Introduction
Lung cancer (LCa) remains one of the primary reasons
of cancer mediated fatalities globally, with 5-year survival rates
of <20% in 2017 (1). In the US
~234,030 new LCa cases were diagnosed and in China ~781,500 new LCa
cases were diagnosed in 2018 (2).
Non-small cell lung cancer (NSCLC) is the most prevalent subtype of
lung cancer, and lung adenocarcinoma (LUAD) is the major subtype of
NSCLC (3). In the United States,
LUAD accounted for ~50% of LC cases (43 and 52% in males and
females, respectively) between 1992–2013 (3). In spite of progress in NSCLC
therapeutics, the molecular mechanisms underlying NSCLC progression
and development are poorly understood (4,5). Further
mechanistic studies to fully understand NSCLC progression are
therefore urgently required.
lncRNAs (long non-coding RNAs) are a group of
sequences that have been used to study several cancers, such as
liver cancer, gastic cancer and colorectal carcinoma (6–8). lncRNAs
are members of a family of >200 nucleotides long linear
transcripts (9). They lack
protein-encoding function and were hence deemed genomic ‘noise’ and
‘junk’ (10). Several recent reports
have implicated lncRNAs in a range of genetic processes for gene
expression modulation at epigenetic, transcriptional and
post-transcriptional levels (11,12). In
NSCLC, lncRNAs are frequently abnormally expressed and are involved
in NSCLC occurrence and development (10–14).
Hence, a detailed study of lncRNAs and their role in NSCLC may lead
to unraveling potential targets to improve the prognosis and
treatment of patients with NSCLC.
Recently lncRNA B4GALT1-AS1 was found as the B4GALT1
antisense counterpart, lncRNA B4GALT1-AS1 shows significant
complementarity with B4GALT1 messenger RNA and enhanced colon
cancer cell stemness and migration (15). Therefore, the activity of B4GALT1-AS1
in NSCLC needed to be investigated. B4GALT1-AS1 was chosen as the
main focus of the present study, which aimed to study the
pathogenicity of NSCLC and offer insights for its therapy.
Materials and methods
Patient details and tissue
specimens
The Ethics Committee of The First Hospital of
Longyan City (Longyan, China; approval no. 2019-015) gave approval
for the present study which was performed according to the
Declaration of Helsinki. Written informed consent was obtained from
all the patients. A total of 56 patients with NSCLC, including 22
men and 34 women (mean age, 53 ± 6.2 years; age range, 32–76 years)
treated by surgical resection in the First Hospital of Longyan City
between January 2013 and December 2016 were recruited in the
present study. The inclusion criteria were as follows: i)
Histologically confirmed as NSCLC; ii) medical information was
completely recorded and iii) informed consent was obtained prior to
the study start. The exclusion criteria were as follows: i)
Combined with other malignancies; ii) combined with chronic
diseases and iii) therapeutic history within 3 months prior to
admission. The surgically resected NSCLC and paracancerous tissues
(at least 5 cm away from the tumor edge) were frozen in liquid
nitrogen and stored at −80°C until further use. The distance
between the NSCLC tissue and normal adjacent tissue was more than 5
cm.
Cell culture
Three NSCLC cell lines (A549, PG49 and H1299) and a
normal human lung cell line MRC-5 were acquired from the American
Type Culture Collection and grown in Dulbecco's modified Eagle's
medium (DMEM) containing 10% heat-inactivated fetal bovine serum
(FBS), 100 mg/ml streptomycin and 100 U/ml penicillin. The DMEM,
FBS and antibiotics were from (Gibco; Thermo Fisher Scientific,
Inc.). All the aforementioned cell lines were grown at 37°C in an
atmosphere with humidity and 5% CO2 for the following
experiments.
Cell transfection
To examine the loss-of-function, siRNA (small
interfering RNA) si-B4GALT1-AS1 (against B4GALT1-AS1) and si-NC
(control, non-targeting siRNA) were purchased from Guangzhou
Ribobio Co., Ltd. The transfection plasmid sequences were as
follows: shB4GALT1-AS1: 5′-GGTTTAGGGCTCCTCTAA-3′; sh-NC:
5′-AATTCTCACGTCACGT-3′. The antagomir-30e used for knockdown of
miR-30e (forward, 5′-CCGCTCGGGAATAGGAAGGTG-3′ and reverse,
5′-GCGAACCTTGGGTAGCCTCCTTGTC-3′) and the negative control
antagomir-NC (forward, 5′-CTTCACAGTGGCTAAGTTCCG-3′ and reverse,
5′-GAATTCCTGGTGCCAAAGCCTTGTC-3′) were synthesized by Shanghai
GenePharma Co., Ltd. The transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in 5×106 cells at a final
concentration of 50 nM, according to the manufacturer's protocol.
After transfection for 24 h, expression of B4GALT1-AS1 or miR-30e
was validated by reverse transcription-quantitative PCR
(RT-qPCR).
