Introduction
Breast cancer is a type of malignant tumor
originating from breast epithelial tissue that accounts for
one-fourth of cancer cases diagnosed in females in 2018 worldwide
and that seriously endangers women's health (1). In recent years, with the increase in
early screening and the improvement of treatment, the survival and
prognosis of breast cancer have improved, but the overall curative
ratio is still not ideal (2–4). Based on statistics, there were ~1.7
million new cases of breast cancer and 520,000 breast
cancer-associated deaths worldwide in 2012 (5), while by 2018, the number of new cases
of breast cancer increased to ~2.1 million, and the number of
deaths was ~630,000 (2). With the
changes in lifestyle and reproductive choices, the incidence and
mortality rate of breast cancer are increasing, and breast cancer
is becoming a serious global health burden (6).
The overactivation of oncogenes and the inactivation
of tumor suppressor genes are important causes of the occurrence
and development of breast cancer (7). Among these genes, forkhead box P3
(FOXP3) plays an important suppressive role in breast cancer. FOXP3
is a specific marker of regulatory T cells, and this gene plays an
important role in the differentiation, development and functional
maintenance of T regulatory cells (8–11). A
large number of studies have shown that FOXP3 is also expressed in
normal breast epithelial cells and is an important X-linked breast
cancer suppressor gene that plays an important role in the
metastatic spread of breast cancer by regulating the expression of
a series of tumor-related genes, including BRCA1/2, HER2,
c-myc and SKP2 (12–14). The
deletion, mutation and change of the cellular localization of FOXP3
are important causes of the occurrence and development of breast
cancer (12). In our previous study,
it was reported that FOXP3 inhibits breast cancer angiogenesis by
regulating the expression of VEGF (15). However, it remains unclear whether
FOXP3 is involved in the regulation of breast cancer cell
apoptosis. The present study aimed to investigate the underlying
genes and networks regulated by FOXP3 in breast cancer.
Materials and methods
Cell lines and culture
The human breast cancer cell lines MCF-7 and
MDA-MB-231 were obtained from The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences. All cell lines were
authenticated by the analysis of short tandem repeat profiles and
100% matched the standard cell lines in the DSMZ data bank. All
cells were negative for the cross-contamination of other human
cells and for mycoplasma contamination. The cells were cultured in
DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) with 10%
estrogen-deprived fetal bovine serum (HyClone; Cyvita) and 100 mg
per ml ampicillin/streptomycin.
Generation of FOXP3-MDA-MB-231
cells
FOXP3- overexpresing lentivirus was constructed and
purchased from GeneChem, Inc. In total, 3×105 MDA-MB-231
cells were seeded onto a six-well culture plate, and the
second-generation lentivirus-containing control vector or a FOXP3
vector (MOI 10) was added to the plate. After 12 h, the medium was
replaced, and cells were cultured in incubator of 37°C for 96 h.
Puromycin (2 µg/ml) was used to select infected cells. MDA-MB-231
breast cancer cells were infected with GFP-labeled
FOXP3-overexpressing adenovirus or control adenovirus, and the
bright field and eGFP expression patterns (representing cells that
were effectively infected with adenovirus) were examined using
fluorescence microscopy at ×20 magnification.
Plasmid construction and RNA
interference
XhoI and KpnI flanked HFOXP3 were synthesized by
TsingKe Biological Technology, and digested HFOXP3 was subcloned
into identically digested pcDNA3.1+ to generated pcDNA3.1-FOXP3
plasmid. The FOXP3 siRNA (sense 5′-GCAGCGGACACUCAAUGAGdTdT-3′;
antisense 5′-CUCUUUGUGUGUCCGCUGCdTdT-3′) (16) and the negative control (sense
5′-GCAGCGGACACUCAAUGAGdTdT-3′; antisense
5′-CUCUUUGUGUGUCCGCUGCdTdT-3′) were purchased from Shanghai
GenePharma Co., Ltd. The pcDNA3.1-FOXP3 plasmid and FOXP3 siRNA
were used to overexpress and knockdown FOXP3 expression,
respectively, in MDA-MB-231and MCF-7 cells. The PDCD4 siRNA (sense,
5′-GCUGCUUUGGACAAGGCUATT-3′; antisense,
5′-UAGCCUUGUCCAAAGCAGCTT-3′) and the negative control (sense,
5′-GCUGCUUTGGACAAGGCUATC-3′; antisense,
5′-UAGCCUAGUCCAAAGCAGCAT-3′) sequences (17) were synthesized by Shanghai GenePharma
Co., Ltd.
Cell transfection
Breast cancer cells MDA-MB-231 and MCF-7 were seeded
in 6-well plate at the density of 2×105 cells/well and
cultured overnight at 37°C. Once cell density reached 70%, the
culture medium was discarded and OPTI-MEM (Gibco; Thermo Fisher
Scientific, Inc.) was added for another 4 h; the pcDNA3.1-FOXP3
plasmid (2.5 µg) and siRNA (MOI 20, 5 µl) were transfected into
MDA-MB-231 and MCF-7 cells, respectively, using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. After 6 h incubation at 37°C, the
medium was replaced and the cells were cultured in appropriate DMEM
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% estrogen-deprived fetal bovine serum (HyClone; Cyvita) for
various time periods. At the same time, cells were transfected with
control siRNA as control group.
Reverse transcription-quantitative
PCR
Total RNA was isolated from cells with RNAIso Plus
(Takara Biotechnology Co., Ltd.), and RNA (100 ng) was reverse
transcribed into single stranded cDNA using a PrimeScript RT
Reagent kit (Takara Biotechnology Co., Ltd.) in 10 µl reaction at
37 C for 15 min, 85°C for 5 min, 4°C for hold. Then, 2 µl cDNA was
used for qPCR using a Prism 7500 real-time thermocycler (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using with SYBR Green
Ex Taq (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions. The reaction protocol was as follows:
30 sec at 95°C, followed by 40 cycles of 5 sec at 95°C and 34 sec
at 60°C. Relative expression level of genes was calculated using
the 2−∆∆Ct method (18).
The sequences of the primers were as follows: FOXP3, forward
5′-CGAAGCTTATGCCCAACCCCAGGCCTG-3′, reverse
5′-CGGGATCCTCAGGGGCCAGGTGTAGGGTTG-3′; PDCD4, forward
5′-TGGATTAACTGTGCCAACCA-3′, reverse 5′-TCTCAAATGCCCTTTCATCC-3′;
SQOR, forward 5′-CACTGGTGGCTGTGGTAT-3′, reverse
5′-CACCCACTTTCCTCTTCAT-3′; PGGHG, forward
5′-GGTGGTCTCAGGAGGATGGA-3, reverse 5′-GGTCGGGTCAGAAGGAAGC-3′; and
GAPDH, forward 5′-GTCAAGGCTGAGAACGGGAA3′ and reverse
5′-AAATGAGCCCCAGCCTTCTC-3′. Each reaction was set up in triplicate.
GAPDH was used as the internal control.
