Introduction
Primary liver cancer is a rapidly progressing
neoplasm with high morbidity and mortality rates (1). Liver cancer ranks fourth among the most
common cancer-related causes of death and sixth in terms of
incidence (2). Worldwide, in 2019,
hepatocellular carcinoma (HCC) accounted for ~80% of all primary
liver cancer cases (1). There are
numerous risk factors for CC, including chronic hepatitis B virus
(HBV) infection, hepatitis C virus (HCV) infection, long-term
intake of aflatoxin, alcoholism and non-alcoholic fatty liver
disease (3). Chronic HBV and HCV
infections are the leading causes of HCC, constituting ~80% of
cases worldwide (1,4), but the mechanism of HCC pathogenesis
has not been fully revealed. The current treatment methods for HCC
have resulted in big improvements in clinical practice, early stage
HCC can be cured via surgical resection, liver transplantation and
radiofrequency ablation (5).
However, the prognosis of HCC is poor, with a 5-year survival rate
of <20% (2). When the location of
a tumor is challenging (such as near the liver capsule,
diaphragmatic top, important bile duct or gastrointestinal tract)
and not suitable for the implementation of the aforementioned
methods, there are certain cell cycle-associated drugs, such as
doxorubicin, cisplatin and mitomycin C, that can be used to
chemoembolize the hepatic artery, in a process known as
transarterial chemoembolization (TACE). TACE is considered as a
remedy for early and very early stage liver cancer (6–11).
However, predictors of early stage HCC remain limited. Ultrasound,
as the common means for screening and monitoring early stage HCC,
can be affected by various factors, such as the skill of the
ultrasound operators, the body habitus and liver nodularity of the
patient, which can sometimes make a big difference in the results
(12,13). Furthermore, with a cut-off value of
20 ng/ml, the specificity of α-fetoprotein (AFP) is 80–90%, but the
sensitivity is 40–60%, and normal AFP levels are detected in ~30%
of patients with HCC (14). AFP
levels can be influenced by cirrhosis or the level of liver
inflammation to a certain extent (12,15).
Therefore, numerous patients with HCC are diagnosed at the advanced
stage, and surgery to remove all the tumor cells is often
difficult, leading to a high recurrence rate and a low 5-year
survival rate (16). Therefore,
early diagnosis and early treatment are of profound importance for
patients with HCC.
Gene variation, deletion and abnormal expression are
closely associated with the occurrence, metastasis and recurrence
of HCC. With the development of molecular biology and genomics,
high-throughput sequencing is expected to provide biomarkers for
screening and monitoring liver cancer (1). Compared with sequencing technologies,
such as gene chip technology, hybridization- or sequence-based
approaches, RNA-sequencing (RNA-seq) has a higher sensitivity,
accuracy, breadth and depth using the ‘next-generation’ sequencing
technology, making the results more comprehensive and repeatable
(17).
The present study aimed to identify the hub genes
and signaling pathways closely associated with HBV-associated early
stage HCC using RNA-seq technology. Additionally, small molecular
compounds that may reverse the altered differentially expressed
genes (DEGs) were identified. These hub genes may be used as
potential diagnostic and prognostic markers, and the small
molecular compounds may be used as promising candidate agents for
HBV-associated early stage HCC.
Materials and methods
Screening dataset and processing raw
data
The study design is presented in the form of a flow
chart (Fig. 1). The Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) is a public genome
database and an online tool for downloading experimental and
curated gene expression raw data. The GSE124535 dataset (18) from the GEO database was used in the
present study, consisting of 35 HBV-associated early stage HCC
tissues and 35 matched normal tissues, which were obtained from
patients in Zhongshan Hospital, Fudan University (Shanghai, China)
and Cancer Hospital & Institute, Peking University (Beijing,
China), with the approval of the Research Ethics Committees of
these two hospitals (18). And its
Sequence Read Archive (SRA) accession number was SRP174991. The SRA
(https://www.ncbi.nlm.nih.gov/sra/) is
a primary archive of the National Institutes of Health regarding
high-throughput sequencing data and is part of the International
Nucleotide Sequence Database Collaboration; it is the largest
publicly available repository of high-throughput sequencing data
and accepts data from all branches of life sciences, as well as
metagenomic and environmental surveys. On June 16, 2020, the total
bases and size of the SRA had reached 4.2182×1016 bases
and 1.52091×1016 bytes, respectively. The SRA stores raw
sequencing data and alignment information to enhance
reproducibility and facilitate novel discoveries through data
analysis. Subsequently, RNA-seq raw data of SRP174991, including 35
early stage HCC tissues and 35 matched normal tissues, were
downloaded from the SRA. Next, the original binary SRA data was
converted into sequencing data as a fastq file. Firstly, reasonable
quality control and filtering were performed on the data. Reads
length, reads quality, uncertain base number and other cut-off
values were set using Cutadapt software (version 2.4) (http://code.google.com/p/cutadapt/); reads and
bases with lower quality were screened out and removed, with wow
quality defined as the length of read is <100 and the quality of
read is <25). Secondly, Hisat2 in the hisat2-build program of
the Hisat2 software and to determine the position of chromosomes,
so as to determine which gene it corresponds to. Thirdly, after the
reads were aligned to the location of the gene in which they were
located, the HTSeq software (19)
(version 0.12.4) so that the gene expression levels in the samples
could be accurately compared. The identification of DEGs was
performed using the ‘limma’ package (20) (version 3.40.4). The cut-off criteria
of |log2 fold-change (FC)|>1 and P<0.01 were
considered to indicate a statistically significant difference.
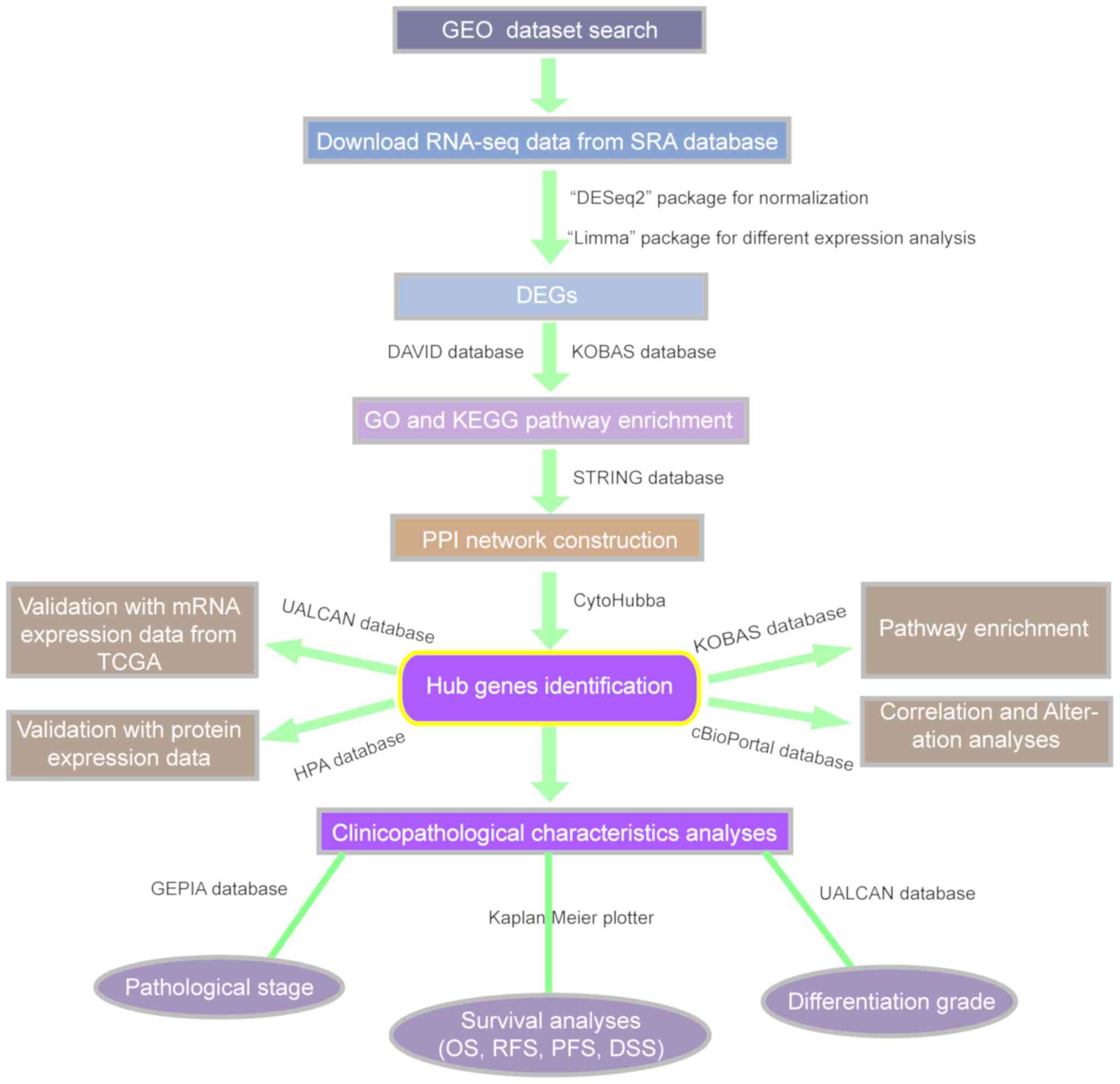 | Figure 1.Flow chart of the study. Gene
expression profile data were extracted from the GEO database, and
DEGs, hub genes and signal pathways were screened by
high-throughput sequencing method, and mRNA and protein expression
of hub genes were verified in UALCAN database and HPA database
respectively. Then, The association between hub genes and the
effects of the abnormal expression of hub genes on the rate of
genetic variation, OS, RFS, PFS and DSS of patients with HCC, as
well as pathological stage and grade, were analyzed using different
databases. GEO, Gene Expression Omnibus; DEG, differentially
expressed gene; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of
Genes and Genomes; PPI, protein-protein interaction; TCGA, The
Cancer Genome Atlas; HPA, Human Protein Atlas; OS, overall
survival; RFS, relapse-free survival; PFS, progression-free
survival; DSS, disease-specific survival; KOBAS, KEGG
Orthology-Based Annotation System; STRING, Search Tool for the
Retrieval of Interacting Genes/Proteins; SRA, Sequence Read
Archive. |
Function and pathway enrichment
analyses of DEGs
To further investigate the identified DEGs, the
Database for Annotation, Visualization and Integrated Discovery
(v6.8; http://david.ncifcrf.gov/) (21) was used for Gene Ontology (GO)
enrichment analysis. GO annotation and functional analyses were
performed for DEGs. GO functional analysis included analysis of
biological processes (BPs), cellular components (CCs) and molecular
functions (MFs). BP refers to pathways and larger processes
comprising the activities of multiple gene products, CC indicates
where within the cell the gene products are active and MF indicates
molecular activities of gene products (21). The Kyoto Encyclopedia of Genes and
Genomes (KEGG) Orthology-Based Annotation System (KOBAS) 3.0
(22) database (http://kobas.cbi.pku.edu.cn/) was used for KEGG
enrichment analysis. KOBAS is a web-based tool for gene functional
enrichment analysis, including corresponding visual pathway
diagrams. P<0.05 was set as the enrichment threshold.