The synthesis of vector pcDNA3.1-SRY-box
transcription factor 9 (pc-SOX9) to overexpress SOX9 and empty
pcDNA3.1 vector was done by Shanghai GenePharma Co., Ltd. One day
prior to transfection, NSCLC cells were seeded into 6-well plates
at a density of 5×106 cells until they reached 70%
confluence. The transfection of the aforementioned molecular
constructs was done transiently into cells using Lipofectamine
2000™ reagent from (Invitrogen; Thermo Fisher Scientific Inc.).
Sox9 expression was detected via reverse-transcription quantitative
(RT-q)PCR analysis 24 h post-transfection.
RT-qPCR
Total RNA was extracted from NSCLC cells and tissues
and isolated using TRIzol® reagent from (Invitrogen;
Thermo Fisher Scientific, Inc.), and a NanoDrop (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.) was used to estimate
its quality and concentration. The expression of miR-30e was done
by reverse transcription using the Mir-X™ miRNA First-Strand
Synthesis kit from Takara Biotechnology, Co., Ltd. and quantitative
evaluation of the synthesized cDNA was done using the Mir-X™ miRNA
RT-qPCR TB Green® kit (Takara Biotechnology, Co., Ltd.).
Small nuclear RNA U6 was used as the endogenous control for the
normalization of the expression of miR-30e. To measure transcript
levels of B4GALT1-AS1 and SOX9, the PrimeScript™ RT Reagent kit
(Takara Biotechnology Co., Ltd.) was used to convert 5 µl total RNA
to cDNA. Then, the amplification of cDNA products was done using
the SYBR-Green PCR Master Mix (Takara Biotechnology Co., Ltd.). The
level of SOX9 and B4GALT1-AS1 transcripts were normalized using
GAPDH as a control. The thermocycling conditions were as follows:
94°C for 30 sec; followed by 45 cycles of 94°C for 5 sec, 60°C for
15 sec and 72°C for 15 sec; dissociation stage. The primer
sequences for miR-30e were as follows: Forward,
5′-TTCACAGAATAATTGC-3′ and reverse, 5′-TTAACACTTTCACGGGATG-3′;
B4GALT1-AS1 forward, 5′-AGCTGAACGTCGAAGCGG-3′ and reverse,
5′-GACACCCTGCGGCCAAAGGC-3′; U6 forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′; and GAPDH forward,
5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse, 5-CAAAGTTGTCATGGA-3′. The
2−ΔΔCq method was used for quantification of gene
expression (16).
Cell counting assay
Assessment of cell proliferation was done using the
Cell Counting Kit-8 assay from Shanghai Haling Biotechnology, Co.,
Ltd (http://haling.bioon.com.cn), according
to the manufacturer's protocol. After incubating the transfected
cells for 24 h, they were collected after trypsinization and seeded
(2×103 cells/well) into 96-well plates. CCK-8 solution
(10 µl) was added per well and kept for 2 h at 37°C. The absorbance
of the mixture was estimated in a microplate reader from Bio-Rad
Laboratories, Inc. at 450 nm.
Colony formation assay
A549 and H1299 cell lines infected with siNC or
si-B4GALT1-AS1 (1×103/well) were seeded into a culture
dish (10 cm) and cultured for 14 days at 37°C in an atmosphere with
humidity and 5% CO2. Finally, colonies were stained with
4% paraformaldehyde for 25 min at 37°C and stained with crystal
violet for 15 min at 37°C followed by the counting of colonies. The
number of colonies (diameter >1 mm) were counted and each
experiment was repeated at least 3 times.
Bioinformatics analysis
Putative B4GALT1-AS1 targets were predicted using
StarBase version 3.0 (http://starbase.sysu.edu.cn/).
RNA immunoprecipitation (RIP)
assay
RIP was performed using the Magna RIP RNA-Binding
Protein Immunopre-cipitation kit (EMD Millipore) to assess
B4GALT1-AS1 and miR-30e interaction in A549 and H1299 cells,
according to the manufacturer's instructions. After treating the
cells in lysis buffer, further incubation of cell lysates was
performed at 4°C overnight with magnetic beads conjugated with
either human protein-argonaute 2 (AGO2) antibody (1:100; cat. no.
ab32381) or IgG (negative control) (1:100; cat. no. ab109489; both
purchased from Abcam). Subsequently, the immunoprecipitated RNA was
isolated after collecting the magnetic beads and RT-qPCR analysis
was conducted to estimate the enrichment of B4GALT1-AS1 and
miR-30e.
Dual-luciferase reporter assay
Design and synthesis of B4GALT1-AS1 fragments
containing binding sites for wild-type (WT) and mutant (MUT) on
miR-30e was done by Shanghai GenePharma Co. Ltd. These were cloned
into the target expression vector pmirGLO dual-luciferase (Promega
Corporation) to get the reporter plasmids WT-B4GALT1-AS1 and
MUT-B4GALT1-AS1. One night prior to transfection, seeding of cells
(60–70% confluent) was done in 24-well plates using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h of transfection, luciferase activity
of the cells was estimated using the Dual-Luciferase Reporter Assay
system (Promega Corporation). The data normalization was done by
comparison with Renilla luciferase activity.