Western blot analysis
Cells (2×105 cells) were collected,
washed twice with pre-cooled PBS and lysed using 100 µl of
pre-cooled RIPA lysis buffer containing protease inhibitor
(Beyotime Institute of Biotechnology for 30 min on ice. Protein
concentration was estimated using BCA protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.). Proteins (30 µg) were separated by
10% SDS-PAGE and were transferred onto PVDF membranes. Membranes
were blocked with 5% skimmed milk in Tris-buffered saline
containing 1% Tween 20 (TBST; pH 7.4) at room temperature for 1 h.
Membranes were incubated with primary antibodies against GAPDH
(ProteinTech Group, Inc.; cat. no. 10494-1-AP; 1:4,000), FOXP3
(Abcam; cat. no. ab22510; 1:500) and PDCD4 (ProteinTech Group,
Inc.; cat. no. 12587-1-AP; 1:1,000) at 4°C overnight. Membranes
were then incubated with horseradish peroxidase-conjugated IgG
secondary antibody (ProteinTech Group, Inc.; cat. no. SA00001-2;
1.400) for 1 h at room temperature. Enhanced chemiluminescence
(Pierce; Thermo Fisher Scientific, Inc.) was used for visualization
of immunoreactive proteins. ChemiDoc™ XRS+ System with Image Lab™
Software 4.1 (Bio-Rad Laboratories, Inc.) was used for densitometry
analysis using GAPDH as internal control.
Flow cytometry
Apoptosis was evaluated by flow cytometry. Cells
were harvested and resuspended in PBS. Cells were stained with
Annexin V-FITC/PI apoptosis kit (cat. no. BD 556547 Annexin V; BD
Biosciences) according to the manufacturers' instructions at room
temperature for 15 min. Apoptotic cells were detected using a FACS
Calibur Flow Cytometer (BD Biosciences) and analyzed using with
FlowJo 10.0 software (FlowJo LLC).
RNA-Seq analysis
Two cell samples, control MDA-MB-231 cells and
FOXP3-MDA-MB-231 cells, were prepared. Then, total RNA was
extracted using an RNA easy Mini kit, and an on-column DNase
digestion in RNase-Free DNase set (both Qiagen GmbH) was used to
avoid contamination by genomic DNA. A sequencing library was built
and sequenced using an Illumina HiSeq 2000 by Gene Denovo
Biotechnology.
Identification of differentially
expressed genes (DEGs)
Differential expression analysis was performed using
the edgeR package in R between two samples (19). Genes with false discovery rates
(FDRs) <0.05 and absolute fold-changes ≥2 were considered
differentially expressed genes.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis
Genes usually interact to participate in certain
biological functions. Pathway-based analysis helps to further
understand the biological functions of genes. KEGG (https://www.genome.jp/kegg/) is a major public
pathway-related database. Pathway enrichment analysis can be used
to identify significantly enriched metabolic pathways or signal
transduction pathways in DEGs compared with the whole-genome
background (20). Here, N is the
number of all genes with a KEGG annotation, n is the number of DEGs
in N, M is the number of all genes annotated to specific pathways,
and m is the number of DEGs in M. The calculated P-value underwent
an FDR correction, and FDR ≤0.05 was set as the threshold. Pathways
meeting this condition were defined as significantly enriched
pathways in DEGs.
Gene set enrichment analysis
(GSEA)
Enrichment analysis was performed using GSEA and
MSigDB software (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp,
v7.1) to determine whether a set of genes in specific Gene Ontoloy
term (http://geneontology.org/) pathways
showed significant differences in two groups (21). Briefly, a gene expression matrix was
constructed and genes were ranked using the signal-to-noise
normalization method. Enrichment scores and P-values were
calculated using the default parameters.
Kaplan-Meier plotter analysis
Survival analysis based on the mRNA expression
levels of PDCD4 in breast cancer was performed using the
Kaplan-Meier plotter website (www.kmplot.com), an online database that can assess
the effect of 54,675 genes on the prognosis of patients with
breast, ovarian, lung and gastric cancer. Briefly, the PDCD4 gene
names were uploaded into the database, and the breast cancer cases
included in the analysis were divided into two cohorts according
the expression level of PDCD4. Patients with PDCD4 expression
higher than the median were pooled into the group with high
expression, while the patients with PDCD4 expression lower than the
median were pooled into the group with low expression. Hazard ratio
(HR), 95% confidence intervals and log-rank P-values were
determined using the database.
Statistical analysis
Statistical analysis between two groups of samples
was performed using unpaired Student's t-test by SPSS version 16
software (SPSS Inc.) and expressed as mean ± SEM from three
independent replicates. A value of P<0.05 was considered to
indicate a statistically significant difference. The statistical
tests were two-sided. The in vitro experiments were repeated
at least three times. Relapse-free survival (RFS) and overall
survival (OS) rates of patients in different cohorts were assessed
by Kaplan Meier plots. The hazard ratio (HR) and log-rank P-values
were calculated using the aforementioned databases. PDCD4 mRNA
levels in different stages (according to the
Scarff-Bloom-Richardson SBR grading system) were determined by
Dunnett's or Tukey's post hoc test following ANOVA. Pearson's
correlation coefficient was used to calculate the correlation
between different genes. Correlation analysis of FOXP3 expression
and PDCD4 were performed by The Cancer Genome Atlas dataset (TCGA;
http://www.cancer.gov/).
