Introduction
Renal cell carcinoma (RCC) is a common renal tumor
in adults, accounting for ~90% of renal tumors and ~3% of all adult
cancer cases, incidence rate of RCC was 5.78 per 100,000 women and
13.14 per 100,000 men in 2013 (1).
The clinical onset of RCC is insidious; patients usually experience
no obvious symptoms in the early stages. This makes RCC a
problematic disease to treat, as the tumor has usually metastasized
before surgery and often relapses following surgery (2). Nevertheless, surgery remains the
primary treatment for RCC, supplemented by other treatments,
including renal artery embolization, chemotherapy, immunotherapy
and molecular targeted therapy (2,3).
Therefore, finding targeted treatments for RCC has great clinical
importance. However, at present, the molecular mechanism underlying
RCC remains unclear (4,5).
The ubiquitin-proteasome system (UPS) exists in
almost all eukaryotic cells. Ubiquitination is reversible, and the
deubiquitination of substrate proteins by deubiquitinases (DUBs)
can regulate the stability of proteins, thereby affecting their
functions (5). Therefore, DUBs serve
an important role in maintaining normal cell functions and
regulating pathological processes. Ubiquitin-specific protease
(USP) is one of the subfamilies of the DUB family. USP family
member USP22 is highly expressed in multiple malignant tumors,
including gastric, colon and prostate cancers (6). The expression level of USP22 is closely
associated with metastasis potential, chemotherapeutic resistance
and prognosis in patients with cancer (7). USP22 deficiency results in myeloid
leukemia via an ETS-family transcription factor pu.1-dependent
mechanism after the activation of carcinogenic Kras (8). However, its function and mechanism in
RCC have yet to be elucidated.
Survivin is a member of the inhibitor of apoptotic
protein family and serves as a subunit of the chromosomal passenger
complex that regulates cell division (9). Survivin expression is associated with a
variety of human cancers and its high expression often indicates a
poor prognosis (10–12). For example, a high expression of
survivin is associated with a poor overall survival and shorter
cancer-specific survival in RCC (13,14).
Survivin protein functions to inhibit caspase activation, thereby
leading to negative regulation of apoptosis or programmed cell
death (15). The stability of
survivin is regulated via the ubiquitin-proteasome pathway
(16). However, whether USP22
influences the ubiquitination-proteasome pathway-dependent
regulation of survivin in the development of RCC has yet to be
elucidated.
The present study hypothesized that USP22 inhibits
the apoptosis of renal carcinoma cells via modulating survivin
level. This was first confirmed by immunohistochemical and western
blotting that the protein levels of USP22 and survivin in RCC were
higher compared with adjacent normal tissues. Then it was
identified that USP22 upregulated survivin protein level and
suppressed cell death via siRNA knockdown and overexpression
experiments. Finally, the interaction between USP22 and survivin
was confirmed, and that USP22 supported survivin stabilization
through deubiquitination. Together, the results of the present
study revealed that USP22 may inhibit the apoptosis of RCC by
deubiquitinating and stabilizing survivin.
Materials and methods
Specimens
Renal carcinoma specimens (n=10) and the
corresponding adjacent normal tissues were obtained by surgical
excision from the Department of Urological Surgery in the Fifth
Hospital of Xiamen (Xiamen, China) between March 2017 and March
2018. The inclusion criteria were as follows, histologically
confirmed renal carcinoma and having adjacent normal tissues as a
control. Patients with prior history of RCC that was resected
within the past 5 years were excluded from the present study.
Patients provided written informed consent in accordance with the
legal and institutional ethical guidelines defined by the hospital
by the Ethics Committee of the Fifth Hospital of Xiamen (Xiamen,
China). The age of patients ranged from 36–85 years, the overall
median age was 60.2 years. Adjacent normal tissues were taken from
at least 2 cm apart from the tumor border.
Cell culture
Human RCC cell lines ACHN and TK-1 (American Type
Culture Collection) were cultured in Dulbecco's modified Eagle's
medium (Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v)
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100
U/ml penicillin (Thermo Fisher Scientific, Inc.) and 100 U/ml
streptomycin (Thermo Fisher Scientific, Inc.) at 37°C in 5%
CO2.
Immunohistochemistry
Tissue sections were dehydrated in gradient
concentration of ethanol solution and fixed with 4%
paraformaldehyde at room temperature for 30 min, and then paraffin
embedded. Consecutive 4-µm sections were used for analysis. Antigen
retrieval was performed by microwaving sections in citrate buffer
(pH 6.0). Subsequently, slides were incubated overnight with
anti-USP22 (cat. no. ab71732; Abcam; 1:1,000) and
anti-SURVIVIN (cat. no. 2808; Cell Signaling Technology,
Inc.; 1:1,000) antibodies at 4°C. Subsequently, slides were block
with goat serum (cat. no. AR0009; Wuhan Boster Biological
Technology, Ltd; 1:20) at room temperature for 1 h and then
incubated with goat anti-rabbit second antibody (cat. no. ab6721;
Abcam; 1:2,000) at RT for 1 h. Streptococcal avidin-biotinylated
peroxidase system (Thermo Fisher Scientific, Inc.) was used to
develop the color according to the manufacturer's instructions.
Tissue sections were visualized using an Olympus microscope IX50 at
×20 magnification and the images were analyzed using ImageJ 1.49
version software (National Institutes of Health). The American
Joint Committee on Cancer (AJCC) Staging Manual system was used to
assess tumor grades (17).
DNA constructs and proliferation
assay
cDNA fragments encoding USP22 were inserted into
pCMV-HA vector to construct an USP22-HA overexpression vector.
Short hairpin (sh)RNA targeting sequences for human USP22 (shRNA1:
5′-AAGTCCTGTATCTGCCATGTC-3′, shRNA2: 5′-GTTTCACAAAGAAGCATATTC-3′)
or scrambled shRNA (sequence: CCTAAGGTTAAGTCGCCCTCG) were inserted
into a pLKO.1-GFP-shRNA construct. ACHN and TK-10 cells were
transfected with USP22 shRNA and ACHN cells were transfected
with USP22-HA vector using Turbofect (Thermo Fisher Scientific,
Inc.). Cell Counting Kit-8 (CCK-8; Thermo Fisher Scientific, Inc.)
was used to estimate cell numbers according to manufacturer's
instructions, 48 h post-transfection. OD values (450 nm) were
measured using a microplate reader (Thermo Fisher Scientific,
Inc.). Cell counting assays were performed using ImageJ software
version 1.49 [National Institutes of Health (NIH)] to establish
growth curves. Each experiment was repeated 4 times in
quintuplicate wells per sample by 2 independent experimenters.
