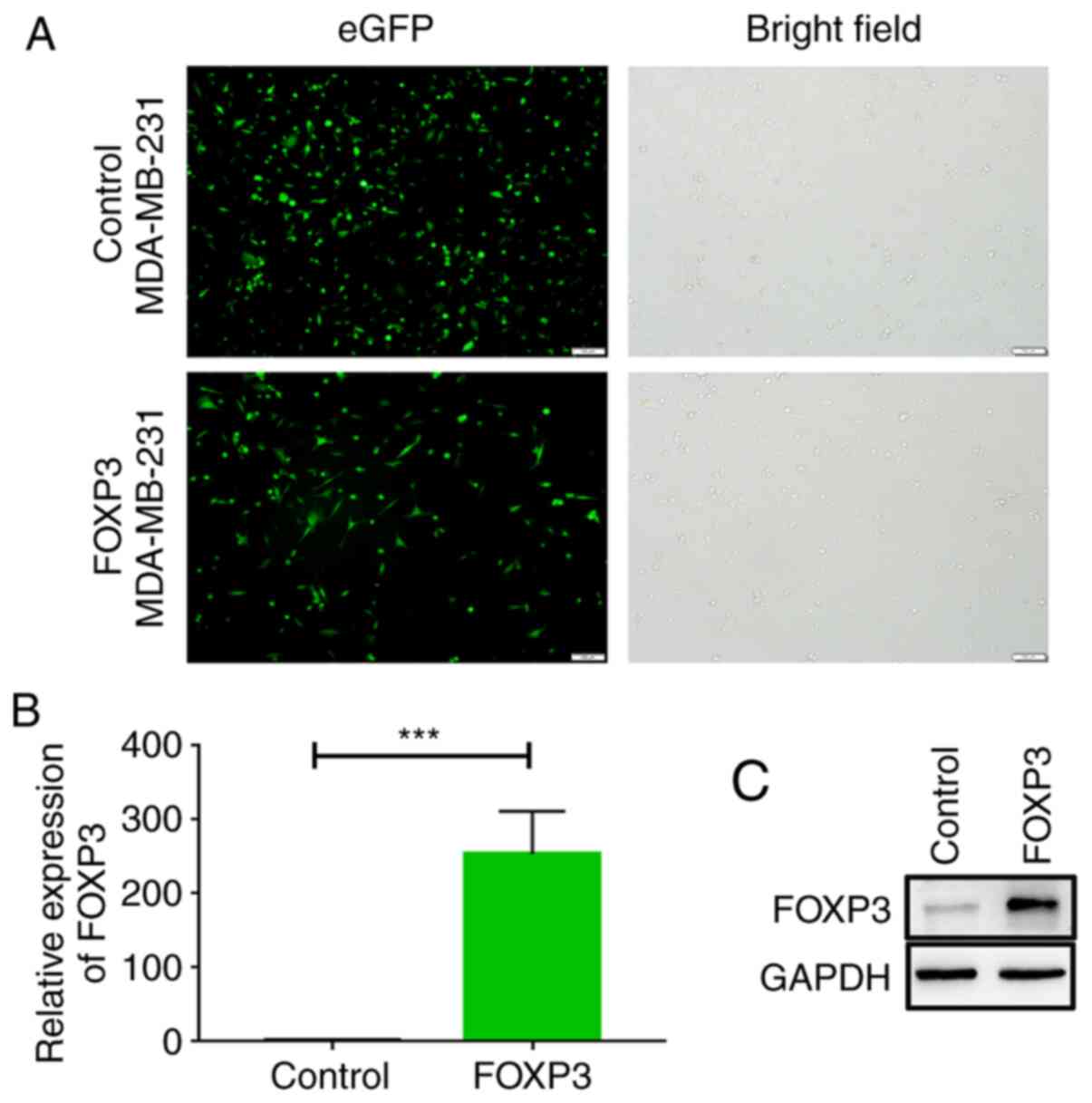
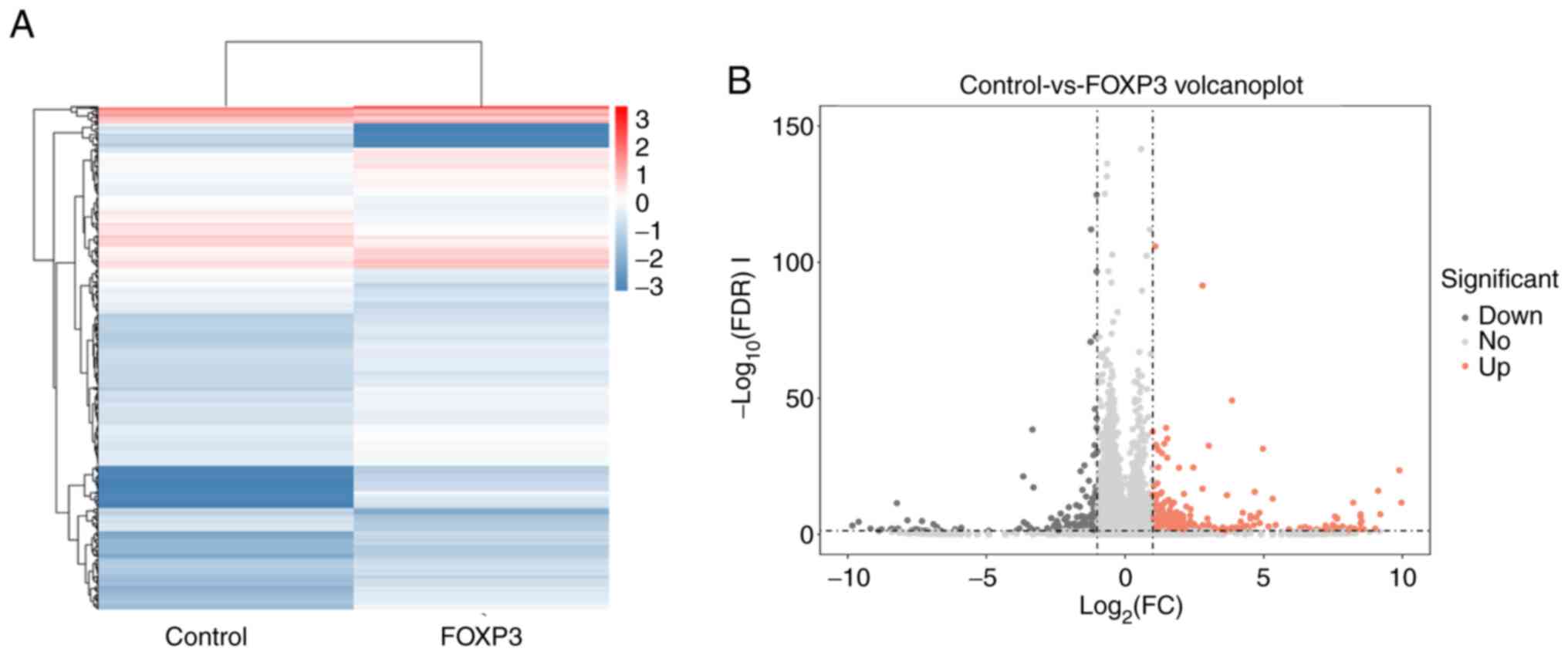