Statistical analyses
All results of independent experiments conducted 3
times are presented as the mean ± SD. Two groups were compared
using paired and unpaired Student's t-test based on dependence and
independence of samples, respectively. Multiple comparisons were
conducted using one-way ANOVA followed by the post hoc Tukey's
test. Pearson correlation coefficients were used for correlation
analysis between B4GALT1-AS1 and Sox9 in NSCLC. P<0.05 was
considered to indicate a statistically significant difference.
Results
B4GALT1-AS1 is increased in NSCLC
tissues and cells
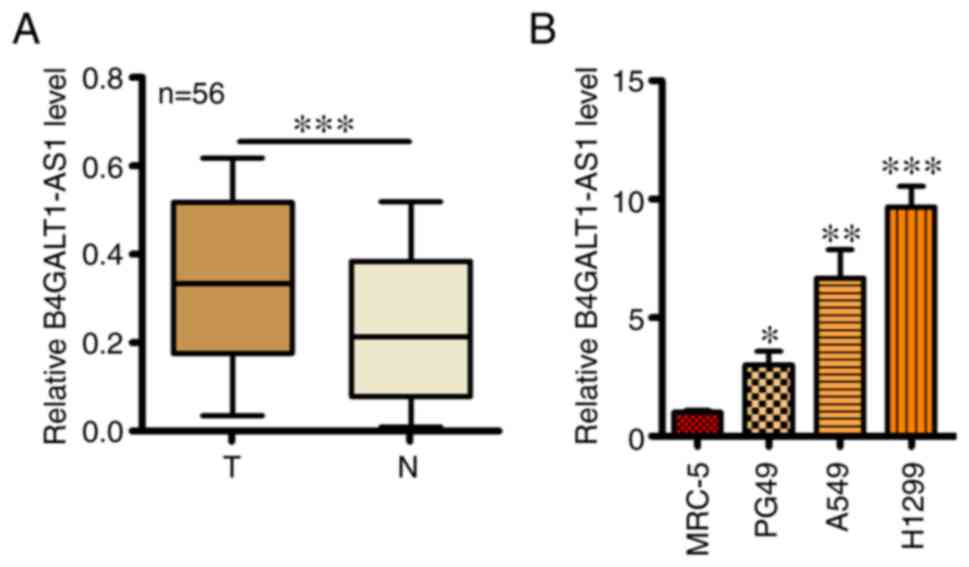
Expression of B4GALT1-AS1 was first assessed by
RT-qPCR in 56 NSCLC tissues and adjacent healthy tissues. An
elevated B4GALT1-AS1 mRNA level was observed in NSCLC tissues
compared with adjacent healthy tissues (Fig. 1A). In addition, RT-qPCR was performed
in 3 NSCLC cell lines (PG49, A549 and H1299) and MRC-5 (human
normal lung cell line) and a significantly higher B4GALT1-AS1 mRNA
expression level was found in NSCLC cell lines compared with MRC-5
cells. Meanwhile, H-1299 cells had the highest level of B4GALT1-AS1
compared with the others cell lines (Fig. 1B).
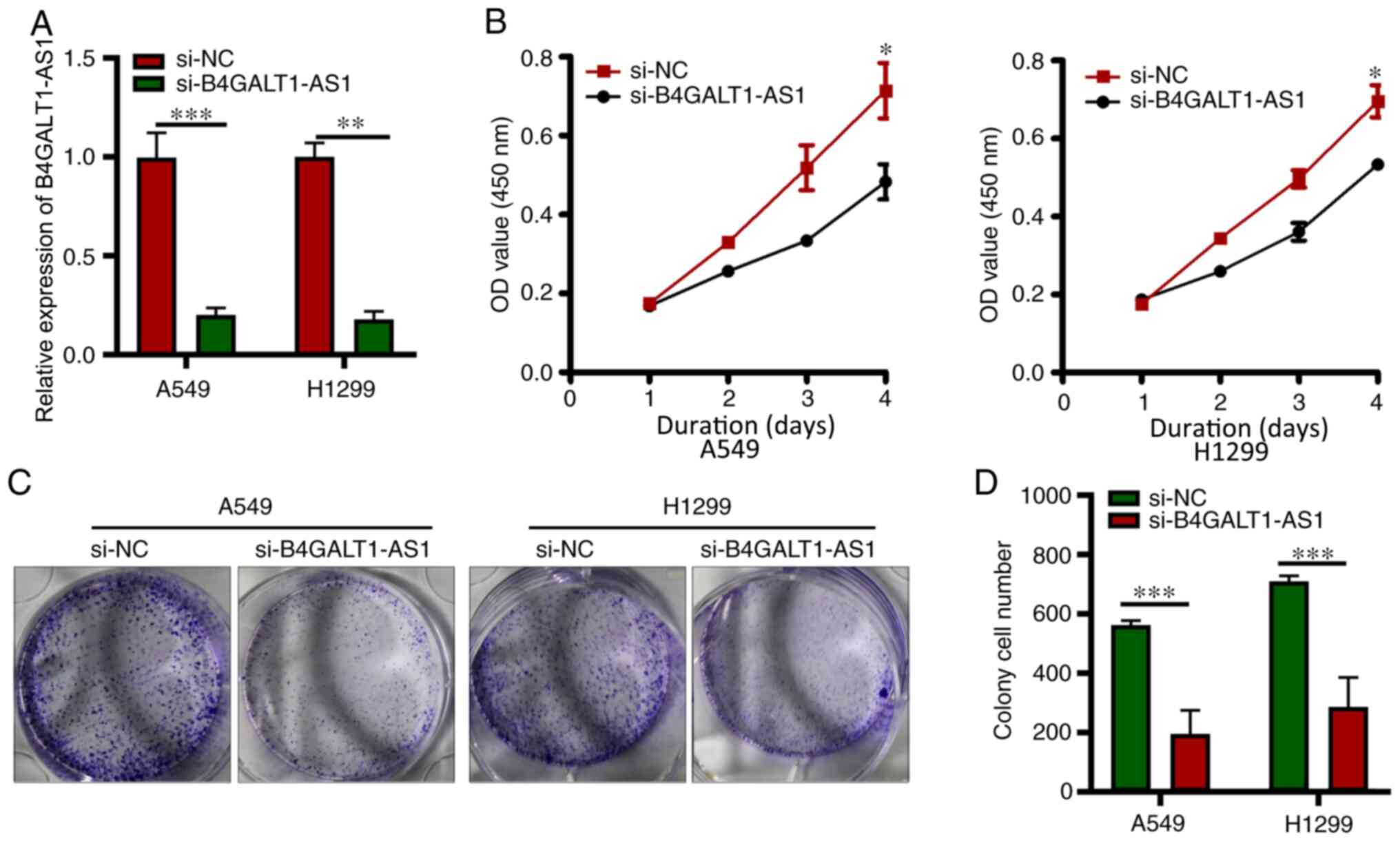
In vitro B4GALT1-AS1 silencing leads
to inhibition of the NSCLC cell malignant phenotype
A higher B4GALT1-AS1 mRNA expression was observed in
A549 and H1299 cells compared with PG49 cells; hence, for
subsequent analysis, H1299 and A549 cells were used. To examine the
function of B4GALT1-AS1 in the progression of NSCLC, si-B4GALT1-AS1
to deplete B4GALT1-AS1 was transfected in A549 and H1299 cells and