Colony formation assay
ACHN cells were seeded in a six-well plate at 800
cells/well, USP22-specific shRNA and negative control shRNA
were transfected into cells using Turbofect (Thermo Fisher
Scientific, Inc.) the next day and the cells were cultured for
another 7 days at 37°C in 5% CO2. Cells were stained
with crystal violet at room temperature for 10 min, the colonies
(>1 mm) in diameter were counted using ImageJ software version
1.49 (NIH) (18).
Western blotting
Kidney tissues (about 100 mg) or ACHN cells
(approximately 3×105 cells) were incubated on ice with
RIPA buffer (Beyotime Institute of Biotechnology) to extract the
total proteins. The protein concentrations were determined with a
bicinchoninic acid assay system (Thermo Fisher Scientific, Inc.).
Protein samples (40 µg/lane) were separated via SDS-PAGE on a 10%
gel (Beijing Solarbio Science & Technology Co., Ltd.), and then
proteins were transferred onto nitrocellulose membranes. Membranes
were then blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) at room
temperature for 1 h and incubated with primary antibodies overnight
at 4°C, and then incubated with HRP-labeled goat anti-rabbit
secondary antibody (cat. no. A0208; Beyotime Institute of
Biotechnology; 1:3,000) or HRP-labeled goat anti-mouse secondary
antibody (cat. no. A0216; Beyotime Institute of Biotechnology;
1:3,000) at room temperature for 1 h. Immunoreactive bands were
visualized using ECL detection reagents (Thermo Fisher Scientific,
Inc.), and protein levels were quantified using ImageJ software
(version 1.49; National Institutes of Health). The antibodies were
USP22 (cat. no. ab71732; Abcam; 1:2,000), SURVIVIN (cat. no.
2808; Cell Signaling Technology, Inc.; 1:2,000), cleaved-caspase-3
(cat. no. 9664; Cell Signaling Technology, Inc.; 1:2,000),
caspase-3 (cat. no. ab197202, Abcam; 1:1,000), phosphorylated
(p-)Akt (cat. no. ab38449, Abcam; 1:1,000), Akt (cat. no. ab8805,
Abcam; 1:2,000), p-Erk (cat. no. ab131438, Abcam; 1:1,000), Erk
(cat. no. ab32537, Abcam; 1:2,000), GAPDH (cat. no. bc002, xmbcss,
www.bcssbio.com; 1:2,000), ubiquitin (cat. no.
sc-8017, Santa Cruz Biotechnology, Inc.; 1:2,000), HA (cat. no.
51064-2-AP, ProteinTech Group, Inc.; HA sequences: YPYDVPDYA;
1:2,000) and β-actin (cat. no. 23660-1-AP; ProteinTech Group, Inc.;
1:2,000).
Co-immunoprecipitation (Co-IP)
The USP22-HA and control plasmid was transfected
into 293T cells and 48 h later, the cells were collected and
dissolved in cold nucleolysis buffer (50 mmol/l Tris-HCl pH 8.0,
150 mmol/l NaCl, 1% NP-40 (Solarbio), 2% v/v complete protease
inhibitor, and NaVO3). Cells were disrupted with a
homogenizer, and the supernatants were collected after 14,000 × g
centrifugation for 15 min at 4°C. The supernatants were incubated
with anti-USP22 antibody (cat. no. ab71732; Abcam; 1:500) or
anti-IgG antibody (cat. no. ab109489; Abcam; 1:500) at 4°C and, 6 h
later, protein A + G agarose beads (Santa Cruz Biotechnology, Inc;
1:200) were added to each sample and the samples were incubated
overnight at 4°C. The beads were then centrifuged at 8,000 × g for
30 sec at 4°C and washed four times with 1 ml cold lysis buffer, as
aforementioned. Then the beads were boiled in 30 µl sample buffer
for 5 min, to release the protein from the beads. Proteins were
detected via western blotting as mentioned above with USP22 (cat.
no. ab71732; Abcam), SURVIVIN (cat. no. 2808; Cell Signaling
Technology, Inc.).
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated from 3×105 cells
using the RNAiso™ Plus kit (Tehermo Fisher Scientific, Inc.).
Reverse transcriptions were conducted using a cDNA synthesis kit
(Toyobo Life Science) in 20 µl reaction system. qPCR was performed
using FastStart Universal SYBR Green Master (Roche, Inc.) in 25 µl
reaction system with thermocycling conditions: Initial denaturation
at 98°C for 30 sec; 35 cycles at 98°C for 10 sec, 60°C for 30 sec,
and 72°C for 30 sec; final extension at 72°C for 2 min. The primer
sequences were as follows: β-actin forward,
5′-AAGGAAGGCTGGAAGAGGTGC-3′ and reverse, 5′-CTGGAGAGAGAGAGAGAAA-3′;
USP22 forward, 5′-GGCGGAGATCACCAGGTAT-3 and reverse,
5′-TTGTGTAGAGACTGTCCGTGGG-3′; and SURVIVIN forward,
5′-TGGCCTTTCAGAGCAGAGTG-3′ and reverse 5′-AAGCCACAGTTAGGGGAACG-3′.
RNA extraction, cDNA synthesis, and qPCR performed according to the
manufacturer's protocols. Normalized relative expression levels
were calculated using the 2−∆∆Cq (cycle threshold)
method (19).
Cycloheximide (CHX) analysis
ACHN cells (approximately 3×105 cells)
were transfected with control or USP22-HA vector. After 48 h, CHX
(500 µM) was added at different time points as indicated, and then
cells were harvested. The expression of Survivin was detected via
western blot analysis and quantification analysis was performed
using ImageJ software (version 1.49; National Institutes of
Health).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 7 software (GraphPad Software, Inc.). All experiments were
conducted in triplicate and repeated at ≥3 times unless otherwise
specified. The data are presented as mean ± standard error of mean.
