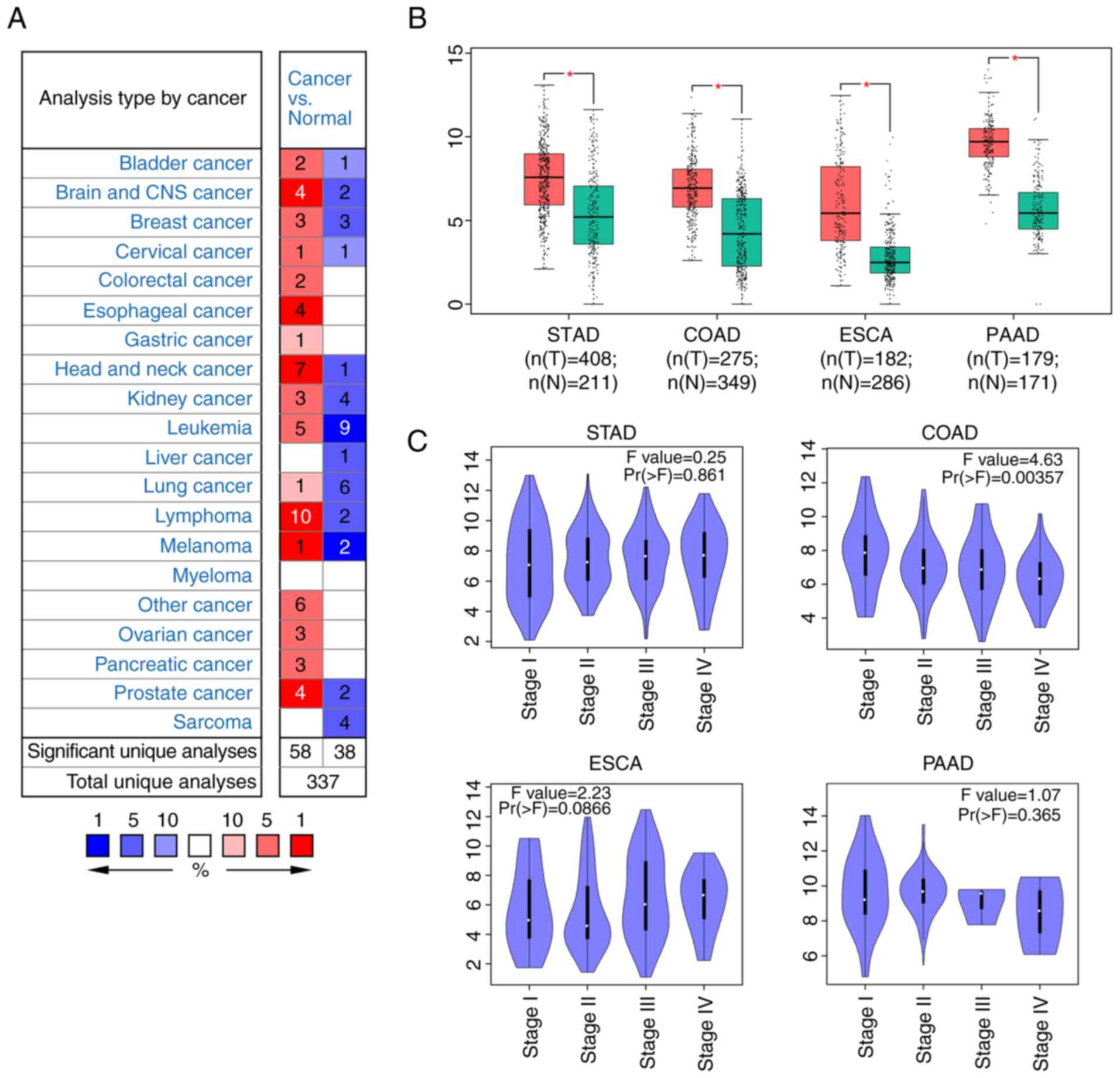
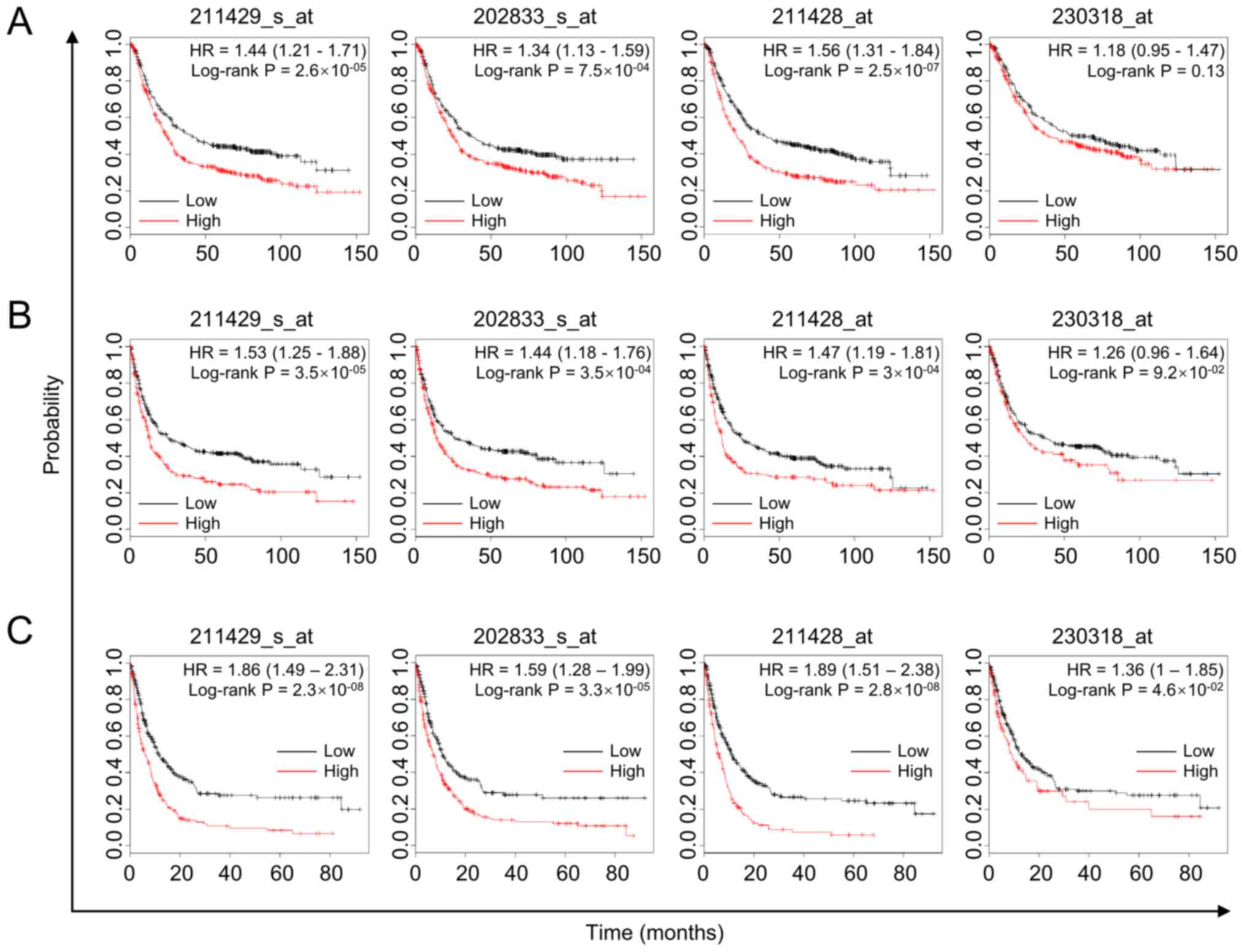
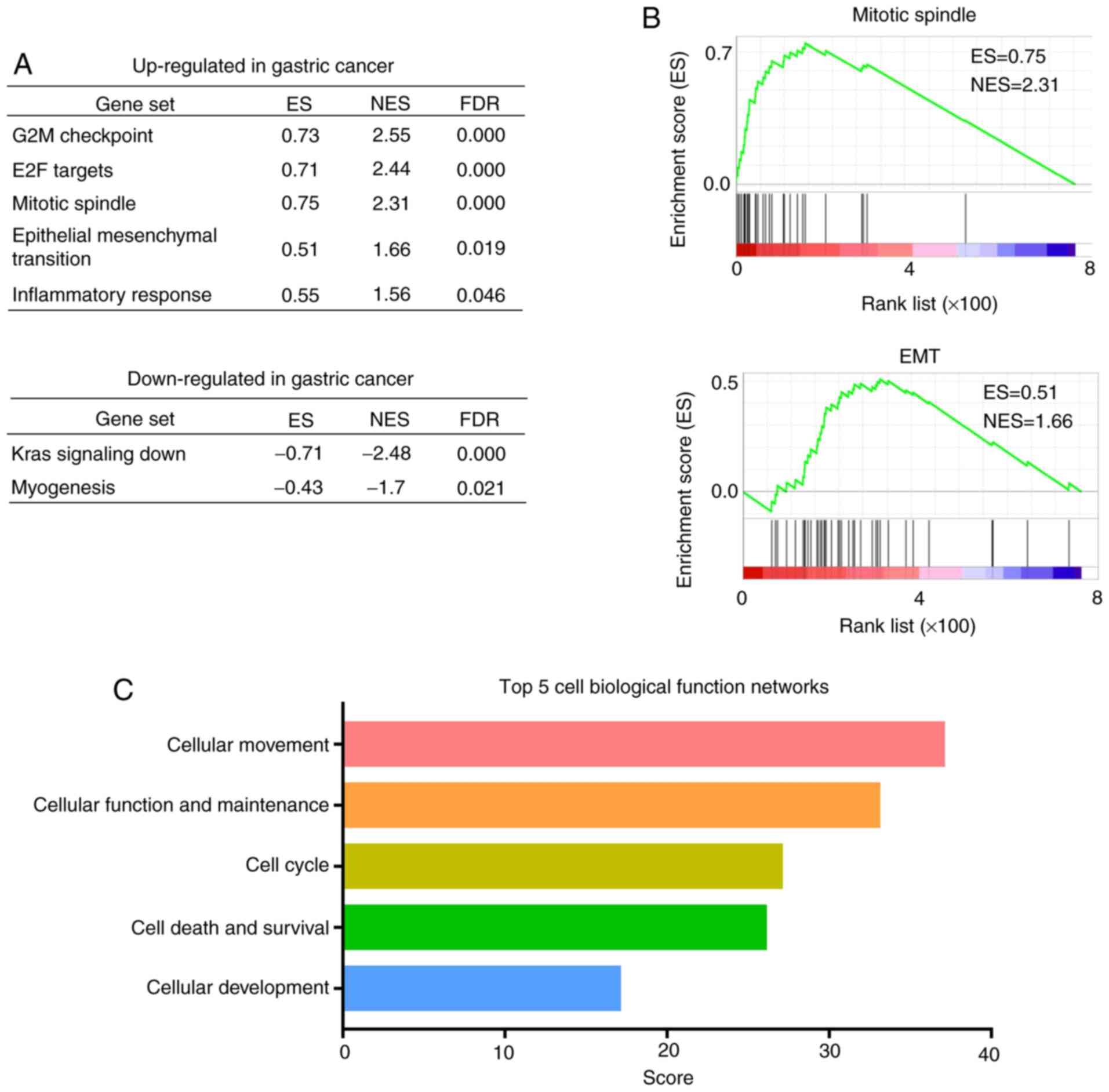
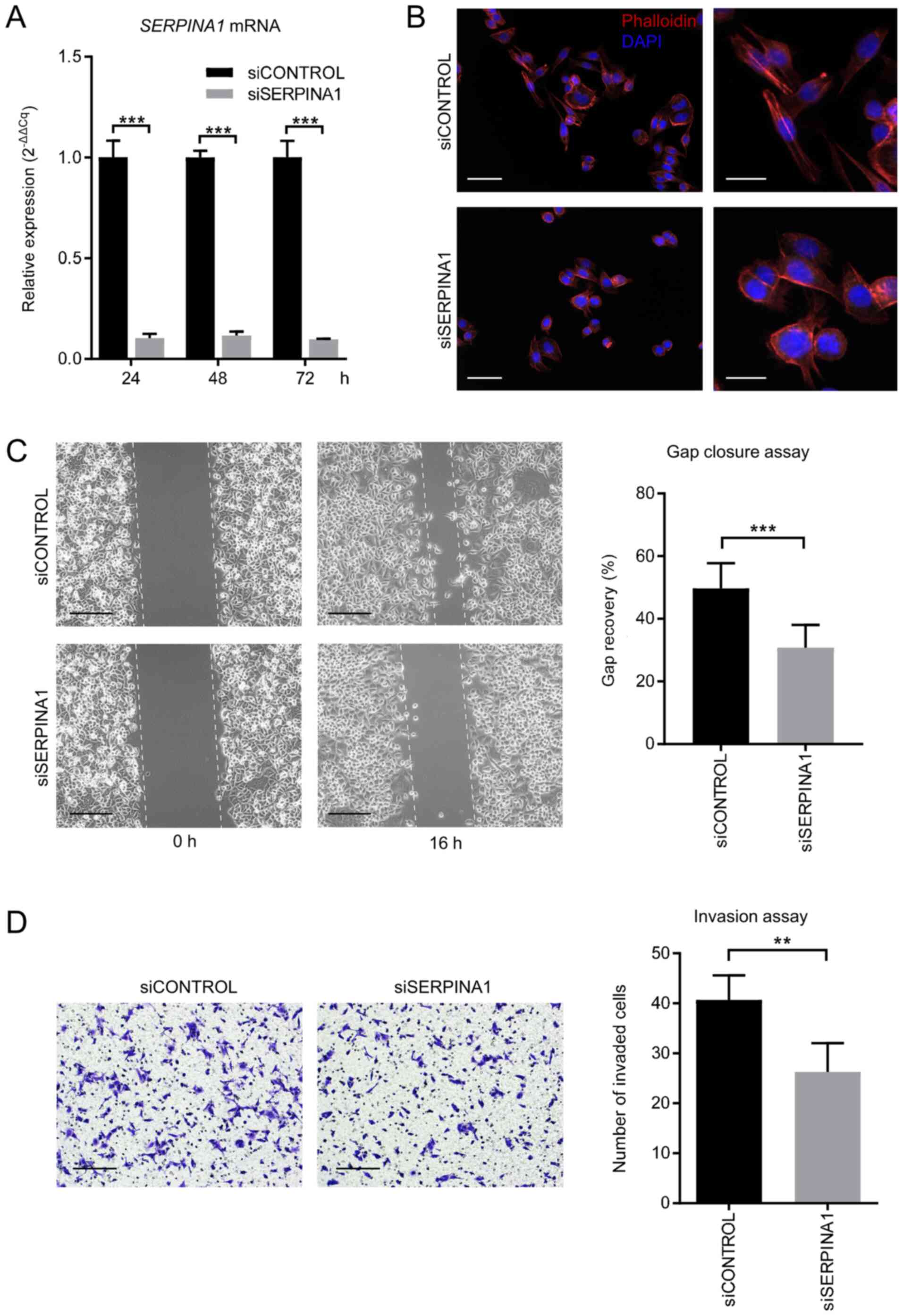
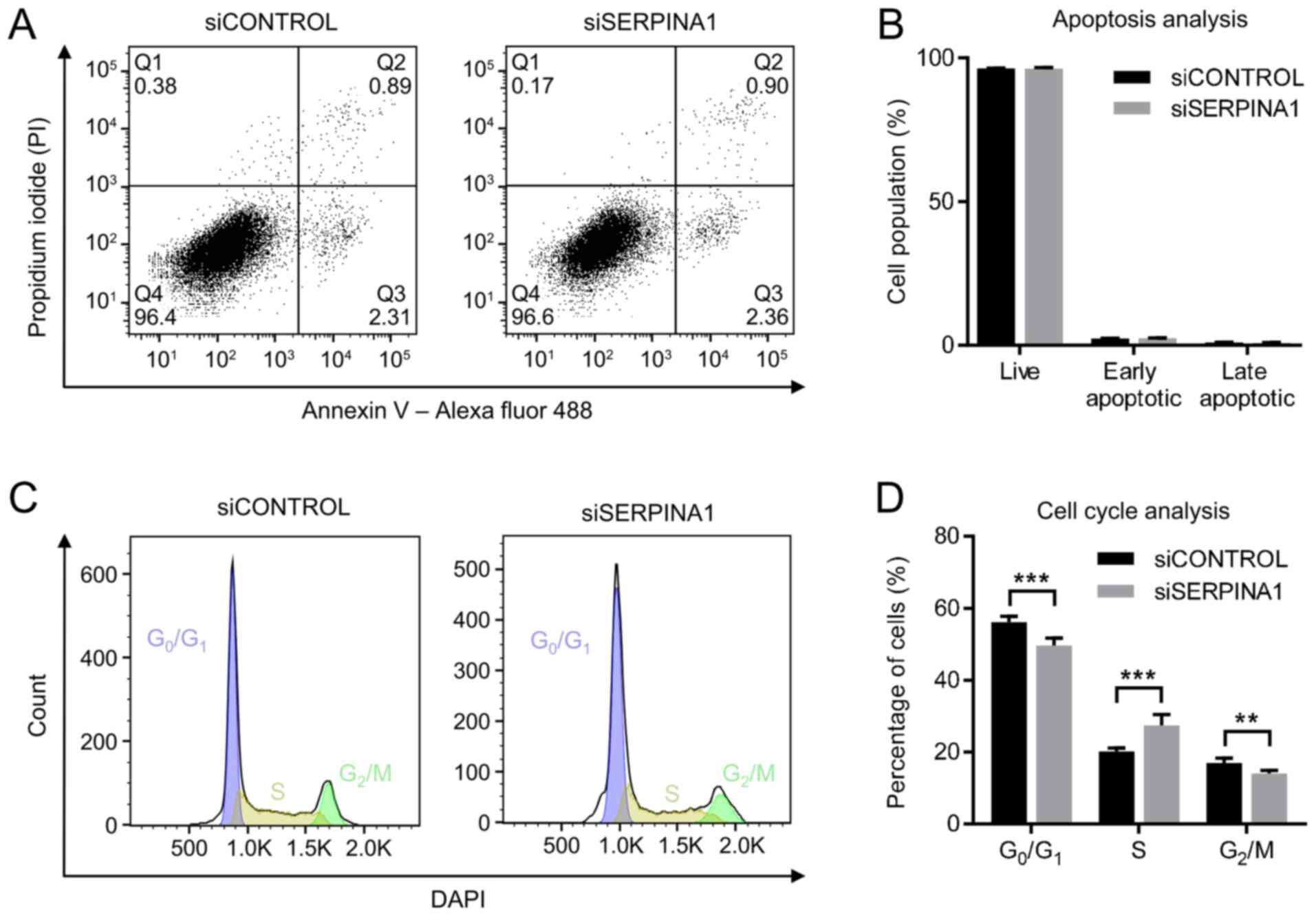
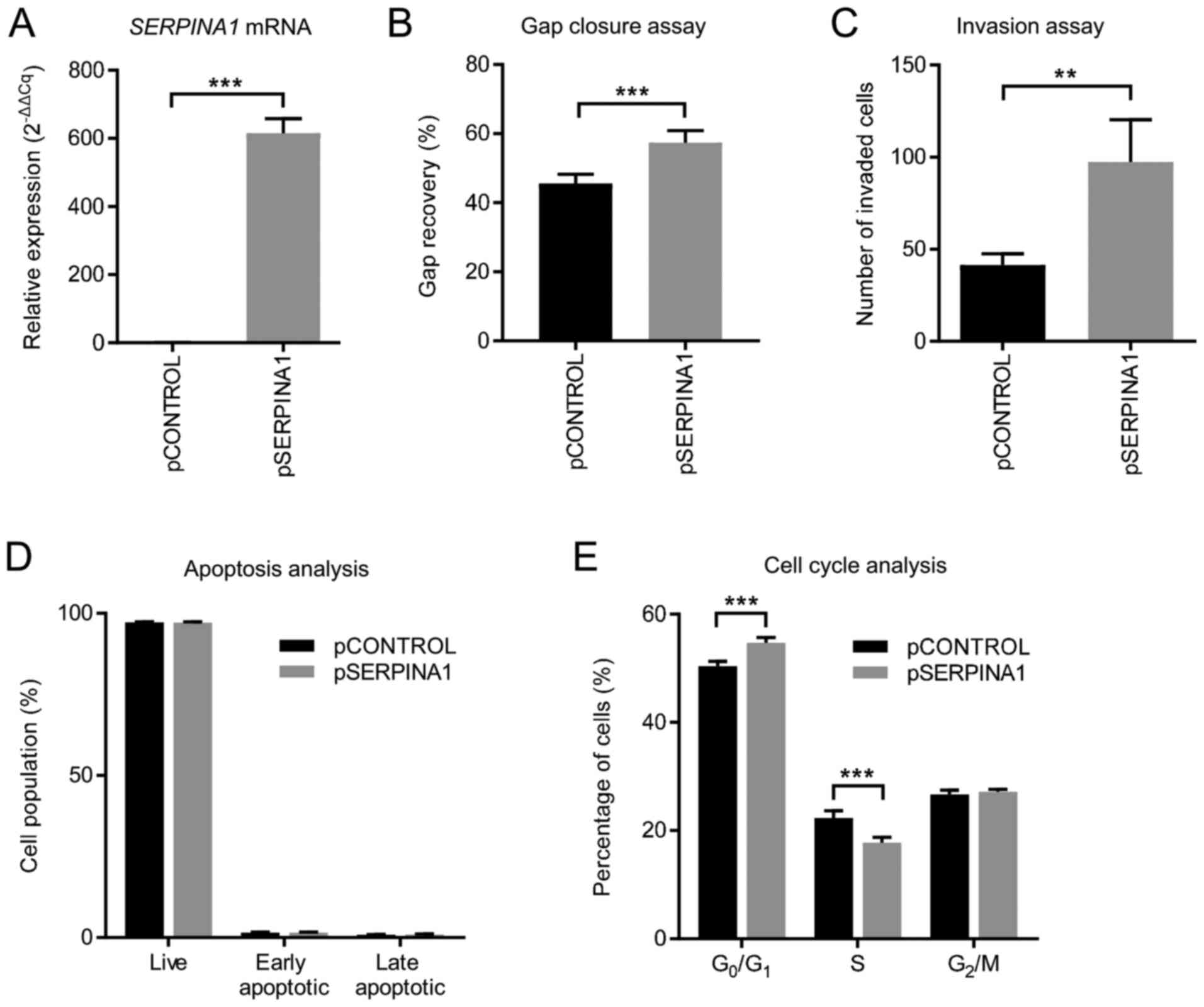
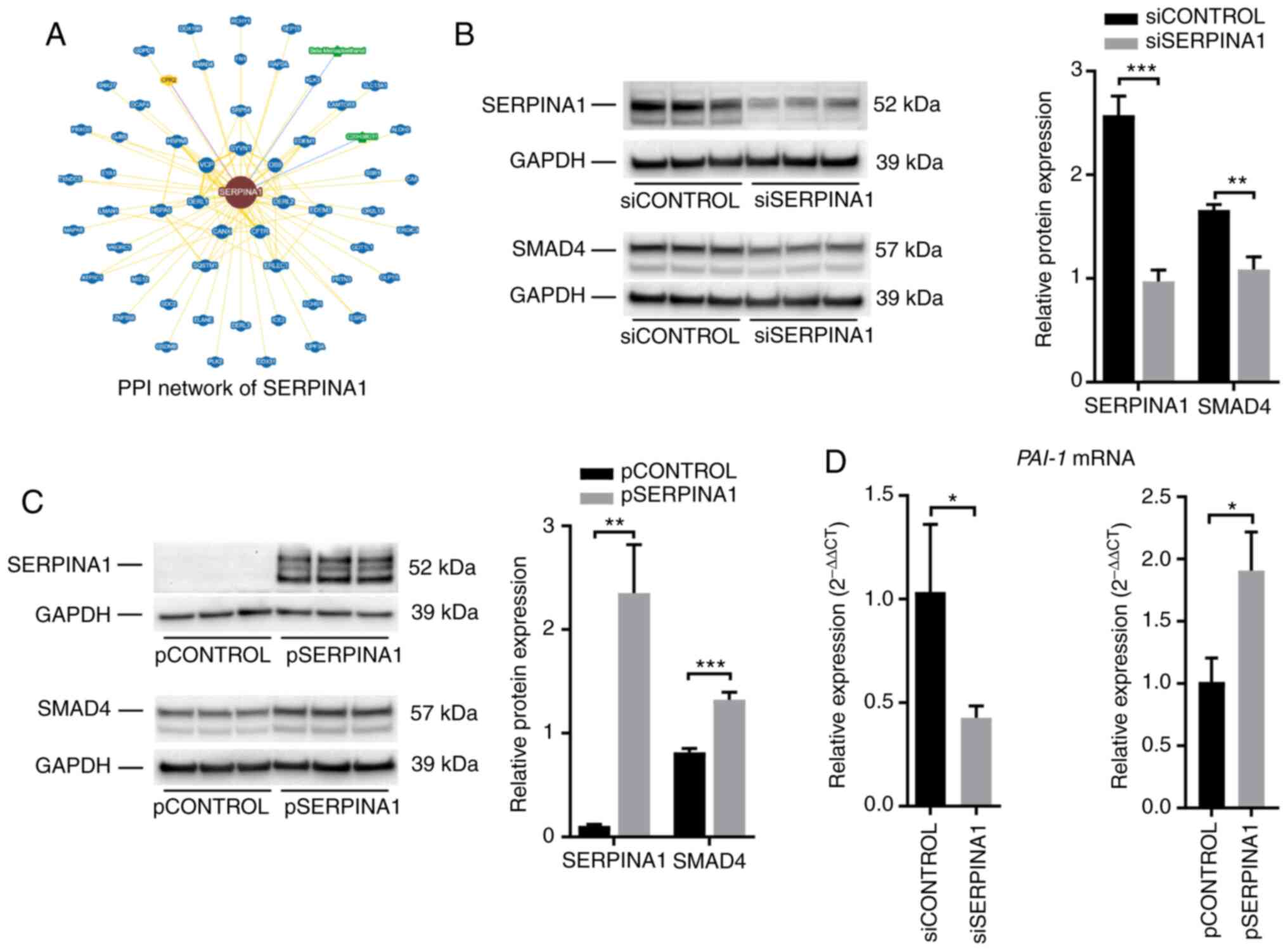
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI
|
|
2
|
Ben Kridis W, Marrekchi G, Mzali R, Daoud
J and Khanfir A: Prognostic factors in metastatic gastric
carcinoma. Exp Oncol. 41:173–175. 2019.PubMed/NCBI
|
|
3
|
Matsuoka T and Yashiro M: Biomarkers of
gastric cancer: Current topics and future perspective. World J
Gastroentero. 24:2818–2832. 2018.
|
|
4
|
Chia NY and Tan P: Molecular
classification of gastric cancer. Ann Oncol. 27:763–769.