si-NC transfected cells were used as the control, and the level of
B4GALT1-AS1 was significantly decreased in A549 and H1299 cells
transfected with si-B4GALT1-AS1 (P<0.05; Fig. 2A). The proliferative ability of both
A549 and H1299 cells harboring si-B4GALT1-AS1 was measured using
the CCK-8 assay and demonstrated inhibition compared with control
cells (P<0.05; Fig. 2B). In
addition, the colony-forming capacity of A549 and H1299 cells
deficient in B4GALT1-AS1 (after transfection with si-B4GALT1-AS1)
was greatly inhibited (P<0.05; Fig.
2C and D). These results strongly indicated the
cancer-enhancing activity of lncRNA B4GALT1-AS1 in NSCLC.
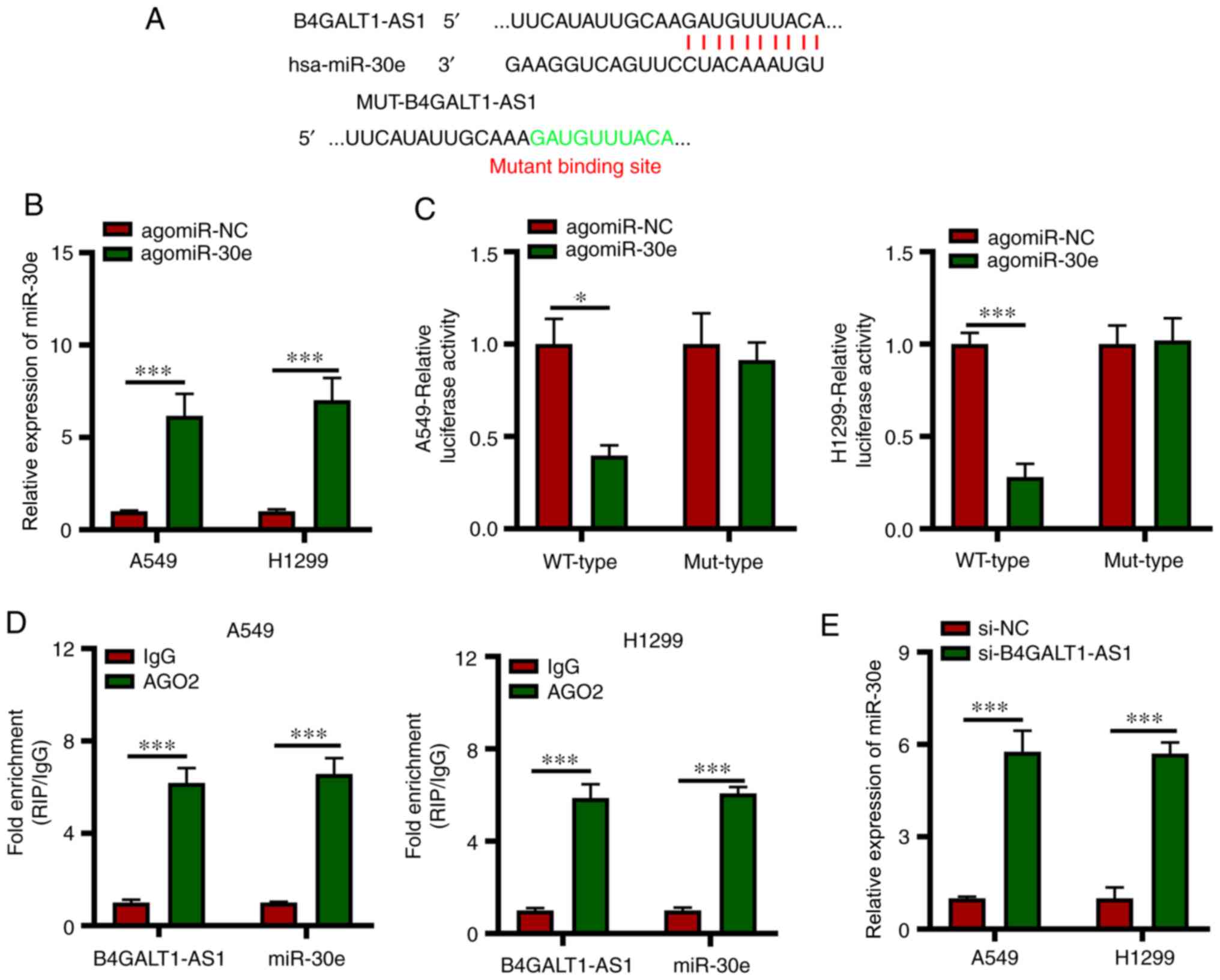
B4GALT1-AS1 interacts directly with
miR-30e in NSCLC cells and acts as a miRNA sponge
lncRNAs participate in several biological processes
by functioning as ceRNA (competing endogenous RNA) against miRNAs
(9). Next, StarBase version 3.0, a
publicly available algorithm was used to predict the miRNAs that
interact directly with B4GALT1-AS1. A binding sites was identified
between B4GALT1-AS1 and miR-30e (Fig.
3A). In addition, miR-30e has also been reported to inhibit
NSCLC progression (17). An increase
in miR-30e expression in A549 and H1299 cells transfected with
si-B4GALT1-AS1 compared with si-NC (Fig.
3B). In addition, the luciferase activity of cells harboring
WT-B4GALT1-AS1 reduced significantly following agomiR-30e
transfection; however, there was no change in MUT-B4GALT1-AS1
activity when miR-30e was overexpressed (Fig. 3C). Next, the RIP assay was conducted
to assess the miR-30e and B4GALT1-AS1 interaction and miR-30e and