The statistically significant differences between two groups were
calculated with unpaired Student's t-test using GraphPad Prism 7
software (GraphPad Software, Inc.). One-way analysis of variance
followed by Tukey's post hoc test were used to compare differences
between multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
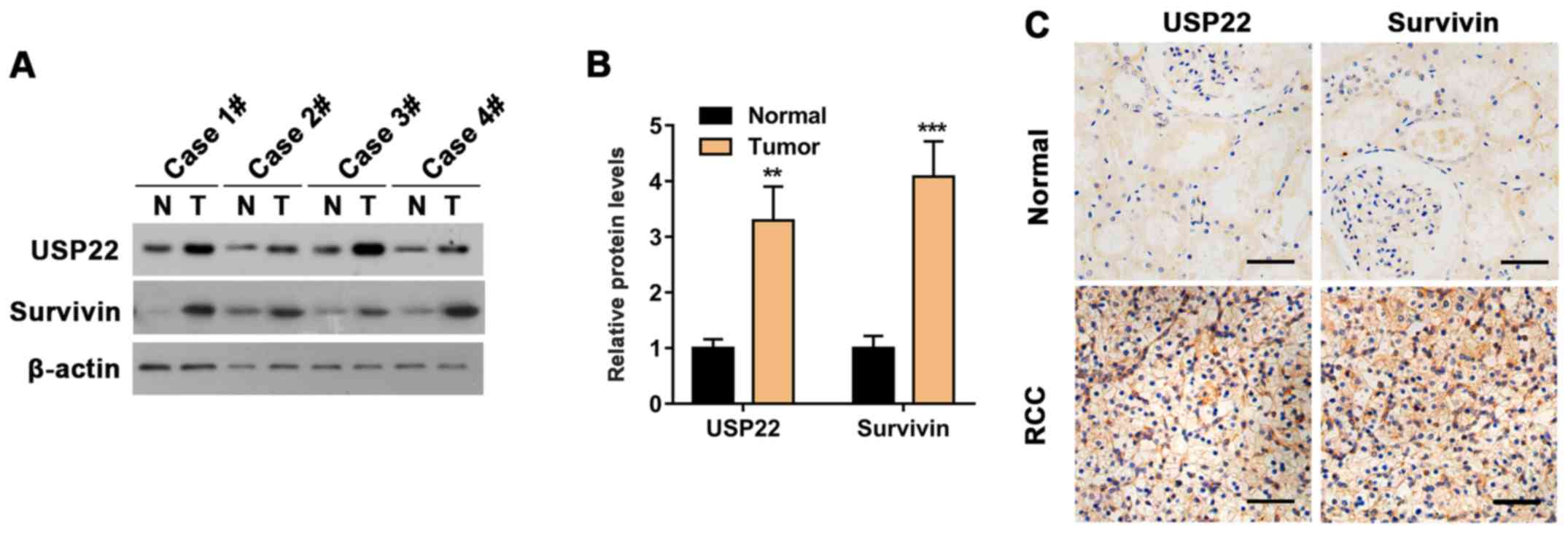
USP22 is highly expressed in RCC and
associated with survivin expression
To determine whether the levels of USP22 and
survivin are clinically correlated with RCC, 10 pairs of tissues
were collected, each consisting of a patient's RCC tissue and an
adjacent normal tissue as the control (Table I). The protein levels of USP22 and
survivin in RCC were quantified with western blotting. Results
indicated that the protein levels of USP22 and survivin in RCC
tissues were significantly higher compared with those in control
tissues (Fig. 1A and B). USP22 and
survivin were also detected in RCC tissues and controls using
immunohistochemistry. Analogous to western blotting,
immunohistochemistry results also suggested upregulation of both
USP22 and survivin in RCC tissues (Fig.
1C). Therefore, it was concluded that USP22 and survivin are
co-expressed in RCC cells and their protein levels are upregulated
in RCC.
 | Table I.Basic clinicopathological data of
renal cell carcinoma patients. |
Table I.
Basic clinicopathological data of
renal cell carcinoma patients.
| Patient | Sex | Age, years | Tumor grade | Treatment |
|---|
| 1 | Male | 54 | II | After radical
resection, regular follow-ups were conducted |
| 2 | Male | 52 | VI | Oral administration
of sorafenib (400 mg bid) |
| 3 | Female | 73 | III | After radical
resection, regular follow-ups were conducted |
| 4 | Female | 56 | IV | Oral administration
of sunitinib (400 mg bid) |
| 5 | Female | 44 | IV | Oral administration
of sorafenib (400 mg bid) |
| 6 | Male | 72 | III | Oral administration
of sorafenib (400 mg bid) |
| 7 | Male | 68 | II | After radical
resection, regular follow-ups were conducted |
| 8 | Male | 62 | III | After radical
resection, regular follow-ups were conducted |