2016.PubMed/NCBI
|
|
5
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013.PubMed/NCBI
|
|
6
|
Chen K, Yang D, Li X, Sun B, Song F, Cao
W, Brat DJ, Gao Z, Li H, Liang H, et al: Mutational landscape of
gastric adenocarcinoma in Chinese: Implications for prognosis and
therapy. Proc Natl Acad Sci USA. 112:1107–1112. 2015.PubMed/NCBI
|
|
7
|
Boku N: HER2-positive gastric cancer.
Gastric Cancer. 17:1–12. 2014.PubMed/NCBI
|
|
8
|
Oh DY and Bang YJ: HER2-targeted
therapies-a role beyond breast cancer. Nat Rev Clin Oncol.
17:33–48. 2020.PubMed/NCBI
|
|
9
|
Pickup M, Novitskiy S and Moses HL: The
roles of TGFβ in the tumour microenvironment. Nat Rev Cancer.
13:788–799. 2013.PubMed/NCBI
|
|
10
|
Batlle E and Massagué J: Transforming
growth factor-β signaling in immunity and cancer. Immunity.
50:924–940. 2019.PubMed/NCBI
|
|
11
|
Zhao M, Mishra L and Deng CX: The role of
TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 14:111–123.
2018.PubMed/NCBI
|
|
12
|
Vincent T, Neve EP, Johnson JR, Kukalev A,
Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL,
et al: A SNAIL1- SMAD3/4 transcriptional repressor complex promotes
TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol.
11:943–950. 2009.PubMed/NCBI
|
|
13
|
Bracken CP, Gregory PA, Kolesnikoff N,
Bert AG, Wang J, Shannon MF and Goodall GJ: A double-negative
feedback loop between ZEB1-SIP1 and the microRNA-200 family
regulates epithelial-mesenchymal transition. Cancer Res.
68:7846–7854. 2008.PubMed/NCBI