B4GALT1-AS1 were found to be enriched in immunoprecipitates
containing AGO2 in comparison with the IgG control in A549 and
H1299 cell lines (Fig. 3D).
Subsequently, it was tested if expression of miR-30e was modulated
by B4GALT1-AS1 in NSCLC cells and it was found to be upregulated in
A549 and H1299 cells due to the silencing of B4GALT1-AS1 (Fig. 3E). The aforementioned findings
indicated the miR-30e quenching function of B4GALT1-AS1 in NSCLC
cells.
B4GALT1-AS1 sequesters miR-30e and
positively modulates the expression of SOX9 in NSCLC cells
miR-30e directly targets the SOX9 gene in NSCLC
cells (17). On observing that
B4GALT1-AS1 sequesters miR-30e, the function of B4GALT1-AS1 in SOX9
regulation in NSCLC cells was next investigated. As shown in
Fig. 4A, B4GALT1-AS1 silencing
markedly reduced the expression of SOX9 at transcript in NSCLC
cells. A significant decline in the level of miR-30e in
antagomiR-30e harboring A549 and H1299 cells was observed compared
with antagomiR-NC (Fig. 4B). In
addition, the decline in the Sox9 transcript because of B4GALT1-AS1
knockdown was reversed by re-introduction of antagomiR-30e in A549
and H1299 cells (P<0.05; Fig.
4C). B4GALT1-AS1 mRNA expression was also positively correlated
with SOX9 mRNA levels in NSCLC tissue (P=0.001; R=0.428; Fig. 4D). Further, rescue experiments
elucidated that B4GALT1-AS1 silencing impeded proliferation of
H1299 and A549 cells and inhibition of miR-30e in these cells
partly neutralized the outcomes of B4GALT1-AS1 silencing
(P<0.05; Fig. 4E). SOX9 was
overexpressed in A549 and H1299 cells transfected with LV-Sox9
compared with LV-NC (P<0.05; Fig.