| 9 | Male | 36 | I | After radical
resection, regular follow-ups were conducted |
| 10 | Female | 85 | I | After radical
resection, regular follow-ups were conducted |
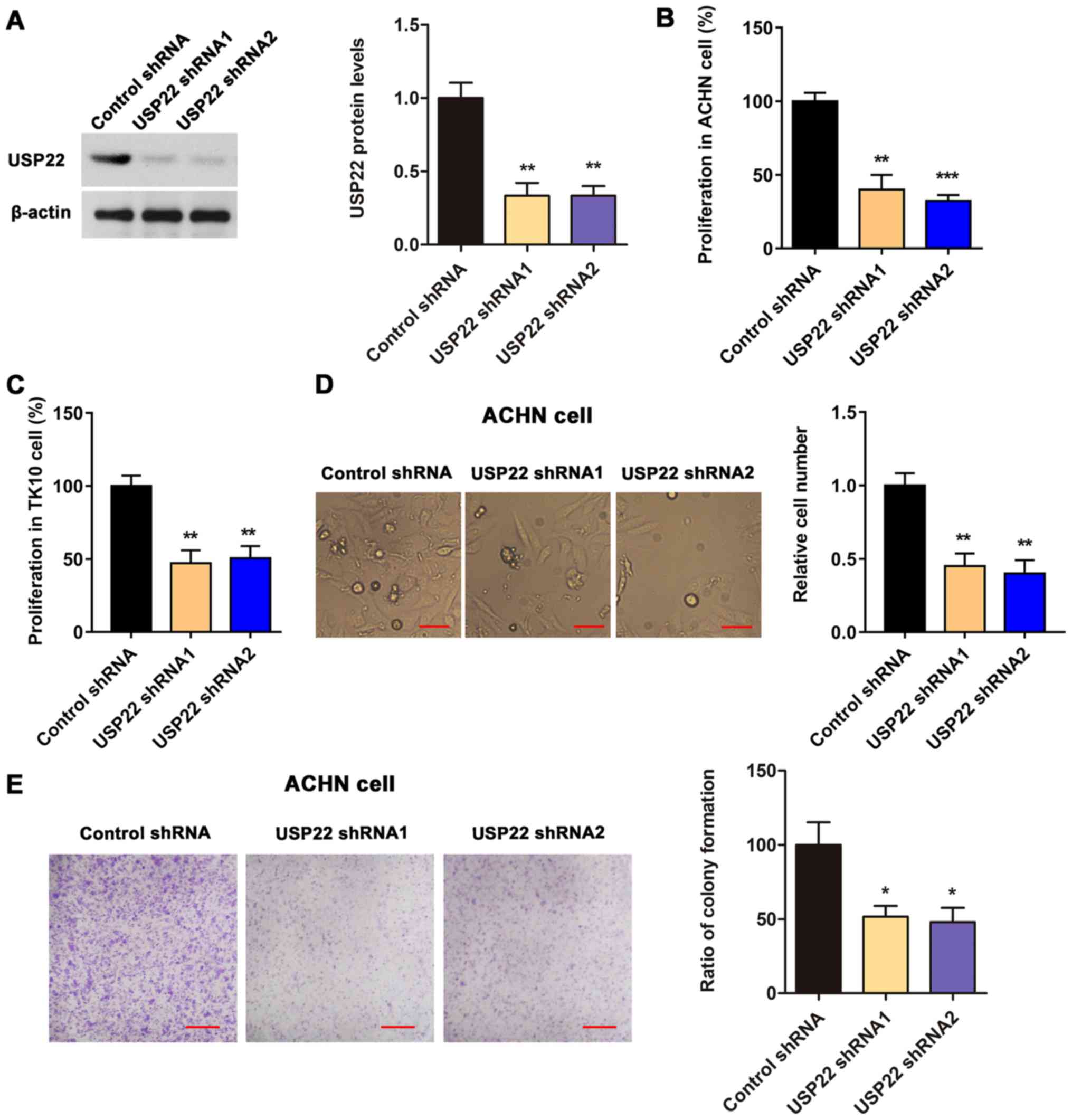
USP22 knockdown inhibits the expansion
and monoclonal colony formation of ACHN cells
To knockdown USP22 in RCC cells, two pairs of
USP22-targeting shRNA were constructed and their knockdown
efficiency was validated via western blotting (Fig. 2A). Next, the role of USP22 in the
proliferation of human renal carcinoma cell line ACHN and TK-10
cells was evaluated with CCK8 assays. The results indicated that
USP22 knockdown significantly inhibited the proliferation of ACHN
and TK-10 cells (Fig. 2B-C).
Although USP22 silencing demonstrated similar inhibitory effects on
both ACHN and TK-1 human RCC cells, only ACHN cells were subjected
to the subsequent experiments as the most commonly utilized cell
line in RCC research.
It was also determined that the cell number and
cloning rate of ACHN cells transfected with USP22 shRNA was
significantly lower compared with the cells transfected with
control shRNA (Fig. 2D-E, P<0.01
in cell number and P<0.05 in cloning rate), indicating that
USP22 may contribute to the proliferation of RCC.
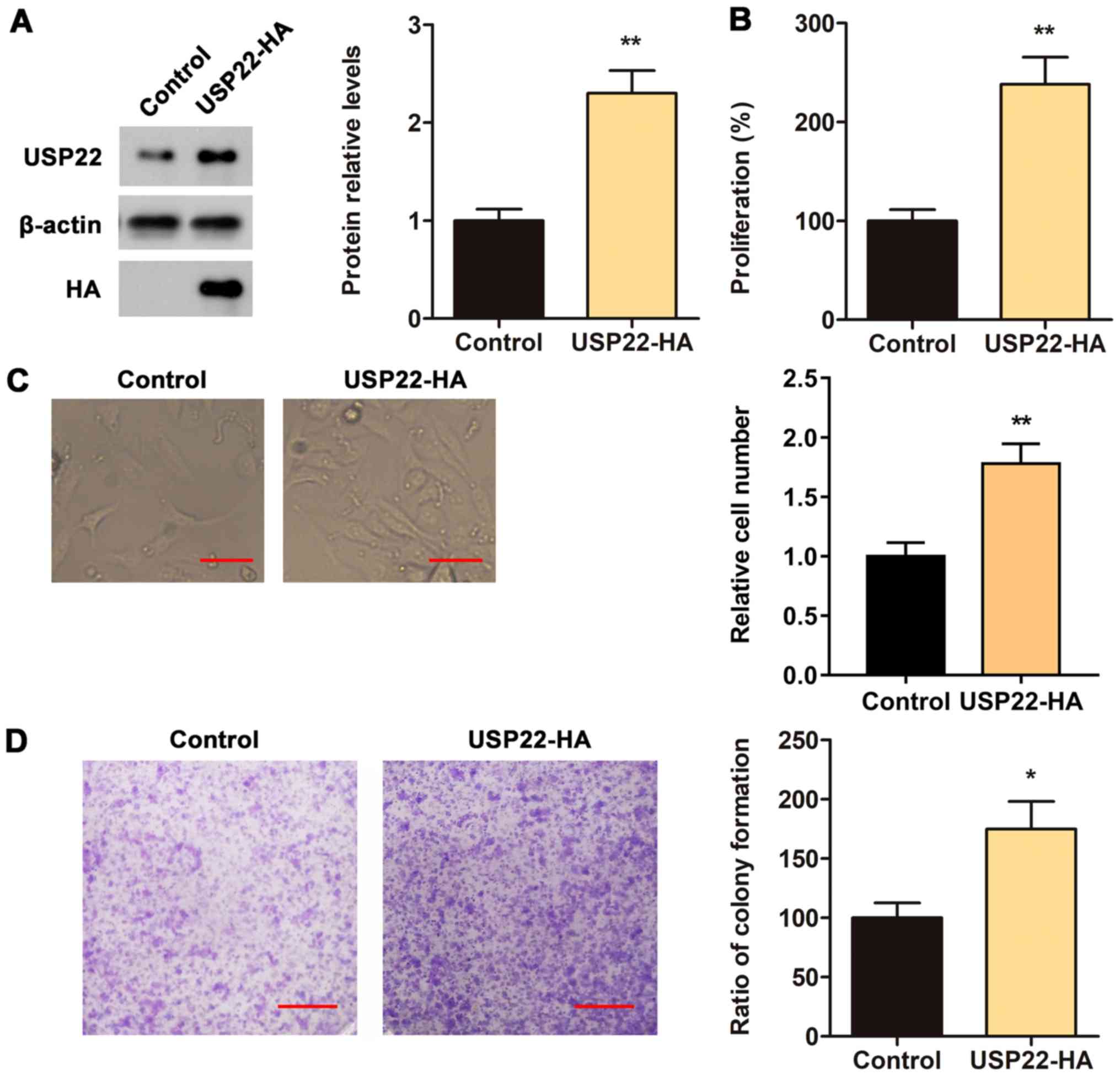
Overexpression of USP22 promotes the
proliferation and monoclonal colony formation of ACHN cells
To confirm the aforementioned findings described, a
USP22-HA vector was constructed to test whether the overexpression
of USP22 promotes the proliferation of ACHN cells. First, the
expression of the USP22-HA vector was validated via western
blotting. The results indicated that USP22-HA plasmids
significantly increased the protein level of USP22 in ACHN cells
(Fig. 3A). CCK8, cell number
counting and monoclonal colony formation assays were performed to
determine the proliferation rate of the cells. The results
demonstrated that the overexpression of USP22 significantly
increased the proliferation rate and the cell cloning rate of ACHN
cells (Fig. 3B-D). These data
further confirmed that USP22 may promote the proliferation of
RCC.