|
|
14
|
Thuault S, Valcourt U, Petersen M,
Manfioletti G, Heldin CH and Moustakas A: Transforming growth
factor-beta employs HMGA2 to elicit epithelial-mesenchymal
transition. J Cell Biol. 174:175–183. 2006.PubMed/NCBI
|
|
15
|
Zhang H, Liu L, Wang Y, Zhao G, Xie R, Liu
C, Xiao X, Wu K, Nie Y, Zhang H and Fan D: KLF8 involves in
TGF-beta-induced EMT and promotes invasion and migration in gastric
cancer cells. J Cancer Res Clin Oncol. 139:1033–1042.
2013.PubMed/NCBI
|
|
16
|
Hawinkels LJ, Verspaget HW, van Duijn W,
van der Zon JM, Zuidwijk K, Kubben FJ, Verheijen JH, Hommes DW,
Lamers CB and Sier CF: Tissue level, activation and cellular
localisation of TGF-beta1 and association with survival in gastric
cancer patients. Br J Cancer. 97:398–404. 2007.PubMed/NCBI
|
|
17
|
Normandin K, Péant B, Le Page C, de
Ladurantaye M, Ouellet V, Tonin PN, Provencher DM and Mes-Masson
AM: Protease inhibitor SERPINA1 expression in epithelial ovarian
cancer. Clin Exp Metastas. 27:55–69. 2010.
|
|
18
|
Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK,
Jo HJ, Kim HS, Oh N, Song GA and Park DY: Snail and serpinA1
promote tumor progression and predict prognosis in colorectal
cancer. Oncotarget. 6:20312–20326. 2015.PubMed/NCBI
|
|
19
|
Ercetin E, Richtmann S, Delgado BM,
Gomez-Mariano G, Wrenger S, Korenbaum E, Liu B, DeLuca D, Kuhnel
MP, Jonigk D, et al: Clinical significance of SERPINA1 gene and its
encoded alpha1-antitrypsin protein in NSCLC. Cancers (Basel).
