Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer in the world, with the second highest mortality rate
among cancers (1). In HCC treatment,
systemic treatment is mostly indispensable, and molecular targeted
therapy is an important part (2).
Targeted drugs, such as sorafenib, can effectively prolong the
survival of patients for several months; however, occasional
treatment resistance can still bring about tumor recurrence or
progression (3). In addition,
immunotherapy has played a key role in the treatment of tumors,
such as melanoma and non-small-cell lung cancer, and has achieved
initial success in the treatment of HCC (4,5).
Increasing evidence indicates that treatment resistance and tumor
response to anti-programmed cell death protein are highly dependent
on tumor heterogeneity, immunogenicity and the lymphocyte
infiltration in immune microenvironment (6–8).
Intratumor heterogeneity (ITH), the result of tumor
cell mutation, is closely associated with neoantigen generation and
tumor immunogenicity (9). The
interaction between tumor cells and the immune microenvironment is
also a reason for ITH (10). Since
ITH represents the cell division and proliferation of tumor cells,
it is also considered a biological process in tumor progression and
is one of the characteristics of malignant tumors (11). Clinical studies have also shown that
ITH has been linked to cancer progression and affects the tumor
immune response and tumor immune escape (12,13).
Mutant-allele tumor heterogeneity (MATH) is an algorithm for
quantifying ITH that was deduced from whole-exome sequencing data
(7). Mroz and Rocco (14) first applied the MATH algorithm to
quantify ITH and reported that the prognosis of neck squamous cell
carcinoma is related to the MATH score. Subsequent studies have
calculated the MATH score in other tumors, and the results verified
the clinical outcome and demonstrated that the MATH score is a
useful biomarker for the metastasis of cancer, such as colon cancer
(15). In a study of breast cancer,
Ma et al (6) reported that a
high MATH score corresponds with a worse prognosis. Wang et
al (16) evaluated ITH in
diffuse large B-cell lymphoma using MATH and demonstrated that a
higher MATH score indicated an increased risk of progression.
Therefore, the MATH score of ITH is a potentially quantitative
indicator for various tumors.
Moreover, the occurrence and development of HCC is
also closely related to tumor immune microenvironment heterogeneity
(TIMH), which is created by the interaction of immune cells,
fibroblasts, endothelial cells, mesenchymal stem cells and
pericytes with tumor cells (17). In
TIMH, cytokines, chemokines and pro-angiogenic and
anti-inflammatory mediators produced in lymphocytes or tumor cells
play a key role in the tumor immune response (18,19). On
the other hand, the recruitment and localization of lymphocytes in
the tumor stroma affects the production of cytokines and immune
factors, which can be used to predict prognosis (20). TIMH is associated with the type and
proportion of lymphocytes in the tumor microenvironment (21). The CIBERSORT method (https://cibersort.stanford.edu/) is an algorithm
used to evaluate gene expression data from tumor RNA sequences and
analyze the distribution of various types of immune cells inside
the sample (22). Based on previous
studies (23,24), the CIBERSORT method was used to
research TIMH in the present study.
To date, the mechanism between ITH and TIMH remains
unclear, and to the best of our knowledge there have been no
reports as to whether the MATH score can be used as a biomarker in
HCC research. Therefore, the present study used the MATH score to
represent ITH and utilized CIBERSORT to calculate the fractions of
22 types of leukocytes in tumor tissues to analyze the intrinsic
relationship between ITH and TIMH.
Materials and methods
Data acquisition and calculation of
MATH score
The TCGA-LIHC (Liver hepatocellular carcinoma)
multi-data were downloaded from The Cancer Genome Atlas (TCGA)
website (https://portal.gdc.cancer.gov/) (accessed on June 20,
2019). From these data, 424 files of RNA-sequencing (RNA-seq) were
obtained, of which 50 were paracancerous samples and 374 were HCC
samples. Next, the gene expression matrix was extracted through R
version 3.5.1 (https://www.r-project.org/). Then, gene symbols were
acquired by matching the Ensembl ID from the Ensembl dataset
(http://asia.ensembl.org/index.html).
The simple nucleotide variation (SNV) data of the 374 patients were
obtained from TCGA as Mutation Annotation Forma (MAF) files, and
the clinical data were acquired as BCR XML files. Here, ‘maftools’
packages of R were applied to analyze the MAF files and to
calculate the MATH score. In this MATH algorithm, a clustering
analysis algorithm was added, and some outliers on the variant
allele frequency were removed based on the conventional algorithm
to increase the accuracy of the algorithm. Finally, the RNA
expression matrix, clinical data and all cohort MATH scores were
acquired.
Exploring oncological features and
tumor immunology
Based on the MATH score, cohort samples were divided
into low and high MATH score groups and the cut-off value was set
as 19.39. Kaplan-Meier survival analysis was used to explore the
relationship between MATH score and clinical prognosis, and the
optimal cut-off value (set as 19.39) was calculated by the
‘survminer’ package.
To explore the oncological features and gene sets
enriched in these two groups, Gene Set Enrichment Analysis (GSEA)
was performed to resolve the underlying molecular mechanisms and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of enriched
gene sets. GSEA was used to investigate the potential mechanisms
involving the Molecular Signatures Database gene sets c2
(c2.cp.kegg.v6.1.symbols and c2.cp.biocarta.v6.1.symbols) and c5
(c5.bp.v6.1.symbols) using the JAVA program (http://software.broadinstitute.org/gsea/index.jsp)
(25). One thousand random sample
permutations were performed, and the significance threshold was set
at P<0.05. After GSEA, the immune-related signaling pathways
were selected from the differential signaling pathways. Next,
immune-related genes were extracted from those immune-related
pathways to explore tumor immunology.
In addition, some immunosuppression-related genes
(indoleamine 2,3-dioxygenase interleukin-10, programmed cell death
1 ligand 1 and transforming growth factor-β) were subjected to
Kaplan-Meier survival analysis and Pearson's correlation analysis
in the group with more immune-related pathways to observe whether
those genes affect prognosis.
Selection of immune- and
survival-related (IS) genes
First, IS genes were selected using Kaplan-Meier and
Cox multivariate regression analysis using the ‘survival’ and
‘survimer’ packages of R software, in which the significance
threshold was set at P<0.05.
Second, coexpression analysis, cluster analysis and
Pearson's correlation coefficient analysis of immune checkpoints
and IS genes were implemented to analyze the positive and negative
regulation of these genes in tumor-associated immunity based on
ITH.
Analysis of TIMH
In this part, Genotype-Tissue Expression (GTEx) data
was downloaded to integrate analysis with TCGA data via UCSC
dataset (http://hgdownload.soe.ucsc.edu/downloads.html). Gene
expression arrays were mined through CIBERSORT to estimate the
fractions of 22 types of leukocyte infiltrates in all samples, and
the algorithm was run with 800 permutations. By analyzing the
difference in the percentage of immune cells between the
paracancerous and tumor tissues, the characteristics of
tumor-infiltrating lymphocytes in HCC tissue can be evaluated
(21). Pearson's correlation
coefficient analysis and coexpression analysis of these immune
cells were performed to determine the interaction of these
lymphocytes. In addition, through Kaplan-Meier and Cox multivariate
regression analyses, the relationship between TIMH and prognosis
was explored. Then, the immune cells associated with overall
survival (OS) time were selected for the next step (P<0.05).
Construction of the immune-related
prognostic model
By incorporating IS genes and immune cells into Cox
multivariate regression analysis, an immune-related model was
established, and the impact of host immunity on OS was further
analyzed. In addition, the receiver operating characteristic (ROC)
curve and the area under the ROC curve (AUC) were used to evaluate
the performance of this model.
The regulatory network of
immune-related genes and immune cells
In the aforementioned steps, IS genes were selected
by analyzing ITH, and immune-related lymphocytes were chosen by
calculating the TIMH. To further investigate the regulatory
relationship between these, a regulatory network was established
linked by miRNAs and their target genes.
First, the HCC miRNA dataset was downloaded from
TCGA, extracted and organized into a miRNA expression matrix.
Through univariate linear regression analysis, miRNAs that were
highly related to immune cell infiltration were selected
(P<0.05). Second, utilizing miRbase (http://www.mirbase.org), the target genes of these
miRNAs were identified, and low-correlation target genes were
filtered out. The Database for Annotation, Visualization and
Integrated Discovery (DAVID) version 6.8 (https://david.ncifcrf.gov/) was used to analyze the
enrichment of the target genes in terms of KEGG pathway and Gene
Ontology (GO) annotation and functional classification. The
high-correlation target genes and the corresponding miRNAs were
used for further analysis.
Finally, the regulatory networks between target
genes, miRNAs, immune-related cells and genes were established by
Cystoscope software version 3.7.1 (26), and the correlation coefficient map
was constructed using R software.
Construction of the network of
weighted coexpressed genes and their associations with potential
molecules
To further analyze the regulatory mechanisms of
these potential molecules (including target genes and miRNAs) in
the tumor microenvironment and the signaling pathways involved, the
Weighted Correlation Network Analysis Model (WGCNA) (27), a systematic biological method
describing the gene correlation pattern between different samples,
was constructed. The molecular mechanism, signaling pathways and
cell functions were elucidated by analyzing the gene modules
corresponding to these potential molecules through WGCNA.
Statistical analysis
Categorized variables were described by proportion
(%) and χ2 tests were used to compare proportions of
lymphocytes between normal and tumor tissues. Associations between
characteristics and overall survival time were evaluated by Cox
proportional hazard models. Kaplan-Meier survival curves and the
log-rank test were used to compare overall survival between the
high and low MATH score groups. A weighted log-rank test (Renyi
test) was performed to generate the P-values when survival curves
crossed over. Mantel test was used to measure the correlation
between key genes and the matrix of miRNAs and target gene, in
which coefficient of correlation (r) described the linear
relationship between the two variables. The ratio of lymphocytes
between normal and tumor tissue were compared using Wilcoxon rank
sum test. The AUC was calculated to assess the predictive ability
of the immune-related model.
All statistical analyses were performed using R
software (https://www.r-project.org/) and
Bioconductor (https://www.bioconductor.org/). All statistical tests
were two-tailed P<0.05 was consider to indicate a statistically
significant difference.