USP22 regulates the protein levels of
survivin and cleaved-caspase-3
A previous report has revealed that USP22 regulates
the proliferation of hepatocellular carcinoma via the survivin
signal pathway (20). In order to
study the mechanism of USP22 in regulating the proliferation of
RCC, the protein level of survivin was detected in ACHN cells
following USP22 knockdown or overexpression. It was revealed
that USP22 knockdown significantly decreased the protein
level of survivin, and that the overexpression of USP22 had
a reverse effect (Fig. 4A).
Furthermore, it was found that cleaved caspase-3, a downstream
protein of survivin, was decreased following USP22 knockdown
and increased by USP22 overexpression (Fig. 4A-D); however, no significant changes
were observed in the signal pathway of cell proliferation,
including p-AKT, AKT, p-Erk, Erk and ratio of p-AKT/AKT and
p-Erk/Erk (Fig. 4A). Notably, it was
demonstrated that the mRNA level of SURVIVIN in ACHN cells
was not changed by either knockdown or overexpression of
USP22 (Fig. 4E). Furthermore,
SURVIVIN overexpression rescued the effect of USP22
shRNA on cell proliferation and monoclonal colony formation of ACHN
cells (Fig. 4F and G). These results
indicate that USP22 regulates ACHN cell expansion via modulating
survivin at the post-transcriptional level.
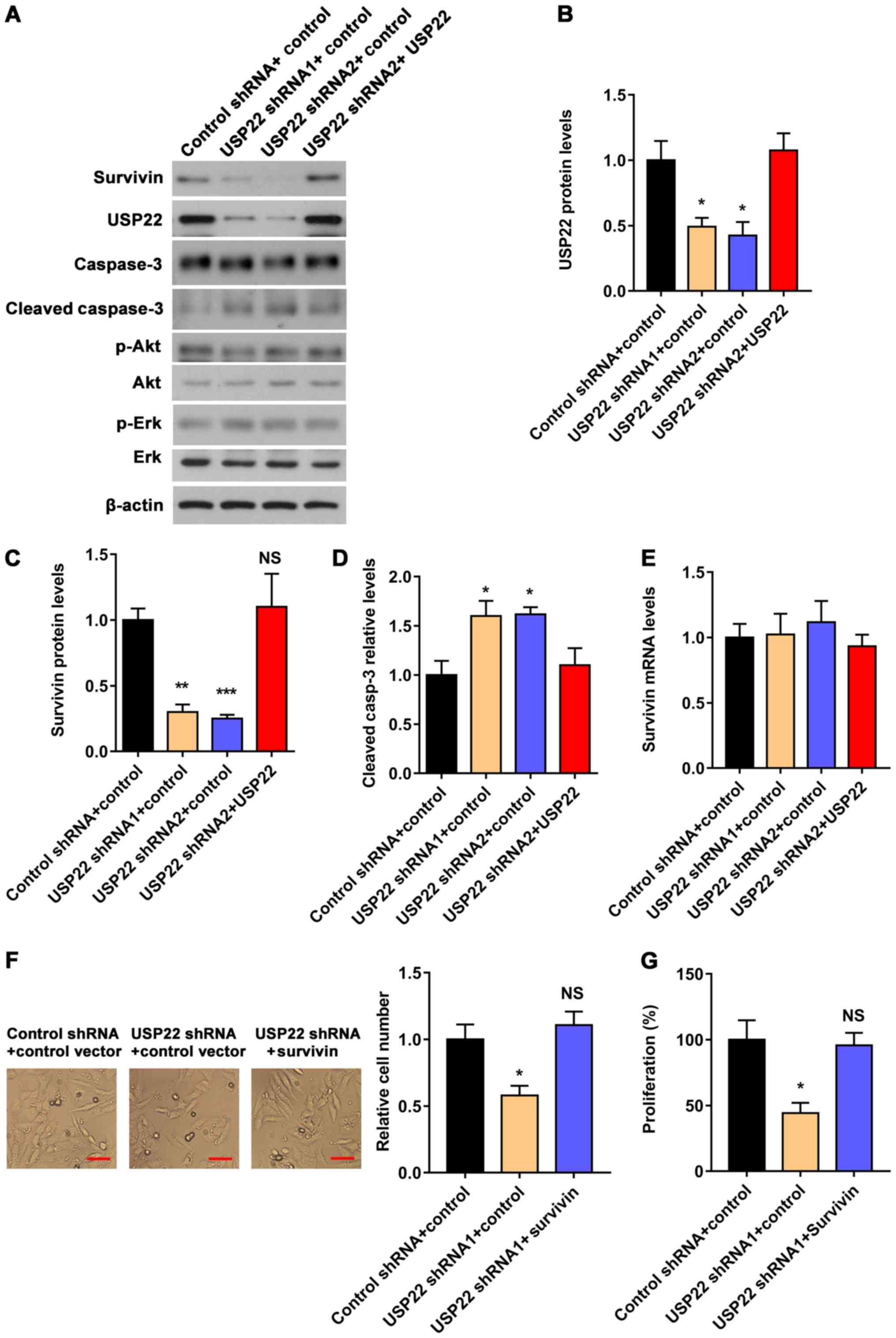 | Figure 4.USP22 regulates the expression of
survivin and the cleavage of caspase-3 but does not affect
SURVIVIN at the mRNA level. (A) Western blot analysis of the
protein levels of survivin, USP22, caspase-3, cleaved caspase-3,
Akt, p-Akt, Erk and p-Erk in ACHN cells transfected with control
shRNA, USP22 shRNA1, USP22 shRNA2 or USP22-HA. (B)
Quantitative analysis of the USP22 protein levels. (C) Quantitative
analysis of the survivin protein levels. (D) Quantitative analysis
of the cleaved caspase-3 levels. (E) Reverse
transcription-quantitative PCR analysis of the mRNA levels of
survivin in ACHN cells transfected with control shRNA, USP22
shRNA1, USP22 shRNA2 or USP22-HA. (F) Effect of
SURVIVIN overexpression on cell numbers in
USP22-knockdown ACHN cells. Scale bar, 20 µm. (G) Effect of
SURVIVIN overexpression on the proliferation of
USP22-knockdown ACHN cells by CCK8 assay. Data are presented