Results
Generation of FOXP3-overexpressing
MDA-MB-231 cells
To explore the function of FOXP3 in breast cancer
cells, MDA-MB-231 breast cancer cells were infected with green
fluorescent protein (GFP)-labeled FOXP3-overexpressing adenovirus,
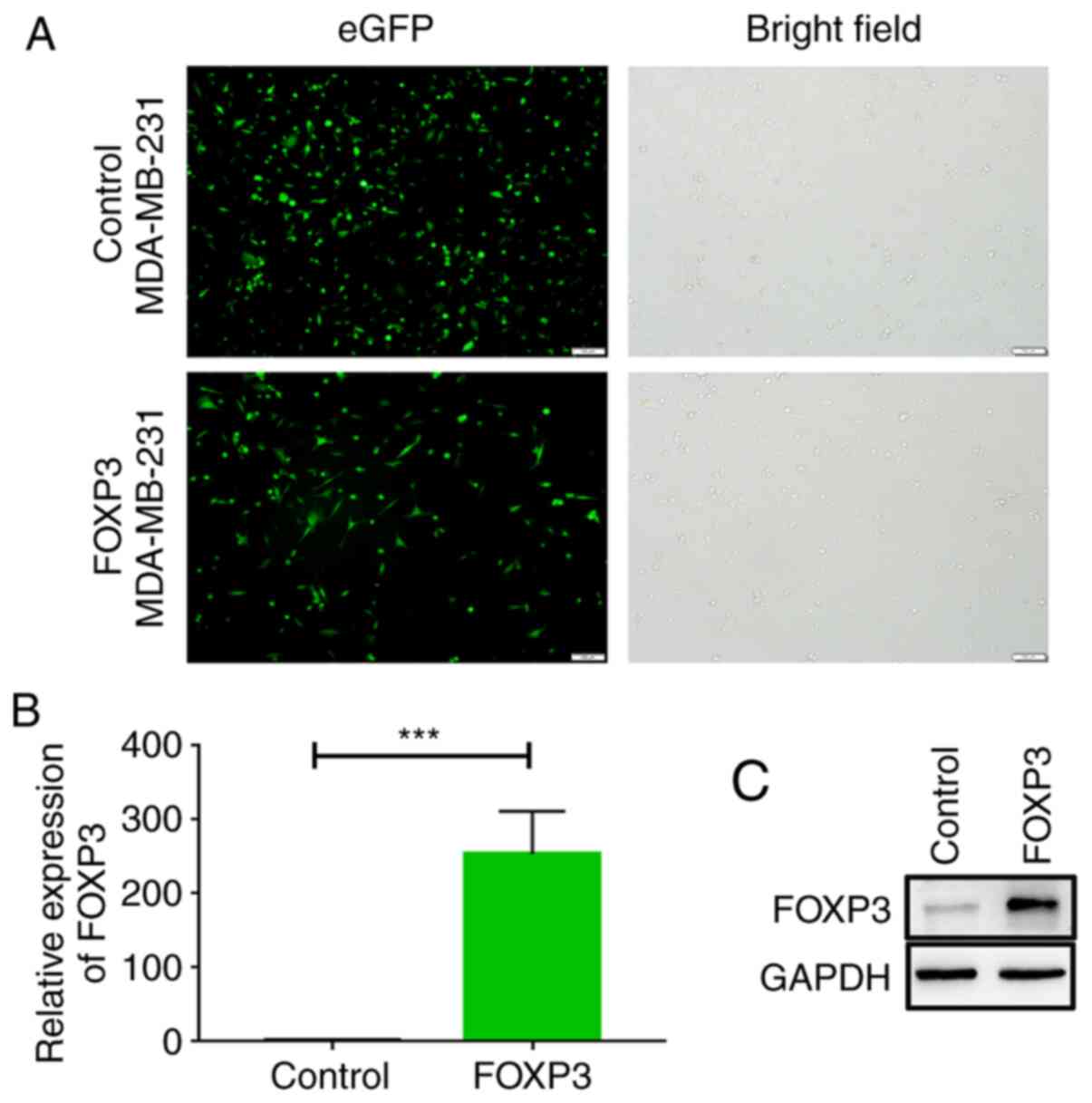
and puromycin was used to select positive cells. As shown in
Fig. 1A, cells expressed GFP, which
indicated that these cells were successfully infected with
adenovirus. RT-qPCR and western blotting was then conducted to
further verify the FOXP3 expression levels. The results
demonstrated that FOXP3 mRNA levels in FOXP3-overexpressing
MDA-MB-231 cells were significantly higher compared with those in
control cells (P<0.001; Fig. 1B).
Moreover, western blot assays showed that control MDA-MB-231 cells
expressed low levels of FOXP3, and FOXP3-MDA-MB-231 cells exhibited
higher FOXP3 expression (Fig. 1C).
Collectively, these results suggested that a FOXP3-overexpressing
MDA-MB-231 cell line was successfully constructed, and this cell
line and the control cells were used in the subsequent studies.
Identification of DEGs between
FOXP3-overexpressing MDA-MB-231 and control MDA-MB-231 cells
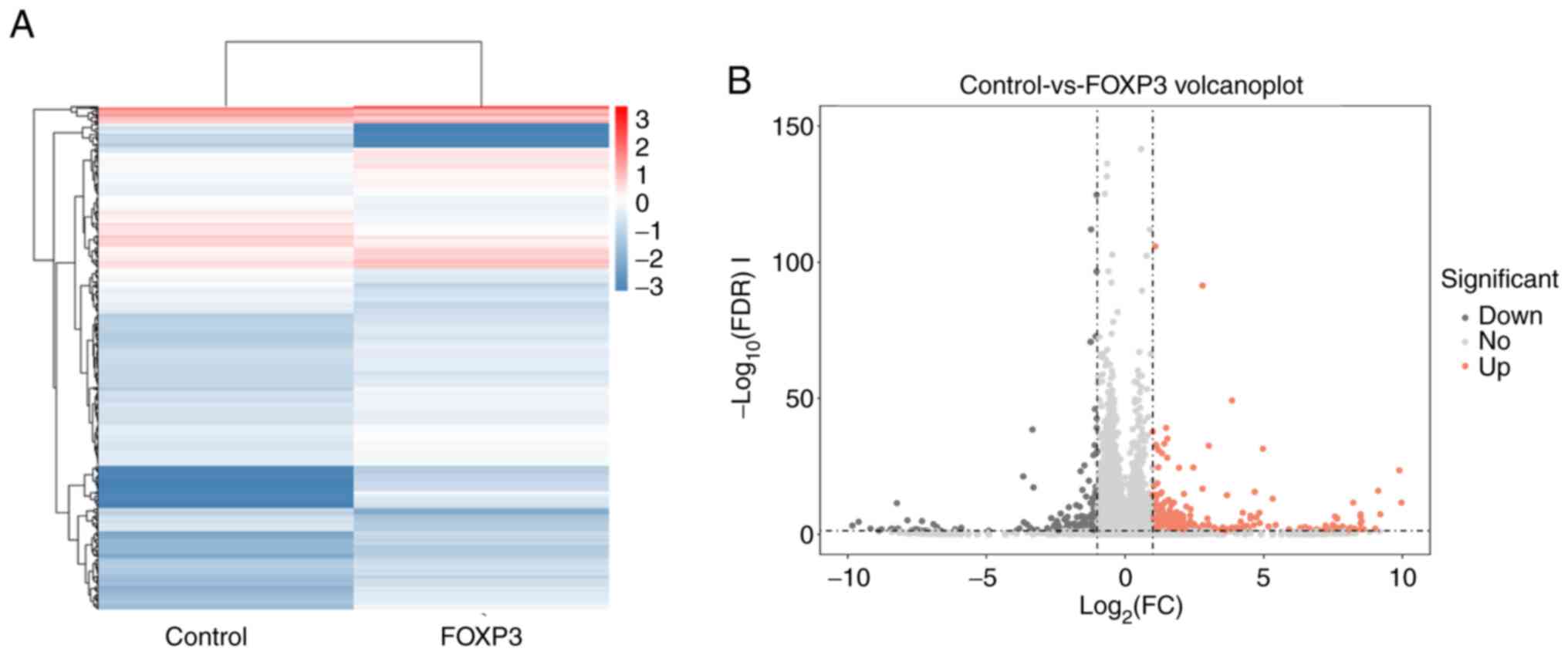
To identify genes regulated by FOXP3, total RNA was
obtained from the FOXP3-overexpressing MDA-MB-231 cells and
wild-type MDA-MB-231 cells and was subjected to RNA-seq. Then,
global gene expression analysis was performed comparing
FOXP3-MDA-MB-231 cells and wild-type cells. A heat map of DEGs was
constructed, and expression changes in genes are shown by
hierarchical cluster analysis (Fig.
