506.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xu D, Jin J, Yu H, Zhao Z, Ma D, Zhang C
and Jiang H: Chrysin inhibited tumor glycolysis and induced
apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J
Exp Clin Cancer Res. 36:442017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xiao W, Dong W, Zhang C, Saren G, Geng P,
Zhao H, Li Q, Zhu J, Li G, Zhang S and Ye M: Effects of the
epigenetic drug MS-275 on the release and function of
exosome-related immune molecules in hepatocellular carcinoma cells.
Eur J Med Res. 18:612013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang B, Yin X and Sui S: Resveratrol
inhibited the progression of human hepatocellular carcinoma by
inducing autophagy via regulating p53 and the phosphoinositide
3kinase/protein kinase B pathway. Oncol Rep. 40:2758–2765.
2018.PubMed/NCBI
|
|
85
|
Park S, Lim J, Kim JR and Cho S:
Inhibitory effects of resveratrol on hepatitis B virus X
protein-induced hepatocellular carcinoma. J Vet Sci. 18:419–429.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liao PC, Ng LT, Lin LT, Richardson CD,
Wang GH and Lin CC: Resveratrol arrests cell cycle and induces
apoptosis in human hepatocellular carcinoma Huh-7 cells. J Med
Food. 13:1415–1423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Notas G, Nifli AP, Kampa M, Vercauteren J,
Kouroumalis E and Castanas E: Resveratrol exerts its
antiproliferative effect on HepG2 hepatocellular carcinoma cells,
by inducing cell cycle arrest, and NOS activation. Biochim Biophys
Acta. 1760:1657–1666. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Bishayee A, Politis T and Darvesh AS:
Resveratrol in the chemoprevention and treatment of hepatocellular
carcinoma. Cancer Treat Rev. 36:43–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li Y, Cheng X, Chen C, Huijuan W, Zhao H,
Liu W, Xiang Z and Wang Q: Apigenin, a flavonoid constituent
derived from P. villosa, inhibits hepatocellular carcinoma cell
growth by CyclinD1/CDK4 regulation via p38 MAPK-p21 signaling.
Pathol Res Pract. 216:1527012020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen
Y, Chen H, Chu F and Zhang Y: Luteolin promotes cell apoptosis by
inducing autophagy in hepatocellular carcinoma. Cell Physiol
Biochem. 43:1803–1812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Fan YP, Liao JZ, Lu YQ, Tian DA, Ye F,
Zhao PX, Xiang GY, Tang WX and He XX: MiR-375 and doxorubicin
Co-delivered by liposomes for combination therapy of hepatocellular
carcinoma. Mol Ther Nucleic Acids. 7:181–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ang C, O'Reilly EM, Carvajal RD, Capanu M,
Gonen M, Doyle L, Ghossein R, Schwartz L, Jacobs G, Ma J, et al: A
nonrandomized, phase II study of sequential irinotecan and
flavopiridol in patients with advanced hepatocellular carcinoma.
Gastrointest Cancer Res. 5:185–189. 2012.PubMed/NCBI
|
|
93
|
Liu L, Chen X, Xie S, Zhang C, Qiu Z and
Zhu F: Variant 1 of KIAA0101, overexpressed in hepatocellular
carcinoma, prevents doxorubicin-induced apoptosis by inhibiting p53
activation. Hepatology. 56:1760–1769. 2012. View Article : Google Scholar : PubMed/NCBI
|