as mean ± standard error of mean. n=4. *P<0.05, **P<0.01 and
***P<0.001 as determined by one-way ANOVA. USP22,
ubiquitin-specific protease 22; sh-, short hairpin. |
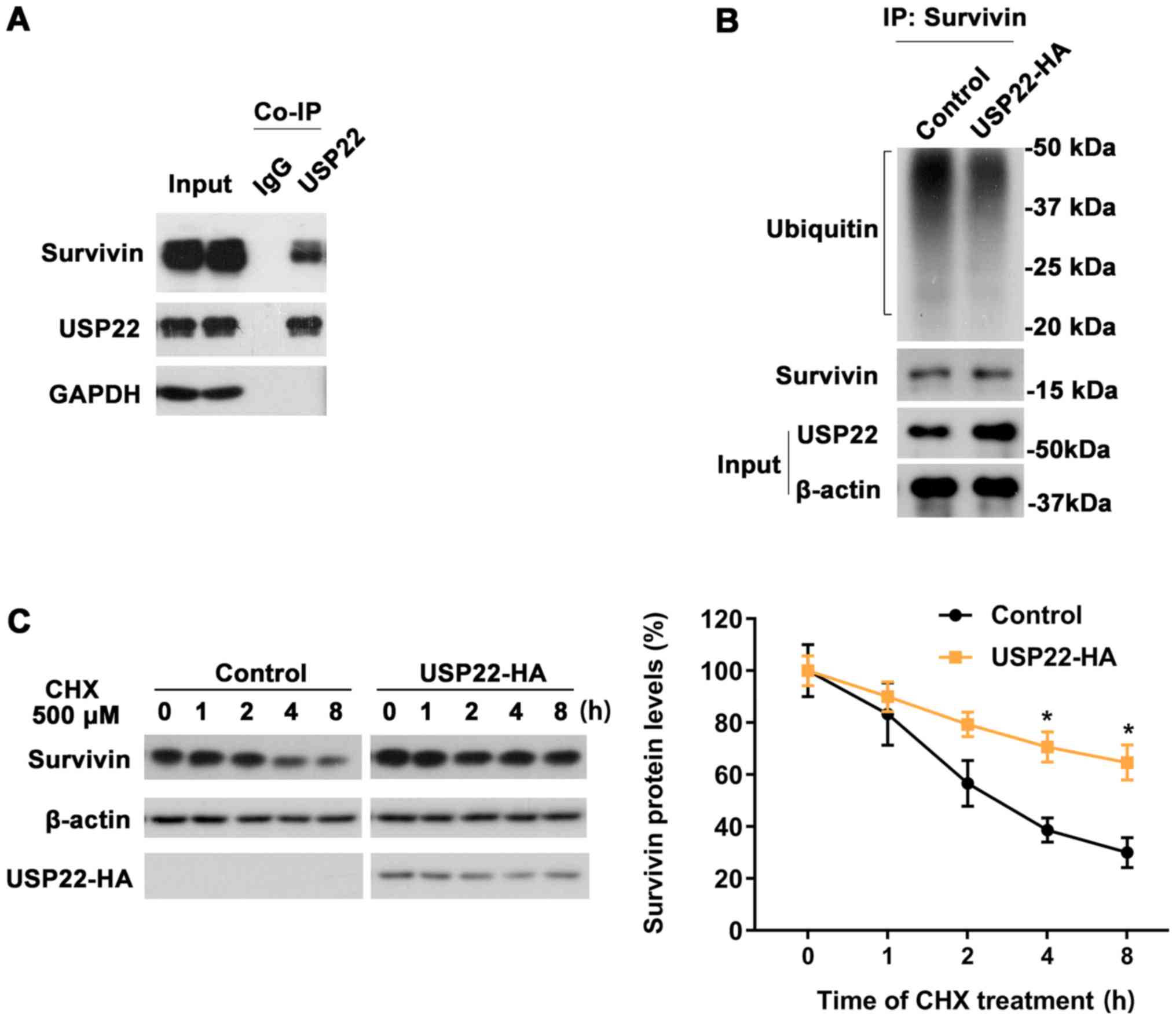
USP22 interacts with survivin and
regulates the ubiquitination of survivin
To further investigate the association between USP22
and survivin, the interaction between USP22 and survivin was
evaluated via co-immunoprecipitation. It was revealed that the
USP22 antibody, but not control IgG, could co-IP with survivin,
suggesting that USP22 interacts with survivin (Fig. 5A). Next, the effect of USP22 on the
ubiquitination of surviving was examined. ACHN cells were
transfected with either USP22-HA or control plasmid. After 48 h of
expression, the two groups were immunoprecipitated with
anti-survivin antibodies and immunoblotted with an anti-ubiquitin
antibody. The results indicated that the ubiquitination level of
survivin was decreased after the overexpression of USP22 (Fig. 5B). Cycloheximide chase assays were
performed in 293T cells and it was revealed that the degradation of
survivin was markedly decreased following overexpression of USP22
(Fig. 5C). Together, these results
indicated that USP22 was associated with survivin and regulated the
stability of survivin via modulating its ubiquitination level
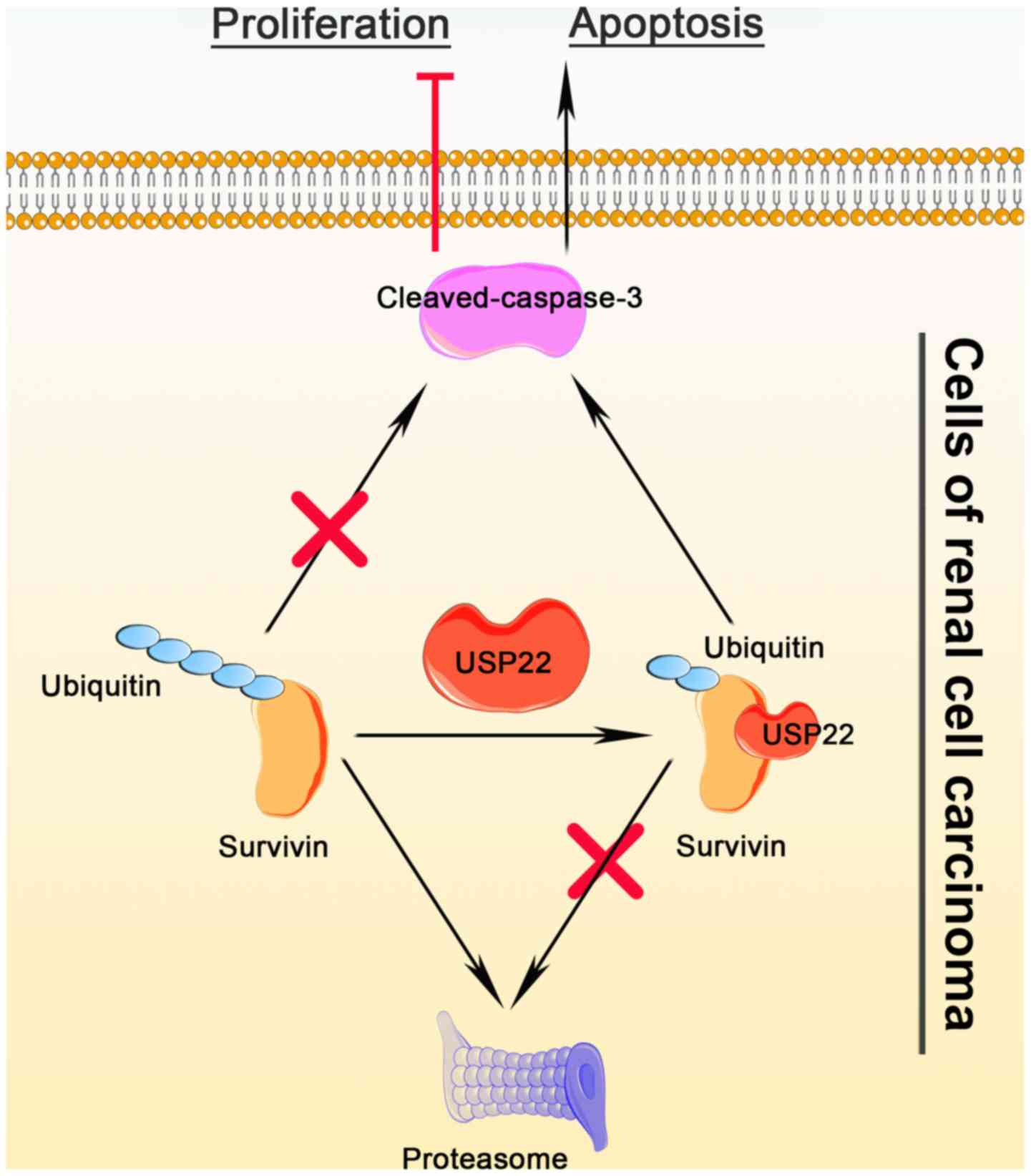
(Fig. 6).
Discussion
The present study reported, for the first time to
the best of the authors' knowledge, that USP22 is highly expressed
in RCC tissues. Upregulation of USP22 has been observed in various
cancers (21–23). Together with the present study, these
findings suggest that a high level of USP22 may be a common
characteristic in cancers. To investigate the roles of USP22 in
RCC, USP22 shRNA and USP22-HA vector were used to manipulate
the expression of USP22 in ACHN cells, and it was revealed
that the proliferation and monoclonal colony formation abilities of
tumor cells were inhibited via USP22 knockdown and promoted
via USP22 overexpression. These in vitro data support
the hypothesis that USP22 may be used as a therapeutic target for
RCC.
Previous studies demonstrate survivin is highly
expressed in numerous tumors, including RCC (14,21–23).
This was confirmed using RCC tissues and it was revealed that