2A). The RT-PCR analysis showed the high expression of three
differentially expressed genes (PGGHG, SQOR and PDCD4 genes) in the
RNA-seq analysis of FOXP3-overexpressing MDA-MB-231 cells (Fig. S1). Furthermore, a volcano plot shows
that 6,285 genes were upregulated and 6,013 genes were
downregulated in FOXP3-MDA-MB-231 cells (Fig. 2B).
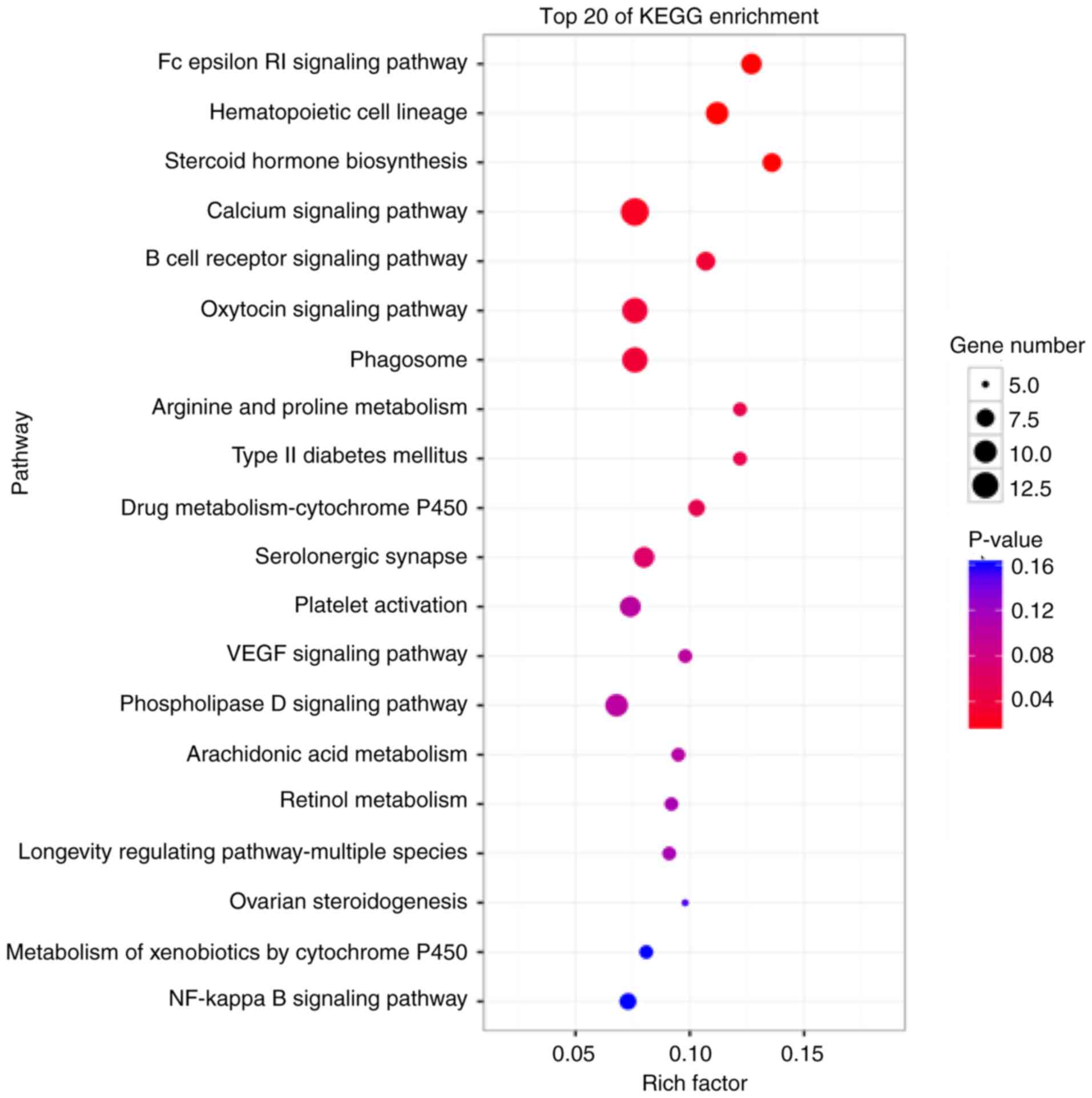
KEGG analysis of DEGs
To further specify the interactions of the pathways
and to clarify the biological functions of FOXP3, KEGG database
analysis, which can be used to find frequently and significantly
enriched pathways, was performed (22). The results demonstrated that the DEGs
were enriched in several signaling pathways, and the main enriched
terms were ‘phagosome’, ‘oxytocin signaling pathway’, ‘serolonergic
synapse’, ‘phospholipase D signaling pathway’, ‘platelet
activation’ and ‘drug metabolism-cytochrome P450’ (Fig. 3).
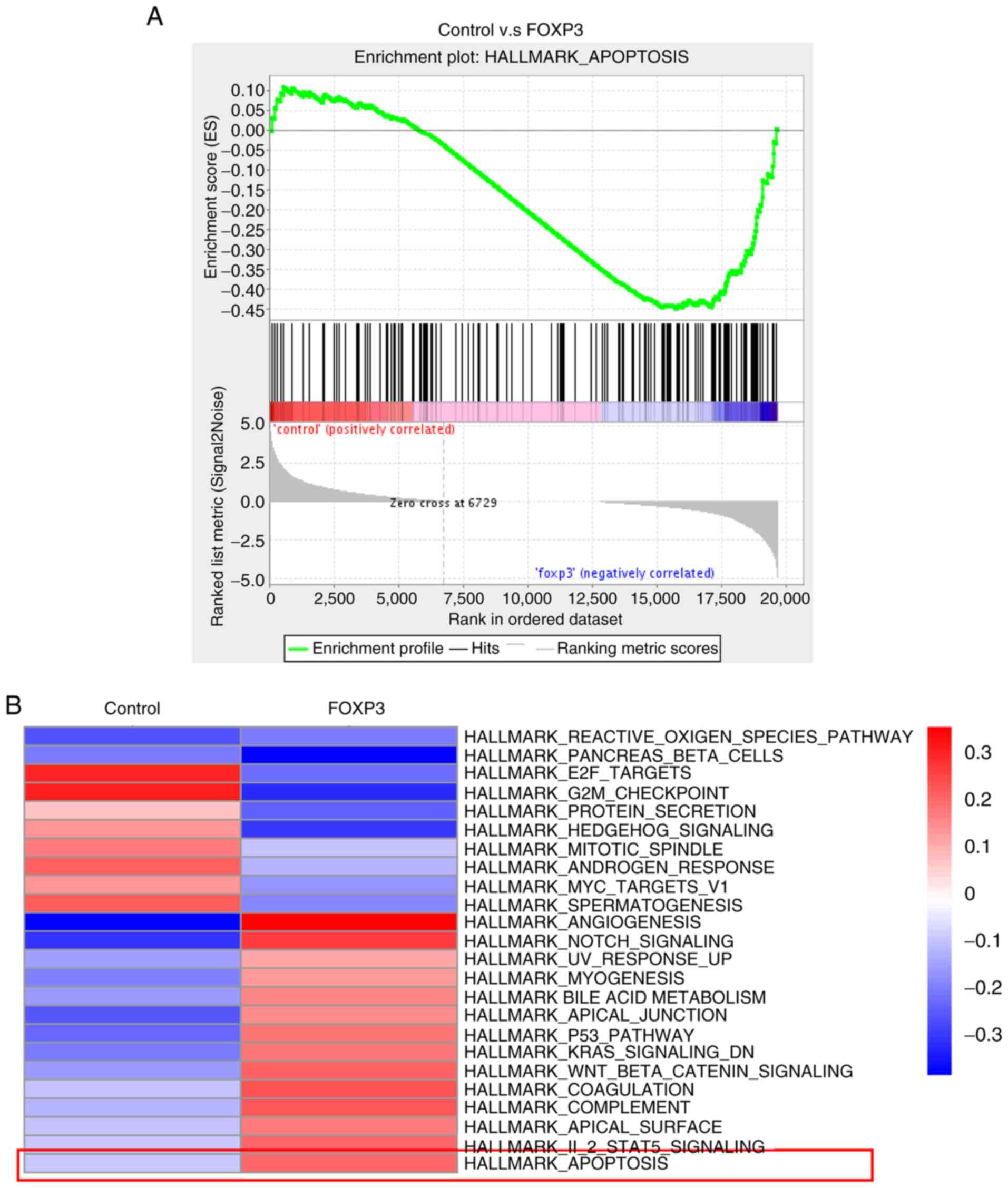
FOXP3 is associated with breast cancer
apoptosis
To determine whether FOXP3 could regulate
apoptosis-related genes, GSEA was conducted. The results revealed
the enrichment of a gene signature related to apoptosis in
FOXP3-overexpressing MDA-MB-231 cells compared with wild-type cells
(Fig. 4A). Functional
enrichment-based clustering analysis was also conducted and it was
demonstrated that, compared with wild-type cells, the upregulated
genes in FOXP3-overexpressing MDA-MB-231 cells were also involved
in apoptosis (Fig. 4B). Taken
together, these data suggested that FOXP3 is associated with breast
cancer apoptosis.
FOXP3 upregulates PDCD4
As aforementioned, FOXP3 expression levels were
associated with apoptosis. To identify the role of FOXP3 in breast
cancer apoptosis, the DEGs in FOXP3-overexpressing MDA-MB-231 cells
and wild-type cells were further analyzed. It was revealed that
PDCD4, a key mediator of apoptosis (23), was upregulated in
FOXP3-overexpressing MDA-MB-231 cells (Fig. 5A). These results indicated that FOXP3
might be a regulator of PDCD4 in breast cancer cells. To further
investigate the regulatory role of FOXP3 in PDCD4 expression in