Protein-protein interaction (PPI)
network construction
To establish the interactions of these DEGs in HCC,
a DEG-associated PPI network was constructed using the Search Tool
for the Retrieval of Interacting Genes/Proteins database
(https://string-db.org.uk/) (23) with a minimum required interaction
score of >0.9. Interactions were subsequently visualized and
analyzed using Cytoscape v3.7.1 (24) after hiding the disconnected
nodes.
Identification of hub genes
CytoHubba (version 0.1) (25), a plug-in of Cytoscape v3.7.1, was
used to screen hub genes. Three topological analysis methods of
CytoHubba, namely degree, closeness and betweenness, were used to
measure each gene in the PPI network. Degree centrality describes
the sum of direct edges each node has, and proteins with higher
degrees are more likely to be essential proteins (25). Closeness centrality is used to
calculate the sum of the distances from one nodes to all other
nodes, and betweenness is utilized to calculate the number of
shortest paths through a node (26).
These three measures are the most commonly used to reveal the
relative importance of nodes in the structure of a network
(26). The top 50 genes in each
analysis method were selected, and the hub genes were filtered from
their intersection.
Validating hub genes with mRNA and
protein expression data
UALCAN (http://ualcan.path.uab.edu/index.html) (27) is an easy to use, interactive
web-portal that can perform in-depth analyses of The Cancer Genome
Atlas (TCGA) Program gene expression data (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
The Human Protein Atlas (HPA) database (http://www.protein atlas.org/) (28) integrates various biotechnologies to
cover the database of proteins in almost all human cells, tissues
and organs. The mRNA expression levels of the hub genes were
verified by the UALCAN online database in 50 normal tissues and 371
HCC tissues from TCGA database. The protein expression levels of
the hub genes were verified using the HPA databases.
Immunohistochemical (IHC) images were downloaded from the HPA
database. The mean integrated optical density (IOD) value of IHC
images was measured using Image-Pro Plus software (v6.0; Media
Cybernetics, Inc.). The higher the total IOD value was, the greater
the expression level was. IHC data were analyzed using an unpaired
Student's t-test using GraphPad Prism® v8.0 (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Pathway analysis correlation and
genetic variation rate of hub genes
The aforementioned KOBAS database was used to
conduct pathway enrichment analysis of hub genes. The cBioPortal
(http://www.cbioportal.org/) (29) for TCGA provides a web resource for
exploring, visualizing and analyzing multidimensional cancer
genomics data (29). TCGA contains
both sequencing and pathological data of 30 different types of
cancer. The HCC study (TCGA, PanCancer Atlas) in the cBioPortal
database, containing 372 samples with mRNA data, was used for
correlation and genetic change analysis of hub genes according to
the online instructions. The data type selected was mRNA, and mRNA
expression z-scores (RNA Seq V2 RSEM) were selected as the mRNA
profile. The correlation between hub genes is represented by
Pearson's correlation coefficient (Pearson's correlation
coefficient is positive for positive correlation; Pearson
correlation coefficient is negative for negative correlation).
Clinical significance of hub
genes
The association between hub genes and
clinicopathological characteristics was analyzed. The
clinicopathological characteristics mainly included the following
aspects: Survival analysis, pathological stage and tumor grade.
Survival analysis consisted of overall survival (OS),
recurrence-free survival (RFS), progression-free survival (PFS) and
disease-specific survival (DSS). The Kaplan-Meier plotter
(http://www.kmplot.com/analysis/index.php?p=background)
(30), and the UALCAN (http://ualcan.path.uab.edu/index.html)
databases were used for survival, pathological stage and grade
analysis, respectively. The follow up threshold of OS and DSS were
set as 60 months. The follow up threshold of RFS and PFS were set
as 36 months. Survival analysis was performed using log-rank tests
(30). Unpaired t-tests (27) were carried out for stage and grade
analysis between groups.
Connectivity map (CMap) analysis
The CMap database (updated on September 12, 2017;
http://portals.broadinstitute.org/cmap/) (31) is a collection of genome-wide
transcriptional expression data from cultured human cells treated
with bioactive small molecules and simple pattern-matching
algorithms that together enable the discovery of functional
connections between drugs, genes and diseases through the
transitory feature of common gene expression changes. CMap analysis
was utilized to predict underlying small molecule compounds that
may reverse the altered expression levels of DEGs in cell lines.
Therefore, small molecule compounds with a prominently negative
association with the HBV-associated early stage HCC signature were
identified and may be potential therapeutic agents for
HBV-associated early stage HCC. Mean <-0.4 and P<0.05 were
set as the threshold values.
Results
Raw data and identification of
DEGs
The GSE124535 dataset was selected, consisting of 35
HBV-associated early stage HCC tissues and 35 matched normal
tissues, which were obtained from patients in Zhongshan Hospital,
Fudan University (Shanghai, China) and Cancer Hospital &
Institute, Peking University (Beijing, China), with the approval of
the Research Ethics Committees of these two hospitals. The clinical
information of the 35 patients is presented in Table I (18). Among these patients, there were 29
males and 6 females, and their average age is 55, and the age range
is 25–80 years. All patients provided early stage (Barcelona Clinic
Liver Cancer stages 0 and A) HCC tissues infected with HBV and
negative for HCV. Additionally, patients had not undergone prior
chemotherapy or radiation. Based on the cut-off criteria, a total
of 1,582 DEGs were identified, among which 530 were upregulated and
1,052 were downregulated. Their distribution was presented using a
volcano plot (Fig. 2A).
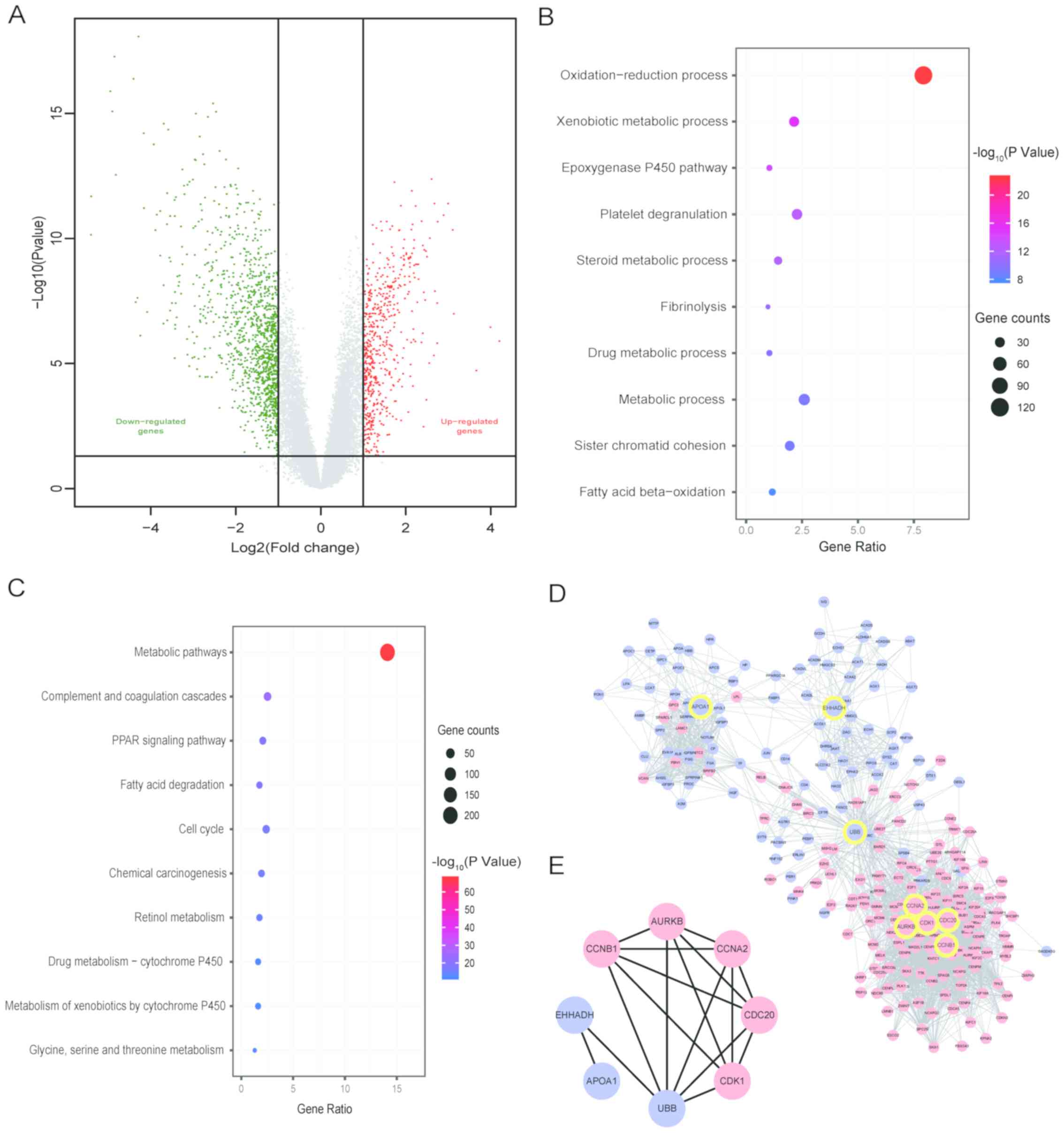 | Figure 2.Distribution of DEGs, GO and KEGG
enrichment and PPI network. (A) Volcano plot of the distribution of
DEGs. (B) Top 10 GO biological processes analysis. (C) Top 10 terms
of KEGG enrichment analysis. (D) PPI network of genes closely
associated with hub genes. The red nodes represent upregulated
genes, the blue nodes represent downregulated genes, the yellow
circles are the positions of the 8 hub genes in the PPI network
diagram. (E) PPI network of hub genes. AURKB, aurora kinase B;
CDK1, cyclin-dependent kinase 1; EHHADH, enoyl-CoA hydratase and
3-hydroxyacyl CoA dehydrogenase; CDC20, cell division cycle 20;
CCNA2/B1, cyclin A2/B1; APOA1, apolipoprotein A1; UBB, ubiquitin B;
DEGs, differentially expressed genes; GO, Gene Ontology; KEGG,
Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein
interaction. |
 | Table I.Clinical information of patients with
HCC (n=35). |
Table I.