Results
The MATH score and GSEA
In the present study, the MATH score was a risk
factor related to prognosis, and the patients had a poor prognosis
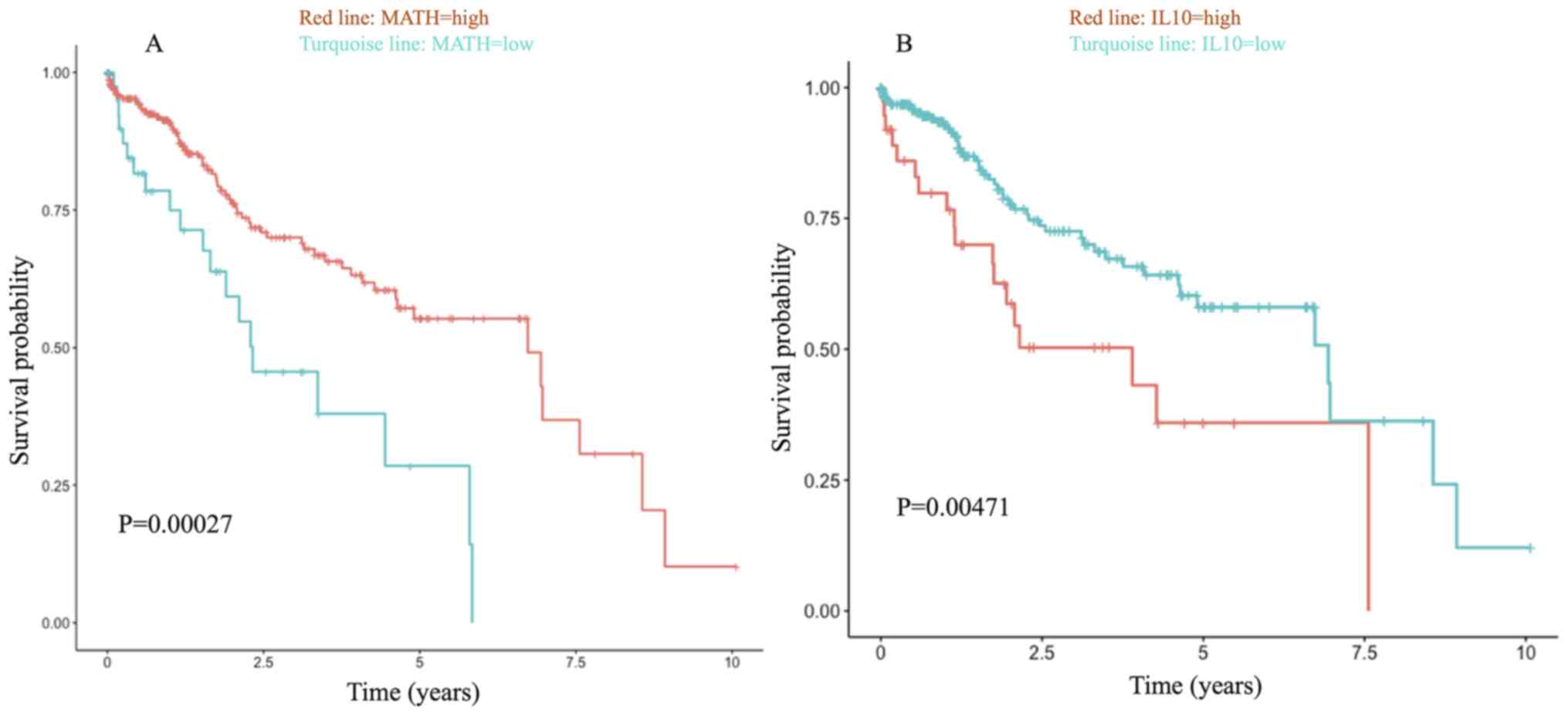
in the MATH-low group (MATH score <19.39) (Fig. 1A). Although the prognosis was
improved in the high MATH score group compared with the low MATH
score group (Fig. 1A), the
immunosuppression pathways involved, such as ‘primary
immunodeficiency’, are detrimental to prognosis (28). Hence, the immunosuppressive factors
in the high-score group were analyzed and it was confirmed that
high IL-10 expression was associated with a worse prognosis
(P=0.00471; Fig. 1B). In the GSEA,
various gene sets were related to the major nutrients, and the bile
acid metabolism gene sets were enriched in the low MATH score group
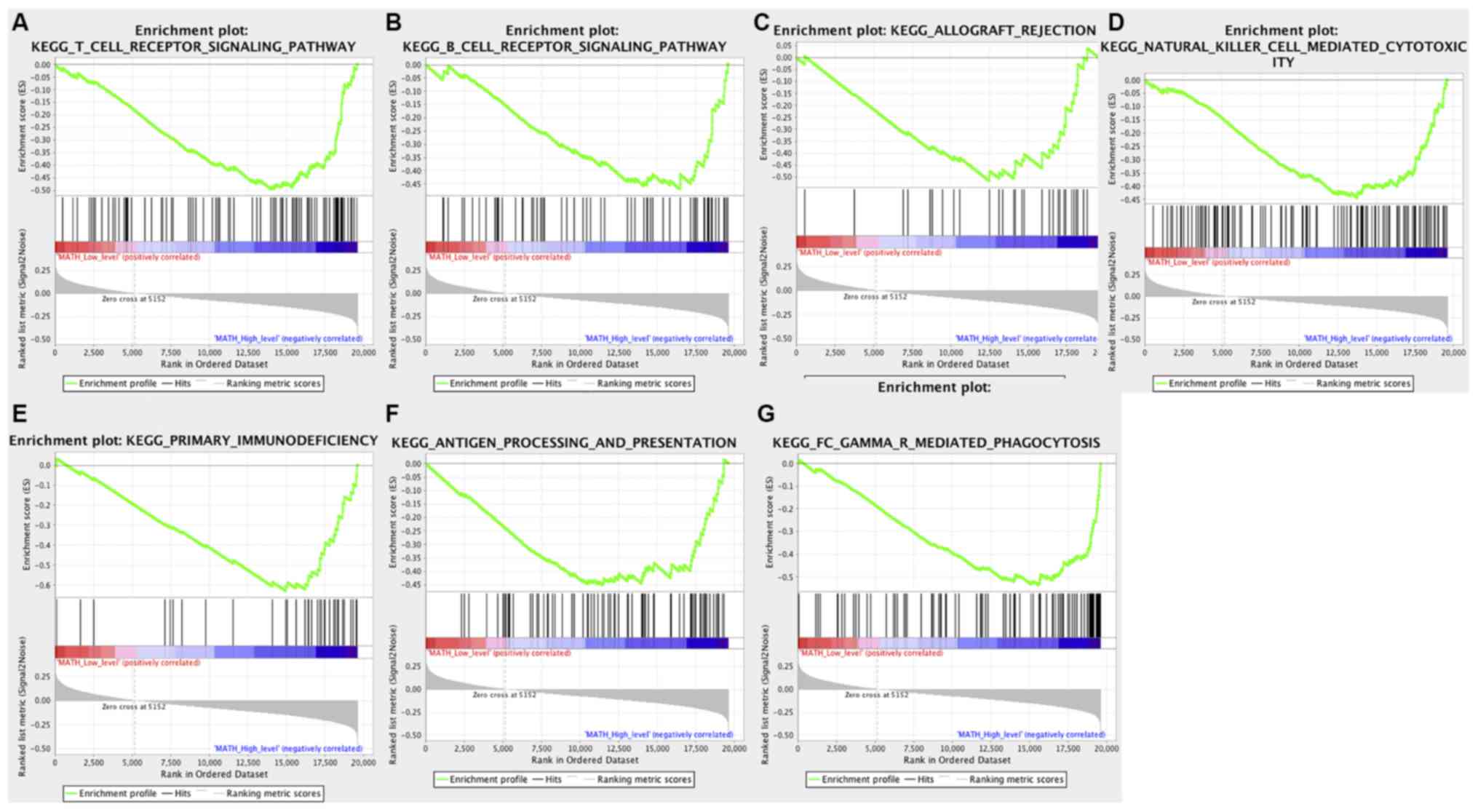
(Table SI). In contrast, seven KEGG
signaling pathways associated with immune responses were found in
the high MATH score group, including ‘allograft rejection’, ‘B
receptor signaling pathway’, ‘antigen processing and presentation’,
‘FC gamma R-mediated phagocytosis’, ‘natural killer cell mediated
cytotoxicity’, ‘primary immunodeficiency’ and ‘T cell receptor
signaling’ (Fig. 2).
Survival and coexpression analysis of
immune-related genes
CD4, CD80, interleukin-1α (IL1A), integrin β-1
(ITGB2), KRAS and PAK4, which were enriched in these seven immune
pathways, were associated with OS. The relationship between these
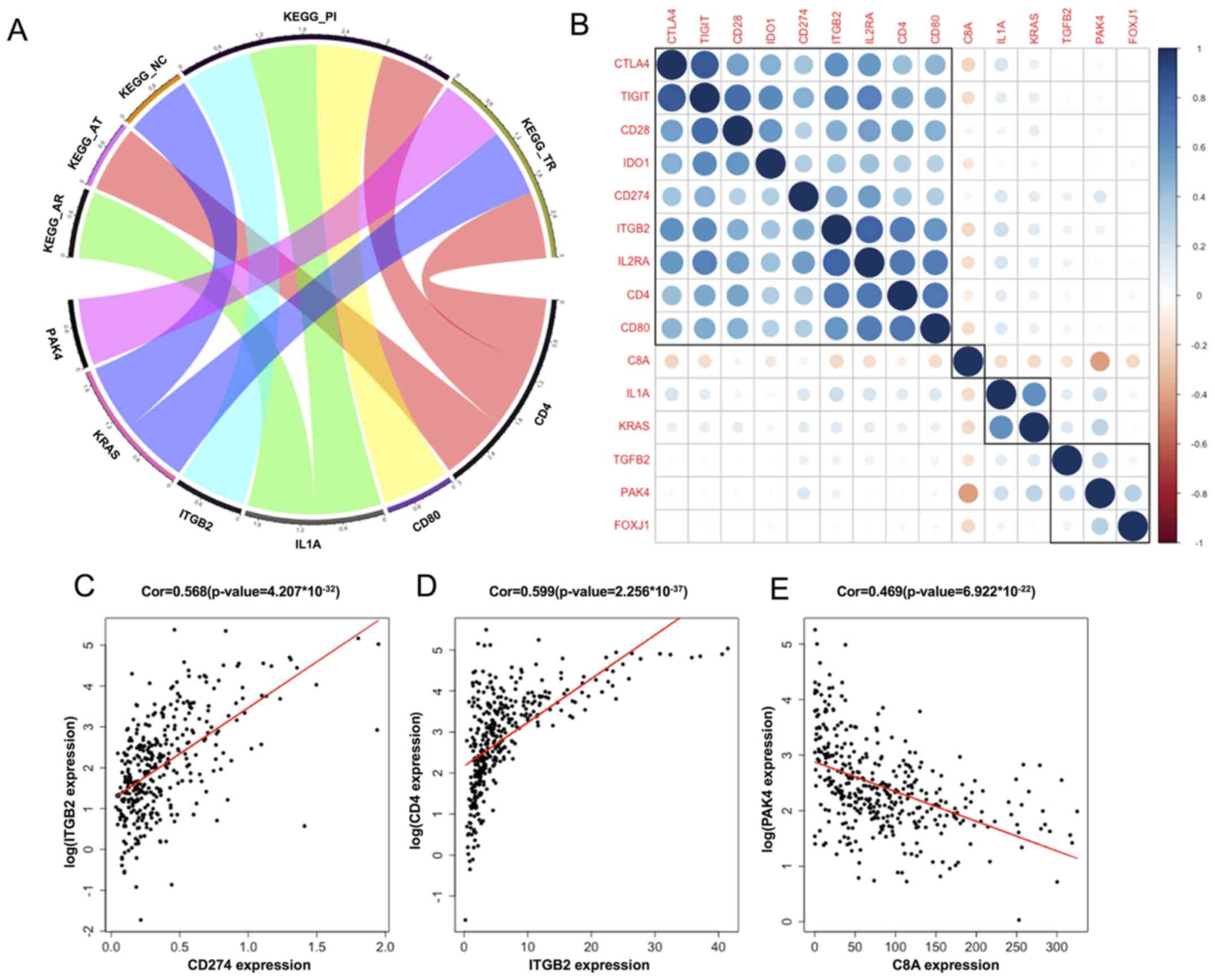
genes and the enriched KEGG pathways is shown in Fig. 3A. ITGB2, IL1A and CD80 were enriched
in the primary immunodeficiency pathway. CD4 was enriched in the
gene set associated with the primary immunodeficiency pathway, as
well as in the T cell receptor and antigen processing cell-related
gene sets.