breast cancer cell lines, RT-qPCR and western blotting were
performed to analyze PDCD4 expression in breast cancer cells that
gained or lost FOXP3 expression. It was reported that the ectopic
expression of FOXP3 in MDA-MB-231 cells upregulated PDCD4
expression and that silencing endogenous FOXP3 in MCF-7 cells
downregulated PDCD4 expression at both the mRNA and protein levels
(Fig. 5B-E). Then, cell apoptosis
was evaluated in FOXP3-overexpressing cells transfected with PDCD4
siRNA or NC siRNA by flow cytometry analysis. The results
demonstrated that, compared with NC siRNA group, knockdown of PDCD4
in FOXP3-overexpressing cells could decrease apoptosis (Fig. 5F). Taken together, these results
indicated that FOXP3 may promote PDCD4 expression in breast
cancer.
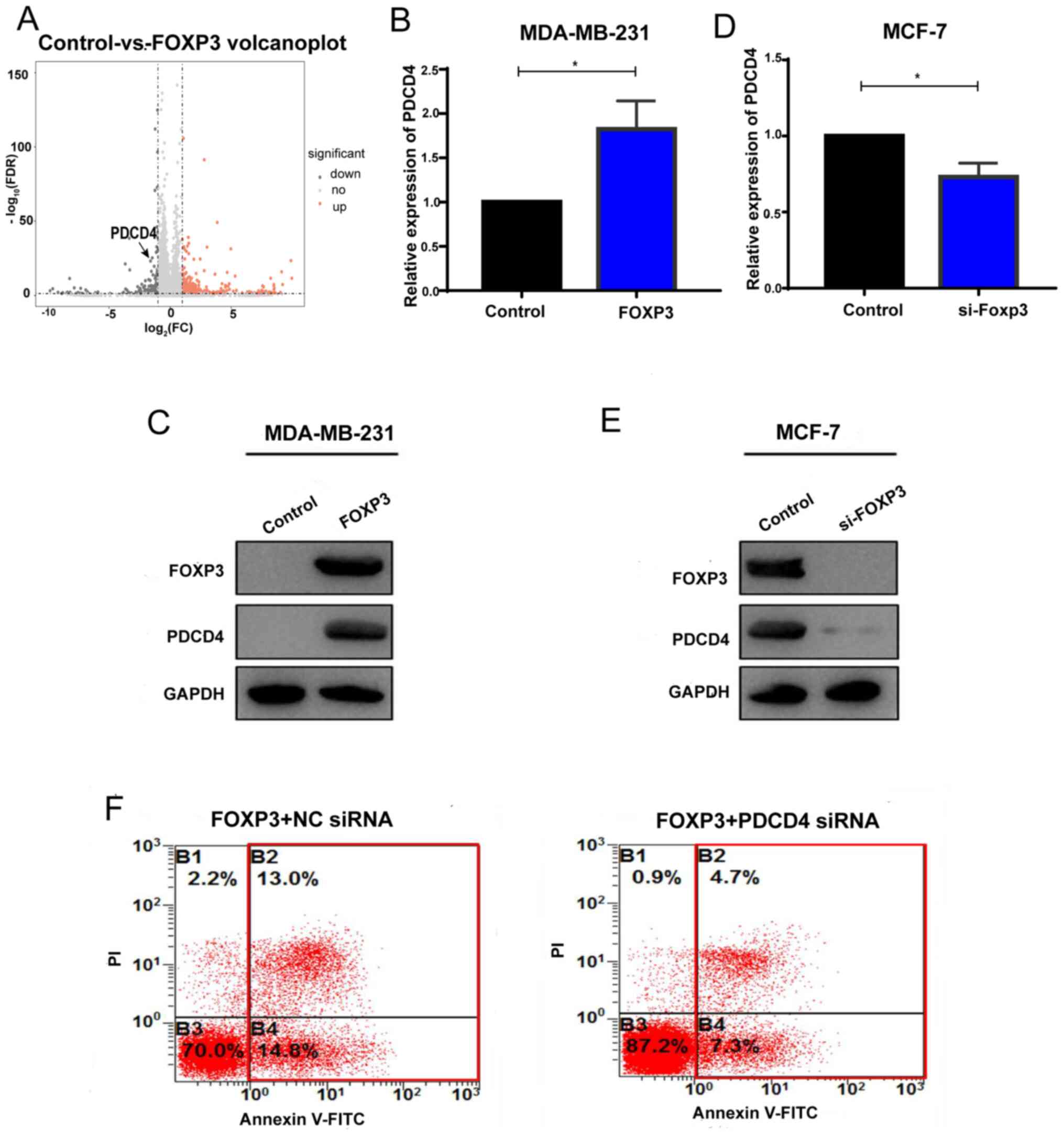 | Figure 5.FOXP3 upregulates PDCD4. (A)
Hierarchical cluster of the differential expression levels between
FOXP3-overexpressing MDA-MB-231 cells and wild-type MDA-MB-231
cells. PDCD4 was included in the upregulated genes. (B and C)
MDA-MB-231 cells were transfected with pcDNA3.1-FOXP3 or a control
vector, and RT-qPCR and western blotting were conducted to detect
the (B) mRNA and (C) protein levels of PDCD4. n=3. (D and E) MCF-7
cells were transfected with FOXP3 siRNA or scrambled RNA, and
RT-qPCR and western blotting were conducted to detect the (D) mRNA
and (E) protein levels of PDCD4. n=3. (F) Apoptosis was evaluated
in FOXP3-overexpressing cells transfected with PDCD4 siRNA or NC
siRNA. *P<0.05. FOXP3, forkhead box P3; PDCD4, programmed cell
death 4; RT-q, reverse transcription-quantitative; si, small
interfering; NC, negative control; FC, fold-change; FDR, false
discovery rate; down, downregulated; no, no difference; up,
upregulated. |
PDCD4 is negatively correlated with
breast cancer progression
To further identify the role of PDCD4 in breast
cancer, the associated between PDCD4 expression and survival was
analyzed for breast cancer samples using Kaplan-Meier plotter. As
shown in Fig. 6A and B, high PDCD4
expression was a protective factor for breast cancer RFS (HR=0.67;
log-rank P<0.001) and OS (HR=0.61; log-rank P<0.001). In