Clinical information of patients with
HCC (n=35).
| Clinical
information | Cases, n |
|---|
| Cirrhosis |
|
|
Yes | 28 |
| No | 7 |
| Number of
tumors |
|
| 1 | 32 |
| 2 | 3 |
| Diameter of tumor,
cma |
|
|
<3 | 9 |
| 35 | 25 |
|
>5 | 1 |
| Lymphatic
metastasis |
|
|
Yes | 0 |
| No | 35 |
| Macrovascular
invasion |
|
|
Yes | 0 |
| No | 35 |
| Microvascular
invasion |
|
|
Yes | 9 |
| No | 26 |
| α-fetoprotein,
ng/ml |
|
|
<20 | 18 |
|
20-400 | 8 |
|
>400 | 9 |
| Cancer
recurrence |
|
|
Yes | 3 |
| No | 32 |
| Died of
recurrence |
|
|
Yes | 0 |
| No | 35 |
GO and KEGG enrichment analyses
The DEGs in the BP category of GO analysis were
mainly enriched in ‘oxidation-reduction process’, ‘xenobiotic
metabolic process’, ‘epoxygenase P450 pathway’, ‘steroid metabolic
process’, ‘metabolic process’ and ‘fatty acid beta-oxidation’
(Fig. 2B). In the KEGG pathway
enrichment analysis, DEGs were significantly enriched in ‘metabolic
pathways’, ‘complement and coagulation cascades’, ‘PPAR signaling
pathway’, ‘fatty acid degradation’, the ‘cell cycle’ and ‘chemical
carcinogenesis’ (Fig. 2C).
PPI network and hub genes
A PPI network of DEGs was established, containing
1,462 nodes, 4,744 edges and an average point degree of 6.49 (data
not shown). The analyses of degree, closeness and betweenness in
the PPI network were performed using CytoHubba. Subsequently, 8 hub
genes were selected from the intersection of the top 50 genes of
the three topological analysis methods using the Cytoscape plug-in
CytoHubba, among which the upregulated hub genes were aurora kinase
B (AURKB), cyclin-dependent kinase 1 (CDK1), cell division cycle 20
(CDC20), cyclin (CCN)B1 and CCNA2, while the downregulated hub
genes were enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase
(EHHADH), apolipoprotein A1 (APOA1) and ubiquitin B (UBB). The PPI
network of genes closely associated with the 8 hub genes, with 244
nodes and 2,781 edges, is presented in Fig. 2D. The PPI network between hub genes
is shown in Fig. 2E.
Validation of hub genes
The UALCAN online database was used to verify the
mRNA expression levels of the 8 hub genes. The protein expression
levels of the hub genes were analyzed using IHC samples from the
HPA online database. As shown in Fig.
3A-C, the mRNA and protein levels of AURKB, CDK1, CDC20, CCNB1
and CCNA2 were upregulated in tumor tissues compared with in normal
tissues, while the mRNA and protein levels of EHHADH, APOA1 and UBB
levels were downregulated in tumor tissues (Fig. 3C), indicating that the mRNA and
protein expression of hub genes were similar in different
databases.
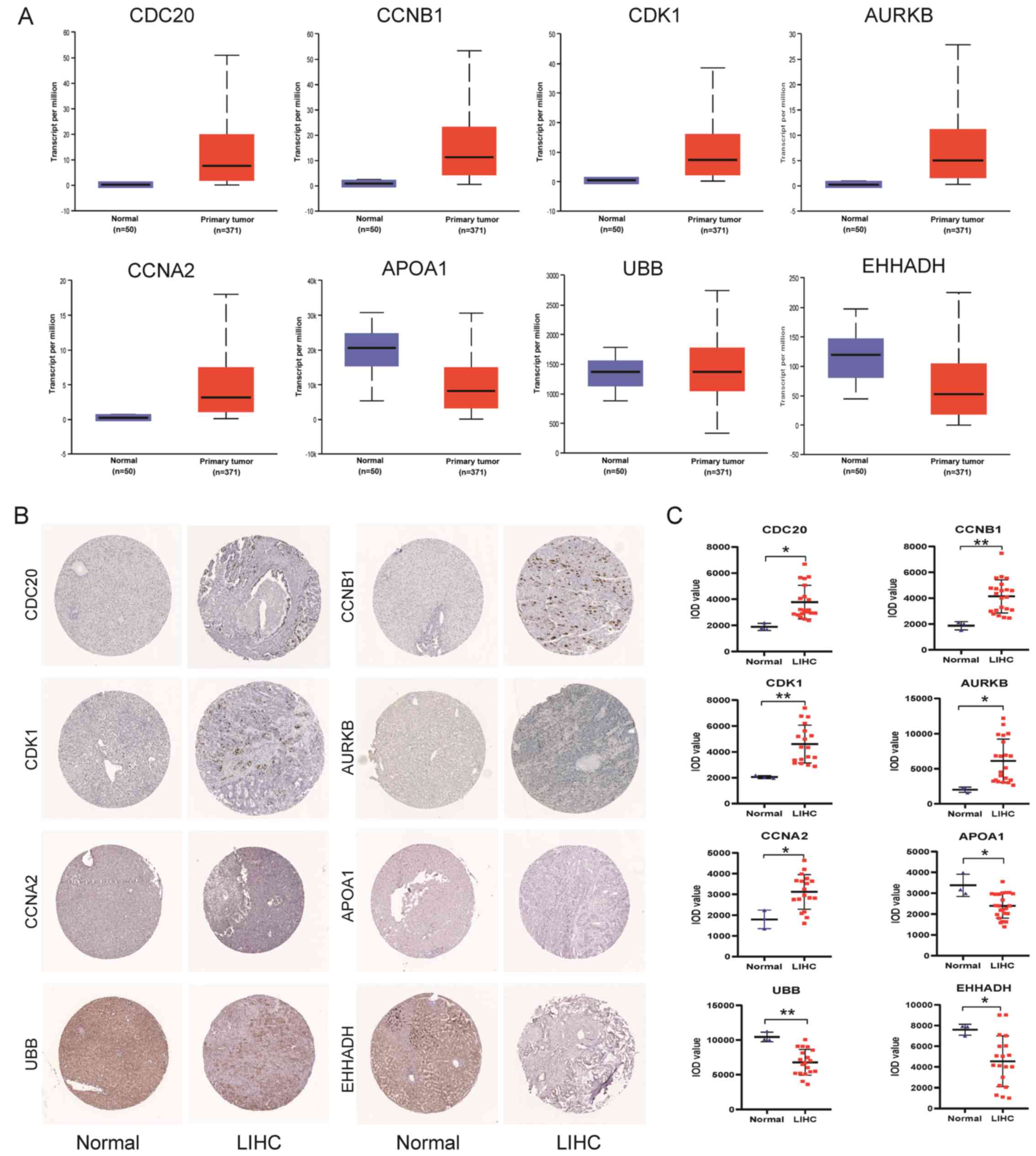 | Figure 3.Validation of hub gene expression.
(A) Validation of hub gene mRNA expression in The Cancer Genome
Atlas database. (B) Validation of protein expression levels of hub
genes in the Human Protein Atlas database. AURKB: https://www.proteinatlas.org/ENSG00000178999-AURKB/tissue/liver#img,
(https://www.proteinatlas.org/ENSG00000178999AURKB/pathology/liver+cancer#img);
CDK1: (https://www.proteinatlas.org/ENSG00000170312-CDK1/tissue/liver#img),
(https://www.proteinatlas.org/ENSG00000170312-CDK1/pathology/liver+cancer#img);
EHHADH: (https://www.proteinatlas.org/ENSG00000113790-EHHADH/tissue#img),
(https://www.proteinatlas.org/ENSG00000113790-EHHADH/pathology/liver+cancer#img);
CDC20: (https://www.proteinatlas.org/ENSG00000117399-CDC20/tissue/liver#img),
(https://www.proteinatlas.org/ENSG00000117399-CDC20/pathology/liver+cancer#img);
CCNA2: (https://www.proteinatlas.org/ENSG00000145386-CCNA2/tissue/liver#img),(https://www.proteinatlas.org/ENSG00000145386--CCNA2/pathology/liver+cancer#img;
CCNB1: (https://www.proteinatlas.org/ENSG00000134057-CCNB1/tissue/liver#img),
(https://www.proteinatlas.org/ENSG00000134057-CCNB1/pathology/liver+cancer#img);
APOA1: (https://www.proteinatlas.org/ENSG00000118137-APOA1/tissue/liver#img),
(https://www.proteinatlas.org/ENSG00000118137APOA1/pathology/liver+cancer#img);
UBB: (https://www.proteinatlas.org/ENSG00000170315-UBB/tissue/liver#img),
(https://www.proteinatlas.org/ENSG00000170315-UBB/pathology/liver+cancer#img).