survivin and USP22 have similar expression patterns. Survivin
regulates cell division and is associated with the development of
tumors (9,24,25). The
present study hypothesized that during the development of RCC,
USP22 upregulates survivin, and thus decreases the apoptosis of RCC
cells. To explore the association between USP22 and survivin, the
present study examined the survivin protein level after
manipulating the expression of USP22 using shRNAs. It was
demonstrated that USP22 knockdown significantly decreased
the protein level of survivin, and USP22 overexpression had
the opposite effect, suggesting USP22 as a novel modulator of
survivin. Notably, the mRNA level of survivin was unaltered,
indicating that USP22 mediates survivin protein at the
post-transcriptional level.
The interactions between USP22 and survivin were
investigated via a Co-IP assay and it was found that USP22 directly
or indirectly binds survivin. This is the first time that USP22 has
been reported to be associated with survivin. USP22 is a
deubiquitinating enzyme involved in regulating the ubiquitination
of many disease-associated proteins (21). It was revealed that USP22 stabilized
survivin via deubiquitination. In addition, the upregulation of the
apoptosis inhibitor survivin was accompanied by a decrease in the
cleaved caspase-3 level, which may explain why the proliferation
and colony formation of RCC cells were enhanced following the
overexpression of survivin. The finding that survivin expression
rescued the effect of USP22 knockdown in cell proliferation also
supported the conclusion that the effect on proliferation of USP22
depends on survivin. Future studies will focus on the research and
development of small-molecule inhibitors for USP22 as a novel
molecular therapeutic approach for RCC.
In conclusion, the present study demonstrated that
USP22 decreased apoptosis in RCC via modulating survivin stability.
The findings indicated that USP22 may be used as a novel
therapeutic target for patients with renal cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Foundation of
the Fifth Hospital of Xiamen (grant no. 2018YJ001), the Fujian
Province Medical Innovation Project (grant no. 2017-CXB-22), the
Fujian Province Natural Science Foundation Project (grant no.