addition, by analyzing the expression of PDCD4 in breast cancer at
different stages, it was demonstrated that PDCD4 was associated
with the breast cancer stage (Fig.
6C). Finally, it was reported that the expression of FOXP3 was
positively correlated with PDCD4 in TCGA dataset (Fig. 6D). Collectively, these results
indicated that PDCD4 may have a suppressive role of breast cancer
progression and that FOXP3 might enhance breast cancer apoptosis by
upregulating PDCD4.
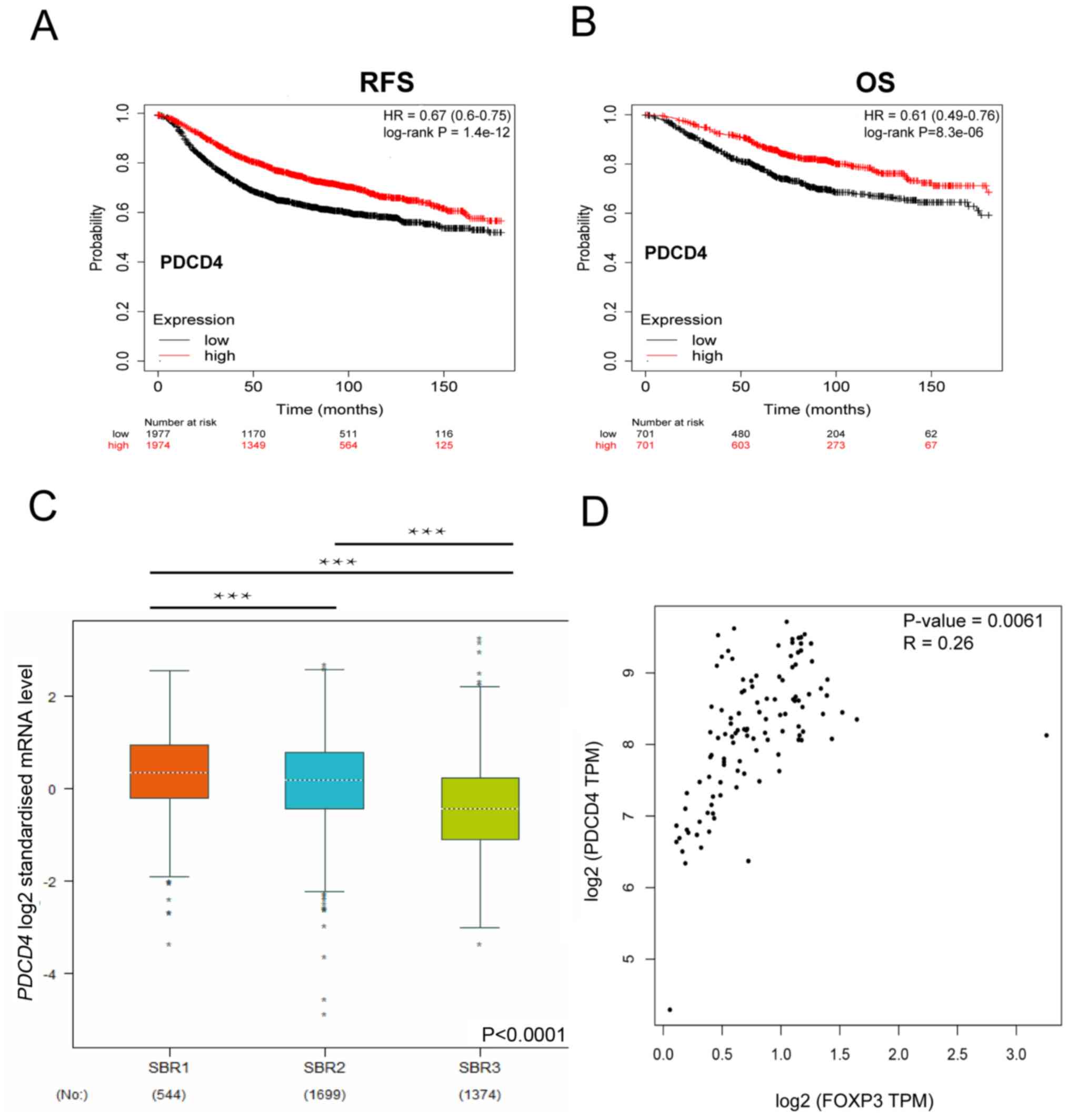 | Figure 6.PDCD4 is associated with breast
cancer progression. (A) Kaplan-Meier survival curves for RFS in
patients with high or low PDCD4. n=3,951. HR=−0.67; log-rank
P=P=1.4×10−12 (B) Kaplan-Meier survival curves for OS of
patients with high and low PDCD4 expression. n=1,402. HR=0.61;
log-rank P=8.3×10−6. (C) PDCD4 mRNA levels in different
stages of breast cancer from a public database. n=3,617 breast
cancer patients. (D) The expression of FOXP3 was positively
correlated with PDCD4 in The Cancer Genome Atlas dataset. Pearson's
correlation coefficient, P=0.0061, R=0.26. ***P<0.0001. RFS,
relapse-free survival; OS, overall survival; TPM, transcripts per
million; SBR, Scarff-Bloom-Richardson; PDCD4, programmed cell death
4; HR, hazard ratio. |
Discussion
Breast cancer is the most frequently diagnosed
cancer in women worldwide. Due to its high invasiveness and
metastasis, breast cancer is the leading cause of cancer-associated
death in women (24,25). In China, the breast cancer is also
the most common cancer in women, and it is occurring with
increasing frequency in younger women (26,27). Our
previous study has shown that FOXP3 is an important breast cancer
suppressor gene (28) and its loss
of function or mutation is closely associated with the development
and prognosis of breast cancer (12,15,29,30). As
a transcription factor, FOXP3 mainly exerts its anticancer function
by regulating the expression of its downstream target genes
(31,32). FOXP3 can inhibit the proliferation of
breast cancer cells by inhibiting the expression of oncogenes, such
as HER2 and S-phase kinase-associated protein 2 (14,33).