(C) IOD level of hub genes in immunohistochemistry sample images.
*P<0.05; **P<0.01 vs. normal tissues. IOD, integrated optical
density; LIHC, liver hepatocellular carcinoma; AURKB, aurora kinase
B; CDK1, cyclin--dependent kinase 1; EHHADH, enoyl-CoA hydratase
and 3-hydroxyacyl CoA dehydrogenase; CDC20, cell division cycle 20;
CCNA2/B1, cyclin A2/B1; APOA1, apolipoprotein A1; UBB, ubiquitin
B. |
Pathway analysis, correlation and
genetic variation rate of hub genes
Hub genes were significantly enriched in signaling
pathways such as the ‘cell cycle’, ‘viral carcinogenesis’, the ‘P53
signaling pathway’ and the ‘PPAR signaling pathway’ (Fig. 4A). Correlation analysis in the
cBioPortal database revealed that most of the hub genes were
correlated with each other when their expression changes were in
the same direction, such as CDC20 with CCNB1, CCNB1 with CDK1, and
CDC20 with AURKB, which were highly and positively correlated with
each other. By contrast, there were significant negative
correlations between all downregulated hub genes and all
upregulated hub genes, such as EHHADH with AURKB, CDK1, CCNB1 and
CDC20 (Fig. 4B). According to the
genetic changes of hub genes, it was revealed that the abnormal
expression of the 8 hub genes were found in ≥30% of all samples
(Fig. 4C and D).
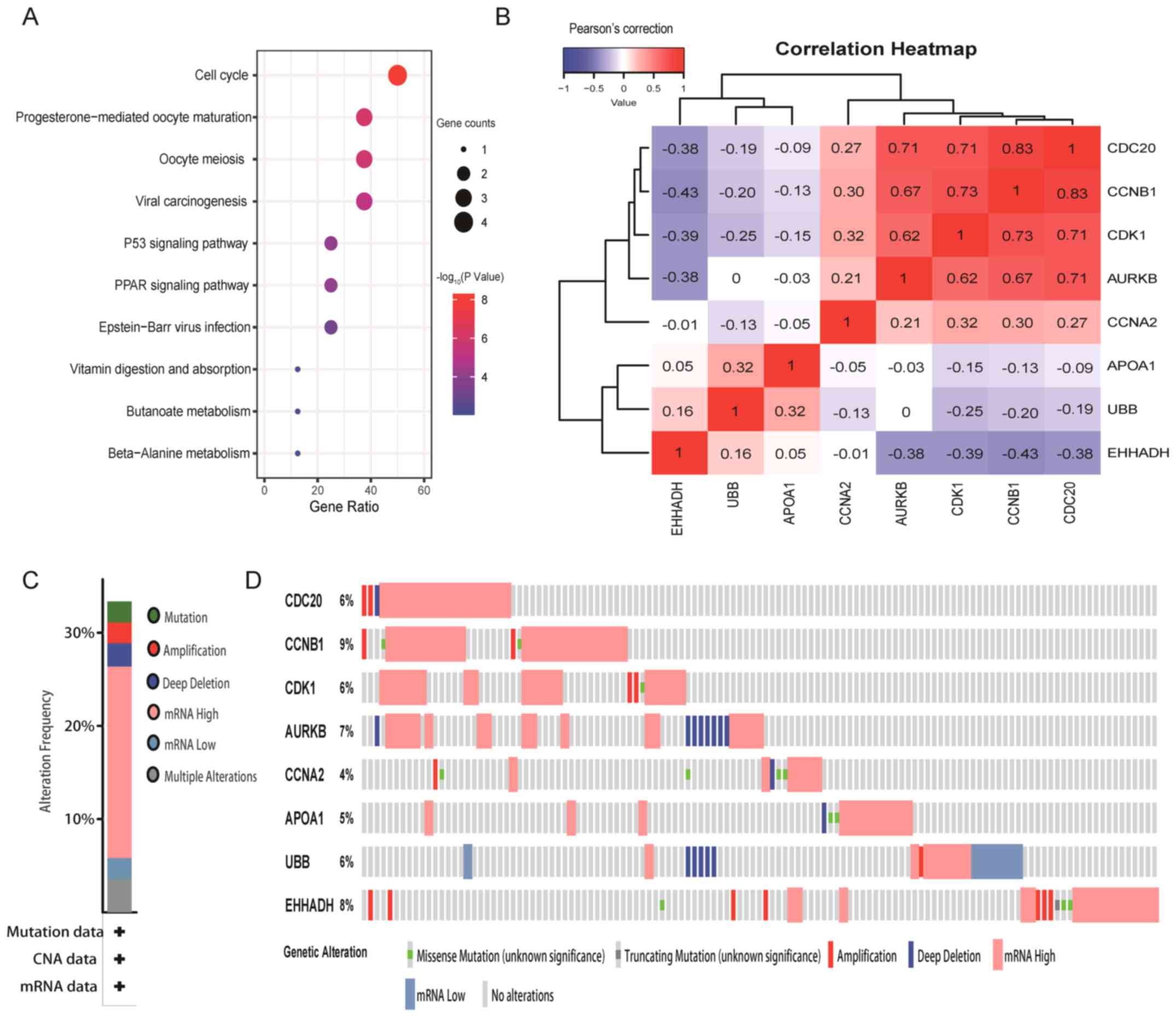 | Figure 4.Pathway enrichment, correlation and
mutation rate of hub genes. (A) Top 10 enriched pathways of hub
genes. (B) Correlation heatmap. Blue indicates that there is a
negative correlation between hub genes, red indicates that there is
a positive correlation between hub genes and the number represents
the Pearson's correlation coefficient. (C and D) Rate of genetic
variation caused by abnormal changes of hub genes in tumor tissues.
(C) The abnormal expression of the eight hub genes were found in
>30% of all samples. (D) Taking CDC20 as an example, the number
of samples with gene CDC20 alterations in all samples accounted for
6% of the total number of samples. Green represents ‘Missense
Mutation’; dark gray represents ‘Truncating Mutation’; red
represents ‘Amplification’; purple represents ‘Deep Deletion’;
flesh color represents ‘mRNA High’; blue represents ‘mRNA Low’;
light gray represents ‘No alterations’. AURKB, aurora kinase B;
CDK1, cyclin-dependent kinase 1; EHHADH, enoyl-CoA hydratase and
3-hydroxyacyl CoA dehydrogenase; CDC20, cell division cycle 20;
CCNA2/B1, cyclin A2/B1; APOA1, apolipoprotein A1; UBB, ubiquitin
B. |
Clinical significance of hub
genes
Kaplan-Meier curves of the 8 hub genes revealed that
the higher the expression levels of CDC20, CCNB1, AURKB, CCNA2 and
CDK1, and the lower the expression levels of APOA1 and UBB, the
poorer the OS, RFS, PFS and DSS of the patients (Fig. 5). In addition, low expression levels
of EHHADH were associated with lower OS, DSS and PFS rates, while
P-values for RFS was not statistically significant, as shown in
Fig. 5. Based on UALCAN online
database, for hub genes CDC20, CDK1, CCNB1, CCNA2 and AURKB, the
higher their expression levels, the higher the stage in HCC
tissues, while for APOA1and EHHADH, the lower its expression
levels, the higher the stage. However, it is worth noting that due
to the small sample size of stage 4, a certain bias in the results
cannot be ruled out. Therefore, stage 4 is not included in the
statistics. Besides, there was no significant relationship between
UBB expression and tumor stage. (Fig.
6A). Furthermore, in the UALCAN online database, for hub genes
CDC20, CDK1, CCNB1, CCNA2 and AURKB, the higher their expression
levels, the higher the grade in HCC tissues, while for EHHADH, the
lower its expression levels, the higher the grade. Besides, there
was no significant relationship between APOA1 and UBB expression
and tumor grade (Fig. 6B).