4F). Subsequently, on performing the CCK-8 assay, pc-SOX9
co-transfection was found to reverse the decline in proliferation
of A549 and H1299 cells caused by the silencing of B4GALT1-AS1
(P<0.05; Fig. 4G). These findings
indicate the oncogenic effect of B4GALT1-AS1 in NSCLC progression
by acting as a ceRNA for miR-30e and consequently enhancing SOX9
expression.
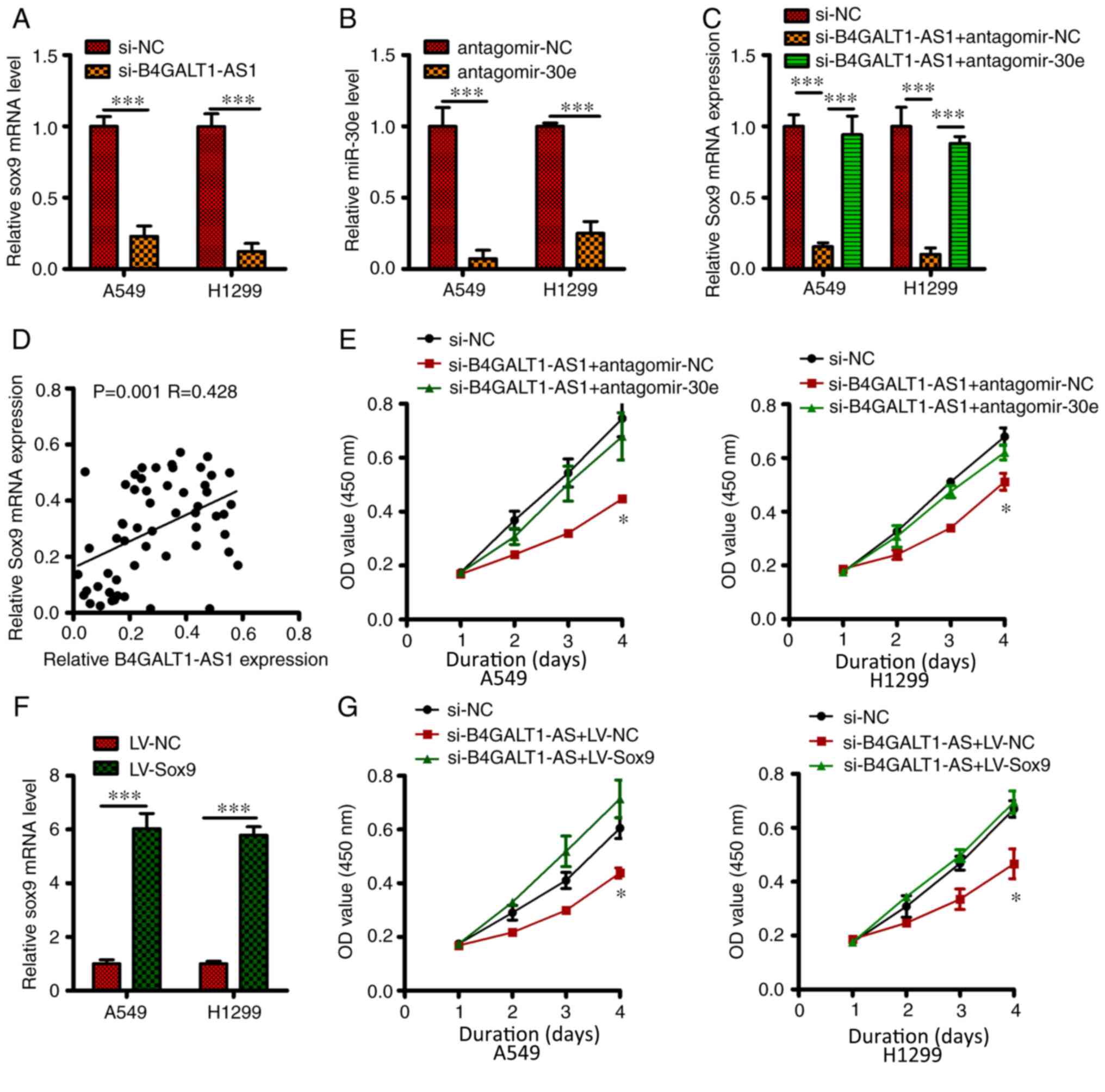 | Figure 4.Role of B4GALT1-AS1 as a competing
endogenous RNA which sequestered miR-30e to regulate the expression
level of SOX9. (A) After transfecting si-B4GALT1-AS1 or si-NC in
A549 and H1299 cells, SOX9 transcript level was quantified by
RT-qPCR. (B) The level of miR-30e was estimated by RT-qPCR after
the transfection of H1299 and A549 cells with antagomiR-30e or
antagomir-NC. (C) Transfection of H1299 and A549 cells with
antagomiR-30e or antagomir-NC in si-B4GALT1-AS1 presence. miR-30e
expression was investigated via RT-qPCR assay. (D) B4GALT1-AS1 and
SOX9 expression was positively correlated in NSCLC tissue. (E)
Co-transfection of A549 and H1299 cells with si-B4GALT1-AS1 and
either antagomiR-30e, or antagomir-NC and assessment of cell
proliferation by the CCK-8 assay. (F) RT-qPCR assay to estimate
SOX9 level in LV-SOX9 or LV-NC transfected A549 and H1299 cells.