Coexpression, correlation and cluster analysis for
those genes and immune checkpoints were implemented to further
study the characteristics of tumor-related immune responses. In the
coexpression study, the expression of ITGB2 was positively
correlated with PD-L1 and CD4, and the expression of PAK4 was
negatively correlated with C8A (P<0.005; Fig. 3C-E). The correlation between these
immune-related genes and the immune checkpoints ranged from weak to
moderate, and they were roughly clustered into four categories by
correlation and cluster analysis (Fig.
3B). These immune-related genes may participate in the
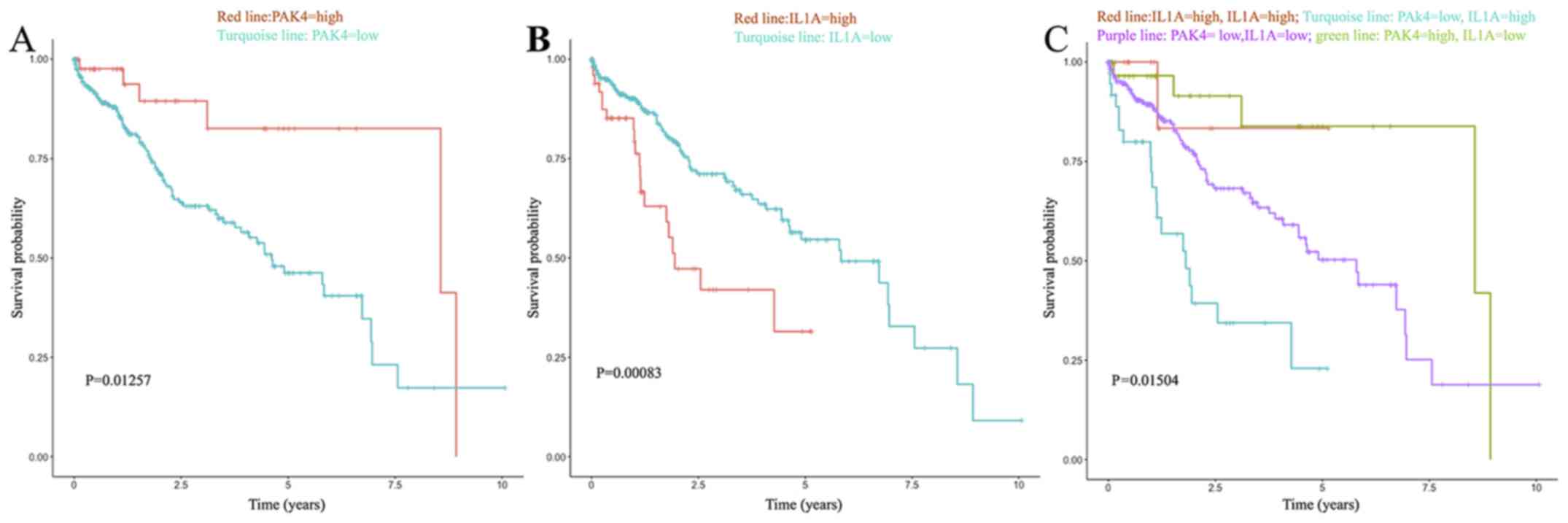
regulation of tumor immune checkpoints. In the multivariate Cox
regression analysis, IL1A and PAK4 were demonstrated to be strong
independent risk factors affecting overall survival (P=0.00083 and
0.01257, respectively; Fig. 4B).
Analysis of immune cells in the tumor
immune microenvironment
Considering that the TCGA-LIHC cohort contains only
50 tumor-adjacent tissues, integrated analysis of TCGA data and
Genotype-Tissue Expression (GTEx) data was applied to enhance the
accuracy of the conclusion. The integrated samples included GTEx
normal liver samples and paracancerous samples. Using CIBERSORT,
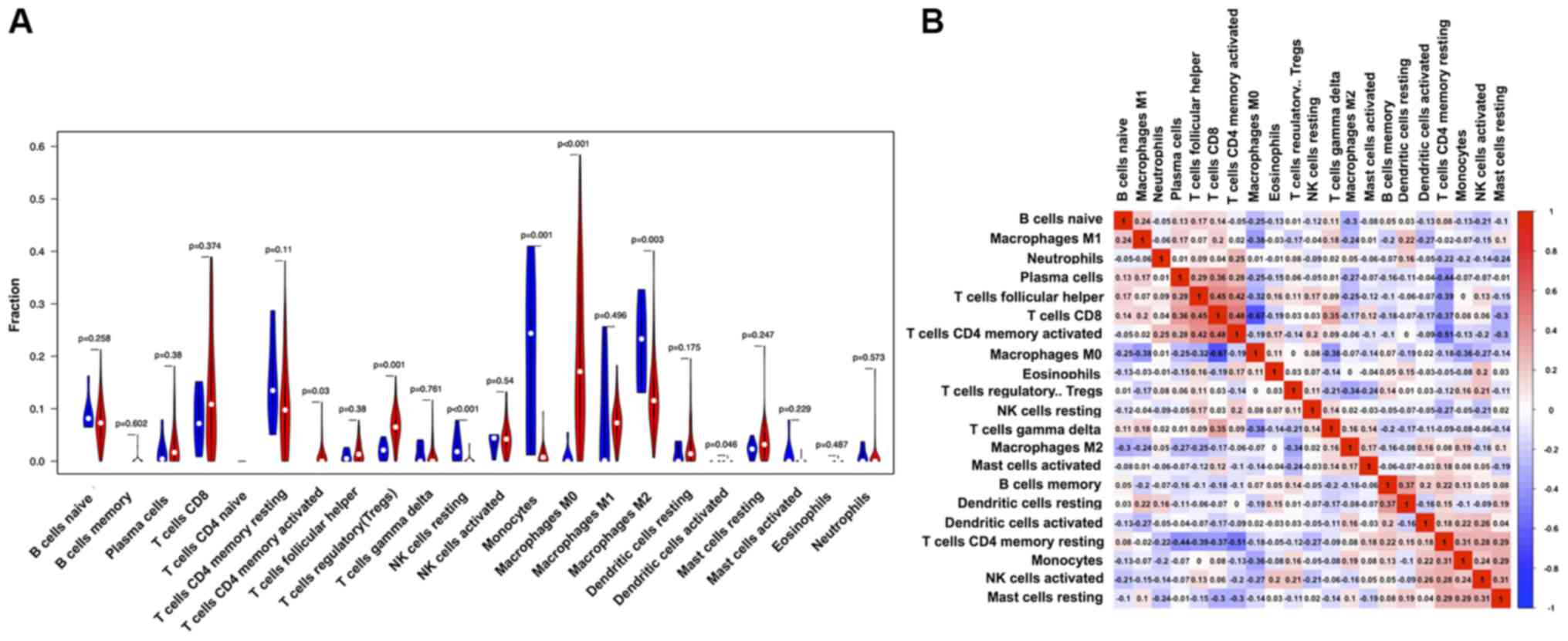
the composition ratio of 22 immune cell types in the HCC samples
was calculated. Accordingly, the difference in the infiltrating
immune cell types between integrated samples and tumor tissue was
estimated. In each tumor tissue, the proportion of different immune
cells varied, indicating TIMH. The proportion of M0 macrophages was
higher in the tumor tissue compared with integrated samples, and
the proportion of M2 macrophages was lower in the tumor tissue
compared with in the integrated samples (Fig. 5A).