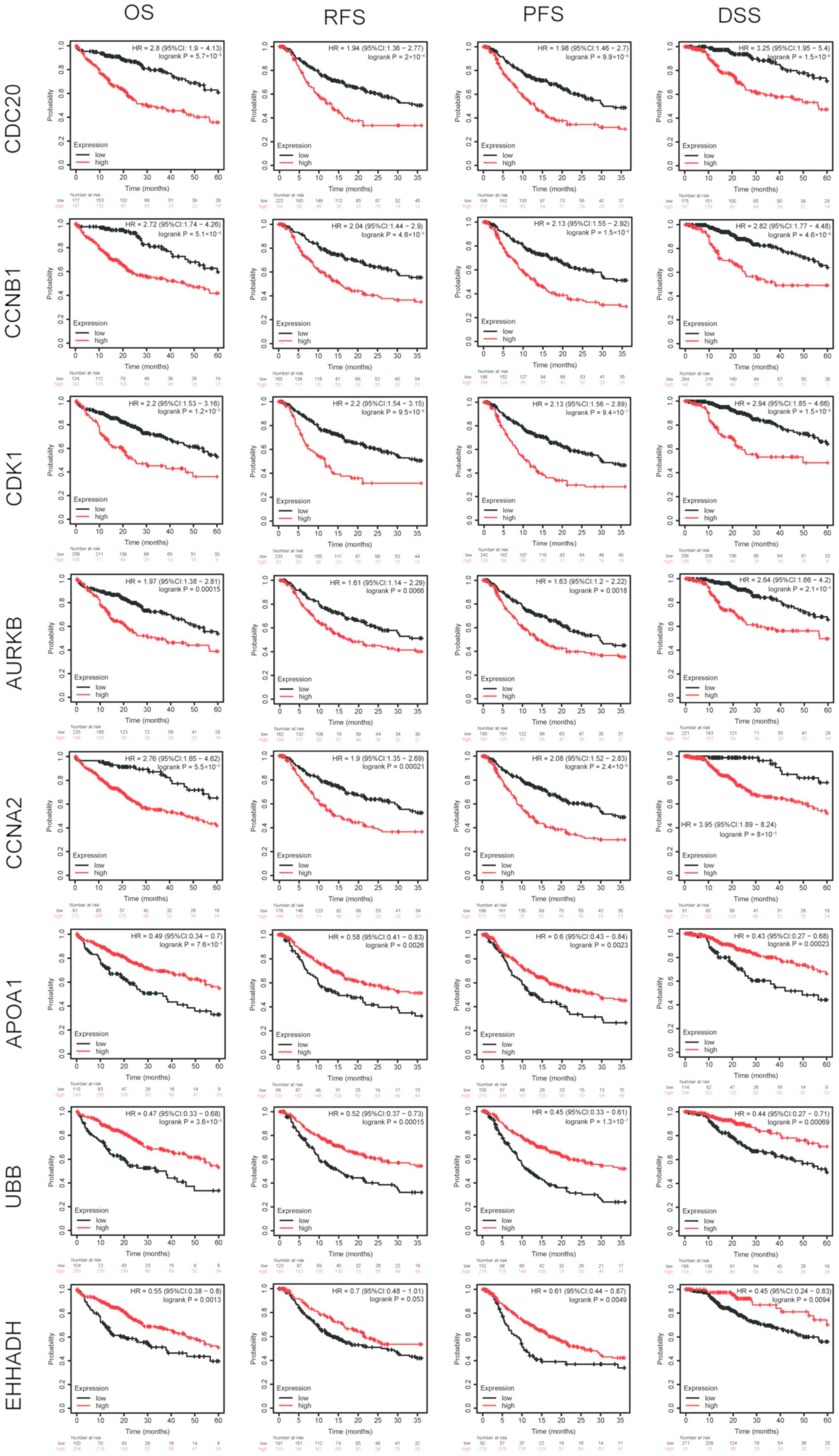 | Figure 5.Survival analysis of hub genes. The
higher the expression levels of CDC20, CCNB1, AURKB, CCNA2, CDK1
and the lower the expression levels of APOA1 and UBB, the poorer
the OS, RFS, PFS and DSS of the patients. In addition, lower levels
of EHHADH resulted in lower OS, DSS and PFS rates, but P-values for
RFS were not statistically significant. AURKB, aurora kinase B;
CDK1, cyclin-dependent kinase 1; EHHADH, enoyl-CoA hydratase and
3-hydroxyacyl CoA dehydrogenase; CDC20, cell division cycle 20;
CCNA2/B1, cyclin A2/B1; APOA1, apolipoprotein A1; UBB, ubiquitin B;
OS, overall survival; RFS, relapse-free survival; PFS,
progression-free survival; DSS, disease-specific survival; HR,
hazard ratio; CI, confidence interval. |
 | Figure 6.Association between hub genes and
pathological stage and grade. (A) For hub genes CDC20, CDK1, CCNB1,
CCNA2 and AURKB, the higher their expression levels, the higher the
stage in HCC tissues, while for APOA1and EHHADH, the lower its
expression levels, the higher the stage (Due to the small sample
size of stage 4, a certain bias in the results cannot be ruled out,
therefore, stage 4 is not included in the statistics). Besides,
there was no significant relationship between UBB expression and
tumor stage. (B) For hub genes CDC20, CCNB1, CDK1 and AURKB, the
higher their expression levels, the higher the grade and therefore
the lower the differentiation level in HCC tissues. For EHHADH, the
lower its expression levels, the higher the grade and the lower the
differentiation level in HCC tissues. For the hub gene CCNA2, the
higher the level of CCNA2 expression in liver cancer tissues, the
lower the differentiation level was, but the difference was not
statistically significant. Comparisons between grade 1/2 and 3/4
for APOA1 and UBB revealed no statistical significance. *P<0.05,
**P<0.01 and ***P<0.001 vs. normal; #P<0.05,
##P<0.01 and ###P<0.001 vs. Grade 1 or
Stage 1; &P<0.05, &&P<0.01
and &&&P<0.001 vs. Grade 2. AURKB, aurora
kinase B; CDK1, cyclin-dependent kinase 1; EHHADH, enoyl-CoA
hydratase and 3-hydroxyacyl CoA dehydrogenase; CDC20, cell division
cycle 20; CCNA2/B1, cyclin A2/B1; APOA1, apolipoprotein A1; UBB,
ubiquitin B; HCC, hepatocellular carcinoma; TMP, transcripts per
million; Pr(>F), P-value. |
CMap analysis
CMap analysis was used to identify potential small
molecular compounds that may reverse the altered expression levels
of DEGs. The most five significant small molecular compounds were
phenoxybenzamine, GW-8510, resveratrol, 0175029-0000 and
daunorubicin (Table II).
 | Table II.CMap analysis of the 20 most
significant small molecular compounds that may reverse the altered
expression levels of the differentially expressed genes. |
Table II.
CMap analysis of the 20 most
significant small molecular compounds that may reverse the altered
expression levels of the differentially expressed genes.
| CMap name | Mean | Enrichment | P-value | Percent
non-null |
|---|
|
Phenoxybenzamine | −0.873 | −0.995 | <0.00001 | 100 |
| GW-8510 | −0.799 | −0.952 | <0.00001 | 100 |
| Resveratrol | −0.781 | −0.827 | <0.00001 | 100 |
| 0175029-0000 | −0.777 | −0.887 | <0.00001 | 100 |
| Daunorubicin | −0.756 | −0.944 | <0.00001 | 100 |
| Apigenin | −0.792 | −0.927 | 0.00002 | 100 |
| Camptothecin | −0.778 | −0.980 | 0.00004 | 100 |
| Thioguanosine | −0.812 | −0.898 | 0.00022 | 100 |
| Irinotecan | −0.763 | −0.953 | 0.00022 | 100 |
| Chrysin | −0.733 | −0.954 | 0.00022 | 100 |
| Luteolin | −0.703 | −0.893 | 0.00026 | 100 |
| Trifluridine | −0.729 | −0.849 | 0.00095 | 100 |
| Alsterpaullone | −0.753 | −0.905 | 0.00162 | 100 |
| Doxorubicin | −0.744 | −0.877 | 0.00375 | 100 |
| Thiostrepton | −0.713 | −0.779 | 0.00493 | 100 |
| Menadione | −0.766 | −0.950 | 0.00545 | 100 |
| Monobenzone | −0.730 | −0.753 | 0.00758 | 100 |
| Ms-275 | −0.729 | −0.931 | 0.01006 | 100 |
| Azacitidine | −0.719 | −0.798 | 0.01683 | 100 |
| 8-azaguanine | −0.753 | −0.683 | 0.02216 | 100 |
Discussion
Primary liver cancer is a rapidly progressing
malignant tumor, and early diagnosis and treatment are of great
importance for patients. Although great progress has been made, the
diagnostic and prognostic biomarkers, and targeted agents of HCC
have not been fully revealed. In the present study, next-generation
high-throughput sequencing technology, namely RNA-seq, was applied
to provide robust raw data for further exploring the critical genes
and pathways associated with HBV-associated early stage HCC. CMap
analysis was applied to identify promising targeted agents against
HBV-associated early stage HCC.
In the present study, a total of 1,582 DEGs were
identified. KEGG enrichment analysis of DEGs revealed that the DEGs
were mostly enriched in metabolic pathways, which was consistent
with previous results showing that the dysfunction of metabolic
pathways was a critical risk factor for HCC, especially for those
infected with HBV or HCV (32–34).
‘Fatty acid degradation’ and ‘PPAR signaling pathway’ in KEGG
analysis, and ‘steroid metabolic process’ and ‘fatty acid
beta-oxidation’ in GO BP analysis, are all strongly associated with
fat metabolism (35), suggesting
that fat metabolism disorders may be closely associated with HCC.
Yamashita et al (36)
revealed that the lipogenic pathway is activated in HCC and can
promote HCC proliferation and may have close association with high
mortality. Jiang et al (18)
demonstrated that when cholesterol homeostasis was impaired,
HBV-infected patients with HCC had the lowest overall survival rate
and a worse prognosis after the initial surgery treatment. In the
present study, the hub gene EHHADH, which is involved in fatty acid
metabolism, was enriched in pathways such as ‘fatty acid
metabolism’, ‘fatty acid beta-oxidation’ and the ‘PPAR signaling
pathway’; the hub gene APOA1, which is associated with tumor
metabolism, was also enriched in the ‘PPAR signaling pathway’.
Additionally, GO analysis of BP provided evidence
that the DEGs were significantly enriched in the
‘oxidation-reduction process’ (redox). To the best of our
knowledge, no reports have confirmed an association between redox
and HCC, but there are studies about oxidative stress and HCC. It
has been demonstrated that oxidative stress can lead to unrepaired
DNA damage, especially of nuclear and mitochondrial DNA, which may
contribute to modifications and mutations of gene expression, and
may subsequently induce the development of HCC (37,38).
Additionally, KEGG analysis of the hub genes in the present study
revealed that the ‘p53 signaling pathway’ was an important pathway.
Previous studies have reported that the p53 signaling pathway
affects the proliferation, metastasis and radiotherapy sensitivity
of HCC (39,40), and is associated with redox (41–43).
Under physiological conditions, cells maintain a balance of redox
reactions by generating and removing reactive oxygen species (ROS)
and reactive nitrogen species (RNS) (44). If the steady state of the redox
reaction is altered, it will induce oxidative stress, which in turn
can lead to excessive production of ROS and RNS (44). Both mild and severe oxidative stress
will activate p53, which will activate different target genes, thus
determining the different fate of the cell (37,44–46).
Under low levels of oxidative stress, activated p53 serves an
anti-oxidant role, reduces the levels of ROS, restores the balance
of redox reactions and enables cells to survive, while under high
levels of oxidative stress, p53 serves a pro-oxidative role,
exacerbating oxidative stress and leading to cell cycle arrest and
apoptosis (37,44–46). ROS
generated under oxidative stress serves an important regulatory
role in the occurrence and development of tumors. Increased ROS
production can induce mutations of tumor suppressor genes, such as
p53, and genetic instability (44).