11:13062019.
|
|
20
|
Janciauskiene S: Conformational properties
of serine proteinase inhibitors (serpins) confer multiple
pathophysiological roles. Biochim Biophys Acta. 1535:221–235.
2001.PubMed/NCBI
|
|
21
|
Farshchian M, Kivisaari A, Ala-aho R,
Riihilä P, Kallajoki M, Grénman R, Peltonen J, Pihlajaniemi T,
Heljasvaara R and Kähäri VM: Serpin peptidase inhibitor Clade A
member 1 (SerpinA1) is a novel biomarker for progression of
cutaneous squamous cell carcinoma. Am J Pathol. 179:1110–1119.
2011.PubMed/NCBI
|
|
22
|
Chan HJ, Li H, Liu Z, Yuan YC, Mortimer J
and Chen S: SERPINA1 is a direct estrogen receptor target gene and
a predictor of survival in breast cancer patients. Oncotarget.
6:25815–25827. 2015.PubMed/NCBI
|
|
23
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004.PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45W:W98–W102. 2017.
|
|
25
|
Bass AJ, Thorsson V, Shmulevich I,
Reynolds SM, Miller M, Bernard B, Hinoue T, Laird PW, Curtis C,
Shen H, et al: Comprehensive molecular characterization of gastric
adenocarcinoma. Nature. 513:202–209. 2014.PubMed/NCBI
|
|
26
|
Muzny DM, Bainbridge MN, Chang K, Dinh HH,
Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF,
et al: Comprehensive molecular characterization of human colon and
rectal cancer. Nature. 487:330–337. 2012.PubMed/NCBI
|
|
27
|
Kim J, Bowlby R, Mungall AJ, Robertson AG,
Odze RD, Cherniack AD, Shih J, Pedamallu CS, Cibulskis C, Dunford
A, et al: Integrated genomic characterization of oesophageal
carcinoma. Nature. 541:169–174. 2017.PubMed/NCBI
|
|
28
|
Raphael BJ, Hruban RH, Aguirre AJ, Moffitt
RA, Yeh JJ, Stewart C, Robertson AG, Cherniack AD, Gupta M, Getz G,
et al: Integrated genomic characterization of pancreatic ductal
adenocarcinoma. Cancer Cell. 32:185–203.e13. 2017.PubMed/NCBI
|
|
29
|
Carithers LJ, Ardlie K, Barcus M, Branton
PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J,
Gelfand ET, et al: A novel approach to high-quality postmortem
tissue procurement: The GTEx project. Biopreserv Biobank.
13:311–319. 2015.PubMed/NCBI
|
|
30
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC cancer staging manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.PubMed/NCBI
|
|
31
|
Szasz AM, Lanczky A, Nagy A, Forster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabo A and Gyorffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016.PubMed/NCBI
|
|
32
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI
|
|
33
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI
|
|
34
|
Kramer A, Green J, Pollard J Jr and
Tugendreich S: Causal analysis approaches in ingenuity pathway
analysis. Bioinformatics. 30:523–530. 2014.PubMed/NCBI
|
|
35
|
Oughtred R, Stark C, Breitkreutz BJ, Rust
J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, et
al: The BioGRID interaction database: 2019 update. Nucleic Acids
Res. 47D:D529–D541. 2019.
|
|
36
|
Orii N and Ganapathiraju MK: Wiki-pi: A
web-server of annotated human protein-protein interactions to aid
in discovery of protein function. PLoS One. 7:e490292012.PubMed/NCBI
|
|
37
|
Kim B: Western blot techniques. Methods
Mol Biol. 1606:133–139. 2017.PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
39
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5:e130912010.PubMed/NCBI
|
|
40
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1263–1275. 2007.PubMed/NCBI
|
|
41
|
Kimchi ET, Posner MC, Park JO, Darga TE,
Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ,
Weichselbaum RR and Khodarev NN: Progression of Barrett's
metaplasia to adenocarcinoma is associated with the suppression of
the transcriptional programs of epidermal differentiation. Cancer
Res. 65:3146–3154. 2005.PubMed/NCBI
|
|
42
|
Kim SM, Park YY, Park ES, Cho JY, Izzo JG,
Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, et al: Prognostic
biomarkers for esophageal adenocarcinoma identified by analysis of
tumor transcriptome. PLoS One. 5:e150742010.PubMed/NCBI
|
|
43
|
Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan
DW, Tang HM and Peng ZH: Upregulated INHBA expression is associated
with poor survival in gastric cancer. Med Oncol. 29:77–83.
2012.PubMed/NCBI
|
|
44
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
45
|
Logsdon CD, Simeone DM, Binkley C,
Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R and Hanash
S: Molecular profiling of pancreatic adenocarcinoma and chronic
pancreatitis identifies multiple genes differentially regulated in
pancreatic cancer. Cancer Res. 63:2649–2657. 2003.PubMed/NCBI
|
|
46
|
Liberzon A, Birger C, Thorvaldsdottir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015.PubMed/NCBI
|
|
47
|
Faulstich H, Zobeley S, Rinnerthaler G and
Small JV: Fluorescent phallotoxins as probes for filamentous actin.
J Muscle Res Cell Motil. 9:370–383. 1988.PubMed/NCBI
|
|
48
|
Deckers M, van Dinther M, Buijs J, Que I,
Lowik C, van der Pluijm G and ten Dijke P: The tumor suppressor
Smad4 is required for transforming growth factor beta-induced
epithelial to mesenchymal transition and bone metastasis of breast
cancer cells. Cancer Res. 66:2202–2209. 2006.PubMed/NCBI
|
|
49
|
Huang G, Du MY, Zhu H, Zhang N, Lu ZW,
Qian LX, Zhang W, Tian X, He X and Yin L: MiRNA-34a reversed
TGF-β-induced epithelial-mesenchymal transition via suppression of
SMAD4 in NPC cells. Biomed Pharmacother. 106:217–224.