The correlation matrix of all 22 lymphocyte types
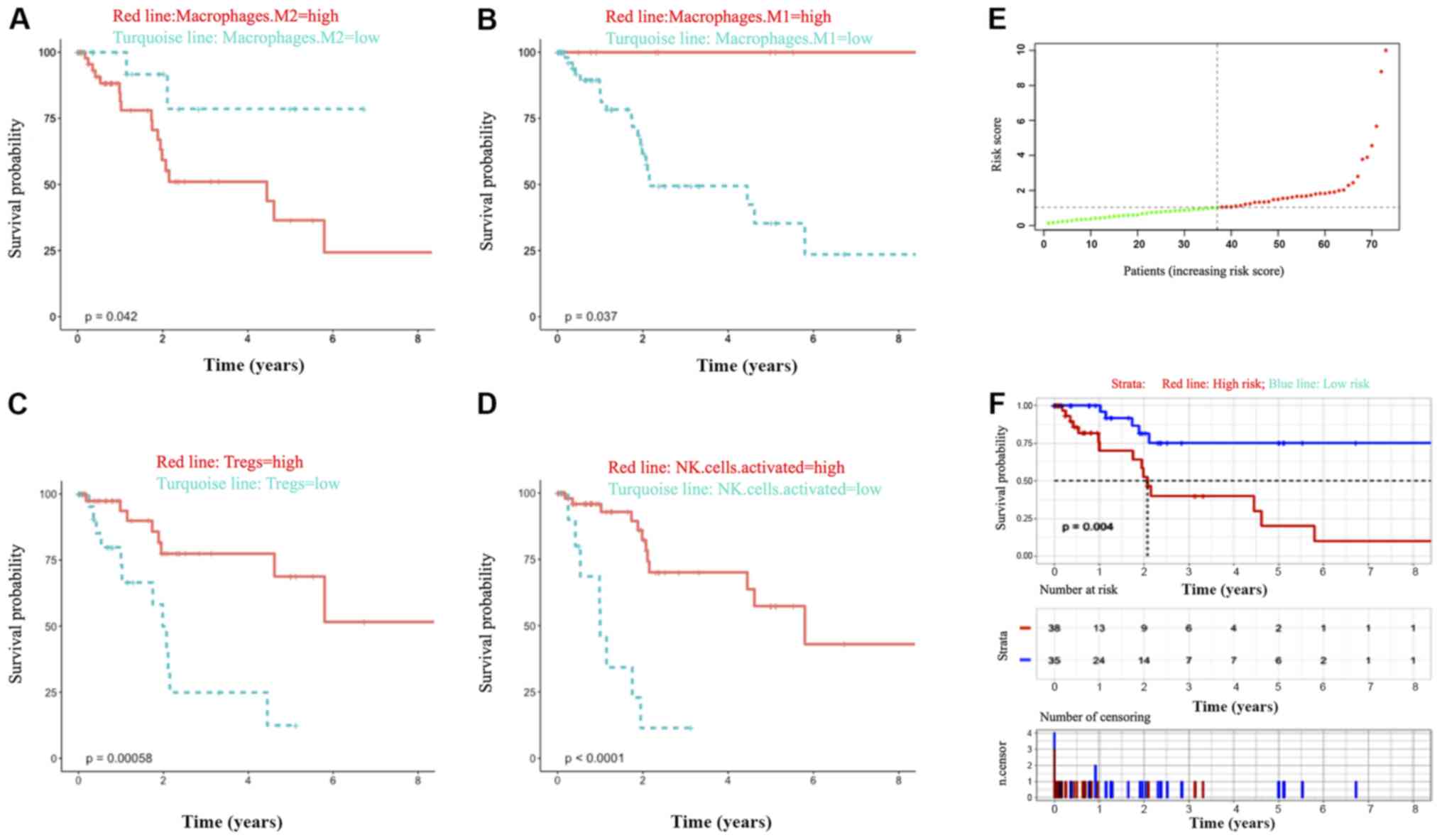
showed their correlation from weak to moderate (Fig. 5B). Univariate and multivariate Cox
analyses of these 22 immune cell types were employed to analyze the
relationship between TIMH and prognosis. Based on the survival
analyses of these immune cells, OS time was extended when the
proportion of M1 macrophages was higher but shortened if the
proportion of M2 macrophages was higher in the tumor tissues. Then,
the phenotype of macrophage polarization was assessed with the
ratio of M1/M2 set as the biomarker (the cut-off value was set as
0.7284), and the results showed that the ratio was better at
predicting survival compared with the proportion of M1 or M2
macrophages alone (Fig. 6A, B and
F). The percentages of NK-activated cells and Tregs were 4.84
and 6.26%, respectively, among all lymphocytes. In addition, a high
proportion of NK activated cells and regulatory T cells worsened
the prognosis of HCC (Fig. 6C and
D). Thus, in the Cox multivariate analysis, the M1/M2 ratio was
confirmed as a strong independent risk factor for prognosis, and OS
was prolonged when the proportion was above 72.83% (Fig. 6F).
Establishing the immune-related
prognosis model
Furthermore, the immune-related prognosis model in
the present study was used to indirectly investigate the effects of
ITH and TIMH on prognosis after integrating IL1A, PAK4 and M1/M2
ratio into the Cox multivariate regression analysis (Fig. 6E and F). After forward stepwise
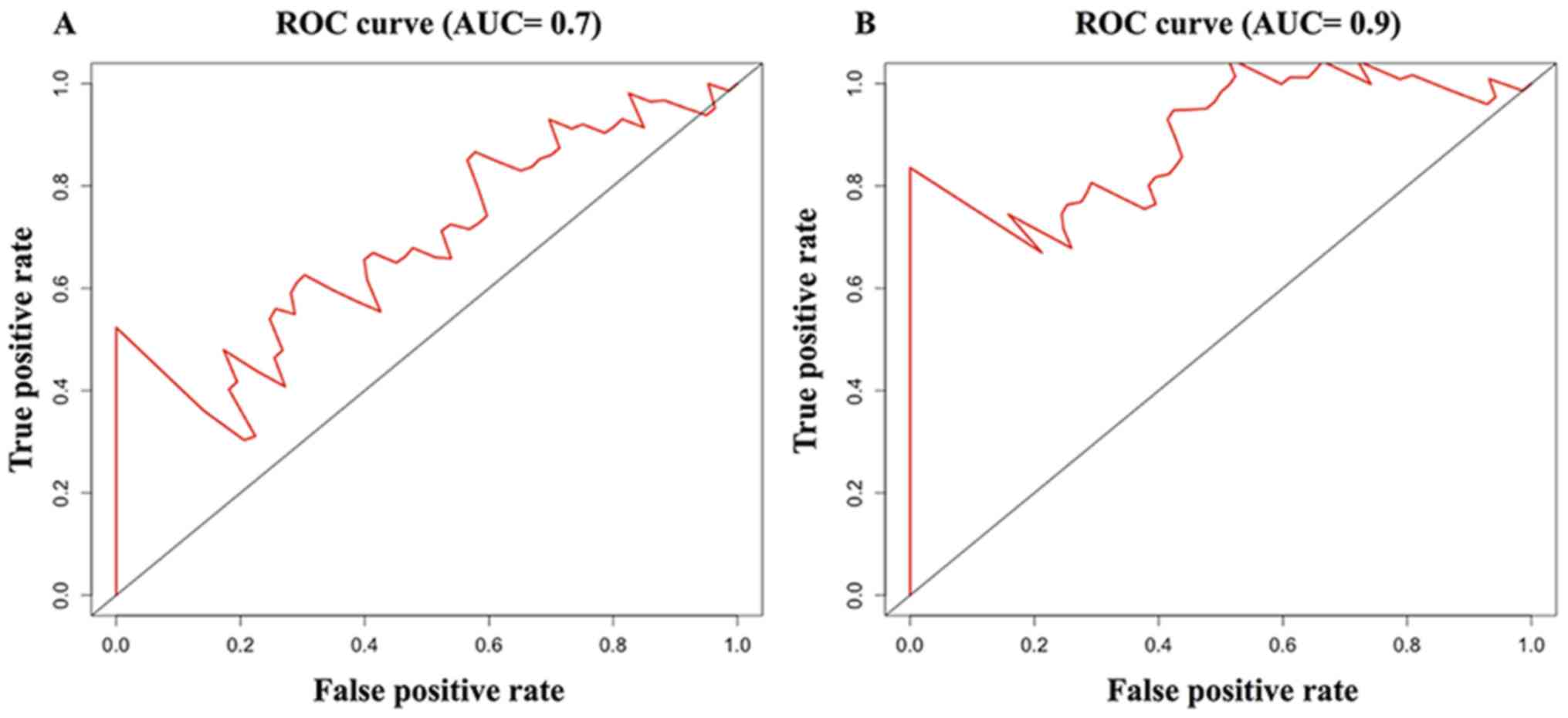
regression, the M1/M2 ratio and IL1A were retained in the model.
The model yielded an AUC of 0.7 in the prediction of 3-year
survival, which was higher compared with that of the IL1A/PAK4
model (AUC=0.6) and the M1/M2 ratio model (AUC=0.651). In the
present model, the ability to predict 5-year survival was even
better, with a higher AUC (0.9) (Fig.
7) compared with the IL1A/PAK4 model and the M1/M2 ratio
model.
The interaction and regulation between
ITH and TIMH
The link between the M1/M2 ratio and immune-related
genes was investigated through examining miRNAs and their
corresponding target genes to study the molecular regulation
mechanisms of macrophage polarization and establish a regulatory
network between IL1A, PAK4 and the M1/M2 ratio. The miRNA
expression matrix was extracted and subjected to regression
analysis with the M1/M2 ratio. In the present study, 19 related
miRNAs were obtained. After excluding the miRNAs with very low
expression levels, the eligible miRNAs that remained were
miRNA-191, −6798, −1269 and −4661. Then, 1,174 target genes of
these miRNAs were identified using the miRBase database, and a
target gene expression matrix was established. In the gene
expression matrix, 92 target genes had a strong linear relationship
with the M1/M2 ratio, and KEGG enrichment analysis of these 92
target genes was performed using the DAVID dataset. The main
enriched KEGG pathways were as follows: ‘MicroRNAs in cancer’;
‘thyroid hormone signaling’; ‘renal cell carcinoma’; ‘tight
junctions’; ‘herpes simplex infection’ and ‘ErbB signaling’
(Table SII).