2018D0022) and the Fujian Science and Technology Plan Guiding
Projects (grant no. 2019D026).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YL, MZ and JH were guarantors of integrity for the
entire study. YL, HZ, BS were responsible for the conception of the
study, and YL, HZ, BS, YP and FL were responsible for the study
design. Definition of intellectual content was conducted by YL, HZ
and BS. Literature research was performed by YL, HZ, BS and YP,
clinical studies by MC, MZ and JH, and experimental studies by YL,
HZ, BS, YP and FL. Data acquisition was conducted by YL, HZ, BS and
YP, and data analysis by YL, HZ, BS and YP. YL, HZ, BS and YP
conducted statistical analyses. YL, MZ and JH prepared the
manuscript and the manuscript was edited by YL, MZ and JH. The
manuscript was reviewed by YL, MZ and JH. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The human studies were approved by the ethical
review board at the Fifth Hospital of Xiamen and patients provided
written informed consent in accordance with the legal and
institutional ethical guidelines defined by the hospital by the
Ethics Committee of the Fifth Hospital of Xiamen.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barata PC and Rini BI: Treatment of renal
cell carcinoma: Current status and future directions. CA Cancer J
Clin. 67:507–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Motzer RJ, Penkov K, Haanen J, Rini B,
Albiges L, Campbell MT, Venugopal B, Kollmannsberger C, Negrier S,
Uemura M, et al: Avelumab plus axitinib versus sunitinib for
advanced renal-cell carcinoma. N Engl J Med. 380:1103–1115. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dabestani S, Marconi L, Hofmann F, Stewart
F, Lam TB, Canfield SE, Staehler M, Powles T, Ljungberg B and Bex
A: Local treatments for metastases of renal cell carcinoma: A
systematic review. Lancet Oncol. 15:e549–e561. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ricketts CJ and Linehan WM: Multi-regional
sequencing elucidates the evolution of clear cell renal cell
carcinoma. Cell. 173:540–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang X, Zang H, Luo Y, Wu J, Fang Z, Zhu W
and Li Y: High expression of USP22 predicts poor prognosis and
advanced clinicopathological features in solid tumors: A
meta-analysis. Onco Targets Ther. 11:3035–3046. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ao N, Wang L and Liu Y: Prognostic and
clinicopathological significance of ubiquitin-specific protease 22
overexpression in cancers: Evidence from a meta-analysis. Onco
Targets Ther. 10:5533–5540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Melo-Cardenas J, Xu Y, Wei J, Tan C, Kong
S, Gao B, Montauti E, Kirsammer G, Licht JD, Yu J, et al: USP22
deficiency leads to myeloid leukemia upon oncogenic Kras activation
through a PU.1-dependent mechanism. Blood. 132:423–434. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim PJ, Plescia J, Clevers H, Fearon ER
and Altieri DC: Survivin and molecular pathogenesis of colorectal
cancer. Lancet. 362:205–209. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arber C, Feng X, Abhyankar H, Romero E, Wu
MF, Heslop HE, Barth P, Dotti G and Savoldo B: Survivin-specific T
cell receptor targets tumor but not T cells. J Clin Invest.
125:157–168. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Campbell CS and Desai A: Tension sensing
by Aurora B kinase is independent of survivin-based centromere
localization. Nature. 497:118–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saenz DT, Fiskus W, Manshouri T, Mill CP,
Qian Y, Raina K, Rajapakshe K, Coarfa C, Soldi R, Bose P, et al:
Targeting nuclear β-catenin as therapy for post-myeloproliferative
neoplasm secondary AML. Leukemia. 33:1373–1386. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krambeck AE, Dong H, Thompson RH, Kuntz
SM, Lohse CM, Leibovich BC, Blute ML, Sebo TJ, Cheville JC, Parker
AS and Kwon ED: Survivin and b7-h1 are collaborative predictors of
survival and represent potential therapeutic targets for patients
with renal cell carcinoma. Clin Cancer Res. 13:1749–1756. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiong C, Liu H, Chen Z, Yu Y and Liang C:
Prognostic role of survivin in renal cell carcinoma: A system
review and meta-analysis. Eur J Intern Med. 33:102–107. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altieri DC: Survivin and apoptosis
control. Adv Cancer Res. 88:31–52. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vong QP, Cao K, Li HY, Iglesias PA and
Zheng Y: Chromosome alignment and segregation regulated by
ubiquitination of survivin. Science. 310:1499–1504. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goldstein NS: The current state of renal
cell carcinoma grading. Union internationale contre le cancer
(UICC) and the American joint committee on cancer (AJCC). Cancer.
80:977–980. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng F, Luo F, Lv S, Zhang H, Cao C, Chen
X, Wang S, Li Z, Wang X, Dou X, et al: A monoclonal antibody
targeting neuropilin-1 inhibits adhesion of MCF7 breast cancer
cells to fibronectin by suppressing the FAK/p130cas signaling
pathway. Anticancer Drugs. 25:663–672. 2014.PubMed/NCBI
|
|
19
|
Cikos S, Bukovska A and Koppel J: Relative
quantification of mRNA: Comparison of methods currently used for
real-time PCR data analysis. BMC Mol Biol. 8:1132007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang B, Liang X, Tang F, Zhang J, Zeng S,
Jin S, Zhou L, Kudo Y and Qi G: Expression of USP22 and survivin is
an indicator of malignant behavior in hepatocellular carcinoma. Int
J Oncol. 47:2208–2216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Melo-Cardenas J, Zhang Y, Zhang DD and
Fang D: Ubiquitin-specific peptidase 22 functions and its
involvement in disease. Oncotarget. 7:44848–44856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ning J, Zhang J, Liu W, Lang Y, Xue Y and
Xu S: Overexpression of ubiquitin-specific protease 22 predicts
poor survival in patients with early-stage non-small cell lung
cancer. Eur J Histochem. 56:e462012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu T, Liu J, Chen Q, Jin S, Mi S, Shao W,
Kudo Y, Zeng S and Qi G: Expression of USP22 and the chromosomal
passenger complex is an indicator of malignant progression in oral
squamous cell carcinoma. Oncol Lett. 17:2040–2046. 2019.PubMed/NCBI
|
|
24
|
Melucci E, Cosimelli M, Carpanese L, Pizzi
G, Izzo F, Fiore F, Golfieri R, Giampalma E, Sperduti I, Ercolani
C, et al: Decrease of survivin, p53 and Bcl-2 expression in
chemorefractory colorectal liver metastases may be predictive of
radiosensivity radiosensivity after radioembolization with
yttrium-90 resin microspheres. J Exp Clin Cancer Res. 32:132013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li G, Xie B, Li X, Chen Y, Wang Q, Xu Y,
Xu-Welliver M and Zou L: Down-regulation of survivin and
hypoxia-inducible factor-1 α by β-elemene enhances the
radiosensitivity of lung adenocarcinoma xenograft. Cancer Biother
Radiopharm. 27:56–64. 2012. View Article : Google Scholar : PubMed/NCBI
|