2018.PubMed/NCBI
|
|
50
|
Yu C, Liu Y, Huang D, Dai Y, Cai G, Sun J,
Xu T, Tian Y and Zhang X: TGF-β1 mediates epithelial to mesenchymal
transition via the TGF-β/Smad pathway in squamous cell carcinoma of
the head and neck. Oncol Rep. 25:1581–1587. 2011.PubMed/NCBI
|
|
51
|
Levy L and Hill CS: Smad4 dependency
defines two classes of transforming growth factor β (TGF-β) target
genes and distinguishes TGF-β-induced epithelial-mesenchymal
transition from its antiproliferative and migratory responses. Mol
Cell Biol. 25:8108–8125. 2005.PubMed/NCBI
|
|
52
|
Bray F, Colombet M, Mery L, Piñeros M,
Znaor A, Zanetti R and Ferlay J: Cancer Incidence in Five
Continents. XI. International Agency for Research on Cancer; Lyon:
2017, http://ci5.iarc.frJuly
20–2020
|
|
53
|
Curado MP, Edwards B, Shin HR, Storm H,
Ferlay J, Heanue M and Boyle P: Cancer Incidence in Five
Continents. IX. International Agency for Research on Cancer; Lyon:
2007, http://ci5.iarc.frJuly
20–2020
|
|
54
|
Heit C, Jackson BC, McAndrews M, Wright
MW, Thompson DC, Silverman GA, Nebert DW and Vasiliou V: Update of
the human and mouse SERPIN gene superfamily. Hum Genomics.
7:222013.PubMed/NCBI
|
|
55
|
El-Akawi ZJ, Al-Hindawi FK and Bashir NA:
Alpha-1 antitrypsin (alpha1-AT) plasma levels in lung, prostate and
breast cancer patients. Neuro Endocrinol Lett. 29:482–484.
2008.PubMed/NCBI
|
|
56
|
Yang J, Xiong X, Wang X, Guo B, He K and
Huang C: Identification of peptide regions of SERPINA1 and ENOSF1
and their protein expression as potential serum biomarkers for
gastric cancer. Tumour Biol. 36:5109–5118. 2015.PubMed/NCBI
|
|
57
|
Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY,
Jo HJ, Kim DH, Kim GH and Park DY: Serpin peptidase inhibitor clade
A member 1 is a biomarker of poor prognosis in gastric cancer. Br J
Cancer. 111:1993–2002. 2014.PubMed/NCBI
|
|
58
|
Pollard TD: The cytoskeleton, cellular
motility and the reductionist agenda. Nature. 422:741–745.
2003.PubMed/NCBI
|
|
59
|
Byon CH, Hardy RW, Ren C, Ponnazhagan S,
Welch DR, McDonald JM and Chen Y: Free fatty acids enhance breast
cancer cell migration through plasminogen activator inhibitor-1 and
SMAD4. Lab Invest. 89:1221–1228. 2009.PubMed/NCBI
|
|
60
|
Hernanda PY, Chen K, Das AM, Sideras K,
Wang W, Li J, Cao W, Bots SJ, Kodach LL, de Man RA, et al: SMAD4
exerts a tumor-promoting role in hepatocellular carcinoma.
Oncogene. 34:5055–5068. 2015.PubMed/NCBI
|
|
61
|
Heldin CH, Miyazono K and Ten Dijke P:
TGF-beta signalling from cell membrane to nucleus through SMAD
proteins. Nature. 390:465–471. 1997.PubMed/NCBI
|
|
62
|
Komiyama T, Gron H, Pemberton PA and
Salvesen GS: Interaction of subtilisins with serpins. Protein Sci.
5:874–882. 1996.PubMed/NCBI
|
|
63
|
Christensson A, Laurell CB and Lilja H:
Enzymatic activity of prostate-specific antigen and its reactions
with extracellular serine proteinase inhibitors. Eur J Biochem.
194:755–763. 1990.PubMed/NCBI
|
|
64
|
Korkmaz B, Attucci S, Hazouard E,
Ferrandiere M, Jourdan ML, Brillard-Bourdet M, Juliano L and
Gauthier F: Discriminating between the activities of human
neutrophil elastase and proteinase 3 using serpin-derived
fluorogenic substrates. J Biol Chem. 277:39074–39081.
2002.PubMed/NCBI
|
|
65
|
Taggart C, Cervantes-Laurean D, Kim G,
McElvaney NG, Wehr N, Moss J and Levine RL: Oxidation of either
methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss
of anti-neutrophil elastase activity. J Biol Chem. 275:27258–27265.
2000.PubMed/NCBI
|
|
66
|
Greenblatt EJ, Olzmann JA and Kopito RR:
Derlin-1 is a rhomboid pseudoprotease required for the dislocation
of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat
Struct Mol Biol. 18:1147–1152. 2011.PubMed/NCBI
|
|
67
|
Hutchins JR, Toyoda Y, Hegemann B, Poser
I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA,
Bulkescher J, et al: Systematic analysis of human protein complexes
identifies chromosome segregation proteins. Science. 328:593–599.
2010.PubMed/NCBI
|
|
68
|
Wang J, Yuan Y, Zhou Y, Guo L, Zhang L,
Kuai X, Deng B, Pan Z, Li D and He F: Protein interaction data set
highlighted with human Ras-MAPK/PI3K signaling pathways. J Proteome
Res. 7:3879–3889. 2008.PubMed/NCBI
|
|
69
|
LaLonde DP and Bretscher A: The UBX
protein SAKS1 negatively regulates endoplasmic reticulum-associated
degradation and p97-dependent degradation. J Biol Chem.
286:4892–4901. 2011.PubMed/NCBI
|
|
70
|
Satoh T, Chen Y, Hu D, Hanashima S,
Yamamoto K and Yamaguchi Y: Structural basis for oligosaccharide
recognition of misfolded glycoproteins by OS-9 in ER-associated
degradation. Mol Cell. 40:905–916. 2010.PubMed/NCBI
|
|
71
|
Christianson JC, Shaler TA, Tyler RE and
Kopito RR: OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the
Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol.
10:272–282. 2008.PubMed/NCBI
|
|
72
|
Fujimori T, Kamiya Y, Nagata K, Kato K and
Hosokawa N: Endoplasmic reticulum lectin XTP3-B inhibits
endoplasmic reticulum-associated degradation of a misfolded
α1-antitrypsin variant. FEBS J. 280:1563–1575. 2013.PubMed/NCBI
|
|
73
|
Schaafhausen A, Rost S, Oldenburg J and
Muller CR: Identification of VKORC1 interaction partners by
split-ubiquitin system and coimmunoprecipitation. Thromb Haemost.