Among the microRNAs in the cancer pathway, PAK4 and
CAT-1 participate in the transformation of normal hepatocytes to
HCC. In the ErbB signaling pathway, PAK4 and JNK participate in
tumor angiogenesis, which is associated with macrophage
polarization (29). Inhibitor of
nuclear factor κ-B kinase (IΚΚΒ), inhibitor of nuclear factor κ-B
kinase α (IKKA) and interferon α-2 are enriched in the ‘herpes
simplex infection pathway’, which suggests that these genes promote
macrophage polarization. In addition, ZO-1 and tight
junction-associated protein 1 were enriched in the ‘cell polarity’,
and ZO-1 and ZONAB were enriched in the ‘cell differentiation’ and
‘reduced cell proliferation’ (Table
SII). These KEGG pathways may be related to macrophage
polarization. In the 92 target genes, seven genes linear to their
corresponding miRNAs were selected as follows: Angiotensin II type
1 receptor (AGTR1), calbindin (CALB1), double cortin
domain-containing protein 1 (DCDC1), glycosaminoglycan
xylosylkinase (FAM20B), neuromodulin (GAP43), heat shock protein
105 kDa (HSPH1) and serine/threonine p21-activated kinases 4
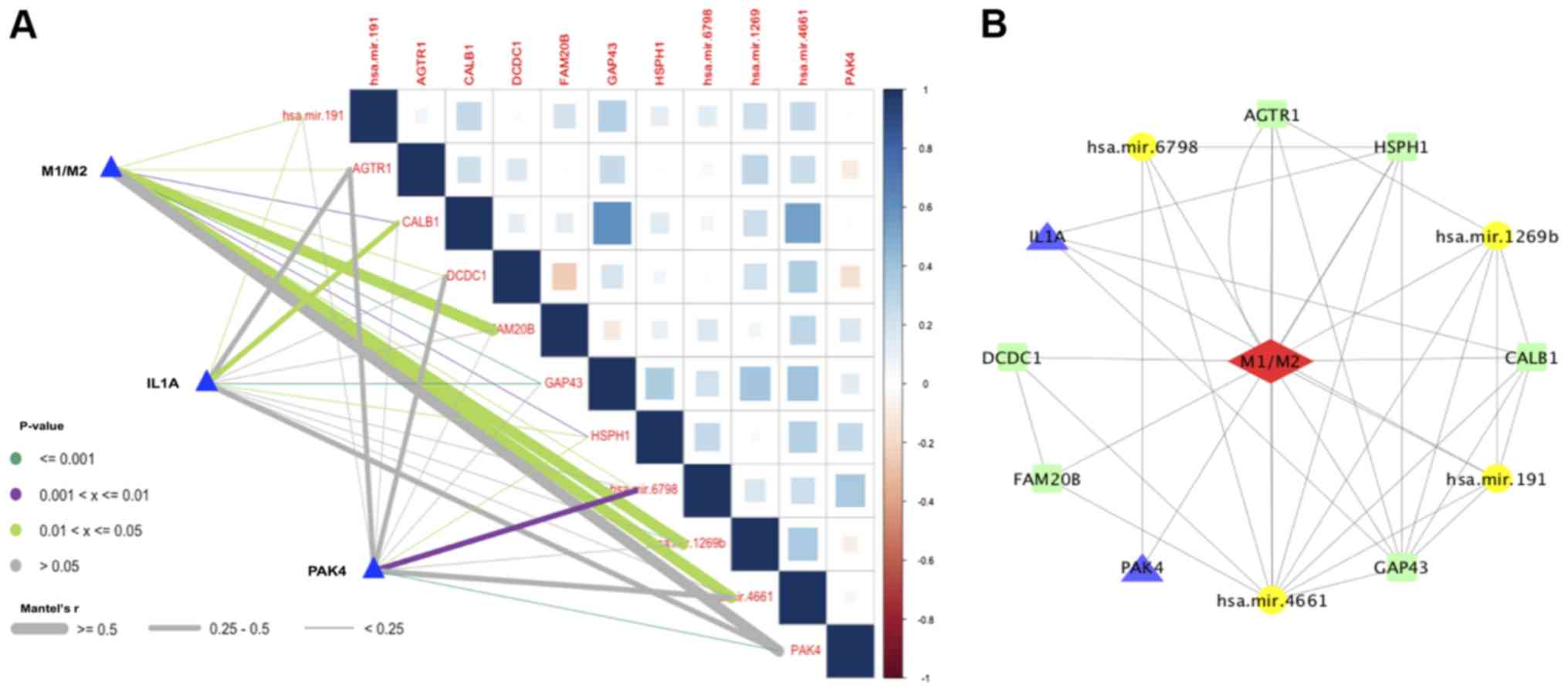
(PAK4). Finally, the correlation matrix map of the seven target
genes and four miRNAs linked with the M1/M2 ratio, IL1A and PAK4
was plotted to show their correlations, which ranged from weak to
strong. The relationship between M1/M2 ratio, IL1A, PAK4, miRNAs
and the target genes is shown in Fig.
8A.
In the final modulation network (Fig. 8B), three pairs of miRNAs and target
genes with linear correlations were obtained as follows:
MiRNA-1269b with GAP43; miR-4661 with HSPH1 and miRNA-6798 with
PAK4. In addition, PAK4 is not only an immune-related gene but is
also a target gene of miRNA-6798. In the regulation network and
correlation coefficient map, it was demonstrated that miRNA-6798
positively regulates the polarization of macrophages and governs
the expression of PAK4. IL1A is coexpressed with CALB1, GAP43 and
HSPH1. Therefore, IL1A and PAK4 indirectly govern the
differentiation of macrophages by coexpression with HSPH1. Finally,
coexpression analysis was applied again to verify the linear
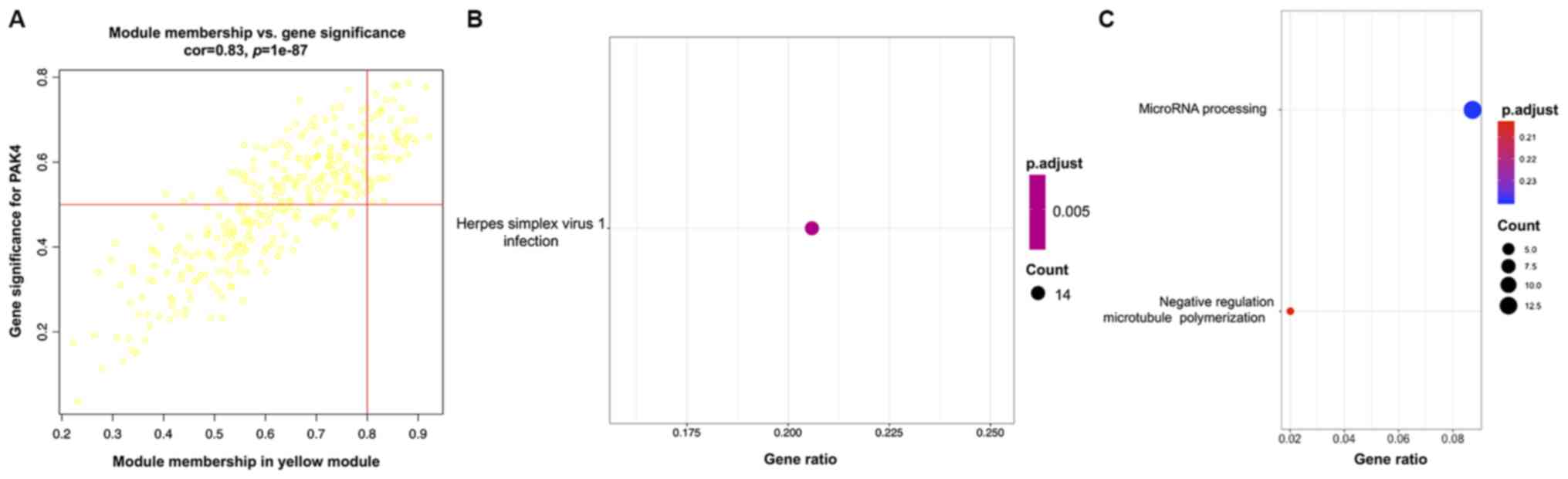
correlation between the expression of PAK4, HSPH1 and MATH scores
(correlation value of PAK4=0.268; P<0.05), which may indicate
the key role of PAK4 in tumor cell differentiation (Fig. S1).
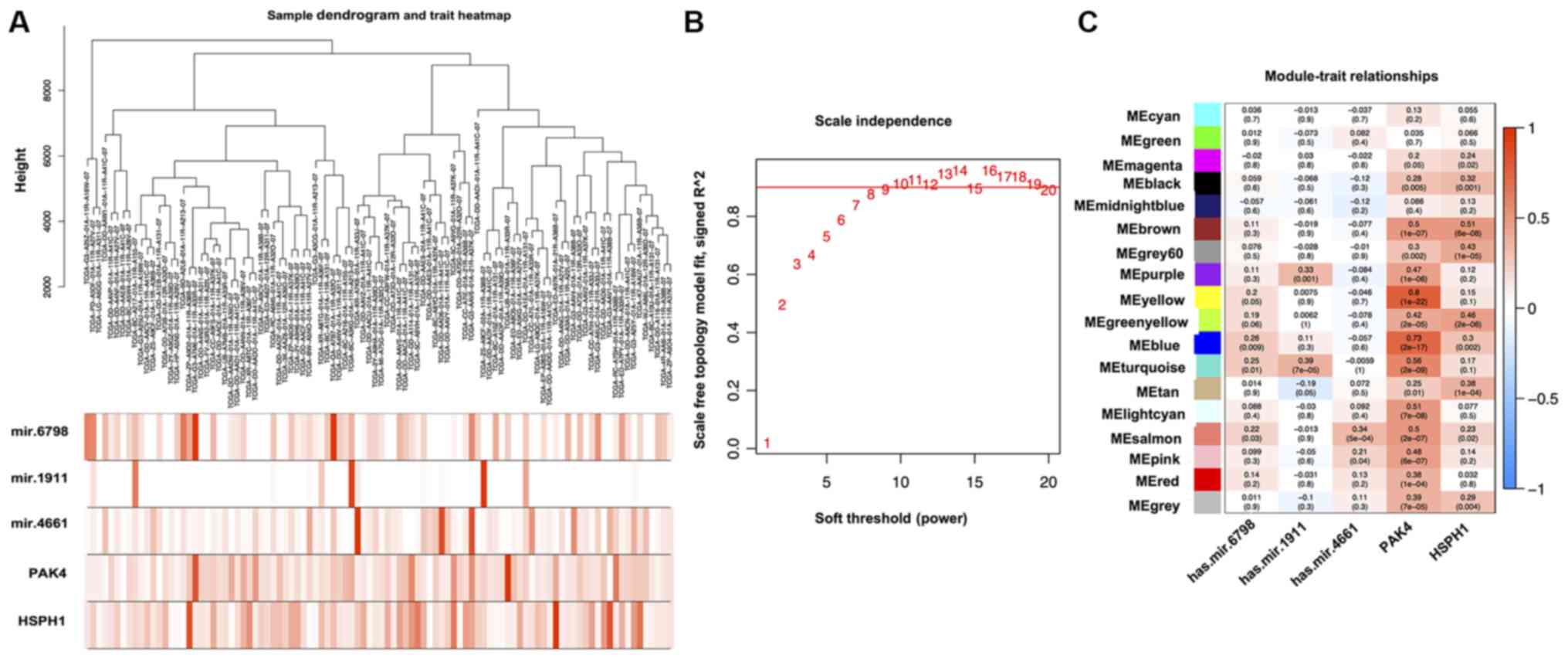
Then, all these coexpressed genes were analyzed with
respect to the expression of those molecules. The yellow consensus
module showed the most significant correlation with the expression
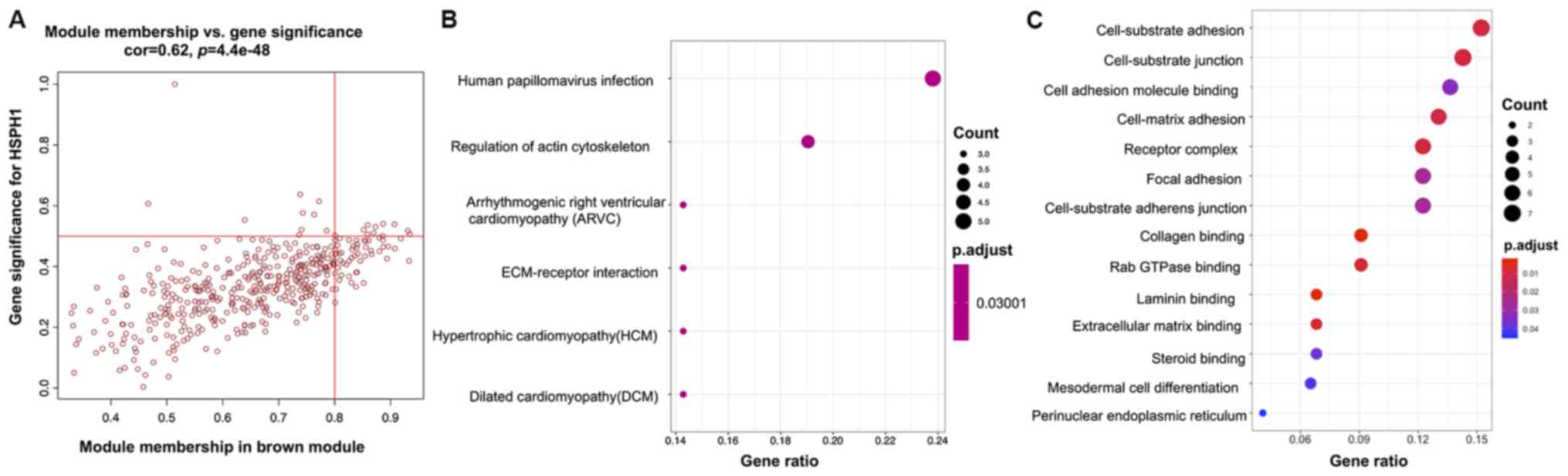
of PAK4 (correlation value=0.8; P<0.001; Fig. 9C). The brown consensus module showed
the most significant correlation with the expression of HSPH1
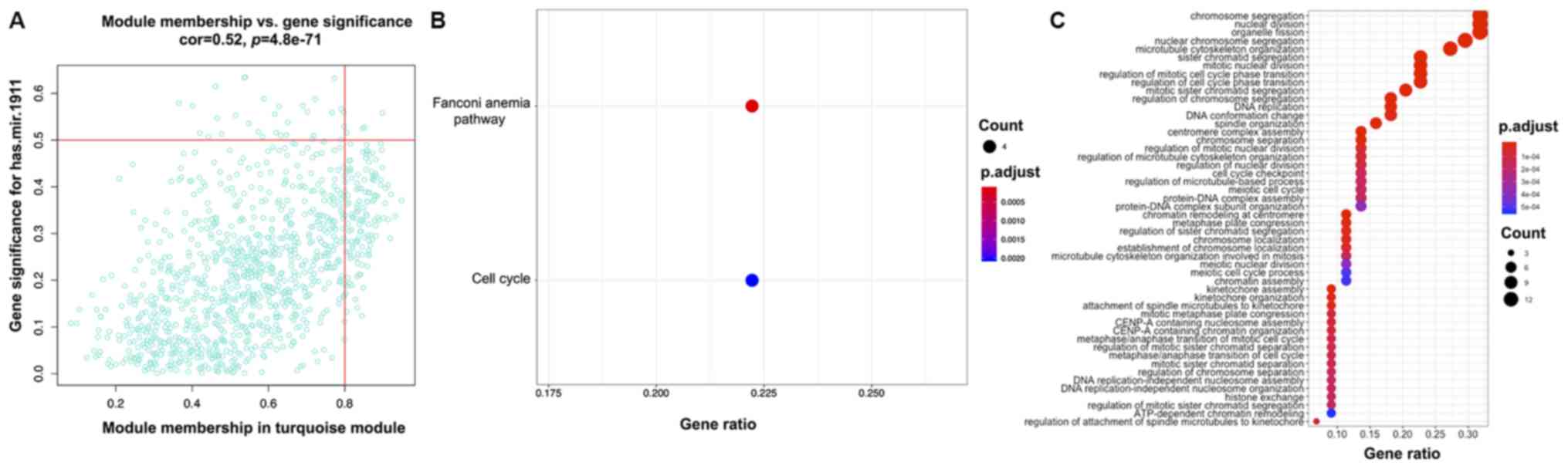
(correlation value=0.51; P<0.001; Fig. 9C). The turquoise consensus module
showed the most significant correlation with the expression of
miRNA-1911 (correlation value=0.39; P<0.001; Fig. 9C). GO analysis indicated enrichment
of mRNA processing in gene sets related to PAK4, cell-substrate
adhesion, cell-substrate junction and cell adhesion binding in gene
sets related to HSPH1, and chromosome segregation, nuclear division
and nuclear chromosome segregation in gene sets related to
miRNA-1911 (Figs. 10C, 11C and 12C, respectively). KEGG pathway analysis
showed the association of ‘herpes simplex virus 1 infection’ with
PAK4; ‘human papillomavirus infection’ with HSPH1 and the ‘Fanconi
anemia pathway’ with miRNA-1911 (Figs.
10B, 11B and 12B).
Discussion
The occurrence of non-synonymous mutations in HCC is
a condition for the production of new antigens that have not
previously been detected by the immune system (30). ITH stems from the production of
neoantigens, which can induce immune surveillance and immune
responses (31). Moreover, the
development of HCC is thought to be associated with the immune
response and the immune microenvironment (18). The immune microenvironment is often
accompanied by chronic liver inflammation in the occurrence of HCC.
The differences in immune cells, immunoregulatory factors and their
gene and protein profiles in the microenvironment of liver cancer
reflect the heterogeneity of the tumor immune microenvironment and
are associated with prognosis, immune response and drug resistance
(22). The present study applied the
MATH algorithm and CIBERSORT method to evaluate the ITH and TMH. By
evaluating the MATH score, it was reported that there was no