Loss of p53 function will increase the production of ROS, so that
cells gradually acquire the ability to continuously proliferate and
invade, which finally leads to the occurrence and development of
cancer (44,47). Maurya and Vinayak (48) revealed that in HepG2 cells, oxidative
stress reduces p53 levels, promotes cell proliferation and reduces
apoptosis by regulating the expression of its target genes.
Additionally, it has been demonstrated that p53 transcription
factor is sensitive to redox and may be upregulated when the DNA is
injured by oxidative stress (37).
Consistently, the hub genes CDK1 and CCNB1, which were enriched in
the p53 signaling pathway, were upregulated in the present
analysis.
In the present study, GO analysis of BP indicated
that the DEGs were associated with ‘sister chromatid cohesion’, and
KEGG analysis provided evidence that the DEGs and hub genes were
all enriched in the ‘cell cycle’ and the ‘p53 signaling pathway’.
These results are consistent with the previous findings that cell
life and death are associated with the cell cycle, and the
disruption of the cell cycle may lead to cell cycle arrest,
dysfunction of transcription and uncontrolled cell growth, which
can be the fundament of tumorigenesis and can affect the prognosis
of cancer (37,49,50). p53
is an important tumor suppressor. p53 and its target genes
constitute a complex p53 signaling pathway that regulates various
biological functions (39). In the
case of DNA damage and oncogene activation, p53 can serve a role in
cell cycle checkpoints to regulate cell cycle arrest, apoptosis and
aging to maintain gene integrity and prevent tumor development
(51). The alternation of p53 and/or
its target genes makes the p53 signaling pathway lose its role of
inhibiting tumorigenesis and obtain carcinogenic functions, such as
promoting the proliferation, metastasis, anti-apoptosis ability and
angiogenesis of tumor cells (39,51,52). Hub
genes such as CDK1 (53), CDC20
(54), CCNB1 (55) and CCNA2 (56), which have been associated with cell
cycle-mediated proliferation and invasion of HCC cells, were all
enriched in the ‘cell cycle’ process, and CDK1 and CCNB1 were
enriched in the ‘p53 signaling pathway’ in the present study.
Additionally, ‘viral carcinogenesis’ and the ‘PPAR
signaling pathway’ are involved in viral infection (57,58),
which is consistent with the present results in the KEGG enrichment
analysis of DEGs and hub genes of HBV-associated early stage HCC.
Overall, the dysfunction of metabolic pathways (especially disorder
of fat metabolism), the cell cycle, oxidation-reduction process and
viral carcinogenesis may serve a critical role in the
carcinogenesis of HBV-associated early stage HCC and may be a
potential therapeutic target.
A total of 8 hub genes were identified in the
present study, among which AURKB, CDK1, CCNA2, CDC20 and CCNB1 were
upregulated, while EHHADH, APOA1 and UBB were downregulated. The
hub genes CDK1, CCNA2, CDC20 and CCNB1 are closely associated with
the cell cycle and are closely connected with each other; for
instance, CCNA2 can activate CDK1 (59), and they can promote the formation of
HCC in combination with other genes (60). Previous studies have demonstrated
that poor clinicopathological characteristics and recurrence after
surgery are closely associated with AURKB upregulation, which may
be used as an independent prognostic predictor and a target for
adjuvant therapy of HCC (61–63). Wu
et al (64) revealed that the
levels of CDK1 are higher in patients with HCC compared with
healthy individuals, which is in accordance with the present study.
Furthermore, Ito et al (53)
reported that CDK1 serves an important regulatory role in the
G2/M phase of the cell cycle and the proliferation of
HCC cells, and is directly associated with portal vein invasion,
distant metastasis and low differentiation. Increasing numbers of
studies have demonstrated that the expression levels of AURKB,
CDK1, CDC20, CCNB1 and CCNA2 in HCC are higher than those in normal
tissues (54,65), which is in accordance with the
present study. Moreover, in the present study, high expression
levels of these genes were associated with poorer OS, RFS, PFS and
DSS rates, indicating that higher levels of these genes may predict
a higher risk of recurrence and metastasis of HCC after surgery,
and a poorer prognosis. Therefore, active monitoring and adjuvant
therapy should be provided to patients with HCC. In addition, the
higher the expression levels of CDC20, CDK1, CCNB1and AURKB, the
higher the grade of HCC cells and the worse the prognosis.
Therefore, these genes may be used for diagnosing the occurrence of
HBV-associated early stage HCC, as well as for predicting the
prognosis of patients and potentially acting as therapeutic
targets.
Among the three downregulated hub genes (EHHADH,
APOA1 and UBB), it has been reported that EHHADH can encode an
enzyme with two functions, which can regulate the metabolism of
fatty acids and nitrogen (66).
Previous studies have demonstrated that APOA1 can inhibit the
progression and metastasis of various types of tumor, such as
breast and ovarian cancer (67,68).
APOA1 does not directly inhibit tumor growth or promote apoptosis
of tumor cells, but acts via indirect effects. On one hand, the
APOA1 mimetic peptide can inhibit the signaling pathway of tumor
vascular epithelial growth factor and basic fibroblast growth
factor, therefore inhibiting tumor-associated angiogenesis
(69); on the other hand,
Zamanian-Daryoush et al (70)
revealed that APOA1 can effectively regulate autogenic and acquired
immune responses, reduce the microenvironment that allows tumor
growth, and increase antitumor macrophages and CD8 T cells, thereby
indirectly inhibiting tumor growth and metabolism.
Based on the UALCAN databases, the present study
revealed that low EHHADH expression was associated with a high
grade of HCC cells, corresponding to a poor prognosis.
Additionally, lower levels of APOA1 were associated with lower OS,
PFS, RFS and DSS rates, while lower levels of EHHADH were
associated with lower OS, DSS and PFS rates. These results
indicated that patients with HCC with low EHHADH and APOA1
expression tended to have rapid tumor growth after surgery, and
higher mortality compared with those with high EHHADH and APOA1
expression, suggesting that EHHADH and APOA1 may be tumor
suppressor genes.
There are different results among previous studies
regarding UBB. Some studies have demonstrated that UBB-knockout can
effectively downregulate ubiquitin and inhibit the growth of tumors
(71), such as non-small cell lung
cancer (72). Additionally, Tian
et al (73) reported that UBB
serves an important regulatory role in maintaining the
characteristics of cancer stem cells and the initiation of cervical
cancer. However, a retrospective study conducted by Valdagni et
al (74) revealed that patients
with prostate cancer who underwent radiotherapy had a lower risk of
rectal bleeding when genes such as UBB were significantly
downregulated. It can be seen that UBB may have different roles in
the diagnosis and treatment of different types of cancer; however,
no studies have yet clarified the role of UBB in HCC.
The present study revealed that UBB downregulation
was closely associated with poor OS, RFS, PFS and DSS rates,
suggesting that in patients with HBV-associated early stage HCC
with low UBB expression, the tumor may be prone to relapse and
rapid growth after surgery, and patients have a high mortality
rate. Therefore, patients should be closely monitored after surgery
and treated swiftly if abnormalities are detected, hoping to
improve their prognosis. Additionally, it could be inferred that
UBB may be a tumor suppressor gene.
In the present study, 20 small molecular compounds
were identified based on the analysis of DEGs, which may be able to
reverse the altered expression of the DEGs; therefore, they may be
promising targeted agents against HBV-associated early stage HCC.
Among them, daunorubicin, irinotecan and doxorubicin have already
been reported to have effectiveness in clinical trials (75–77).
Additionally, resveratrol, apigenin, luteolin and menadione have
been demonstrated to display good therapeutic effects in animal
models (45,78–80).
Camptothecin, chrysin and MS-275 have only been studied in
vivo or in cell experiments, but may have potential to treat
HCC (81–83). Other small molecular compounds have
not yet been demonstrated to have effectiveness against HCC, and
therefore further research is required.
Previous studies have revealed that some of these
compounds act on important signaling pathways that have been
identified in the present study, which further supports our views
on the mechanism of HBV-associated HCC formation. Lin et al
(78) reported that resveratrol can
promote the recovery of the fatty liver and effectively decrease
the incidence of HCC caused by the HBV X protein (HBX) in HBX
transgenic mice via downregulating lipogenesis, promoting transient
liver regeneration and stimulating antioxidant activity.
Additionally, it has been demonstrated that resveratrol has
therapeutic effects on HCC through the p53 (84), cell cycle (85–87). A
previous study revealed that menadione kills tumor cells by
influencing their redox cycle (45).
In addition, other small molecular compounds that act on the cell
cycle are apigenin, luteolin and doxorubicin, while further small
molecular compounds that are associated with the p53 pathway are
irinotecan and doxorubicin (89–93).
Given the broad spectrum of these small molecular compounds, it is
not surprising that these compounds may act as promising
therapeutic targets for HBV-associated early stage HCC.
In conclusion, the present study revealed eight hub
genes, important signaling pathways and potential targeted agents
based on the high-throughput sequencing method. These may provide
clues for revealing the molecular mechanism of early stage HCC
caused by HBV infection and offer potential therapeutic, diagnostic
and prognostic biomarkers. Furthermore, these compounds may
represent promising targeted therapeutic agents against
HBV-associated early stage HCC. However, in order to verify the
predictions of the present study, further research is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by the High-level
University Construction of Guangzhou University of Chinese Medicine
(grant no. A1-AFD018181A29) and Project of Administration of
Traditional Chinese Medicine of Guangdong Province of China
(Project No. 20201109).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GSE124535 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124535)
from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
ZZ, ZC and YT conceived and designed the study. ZZ
performed the bioinformatics analysis. All authors contributed to
data collection and analysis. ZZ, YT and ZC contributed to data
visualization. ZZ wrote the manuscript. ZC and YT reviewed and
edited the manuscript. All authors contributed to the manuscript
and agreed to be responsible for all aspects of the work.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
differentially expressed genes
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
AURKB
|
aurora kinase B
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
CDC20
|
cell division cycle 20
|
|
CCNB1/A2
|
cyclin B1/A2
|
|
EHHADH
|
enoyl-CoA hydratase and 3-hydroxyacyl
CoA dehydrogenase
|
|
APOA1
|
apolipoprotein A1
|
|
UBB
|
ubiquitin B
|
|
HBV
|
hepatitis B virus
|
|
HCC
|
hepatocellular carcinoma
|
|
RNA-seq
|
RNA sequencing
|
|
OS
|
overall survival
|
|
RFS
|
relapse-free survival
|
|
PFS
|
progression-free survival
|
|
DSS
|
disease-specific survival
|
References
|
1
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bruix J and Sherman M; Practice Guidelines
Committee, American Association for the Study of Liver Diseases, :
Management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vogel A, Cervantes A, Chau I, Daniele B,
Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, et al:
Hepatocellular carcinoma: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 29 (Suppl
4):iv238–iv255. 2018. View Article : Google Scholar
|
|
7
|
Bruix J and Sherman M; American
Association for the Study of Liver Diseases, : Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Varela M, Reig M, de la Mata M, Matilla A,
Bustamante J, Pascual S, Turnes J, Aracil C, Del Val A, Pascasio
JM, et al: Treatment approach of hepatocellular carcinoma in Spain.