105:285–294. 2011.PubMed/NCBI
|
|
74
|
Mikami K, Yamaguchi D, Tateno H, Hu D, Qin
SY, Kawasaki N, Yamada M, Matsumoto N, Hirabayashi J, Ito Y and
Yamamoto K: The sugar-binding ability of human OS-9 and its
involvement in ER-associated degradation. Glycobiology. 20:310–321.
2010.PubMed/NCBI
|
|
75
|
Yamaguchi D, Hu D, Matsumoto N and
Yamamoto K: Human XTP3-B binds to alpha1-antitrypsin variant null
(Hong Kong) via the C-terminal MRH domain in a glycan-dependent
manner. Glycobiology. 20:348–355. 2010.PubMed/NCBI
|
|
76
|
Nagasawa K, Higashi T, Hosokawa N, Kaufman
RJ and Nagata K: Simultaneous induction of the four subunits of the
TRAP complex by ER stress accelerates ER degradation. EMBO Rep.
8:483–489. 2007.PubMed/NCBI
|
|
77
|
Moussavi-Harami SF, Annis DS, Ma W, Berry
SM, Coughlin EE, Strotman LN, Maurer LM, Westphall MS, Coon JJ,
Mosher DF and Beebe DJ: Characterization of molecules binding to
the 70 K N-terminal region of fibronectin by IFAST purification
coupled with mass spectrometry. J Proteome Res. 12:3393–3404.
2013.PubMed/NCBI
|
|
78
|
Huttlin EL, Ting L, Bruckner RJ, Gebreab
F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al:
The BioPlex network: A systematic exploration of the human
interactome. Cell. 162:425–440. 2015.PubMed/NCBI
|
|
79
|
Jung CH, Kim YH, Lee K and Im H: Retarded
protein folding of the human Z-type α1-antitrypsin
variant is suppressed by Cpr2p. Biochem Biophys Res Commun.
445:191–195. 2014.PubMed/NCBI
|
|
80
|
Huang CH, Hsiao HT, Chu YR, Ye Y and Chen
X: Derlin2 protein facilitates HRD1-mediated retro-translocation of
sonic hedgehog at the endoplasmic reticulum. J Biol Chem.
288:25330–25339. 2013.PubMed/NCBI
|
|
81
|
Fan W, Cheng J, Zhang S and Liu X: Cloning
and functions of the HBxAg-binding protein XBP1. Mol Med Rep.
7:618–622. 2013.PubMed/NCBI
|
|
82
|
Wang J, Huo K, Ma L, Tang L, Li D, Huang
X, Yuan Y, Li C, Wang W, Guan W, et al: Toward an understanding of
the protein interaction network of the human liver. Mol Syst Biol.
7:5362011.PubMed/NCBI
|
|
83
|
Feng L, Zhang J, Zhu N, Ding Q, Zhang X,
Yu J, Qiang W, Zhang Z, Ma Y, Huang D, et al: Ubiquitin ligase
SYVN1/HRD1 facilitates degradation of the SERPINA1 Z
variant/α-1-antitrypsin Z variant via SQSTM1/p62-dependent
selective autophagy. Autophagy. 13:686–702. 2017.PubMed/NCBI
|
|
84
|
Huttlin EL, Bruckner RJ, Paulo JA, Cannon
JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et
al: Architecture of the human interactome defines protein
communities and disease networks. Nature. 545:505–509.
2017.PubMed/NCBI
|
|
85
|
Wen JH, Wen H, Gibson-Corley KN and Glenn
KA: FBG1 is the final arbitrator of A1AT-Z degradation. PLoS One.
10:e1355912015.
|
|
86
|
Dersh D, Jones SM, Eletto D, Christianson
JC and Argon Y: OS-9 facilitates turnover of nonnative GRP94 marked
by hyperglycosylation. Mol Biol Cell. 25:2220–2234. 2014.PubMed/NCBI
|
|
87
|
Khodayari N, Wang RL, Marek G, Krotova K,
Kirst M, Liu C, Rouhani F and Brantly M: SVIP regulates Z variant
alpha-1 antitrypsin retro-translocation by inhibiting ubiquitin
ligase gp78. PLoS One. 12:e1729832017.
|
|
88
|
Xiao Y, Han J, Wang Q, Mao Y, Wei M, Jia W
and Wei L: A novel interacting protein SERP1 regulates the N-Linked
glycosylation and function of GLP-1 receptor in the liver. J Cell
Biochem. 118:3616–3626. 2017.PubMed/NCBI
|
|
89
|
Kadowaki H, Nagai A, Maruyama T, Takami Y,
Satrimafitrah P, Kato H, Honda A, Hatta T, Natsume T, Sato T, et
al: Pre-emptive quality control protects the ER from protein
overload via the proximity of ERAD components and SRP. Cell Rep.
13:944–956. 2015.PubMed/NCBI
|
|
90
|
Pankow S, Bamberger C, Calzolari D,
Martinez-Bartolome S, Lavallee-Adam M, Balch WE and Yates JR III:
∆F508 CFTR interactome remodelling promotes rescue of cystic
fibrosis. Nature. 528:510–516. 2015.PubMed/NCBI
|
|
91
|
Kadowaki H, Satrimafitrah P, Takami Y and
Nishitoh H: Molecular mechanism of ER stress-induced pre-emptive
quality control involving association of the translocon, Derlin-1,
and HRD1. Sci Rep. 8:73172018.PubMed/NCBI
|
|
92
|
Giurato G, Nassa G, Salvati A, Alexandrova
E, Rizzo F, Nyman TA, Weisz A and Tarallo R: Quantitative mapping
of RNA-mediated nuclear estrogen receptor β interactome in human
breast cancer cells. Sci Data. 5:1800312018.PubMed/NCBI
|
|
93
|
Yu S, Ito S, Wada I and Hosokawa N:
ER-resident protein 46 (ERp46) triggers the mannose-trimming
activity of ER degradation-enhancing α-mannosidase-like protein 3
(EDEM3). J Biol Chem. 293:10663–10674. 2018.PubMed/NCBI
|
|
94
|
Lamriben L, Oster ME, Tamura T, Tian W,
Yang Z, Clausen H and Hebert DN: EDEM1's mannosidase-like domain
binds ERAD client proteins in a redox-sensitive manner and
possesses catalytic activity. J Biol Chem. 293:13932–13945.
2018.PubMed/NCBI
|