difference in OS when grouped by the median score. However, when
using 19.39 as the optimal cutoff value, the overall survival in
the low MATH score group was lower compared with that of the high
MATH score group, which was different from previous reports which
demonstrated that high MATH scores indicate poor prognosis in colon
cancer and diffuse large B-cell lymphoma (15,16). The
low MATH score group accounted for only 13% of the cohort; however,
it is still necessary to resolve the reasons for the differences
observed.
In the GSEA, three major nutrient metabolism
pathways and the bile acid metabolism pathway were enriched in the
low MATH score group, while seven immune-related gene sets were
enriched in the high MATH score group. Therefore, it can be
concluded that extremely low ITH indicates lower tumor
immunogenicity, which protects tumor cells from being recognized by
the immune system and avoids the activation of anticancer immunity
(30). This also explains the poor
prognosis phenomenon in the low MATH score group. However, compared
with the overall survival of the total study cohort, there was no
difference in the overall survival of the high MATH score group. As
shown in the results, the enriched immunosuppressive pathway may
influence prognosis in the high MATH score group. The
overexpression of the immunosuppressive factor IL10 indicated
shortened survival time in the high MATH score group. The
aforementioned results indicated the paradoxical role of cytokines
in tumor immunity. The tumor immune response is highly related to a
complex regulatory process of ITH (32,33).
Next, the genes enriched in the immune-related
pathway were identified in the high MATH score group and confirmed
to be associated with OS. ITGB2 was positively correlated with CD4
and PD-L1, and when ITGB2 was upregulated, the survival time is
shortened. The correlation between these genes and immune
checkpoints ranged from weak to moderate. IL1A and PAK4 were also
found to be strongly independent risk factors for survival through
multivariate analysis. IL1A was enriched in the ‘primary
immunodeficiency’ and ‘allograft rejection’ pathways, and PAK4 was
also enriched in the ‘T cell receptor signaling pathway’. The IL1A
cytokine is produced by macrophages and monocytes and is involved
in various immune responses and inflammatory processes, such as
stimulated the production of chemokines resulting in the
infiltration of neutrophils (34).
IL1A affects various stages of carcinogenesis, tumor growth and
tumor cell invasiveness and also the pattern of interactions
between tumor cells, the host immune system and the immune
microenvironment, in which IL-1 may also enhance the invasiveness
of tumor cells by the induction of heparanase, chemokines or
integrins on the malignant cells or endothelial cells (35). PAK4 is upregulated in tumor tissue,
especially pancreatic cancer and oral squamous cell carcinoma. The
amplification of PAK4 plays an important role in tumor invasion
associated with poor prognosis (36). It has been reported that the growth
of breast cancer is suppressed when the PAK4 pathway is inhibited
in in vitro experiments (37). In addition, miRNA-199a-regulated PAK4
promotes HCC occurrence, and PAK4-regulated TP53 promotes HCC
progression in in vivo experiments (38). In the T cell receptor signaling
pathway, PAK4 acts as an inhibitor of the regulation of the actin
cytoskeleton and effectively protects T cells from the host immune
response (39).
The CIBERSORT algorithm was employed to determine
the percentage of lymphocytes in a bulk of tumor transcriptomes to
explore the tumor immune mechanism and TIMH from leukocyte
infiltration in the HCC immune microenvironment. Previous studies
have conducted a similar analysis of immune cell infiltration in
HCC tissues and the impact of immune cells, such as macrophages and
Tregs on prognosis (40,41). One of the studies concluded that a
higher ratio of M1 macrophages indicates an improved prognosis
(40), while another study
illustrates that the total number of macrophages was negatively
correlated with OS (41). To gain
more insight into the relationship between the ratio of different
types of macrophages and prognosis, the present study analyzed the
relationship between the content of three types of macrophages and
prognosis separately and also clarified the effect of the M1/M2
ratio on prognosis. The percentage of M0 macrophages in tumor
tissues was higher compared with that in normal tissues.