(G) Co-transfection of H1299 and A549 cells with si-B4GALT1-AS1
along with either pc-SOX9 or pcDNA3.1. CCK-8 assay was performed to
estimate cellular proliferation. ***P<0.001 and *P<0.05. AS1,
antisense RNA; RT-q, reverse-transcription quantitative; NC,
negative control; si, small interfering; NSCLC, non-small cell lung
carcinoma; miR, micro RNA; LV, lentiviral; OD, optical density; SOX
9, SRY-box transcription factor 9. |
Discussion
lncRNAs have attracted a lot of interest because of
their vital function in the progression of cancer (18,19). It
has been reported that aberrant expression of lncRNAs contributed
to the progression of NSCLC (20).
Hence, potential treatment targets may be revealed by studying the
roles of lncRNAs in NSCLC tumorigenesis. To the best of our
knowledge, until recently the function of B4GALT1-AS1 in the
malignancy of NSCLC has not been studied in detail. Hence, the
present study was firstly evaluated the B4GALT1-AS1 expression
level and its specific role in the malignancy of NSCLC cells and
elucidated the tumor-enhancing function of B4GALT1 in NSCLC. The
present study found a high expression of B4GALT1-AS1 in NSCLC
tissue samples compared with adjacent normal tissues as well as the
NSCLC cell lines compared with the normal lung cell line. The
functional experiments conducted in the present study revealed
inhibition of NSCLC cell proliferative and colony-forming
capacities as a result of B4GALT1-AS1 silencing.
Notably, lncRNAs may play key role as competing
endogenous RNAs (ceRNAs) to regulate miRNAs expression (21). In the present study, after revealing
the tumor-enhancing role of B4GALT1-AS1 in NSCLC. Based on the
results from bioinformatics analysis in the present study, it was
predicted that miR-30e possesses a binding site complementary to
that on B4GALT1-AS1. To further confirm the relationship between
miR-30e and B4GALT1-AS1, luciferase reporter and RIP assays were
conducted, which identified that miR-30e was the direct target of
B4GALT1-AS1. In addition, the silencing of B4GALT1-AS1 enhanced
miR-30e level and led to a decline in the expression of SOX9. In
the present study, the outcomes of B4GALT1-AS1 silencing on
malignant phenotypes of NSCLC could be abrogated by miR-30e
inhibition or SOX9 restoration. Taken together, the findings of the
present study indicate a ceRNA model that includes B4GALT1-AS1,
SOX9 and miR-30e in NSCLC cells.
In multiple potential target genes, SOX9 was
selected for further validation since SOX9 has been demonstrated to
be involved in NSCLC carcinogenesis and progression (22). In addition, SOX9 was identified as a
direct target of many diverse miRNAs in NSCLC, including miR-124
(23), miR-206 (24), and miR-32 (25). The results of the present study
verified that SOX9 was a direct and functional target gene of
miR-30e. In the present study, the upregulation of SOX9 targeted by
miR-30e in NSCLC cells was observed and for the first time to the
best of our knowledge, an upstream mechanism modulating the axis of
miR-30e/SOX9 in NSCLC cells in vitro was identified. The
present study indicated that B4GALT1-AS1 possesses a miR-30e
binding site, and acts as a ceRNA and sequesters miR-30e in NSCLC
cells, leading to an enhanced level of SOX9.
To the best of our knowledge, the present study
identified the mode of action of B4GALT1-AS1 in enhancing NSCLC
progression for the first time. B4GALT1-AS1 silencing enhanced the
malignancy of NSCLC cells. B4GALT1-AS1 positively regulated SOX9
expression by sequestering miR-30e in NSCLC cells. Further studies
are needed to focus on the in vivo function of B4GALT1-AS1
in NSCLC. Identification of the regulatory network of
B4GALT1-AS1/miR-30e/SOX9 will potentially aid in fully determining
the stage of NSCLC and provide possible targets for the therapy of
patients with NSCLC. Future studies will focus on seeking out
suitable online databases to confirm the correlation analysis
between B4GALT1-AS1 and Sox9 in lung cancer. The sample size of
this study was relatively small. In the future, the biological
potential of B4GALT1-AS1 as a biomarker of NSCLC needs to be
validated in multi-center studies with a large sample size.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JHL and JM wrote the manuscript and contributed to
the conception of the study. JHL, FNC and CXW performed the
experiments and data analysis. SQH contributed to data acquisition
and analysis and revised the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of The First Hospital of Longyan City (Longyan,
China; approval no. 2019-015) and performed in accordance with the
Declaration of Helsinki. All the patients provided written informed
consent prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI
|
|
2
|
Zhao J, Mao Z, Fedewa SA, Nogueira L,
Yabroff KR, Jemal A and Han X: The affordable care act and access
to care across the cancer control continuum: A review at 10 years.