FOXP3 can also inhibit the metastasis of breast cancer by
inhibiting the expression of CD44 and C-X-C chemokine receptor type
4 (16,34). In addition, it has been reported that
FOXP3 can inhibit breast cancer angiogenesis by downregulating VEGF
(15). Therefore, the confirmation
and increased understanding of the target genes regulated by FOXP3
in breast cancer is critical, as it will be helpful to improve our
understanding of the anticancer function of FOXP3 and its clinical
application.
To further investigate the role of FOXP3 in breast
cancer progression, the present study constructed a
FOXP3-overexpressing MDA-MB-231 cell line. RNA-seq revealed that
apoptosis-related genes were significantly enriched in
FOXP3-overexpressing MDA-MB-231 cells, which suggested that FOXP3
may play a role in breast cancer cell apoptosis.
The balance of cell proliferation and death is the
key to the maintenance of homeostasis (35,36).
Abnormal cell proliferation and apoptosis are important causes of
numerous diseases such as Hirschsprung's disease and cancers
(37). PDCD4 is a key molecule in
apoptosis, which can inhibit cell growth, promoted by
downregulating the expression of MAP4K1 (23) or by inhibiting the interaction
between eukaryotic initiation factor 4A-I (EIF)4A1 and EIF4G
(38,39). Studies have revealed that PDCD4 is a
tumor suppressor gene that can inhibit tumor progression by
promoting apoptosis (40,41). Liu et al (42) first reported that FOXP3 could promote
breast cancer apoptosis by inducing microRNA-146 expression, which
inhibited NF-κB activation. The present study reported another
molecular mechanism by which FOXP3 induces apoptosis. Sequencing
analysis demonstrated that PDCD4 is highly expressed in
FOXP3-overexpressing MDA-MB-231 cells, and that FOXP3 promotes
breast cancer apoptosis by upregulating PDCD4 expression. The
current study revealed that FOXP3 can promote the expression of
PDCD4; however, the specific mechanism by which FOXP3 upregulates
PDCD4 requires further study.
The limitation of the present study was that RNA-seq
data was performed with one sample per group; however, PCR was
performed to further confirm the results of the existing RNA-seq
analysis. The qPCR results confirmed that PDCD4 was upregulated in
FOXP3-overexpressing MDA-MB-231 cells. In addition, the
overexpression and knockdown of FOXP3 was not performed in a same
breast cancer cell line. However, our previously studies confirmed
that MDA-MB-231 is a FOXP3 negative cell line, and MCF-7 is
FOXP3-positive cell line (14,16,33).
Therefore, the present study overexpressed FOXP3 in MDA-MB-231 and
knocked down FOXP3 expression in MCF-7 cells to explore the effect
of FOXP3 on PDCD4 expression. The results indicated that
overexpressing FOXP3 in MDA-MB-231 cells resulted in the
upregulation of PDCD4 at both the mRNA and protein levels, while
siRNA-mediated silencing of endogenous FOXP3 in MCF-7 cells
resulted in the downregulation of PDCD4, which indicated that FOXP3
can promote PDCD4 expression in breast cancer cells.
In conclusion, DEGs were identified in FOXP3-
overexpressing MDA-MB-231 cells compared with wild-type MDA-MB-231
cells using RNA-Seq analysis, and KEGG pathway analysis was
performed to examine the roles of FOXP3 in breast cancer. Notably,
it was demonstrated that the expression level of FOXP3 is closely
associated with apoptosis using bioinformatics analysis.
Furthermore, it was confirmed that PDCD4 expression, a key molecule
of apoptosis (23), can be promoted
by FOXP3. Therefore, the present study provides insights into the
roles of FOXP3 in breast cancer, suggesting that FOXP3 can induce
breast cancer apoptosis by promoting the expression of PDCD4.
Ultimately, this provides novel insights into the anticancer
function of FOXP3.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Guangzhou Gene
Denovo Biotechnology Co., Ltd for assisting in sequencing.
Funding
This work was funded by the National Natural Science
Foundation of China (grant nos. 81902678, 81672864, 81702590,
81802632, 81672800 and 81673020) and the Key Research and
Development Program of Shaanxi Province (grant nos.
2017ZDCXL-SF-01-03 and 2017SF-149).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contribution
JL and SW conceived and designed the study. YX and
DF drafted the manuscript. JL and CZ contributed to the later
revision and finalization of this paper. YX and CZ constructed the
FOXP3 transgenic MDA-MB-231 cell line. DF, JH and LZ performed the
molecular biological experiments. YX, CZ, SW and HZ contributed to
the bioinformatics analysis. YZ contributed to data collection
preliminary analysis. YZ and CZ reviewed and edited the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Deng C, Zhang Q, Jia M, Zhao J, Sun X,
Gong T and Zhang Z: Tumors and their microenvironment
dual-targeting chemotherapy with local immune adjuvant therapy for
effective antitumor immunity against breast cancer. Adv Sci
(Weinh). 6:18018682019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu
ZY, Shi W, Jiang J, Yao PP and Zhu HP: Risk factors and preventions
of breast cancer. Int J Biol Sci. 13:1387–1397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pranavathiyani G, Thanmalagan RR,
Leimarembi Devi N and Venkatesan A: Integrated transcriptome
interactome study of oncogenes and tumor suppressor genes in breast
cancer. Genes Dis. 6:78–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brunkow ME, Jeffery EW, Hjerrild KA,
Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF
and Ramsdell F: Disruption of a new forkhead/winged-helix protein,
scurfin, results in the fatal lymphoproliferative disorder of the
scurfy mouse. Nat Genet. 27:68–73. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rudensky AY: Regulatory T cells and Foxp3.