Tumor-associated macrophages are important components of the HCC
immune microenvironment (42). In
the present study, among the 22 types of infiltrating lymphocyte,
M0, M1 and M2 accounted for 20.05, 7.76 and 13.24%, respectively.
In the HCC microenvironment, M2 macrophages are the characteristic
phenotype of tumor-associated macrophages, which promote
angiogenesis to support tumor cell invasion and metastasis
(43,44). Based on previous research, M2
macrophages are associated with poor clinical prognosis, while M1
macrophages are considered to inhibit tumors growth and are
tumoricidal (45). However, studies
have also shown that M1 macrophages can induce epithelial stromal
cell transformation of pancreatic ductal adenocarcinoma (46), activate hepatoma cells (47) and induce PD-L1 expression (42). In the present study, the survival
time was prolonged when the M1 percentage was high, while the
survival time was shortened when the M2 percentage was high. The
cut-off value for the M1/M2 ratio was set as 0.7284, and the HCC
cohort was divided into high-risk and a low-risk groups. The
survival of the two groups was significantly different (P=0.004),
which further indicated that the polarization of macrophages into
M1 can bring about good prognosis, while M2 usually predicts a poor
prognosis. The balance of M1-M2 macrophages is related to various
cancer and inflammatory diseases, such as melanoma, lung cancer and
asthma (48–50), and the ratio of M1/M2 also serves as
a risk factor for survival in the present study. Furthermore, the
proportion of NK-activated cells and Tregs cells was also
associated with clinical prognosis. The percentages of NK-activated
cells and Tregs were 4.84 and 6.26%, respectively, among all
lymphocytes. This was consistent with a previous study that
demonstrated that tumor-infiltrating NK-activated cells play a role
in immune surveillance and killing tumor cells by natural
cytotoxicity (51). In multivariate
Cox regression analysis, the M1/M2 ratio acted as the best
biomarker for survival prediction. Next, IL1A and the M1/M2 ratio
were integrated into the final model by multivariate Cox regression
analysis. The predictive ability of the model was judged by the AUC
value, wherein the 3-year prediction accuracy was 0.651 and the
5-year prediction accuracy was higher with an average value of 0.9.
Moreover, the heat map of the risk score for immune cells and the
gene model showed that the ratio of the M1/M2 ratio was notably
different in the high- and low-risk groups compared with the IL1A
group. Therefore, the M1/M2 ratio is a critical factor in the model
risk score.
In a previous study, macrophage polarization
modulated by miRNAs has been studied (48). In the present study, the expression
of miRNA-1269b, −6798, −191 and −4661 was linearly related to the
M1/M2 ratio, indicating that these miRNAs affect macrophage
polarization. It has also been demonstrated that miRNA-4661 can
regulate the immune response through the expression of IL10
(52), and miRNA-191 is highly
expressed in HCC and involved in promoting the cell cycle and tumor
cell proliferation (53). Activation
of the NF-κB pathway promotes the expression of miRNA-1269b,
inducing the development of liver cancer (54). The seven target genes (AGTR1,
HSPH1, CALB1, GAP43, FAM20B, DCDC1 and PAK4) were also
related to the M1/M2 ratio. Furthermore, IL1A was highly correlated
with HSPH1, CALB1, miRNA-191 and GAP43. PAK4 was
highly correlated with HSPH1. The relationship network of these
miRNAs, target genes, immune-related genes and the M1/M2 ratio
underpin the regulatory relationship between IL1A and PAK4 and the
M1/M2 ratio. As shown in the results, both IL1A and PAK4 regulate
macrophage polarization through HSPH1, which may indicate that
HSPH1 is an essential factor in macrophage polarization regulation
and in the immune microenvironment of liver cancer. In the colon
cancer immune response, HSPH1 modulates macrophage polarization
(55) and is associated with
chemotherapy sensitivity (56) and
immunogenicity (57). The present
study also indicated that HSPH1 may play a role in regulating
macrophage polarization and tumor immune response in liver cancer.
By establishing the regulatory network of IL1A, PAK4, HSPH1 and
M1/M2 ratio, it was clarified how the tumor immune response
mediated by ITH alters tumor-infiltrating immune cells and thus
affects TIMH in HCC. On the other hand, PAK4 not only plays a
regulatory role in the development of HCC but also plays a marked
role in mutation, neoantigen production and regulation of tumor
immunity. Moreover, HSPH1 is a mediator in the network that
regulates ITH and TIMH in HCC. In the WGCNA, the gene sets were
enriched in ‘herpes simplex virus 1 infection’, ‘human
papillomavirus infection’ and ‘Fanconi anemia’, which were in turn
related to immune responses and macrophage differentiation
(58) illustrating the potential of
these molecules to regulate the tumor immune microenvironment. It
has been reported that DNA-dependent protein kinase/Akt-mTORC1,
Toll-like receptor/nitric oxide and DNA-dependent protein kinase
(DNA-PK) /Rac1 signaling pathways are associated with the
immune-related KEGG pathway, therefore these may have a role in the
altered microenvironment of HCC (59–61).
Overall, the present study explored the potential
relationship between HCC ITH and TIMH via bioinformatics analysis.
According to the multi-omics analysis, IL1A, PAK4 and
HSPH1 may be key genes in tumor evolution, and liver cancer
immune-related signaling pathways were identified. Furthermore,
miRNA-1269b, −6798, −191 and −4661 are involved in the regulation
of the tumor microenvironment and may play an important role in
cross-talk between tumor cells and immune cells. Owing to the
experimental limitation, the majority of the present study focused
on multi-omics analysis of bioinformatics, whereas discussion on
other tumor immune microenvironment factors, such as fibroblasts,
transforming growth factors, chemokines were not included.
Considering that the immune microenvironment of liver cancer is
extremely complex, further exploration is needed in future studies,
such as the effect of glycolytic metabolites on the polarization of
macrophages and the molecular interaction mechanism between immune
cells and tumor cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was funded by a Grant-In-Aid for
Scientific Research from the National Natural Science Foundation
for the Youth of China (grant no. 81704042).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in The Cancer Genome Atlas repository,
(https://portal.gdc.cancer.gov/) and
Genotype-Tissue Expression repository (https://xenabrowser.net/datapages).
Authors' contributions
LBL designed and conceived the study. LY collected
the MAF files in the TCGA-LIHC database and calculated the MATH
score to analyzed the association between MATH score and survival
time. GQX collected the RNA seq files to explore the oncological
features of immune genes via GSEA and KEGG pathways. XCZ collected
and integrate analysis Genotype Tissue Expression with TCGA dataset
to analyze the difference in the percentage of immune cells by
CIBERSORT algorithm. QLX collected and analyzed TCGA-LIHC miRNA
dataset to establish the regulatory networks between target genes
and miRNAs. XBS organizes TCGA RNA-seq and miRNA dataset, and
analyzes the correlation of immune-related genes and draws relevant
figures. XDG made substantial contributions to conception and
design of this study and participated in drafting the manuscript.
ZYM polished the language and performed the data analyses. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nault JC, Ningarhari M, Rebouissou S and
Zucman-Rossi J: The role of telomeres and telomerase in cancer. Nat
Rev Gastroenterol Hepatol. 16:544–558. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kudo M: Systemic therapy for
hepatocellular carcinoma: Latest advances. Cancers (Basel).
10:4122018. View Article : Google Scholar
|
|
3
|
Ricke J, Klumpen HJ, Amthauer H,
Bargellini I, Bartenstein P, de Toni EN, Gasbarrini A, Pech M,
Peck-Radosavljevic M, Popovič P, et al: Impact of combined
selective internal radiation therapy and sorafenib on survival in
advanced hepatocellular carcinoma. J Hepatol. 71:1164–1174. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moehler M, Heo J, Lee HC, Tak WY, Chao Y,
Paik SW, Yim HJ, Byun KS, Baron A, Ungerechts G, et al:
Vaccinia-based oncolytic immunotherapy Pexastimogene Devacirepvec
in patients with advanced hepatocellular carcinoma after sorafenib
failure: A randomized multicenter Phase IIb trial (TRAVERSE).
Oncoimmunology. 8:16158172019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Sosman JA,
Atkins MB, Leming PD, et al: Five-year survival and correlates
among patients with advanced melanoma, renal cell carcinoma, or
Non-small cell lung cancer treated with nivolumab. JAMA Oncol.
5:1411–1420. 2019. View Article : Google Scholar
|
|
6
|
Qazi MA, Vora P, Venugopal C, Sidhu SS,
Moffat J, Swanton C and Singh SK: Intratumoral heterogeneity:
Pathways to treatment resistance and relapse in human glioblastoma.
Ann Oncol. 28:1448–1456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McDonald KA, Kawaguchi T, Qi Q, Peng X,
Asaoka M, Young J, Opyrchal M, Yan L, Patnaik S, Otsuji E and
Takabe K: Tumor heterogeneity correlates with less immune response
and worse survival in breast cancer patients. Ann Surg Oncol.
26:2191–2199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Z, Chen J, Hu J, Zhang H, Xu F, He W,
Wang X, Li M, Lu W, Zeng G, et al: cGAS/STING axis mediates a
topoisomerase II inhibitor-induced tumor immunogenicity. J Clin
Invest. 129:4850–4862. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lindström LS, Yau C, Czene K, Thompson CK,
Hoadley KA, Van't Veer LJ, Balassanian R, Bishop JW, Carpenter PM,
Chen YY, et al: Intratumor heterogeneity of the estrogen receptor
and the long-term risk of fatal Breast cancer. J Natl Cancer Inst.
110:726–733. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nassar D and Blanpain C: Cancer stem
cells: Basic concepts and therapeutic implications. Annu Rev
Pathol. 11:47–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Fujimoto J, Zhang J, Wedge DC,
Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al: Intratumor
heterogeneity in localized lung adenocarcinomas delineated by
multiregion sequencing. Science. 346:256–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jackson CM, Choi J and Lim M: Mechanisms
of immunotherapy resistance: Lessons from glioblastoma. Nat
Immunol. 20:1100–1109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Lou Y, Yang J, Wang J, Feng J,
Zhao Y, Wang L, Huang X, Fu Q, Ye M, et al: Integrated multiomic
analysis reveals comprehensive tumour heterogeneity and novel
immunophenotypic classification in hepatocellular carcinomas. Gut.