CA Cancer J Clin. 70:165–181. 2020.PubMed/NCBI
|
|
3
|
Noone AM, Cronin KA, Altekruse SF,
Howlader N, Lewis DR, Petkov VI and Penberthy L: Cancer incidence
and survival trends by subtype using data from the surveillance
epidemiology and end results program, 1992–2013. Cancer Epidemiol
Biomarkers Prev. 26:632–641. 2017.PubMed/NCBI
|
|
4
|
Singh M and Jadhav HR: Targeting non-small
cell lung cancer with small-molecule EGFR tyrosine kinase
inhibitors. Drug Discov Today. 23:745–753. 2018.PubMed/NCBI
|
|
5
|
Hill A, Gupta R, Zhao D, Vankina R, Amanam
I and Salgia R: Targeted therapies in non-small-cell lung cancer.
Cancer Treat Res. 178:3–43. 2019.PubMed/NCBI
|
|
6
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013.PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009.PubMed/NCBI
|
|
8
|
Hauptman N and Glavac D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013.PubMed/NCBI
|
|
9
|
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q and
Dong D: lncRNADisease 2.0: An updated database of long non-coding
RNA-associated diseases. Nucleic Acids Res. 47:D1034–D1037.
2019.PubMed/NCBI
|
|
10
|
Han S, Cao D, Sha J, Zhu X and Chen D:
lncRNA ZFPM2-AS1 promotes lung adenocarcinoma progression by
interacting with UPF1 to destabilize ZFPM2. Mol Oncol.
14:1074–1088. 2020.PubMed/NCBI
|
|
11
|
Yang S, Liu T, Sun Y and Liang X: The long
noncoding RNA LINC00483 promotes lung adenocarcinoma progression by
sponging miR-204-3p. Cell Mol Biol Lett. 24:702019.PubMed/NCBI
|
|
12
|
Zhao M, Xin XF, Zhang JY, Dai W, Lv TF and
Song Y: lncRNA GMDS-AS1 inhibits lung adenocarcinoma development by
regulating miR-96-5p/CYLD signaling. Cancer Med. 9:1196–1208.
2020.PubMed/NCBI
|
|
13
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30:6942013.PubMed/NCBI
|
|
14
|
Luo Z, Han Z, Shou F, Li Y and Chen Y:
LINC00958 accelerates cell proliferation and migration in non-small
cell lung cancer through JNK/c-JUN signaling. Hum Gene Ther
Methods. 30:226–234. 2019.PubMed/NCBI
|
|
15
|
Zhang Y, Fang Z, Guo X, Dong H, Zhou K,
Huang Z and Xiao Z: lncRNA B4GALT1-AS1 promotes colon cancer cell
stemness and migration by recruiting YAP to the nucleus and
enhancing YAP transcriptional activity. J Cell Physiol.
234:18524–18534. 2019.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
17
|
Liu C, Yang Z, Deng Z, Zhou Y, Gong Q,
Zhao R and Chen T: Downregulated miR-30e contributes to progression
of lung adenocarcinoma through elevating the expression of SOX9.
Cancer Med. 7:5554–5566. 2018.PubMed/NCBI
|
|
18
|
Chu F, Xue L and Miao H: Long noncoding
RNA TP73-AS1 in human cancers. Clin Chim Acta. 500:104–108.
2020.PubMed/NCBI
|
|
19
|
Gourvest M, Brousset P and Bousquet M:
Long noncoding RNAs in acute myeloid leukemia: Functional
characterization and clinical relevance. Cancers (Basel).
11:16382019.
|
|
20
|
Lu T, Wang Y, Chen D, Liu J and Jiao W:
Potential clinical application of lncRNAs in non-small cell lung
cancer. Onco Targets Ther. 11:8045–8052. 2018.PubMed/NCBI
|
|
21
|
Abdollahzadeh R, Daraei A, Mansoori Y,
Sepahvand M, Amoli MM and Tavakkoly-Bazzaz J: Competing endogenous
RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A
new look at hallmarks of breast cancer. J Cell Physiol.
234:10080–10100. 2019.PubMed/NCBI
|
|
22
|
Wang X, Ju Y, Zhou MI, Liu X and Zhou C:
Upregulation of SOX9 promotes cell proliferation, migration and
invasion in lung adenocarcinoma. Oncol Lett. 10:990–994.
2015.PubMed/NCBI
|
|
23
|
Wang X, Liu Y, Liu X, Yang J, Teng G,
Zhang L and Zhou C: miR-124 inhibits cell proliferation, migration
and invasion by directly targeting SOX9 in lung adenocarcinoma.
Oncol Rep. 35:3115–3121. 2016.PubMed/NCBI
|
|
24
|
Zhang YJ, Xu F, Zhang YJ, Li HB, Han JC
and Li L: miR-206 inhibits non small cell lung cancer cell
proliferation and invasion by targeting SOX9. Int J Clin Exp Med.
8:9107–9113. 2015.PubMed/NCBI
|
|
25
|
miR-32 functions as a tumor suppressor and
directly targets SOX9 in human non-small cell lung cancer
[Retraction]. Onco Targets Ther. 9:63792016.PubMed/NCBI
|