Immunol Rev. 241:260–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gregorczyk I and Maslanka T: Significant
expression of Foxp3 in murine extrathymic CD4+CD8+ double positive
T cells. Pol J Vet Sci. 20:815–817. 2017.PubMed/NCBI
|
|
11
|
Pierini A, Nishikii H, Baker J, Kimura T,
Kwon HS, Pan Y, Chen Y, Alvarez M, Strober W, Velardi A, et al:
Foxp3+ regulatory T cells maintain the bone marrow
microenvironment for B cell lymphopoiesis. Nat Commun. 8:150682017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Douglass S, Ali S, Meeson AP, Browell D
and Kirby JA: The role of FOXP3 in the development and metastatic
spread of breast cancer. Cancer Metastasis Rev. 31:843–854. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karanikas V, Speletas M, Zamanakou M,
Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI and
Germenis AE: Foxp3 expression in human cancer cells. J Transl Med.
6:192008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zuo T, Wang L, Morrison C, Chang X, Zhang
H, Li W, Liu Y, Wang Y, Liu X, Chan MW, et al: FOXP3 is an X-linked
breast cancer suppressor gene and an important repressor of the
HER-2/ErbB2 oncogene. Cell. 129:1275–1286. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Gao Y, Li J, Zhang K, Han J, Li W,
Hao Q, Zhang W, Wang S, Zeng C, et al: FOXP3 inhibits angiogenesis
by downregulating VEGF in breast cancer. Cell Death Dis. 9:7442018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang C, Xu Y, Hao Q, Wang S, Li H, Li J,
Gao Y, Li M, Li W, Xue X, et al: FOXP3 suppresses breast cancer
metastasis through downregulation of CD44. Int J Cancer.
137:1279–1290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan H, Xin S, Huang Y, Bao Y, Jiang H,
Zhou L, Ren X, Li L, Wang Q and Zhang J: Downregulation of PDCD4 by
miR-21 suppresses tumor transformation and proliferation in a nude
mouse renal cancer model. Oncol Lett. 14:3371–3378. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang HS, Matthews CP, Clair T, Wang Q,
Baker AR, Li CC, Tan TH and Colburn NH: Tumorigenesis suppressor
Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase
kinase 1 expression to suppress colon carcinoma cell invasion. Mol
Cell Biol. 26:1297–1306. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ghislain I, Zikos E, Coens C, Quinten C,
Balta V, Tryfonidis K, Piccart M, Zardavas D, Nagele E,
Bjelic-Radisic V, et al: Health-related quality of life in locally
advanced and metastatic breast cancer: Methodological and clinical
issues in randomised controlled trials. Lancet Oncol. 17:e294–e304.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Y, Wang Z, Hao Q, Li W, Xu Y, Zhang J,
Zhang W, Wang S, Liu S, Li M, et al: Loss of ERα induces
amoeboid-like migration of breast cancer cells by downregulating
vinculin. Nat Commun. 8:144832017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qin H, Yu H, Sheng J, Zhang D, Shen N, Liu
L, Tang Z and Chen X: PI3Kgamma inhibitor attenuates
immunosuppressive effect of poly(l-Glutamic Acid)-combretastatin A4
conjugate in metastatic breast cancer. Adv Sci (Weinh).
6:19003272019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao Y, Li X, Shu Z, Zhang K, Xue X, Li W,
Hao Q, Wang Z, Zhang W, Wang S, et al: Nuclear galectin-1-FOXP3
interaction dampens the tumor-suppressive properties of FOXP3 in
breast cancer. Cell Death Dis. 9:4162018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tian T, Wang M, Zheng Y, Yang T, Zhu W, Li
H, Lin S, Liu K, Xu P, Deng Y, et al: Association of two FOXP3
polymorphisms with breast cancer susceptibility in Chinese Han
women. Cancer Manag Res. 10:867–872. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Recouvreux MS, Grasso EN, Echeverria PC,
Rocha-Viegas L, Castilla LH, Schere-Levy C, Tocci JM, Kordon EC and
Rubinstein N: RUNX1 and FOXP3 interplay regulates expression of
breast cancer related genes. Oncotarget. 7:6552–6565. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Katoh H, Zheng P and Liu Y: Signalling
through FOXP3 as an X-linked tumor suppressor. Int J Biochem Cell
Biol. 42:1784–1787. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lopes JE, Torgerson TR, Schubert LA,
Anover SD, Ocheltree EL, Ochs HD and Ziegler SF: Analysis of FOXP3
reveals multiple domains required for its function as a
transcriptional repressor. J Immunol. 177:3133–3142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zuo T, Liu R, Zhang H, Chang X and Liu Y,
Wang L, Zheng P and Liu Y: FOXP3 is a novel transcriptional
repressor for the breast cancer oncogene SKP2. J Clin Invest.
117:3765–3773. 2007.PubMed/NCBI
|
|
34
|
Douglass S, Meeson AP, Overbeck-Zubrzycka
D, Brain JG, Bennett MR, Lamb CA, Lennard TW, Browell D, Ali S and
Kirby JA: Breast cancer metastasis: Demonstration that FOXP3
regulates CXCR4 expression and the response to CXCL12. J Pathol.
234:74–85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abraha AM and Ketema EB: Apoptotic
pathways as a therapeutic target for colorectal cancer treatment.
World J Gastrointest Oncol. 8:583–591. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hipfner DR and Cohen SM: Connecting
proliferation and apoptosis in development and disease. Nat Rev Mol
Cell Biol. 5:805–815. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gudipaty SA, Conner CM, Rosenblatt J and
Montell DJ: Unconventional ways to live and die: Cell death and
survival in development, homeostasis, and disease. Annu Rev Cell
Dev Biol. 34:311–332. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Modelska A, Turro E, Russell R, Beaton J,
Sbarrato T, Spriggs K, Miller J, Graf S, Provenzano E, Blows F, et
al: The malignant phenotype in breast cancer is driven by
eIF4A1-mediated changes in the translational landscape. Cell Death
Dis. 6:e16032015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goggin MM, Nelsen CJ, Kimball SR,
Jefferson LS, Morley SJ and Albrecht JH: Rapamycin-sensitive
induction of eukaryotic initiation factor 4F in regenerating mouse
liver. Hepatology. 40:537–544. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Song X, Zhang X, Wang X, Zhu F, Guo C,
Wang Q, Shi Y, Wang J, Chen Y and Zhang L: Tumor suppressor gene
PDCD4 negatively regulates autophagy by inhibiting the expression
of autophagy-related gene ATG5. Autophagy. 9:743–755. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Li Y, Wan L, Liu Y, Sun Y, Liu Y,
Shi Y, Zhang L, Zhou H, Wang J, et al: Downregulation of PDCD4
induced by progesterone is mediated by the PI3K/AKT signaling
pathway in human endometrial cancer cells. Oncol Rep. 42:849–856.
2019.PubMed/NCBI
|
|
42
|
Liu R, Liu C, Chen D, Yang WH, Liu X, Liu
CG, Dugas CM, Tang F, Zheng P, Liu Y and Wang L: FOXP3 controls an
miR-146/NF-KB negative feedback loop that inhibits apoptosis in
breast cancer cells. Cancer Res. 75:1703–1713. 2015. View Article : Google Scholar : PubMed/NCBI
|