68:2019–2031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mroz EA and Rocco JW: MATH, a novel
measure of intratumor genetic heterogeneity, is high in
poor-outcome classes of head and neck squamous cell carcinoma. Oral
Oncol. 49:211–215. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rajput A, Bocklage T, Greenbaum A, Lee JH
and Ness SA: Mutant-allele tumor heterogeneity scores correlate
with risk of metastases in colon cancer. Clin Colorectal Cancer.
16:e165–e170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Feng W and Liu P: Genomic pattern
of intratumor heterogeneity predicts the risk of progression in
early stage diffuse large B-cell lymphoma. Carcinogenesis.
40:1427–1434. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yarchoan M, Xing D, Luan L, Xu H, Sharma
RB, Popovic A, Pawlik TM, Kim AK, Zhu Q, Jaffee EM, et al:
Characterization of the immune microenvironment in hepatocellular
carcinoma. Clin Cancer Res. 23:7333–7339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Atretkhany KSN, Drutskaya MS, Nedospasov
SA, Grivennikov SI and Kuprash DV: Chemokines, cytokines and
exosomes help tumors to shape inflammatory microenvironment.
Pharmacol Ther. 168:98–112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alifano M, Mansuet-Lupo A, Lococo F, Roche
N, Bobbio A, Canny E, Schussler O, Dermine H, Régnard JF, Burroni
B, et al: Systemic inflammation, nutritional status and tumor
immune microenvironment determine outcome of resected non-small
cell lung cancer. PLoS One. 9:e1069142014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang L, Zhu B, Zhang M and Wang X: Roles
of immune microenvironment heterogeneity in therapy-associated
biomarkers in lung cancer. Semin Cell Dev Biol. 64:90–97. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye L, Zhang T, Kang Z, Guo G, Sun Y, Lin
K, Huang Q, Shi X, Ni Z, Ding N, et al: Tumor-infiltrating immune
cells act as a marker for prognosis in colorectal cancer. Front
Immunol. 10:23682019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen H, Chong W, Teng C, Yao Y, Wang X and
Li X: The immune response-related mutational signatures and driver
genes in non-small-cell lung cancer. Cancer Sci. 110:2348–2356.
2019.PubMed/NCBI
|
|
25
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation. network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu K and Chen F: Identification of
significant pathways in gastric cancer based on protein-protein
interaction networks and cluster analysis. Genet Mol Biol.
35:701–708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Komohara Y, Fujiwara Y, Ohnishi K and
Takeya M: Tumor-associated macrophages: Potential therapeutic
targets for anti-cancer therapy. Adv Drug Deliv Rev. 99:180–185.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prieto J, Melero I and Sangro B:
Immunological landscape and immunotherapy of hepatocellular
carcinoma. Nat Rev Gastroenterol Hepatol. 12:681–700. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McGranahan N, Furness AJ, Rosenthal R,
Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak
NJ, Hiley CT, et al: Clonal neoantigens elicit T cell
immunoreactivity and sensitivity to immune checkpoint blockade.
Science. 351:1463–1469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mannino MH, Zhu Z, Xiao H, Bai Q,
Wakefield MR and Fang Y: The paradoxical role of IL-10 in immunity
and cancer. Cancer Lett. 367:103–107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lippitz BE: Cytokine patterns in patients
with cancer: A systematic review. Lancet Oncol. 14:e218–e228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dinarello CA: Overview of the IL-1 family
in innate inflammation and acquired immunity. Immunol Rev.
281:8–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Apte RN, Dotan S, Elkabets M, White MR,
Reich E, Carmi Y, Song X, Dvozkin T, Krelin Y and Voronov E: The
involvement of IL-1 in tumorigenesis, tumor invasiveness,
metastasis and tumor-host interactions. Cancer Metastasis Rev.
25:387–408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Jia S and Dai W: Fisetin modulates
human oral squamous cell carcinoma proliferation by Blocking
PAK4 signaling pathways. Drug Des Devel Ther.
14:773–782. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Costa TDF, Zhuang T, Lorent J, Turco E,
Olofsson H, Masia-Balague M, Zhao M, Rabieifar P, Robertson N,
Kuiper R, et al: PAK4 suppresses RELB to prevent senescence-like
growth arrest in breast cancer. Nat Commun. 10:35892019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu HT, Lai WL, Liu HF, Wong LLY, Ng IOL
and Ching YP: PAK4 phosphorylates p53 at serine 215 to promote
liver cancer metastasis. Cancer Res. 76:5732–5742. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Ke Q, Li Y, Liu F, Zhu G and Li F:
DGCR6L, a novel PAK4 interaction protein, regulates PAK4-mediated
migration of human gastric cancer cell via LIMK1. Int J Biochem
Cell Biol. 42:70–79. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rohr-Udilova N, Klinglmuller F,
Schulte-Hermann R, Stift J, Herac M, Salzmann M, Finotello F,
Timelthaler G, Oberhuber G, Pinter M, et al: Deviations of the
immune cell landscape between healthy liver and hepatocellular
carcinoma. Sci Rep. 8:62202018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Foerster F, Hess M, Gerhold-Ay A,
Marquardt JU, Becker D, Galle PR, Schuppan D, Binder H and Bockamp
E: The immune contexture of hepatocellular carcinoma predicts
clinical outcome. Sci Rep. 8:53512018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zong Z, Zou J, Mao R, Ma C, Li N, Wang J,
Wang X, Zhou H, Zhang L and Shi Y: M1 Macrophages induce PD-L1
expression in hepatocellular carcinoma cells through IL-1β
signaling. Front Immunol. 10:16432019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu
L, Grzesik DA, Qian H, Xue XN and Pollard JW: Macrophages regulate
the angiogenic switch in a mouse model of breast cancer. Cancer
Res. 66:11238–11246. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qian B, Deng Y, Im JH, Muschel RJ, Zou Y,
Li J, Lang RA and Pollard JW: A distinct macrophage population
mediates metastatic breast cancer cell extravasation, establishment
and growth. PLoS One. 4:e65622009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wanderley CW, Colón DF, Luiz JP, Oliveira
FF, Viacava PR, Leite CA, Pereira JA, Silva CM, Silva CR, Silva RL,
et al: Paclitaxel reduces tumor growth by reprogramming
tumor-associated macrophages to an M1 profile in a TLR4-dependent
manner. Cancer Res. 78:5891–5900. 2018.PubMed/NCBI
|
|
46
|
Helm O, Held-Feindt J, Grage-Griebenow E,
Reiling N, Ungefroren H, Vogel I, Krüger U, Becker T, Ebsen M,
Röcken C, et al: Tumor-associated macrophages exhibit pro- and
anti-inflammatory properties by which they impact on pancreatic
tumorigenesis. Int J Cancer. 135:843–861. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang H, Wang X, Li X, Fan Y, Li G, Guo C,
Zhu F, Zhang L and Shi Y: CD68+HLA-DR+ M1-like macrophages promote
motility of HCC cells via NF-κB/FAK pathway. Cancer Lett.
345:91–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang N, Liang H and Zen K: Molecular
mechanisms that influence the macrophage M1-M2 polarization
balance. Front Immunol. 5:6142014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC,
Chen YY, Liu YC, Hong TH, Yu SL, Chen JJ and Yang PC: Opposite
effects of M1 and M2 macrophage subtypes on lung cancer
progression. Sci Rep. 5:142732015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bardi GT, Smith MA and Hood JL: Melanoma
exosomes promote mixed M1 and M2 macrophage polarization. Cytokine.
105:63–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Imai K, Matsuyama S, Miyake S, Suga K and
Nakachi K: Natural cytotoxic activity of peripheral-blood
lymphocytes and cancer incidence: An 11-year follow-up study of a
general population. Lancet. 356:1795–1799. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Quinn SR and O'Neill LA: The role of
microRNAs in the control and mechanism of action of IL-10. Curr Top
Microbiol Immunol. 380:145–155. 2014.PubMed/NCBI
|
|
53
|
Tian F, Yu C, Wu M, Wu X, Wan L and Zhu X:
MicroRNA-191 promotes hepatocellular carcinoma cell proliferation
by has_circ_0000204/miR-191/KLF6 axis. Cell Prolif. 52:e126352019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kong XX, Lv YR, Shao LP, Nong XY, Zhang
GL, Zhang Y, Fan HX, Liu M, Li X and Tang H: HBx-induced MiR-1269b
in NF-κB dependent manner upregulates cell division cycle 40
homolog (CDC40) to promote proliferation and migration in hepatoma
cells. J Transl Med. 14:1892016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Berthenet K, Boudesco C, Collura A, Svrcek
M, Richaud S, Hammann A, Causse S, Yousfi N, Wanherdrick K, Duplomb
L, et al: Extracellular HSP110 skews macrophage polarization in
colorectal cancer. Oncoimmunology. 5:e11702642016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dorard C, de Thonel A, Collura A, Marisa
L, Svrcek M, Lagrange A, Jego G, Wanherdrick K, Joly AL, Buhard O,
et al: Expression of a mutant HSP110 sensitizes colorectal cancer
cells to chemotherapy and improves disease prognosis. Nat Med.
17:1283–1289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang XY, Li Y, Manjili MH, Repasky EA,
Pardoll DM and Subjeck JR: Hsp110 over-expression increases the
immunogenicity of the murine CT26 colon tumor. Cancer Immunol
Immunother. 51:311–319. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lu WT, Lemonidis K, Drayton RM and
Nouspikel T: The Fanconi anemia pathway is downregulated upon
macrophage differentiation through two distinct mechanisms. Cell
Cycle. 10:3300–3310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li J, Rao H, Jin C and Liu J: Involvement
of the Toll-like receptor/Nitric oxide signaling pathway in the
pathogenesis of cervical cancer caused by high-risk human
papillomavirus infection. Biomed Res Int.
2017:78302622017.PubMed/NCBI
|
|
60
|
Hu HL, Shiflett LA, Kobayashi M, Chao MV,
Wilson AC, Mohr I and Huang TT: TOP2β-dependent nuclear DNA damage
shapes extracellular growth factor responses via dynamic AKT
phosphorylation to control virus latency. Mol Cell. 74:466–480.e4.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Romick-Rosendale LE, Hoskins EE, Privette
Vinnedge LM, Foglesong GD, Brusadelli MG, Potter SS, Komurov K,
Brugmann SA, Lambert PF, Kimple RJ, et al: Defects in the fanconi
anemia pathway in head and neck cancer cells stimulate tumor cell
invasion through DNA-PK and Rac1 signaling. Clin Cancer Res.
22:2062–2073. 2016. View Article : Google Scholar : PubMed/NCBI
|