Analysis of 705 patients from 62 centers. Med Clin (Barc).
134:569–576. 2010.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bargellini I, Sacco R, Bozzi E, Bertini M,
Ginanni B, Romano A, Cicorelli A, Tumino E, Federici G, Cioni R, et
al: Transarterial chemoembolization in very early and early-stage
hepatocellular carcinoma patients excluded from curative treatment:
A prospective cohort study. Eur J Radiol. 81:1173–1178. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song YG, Shin SW, Cho SK, Choi D, Rhim H,
Lee MW, Kim YS, Park KB, Park HS, Choo SW, et al: Transarterial
chemoembolization as first-line therapy for hepatocellular
carcinomas infeasible for ultrasound-guided radiofrequency
ablation: A retrospective cohort study of 116 patients. Acta
Radiol. 56:70–77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim JW, Kim JH, Sung KB, Ko HK, Shin JH,
Kim PN, Choi HK, Ko GY, Yoon HK, Chun SY and Gwon DI: Transarterial
chemoembolization vs. radiofrequency ablation for the treatment of
single hepatocellular carcinoma 2 cm or smaller. Am J
Gastroenterol. 109:1234–1240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gopal P, Yopp AC, Waljee AK, Chiang J,
Nehra M, Kandunoori P and Singal AG: Factors that affect accuracy
of α-fetoprotein test in detection of hepatocellular carcinoma in
patients with cirrhosis. Clin Gastroenterol Hepatol. 12:870–877.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang TS, Wu YC, Tung SY, Wei KL, Hsieh
YY, Huang HC, Chen WM, Shen CH, Lu CH, Wu CS, et al:
Alpha-fetoprotein measurement benefits hepatocellular carcinoma
surveillance in patients with cirrhosis. Am J Gastroenterol.
110:836–845. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou J, Sun HC, Wang Z, Cong WM, Wang JH,
Zeng MS, Yang JM, Bie P, Liu LX, Wen TF, et al: Guidelines for
diagnosis and treatment of primary liver cancer in China (2017
edition). Liver Cancer. 7:235–260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takikawa Y and Suzuki K: Is AFP a new
reliable marker of liver regeneration in acute hepatic failure? J
Gastroenterol. 37:681–682. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Minguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Sun A, Zhao Y, Ying W, Sun H,
Yang X, Xing B, Sun W, Ren L, Hu B, et al: Proteomics identifies
new therapeutic targets of early-stage hepatocellular carcinoma.
Nature. 567:257–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anders S, Pyl PT and Huber W: HTSeq-a
python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bringmann LF, Elmer T, Epskamp S, Krause
RW, Schoch D, Wichers M, Wigman JTW and Snippe E: What do
centrality measures measure in psychological networks? J Abnorm
Psychol. 128:892–903. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Subramanian A, Narayan R, Corsello SM,
Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK,
et al: A next generation connectivity map: L1000 platform and the
first 1,000,000 profiles. Cell. 171:1437–1452 e17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu SC, Huang YW, Wang TC, Hu JT, Chen DS
and Yang SS: Increased risk of hepatocellular carcinoma in chronic
hepatitis B patients with new onset diabetes: A nationwide cohort
study. Aliment Pharmacol Ther. 41:1200–1209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang YW, Wang TC, Yang SS, Lin SY, Fu SC,
Hu JT, Liu CJ, Kao JH and Chen DS: Increased risk of hepatocellular
carcinoma in chronic hepatitis C patients with new onset diabetes:
A nation-wide cohort study. Aliment Pharmacol Ther. 42:902–911.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xia H, Chen J, Sekar K, Shi M, Xie T and
Hui KM: Clinical and metabolomics analysis of hepatocellular
carcinoma patients with diabetes mellitus. Metabolomics.
15:1562019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou QC, Shi B, Jiao LF, Jin M, Sun P,
Ding LY and Yuan Y: Hepatopancreas and ovarian transcriptome
response to different dietary soybean lecithin levels in portunus
trituberculatus. Comp Biochem Physiol Part D Genomics Proteomics.
31:1006002019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamashita T, Honda M, Takatori H, Nishino
R, Minato H, Takamura H, Ohta T and Kaneko S: Activation of
lipogenic pathway correlates with cell proliferation and poor
prognosis in hepatocellular carcinoma. J Hepatol. 50:100–110. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li N, Li L and Chen Y: The identification
of core gene expression signature in hepatocellular carcinoma. Oxid
Med Cell Longev. 2018:34783052018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong H, Xiao M, Zarkovic K, Zhu M, Sa R,
Lu J, Tao Y, Chen Q, Xia L, Cheng S, et al: Mitochondrial control
of apoptosis through modulation of cardiolipin oxidation in
hepatocellular carcinoma: A novel link between oxidative stress and
cancer. Free Radic Biol Med. 102:67–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J, Zhang C and Feng Z: Tumor
suppressor p53 and its gain-of-function mutants in cancer. Acta
Biochim Biophys Sin (Shanghai). 46:170–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rebouissou S and Nault JC: Advances in
molecular classification and precision oncology in hepatocellular
carcinoma. J Hepatol. 72:215–229. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shao Y, Song X, Jiang W, Chen Y, Ning Z,
Gu W and Jiang J: MicroRNA-621 acts as a tumor radiosensitizer by
directly targeting SETDB1 in hepatocellular carcinoma. Mol Ther.
27:355–364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pan YH, Yang M, Liu LP, Wu DC, Li MY and
Su SG: UBE2S enhances the ubiquitination of p53 and exerts
oncogenic activities in hepatocellular carcinoma. Biochem Biophys
Res Commun. 503:895–902. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun G, Sui X, Han D, Gao J, Liu Y and Zhou
L: TRIM59 promotes cell proliferation, migration and invasion in
human hepatocellular carcinoma cells. Pharmazie. 72:674–679.
2017.PubMed/NCBI
|
|
44
|
Trachootham D, Lu W, Ogasawara MA, Nilsa
RD and Huang P: Redox regulation of cell survival. Antioxid Redox
Signal. 10:1343–1374. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Oztopcu-Vatan P, Sayitoglu M, Gunindi M
and Inan E: Cytotoxic and apoptotic effects of menadione on rat
hepatocellular carcinoma cells. Cytotechnology. 67:1003–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu D and Xu Y: p53, oxidative stress, and
aging. Antioxid Redox Signal. 15:1669–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
D'Souza LC, Mishra S, Chakraborty A,
Shekher A, Sharma A and Gupta SC: Oxidative stress and cancer
development: Are noncoding RNAs the missing links? Antioxid Redox
Signal. Jan 24–2020.(Epub ahead of print). doi:
10.1089/ars.2019.7987. View Article : Google Scholar
|
|
48
|
Maurya AK and Vinayak M: Anticarcinogenic
action of quercetin by downregulation of phosphatidylinositol
3-kinase (PI3K) and protein kinase C (PKC) via induction of p53 in
hepatocellular carcinoma (HepG2) cell line. Mol Biol Rep.
42:1419–1429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu S, Yang TB, Nan YL, Li AH, Pan DX, Xu
Y, Li S, Li T, Zeng XY and Qiu XQ: Genetic variants of cell cycle
pathway genes predict disease-free survival of hepatocellular
carcinoma. Cancer Med. 6:1512–1522. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Maddika S, Ande SR, Panigrahi S,
Paranjothy T, Weglarczyk K, Zuse A, Eshraghi M, Manda KD, Wiechec E
and Los M: Cell survival, cell death and cell cycle pathways are
interconnected: Implications for cancer therapy. Drug Resist Updat.
10:13–29. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Giono LE and Manfredi JJ: The p53 tumor
suppressor participates in multiple cell cycle checkpoints. J Cell
Physiol. 209:13–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen J: The cell-cycle arrest and
apoptotic functions of p53 in tumor initiation and progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ito Y, Takeda T, Sakon M, Monden M,
Tsujimoto M and Matsuura N: Expression and prognostic role of
cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma.
Oncology. 59:68–74. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gu J, Liu X, Li J and He Y: MicroRNA-144
inhibits cell proliferation, migration and invasion in human
hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int.
19:152019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bayard Q, Meunier L, Peneau C, Renault V,
Shinde J, Nault JC, Mami I, Couchy G, Amaddeo G, Tubacher E, et al:
Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass
with a rearrangement signature of replication stress. Nat Commun.
9:52352018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shi Y, Li Y, Huang C, Ying L, Xue J, Wu H,
Chen Z and Yang Z: Resveratrol enhances HBV replication through
activating Sirt1-PGC-1α-PPARα pathway. Sci Rep. 6:247442016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang JY, Chou SF, Lee JW, Chen HL, Chen
CM, Tao MH and Shih C: MicroRNA-130a can inhibit hepatitis B virus
replication via targeting PGC1α and PPARγ. RNA. 21:385–400. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kanakkanthara A, Jeganathan KB, Limzerwala
JF, Baker DJ, Hamada M, Nam HJ, van Deursen WH, Hamada N, Naylor
RM, Becker NA, et al: Cyclin A2 is an RNA binding protein that
controls Mre11 mRNA translation. Science. 353:1549–1552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yan H, Li Z, Shen Q, Wang Q, Tian J, Jiang
Q and Gao L: Aberrant expression of cell cycle and material
metabolism related genes contributes to hepatocellular carcinoma
occurrence. Pathol Res Pract. 213:316–321. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW,
Lai PL, Lee PH, Cheng AL and Hsu HC: Significance of Aurora B
overexpression in hepatocellular carcinoma. Aurora B overexpression
in HCC. BMC Cancer. 10:4612010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yasen M, Mizushima H, Mogushi K, Obulhasim
G, Miyaguchi K, Inoue K, Nakahara I, Ohta T, Aihara A, Tanaka S, et
al: Expression of Aurora B and alternative variant forms in
hepatocellular carcinoma and adjacent tissue. Cancer Sci.
100:472–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tanaka S, Arii S, Yasen M, Mogushi K, Su
NT, Zhao C, Imoto I, Eishi Y, Inazawa J, Miki Y and Tanaka H:
Aurora kinase B is a predictive factor for the aggressive
recurrence of hepatocellular carcinoma after curative hepatectomy.
Br J Surg. 95:611–619. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY,
Chan ACY, Ma KW, Xia W and Cheung TT: Blocking CDK1/PDK1/β-Catenin
signaling by CDK1 inhibitor RO3306 increased the efficacy of
sorafenib treatment by targeting cancer stem cells in a preclinical
model of hepatocellular carcinoma. Theranostics. 8:3737–3750. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang W, Zhang L, Guo Q, Wang H, Ma M, Sun
J and Chen C: Identification of the pathogenic biomarkers for
hepatocellular carcinoma based on RNA-seq analyses. Pathol Oncol
Res. 25:1207–1213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ganapathy E, Su F, Meriwether D, Devarajan
A, Grijalva V, Gao F, Chattopadhyay A, Anantharamaiah GM, Navab M,
Fogelman AM, et al: D-4F, an apoA-I mimetic peptide, inhibits
proliferation and tumorigenicity of epithelial ovarian cancer cells
by upregulating the antioxidant enzyme MnSOD. Int J Cancer.
130:1071–1081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cedó L, García-León A, Baila-Rueda L,
Santos D, Grijalva V, Martínez-Cignoni MR, Carbó JM, Metso J,
López-Vilaró L, Zorzano A, et al: ApoA-I mimetic administration,
but not increased apoA-I-containing HDL, inhibits tumour growth in
a mouse model of inherited breast cancer. Sci Rep. 6:363872016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gao F, Vasquez SX, Su F, Roberts S, Shah
N, Grijalva V, Imaizumi S, Chattopadhyay A, Ganapathy E, Meriwether
D, et al: L-5F, an apolipoprotein A-I mimetic, inhibits tumor
angiogenesis by suppressing VEGF/basic FGF signaling pathways.
Integr Biol (Camb). 3:479–489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zamanian-Daryoush M, Lindner D, Tallant
TC, Wang Z, Buffa J, Klipfell E, Parker Y, Hatala D,
Parsons-Wingerter P, Rayman P, et al: The cardioprotective protein
apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol
Chem. 288:21237–21252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Oh C, Park S, Lee EK and Yoo YJ:
Downregulation of ubiquitin level via knockdown of polyubiquitin
gene Ubb as potential cancer therapeutic intervention. Sci Rep.
3:26232013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tang Y, Geng Y, Luo J, Shen W, Zhu W, Meng
C, Li M, Zhou X, Zhang S and Cao J: Downregulation of ubiquitin
inhibits the proliferation and radioresistance of non-small cell
lung cancer cells in vitro and in vivo. Sci Rep. 5:94762015.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tian Y, Ding W, Wang Y, Ji T, Sun S, Mo Q,
Chen P, Fang Y, Liu J, Wang B, et al: Ubiquitin B in cervical
cancer: Critical for the maintenance of cancer stem-like cell
characters. PLoS One. 8:e844572013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Valdagni R, Rancati T, Ghilotti M,
Cozzarini C, Vavassori V, Fellin G, Fiorino C, Girelli G, Barra S,
Zaffaroni N, et al: To bleed or not to bleed. A prediction based on
individual gene profiling combined with dose-volume histogram
shapes in prostate cancer patients undergoing three-dimensional
conformal radiation therapy. Int J Radiat Oncol Biol Phys.
74:1431–1440. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Daniele B, De Vivo R, Perrone F, Lastoria
S, Tambaro R, Izzo F, Fiore F, Vallone P and Pignata S: Phase I
clinical trial of liposomal daunorubicin in hepatocellular
carcinoma complicating liver cirrhosis. Anticancer Res.
20:1249–1251. 2000.PubMed/NCBI
|
|
76
|
Brandi G, Biasco G, Mirarchi MG, Golfieri
R, Di Paolo A, Borghi A, Fanello S, Derenzini E, Agostini V,
Giampalma E, et al: A phase I study of continuous hepatic arterial
infusion of Irinotecan in patients with locally advanced
hepatocellular carcinoma. Dig Liver Dis. 43:1015–1021. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Tak WY, Lin SM, Wang Y, Zheng J, Vecchione
A, Park SY, Chen MH, Wong S, Xu R, Peng CY, et al: Phase III HEAT
study adding lyso-thermosensitive liposomal doxorubicin to
radiofrequency ablation in patients with unresectable
hepatocellular carcinoma lesions. Clin Cancer Res. 24:73–83. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lin HC, Chen YF, Hsu WH, Yang CW, Kao CH
and Tsai TF: Resveratrol helps recovery from fatty liver and
protects against hepatocellular carcinoma induced by hepatitis B
virus X protein in a mouse model. Cancer Prev Res (Phila).
5:952–962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bhattacharya S, Mondal L, Mukherjee B,
Dutta L, Ehsan I, Debnath MC, Gaonkar RH, Pal MM and Majumdar S:
Apigenin loaded nanoparticle delayed development of hepatocellular
carcinoma in rats. Nanomedicine. 14:1905–1917. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Balamurugan K and Karthikeyan J:
Evaluation of luteolin in the prevention of
N-nitrosodiethylamine-induced Hepatocellular carcinoma using animal
model system. Indian J Clin Biochem. 27:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen F, Wang H, Zhu J, Zhao R, Xue P,
Zhang Q, Bud Nelson M, Qu W, Feng B and Pi J: Camptothecin
suppresses NRF2-ARE activity and sensitises hepatocellular
carcinoma cells to anticancer drugs. Br J Cancer. 117:1495–1506.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xu D, Jin J, Yu H, Zhao Z, Ma D, Zhang C
and Jiang H: Chrysin inhibited tumor glycolysis and induced
apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J
Exp Clin Cancer Res. 36:442017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xiao W, Dong W, Zhang C, Saren G, Geng P,
Zhao H, Li Q, Zhu J, Li G, Zhang S and Ye M: Effects of the
epigenetic drug MS-275 on the release and function of
exosome-related immune molecules in hepatocellular carcinoma cells.
Eur J Med Res. 18:612013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang B, Yin X and Sui S: Resveratrol
inhibited the progression of human hepatocellular carcinoma by
inducing autophagy via regulating p53 and the phosphoinositide
3kinase/protein kinase B pathway. Oncol Rep. 40:2758–2765.
2018.PubMed/NCBI
|
|
85
|
Park S, Lim J, Kim JR and Cho S:
Inhibitory effects of resveratrol on hepatitis B virus X
protein-induced hepatocellular carcinoma. J Vet Sci. 18:419–429.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liao PC, Ng LT, Lin LT, Richardson CD,
Wang GH and Lin CC: Resveratrol arrests cell cycle and induces
apoptosis in human hepatocellular carcinoma Huh-7 cells. J Med
Food. 13:1415–1423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Notas G, Nifli AP, Kampa M, Vercauteren J,
Kouroumalis E and Castanas E: Resveratrol exerts its
antiproliferative effect on HepG2 hepatocellular carcinoma cells,
by inducing cell cycle arrest, and NOS activation. Biochim Biophys
Acta. 1760:1657–1666. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Bishayee A, Politis T and Darvesh AS:
Resveratrol in the chemoprevention and treatment of hepatocellular
carcinoma. Cancer Treat Rev. 36:43–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li Y, Cheng X, Chen C, Huijuan W, Zhao H,
Liu W, Xiang Z and Wang Q: Apigenin, a flavonoid constituent
derived from P. villosa, inhibits hepatocellular carcinoma cell
growth by CyclinD1/CDK4 regulation via p38 MAPK-p21 signaling.
Pathol Res Pract. 216:1527012020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen
Y, Chen H, Chu F and Zhang Y: Luteolin promotes cell apoptosis by
inducing autophagy in hepatocellular carcinoma. Cell Physiol
Biochem. 43:1803–1812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Fan YP, Liao JZ, Lu YQ, Tian DA, Ye F,
Zhao PX, Xiang GY, Tang WX and He XX: MiR-375 and doxorubicin
Co-delivered by liposomes for combination therapy of hepatocellular
carcinoma. Mol Ther Nucleic Acids. 7:181–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ang C, O'Reilly EM, Carvajal RD, Capanu M,
Gonen M, Doyle L, Ghossein R, Schwartz L, Jacobs G, Ma J, et al: A
nonrandomized, phase II study of sequential irinotecan and
flavopiridol in patients with advanced hepatocellular carcinoma.
Gastrointest Cancer Res. 5:185–189. 2012.PubMed/NCBI
|
|
93
|
Liu L, Chen X, Xie S, Zhang C, Qiu Z and
Zhu F: Variant 1 of KIAA0101, overexpressed in hepatocellular
carcinoma, prevents doxorubicin-induced apoptosis by inhibiting p53
activation. Hepatology. 56:1760–1769. 2012. View Article : Google Scholar : PubMed/NCBI
|




