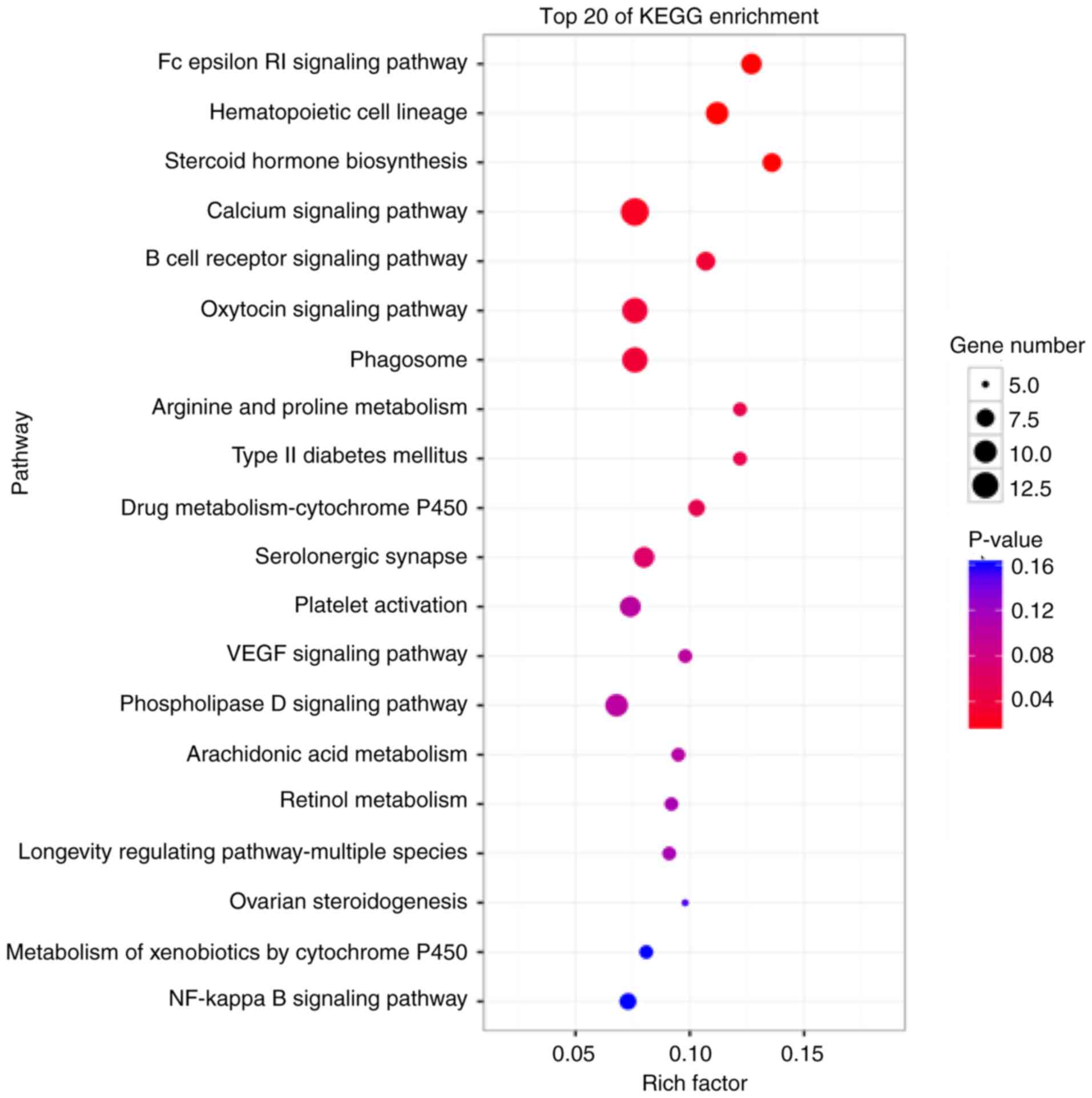
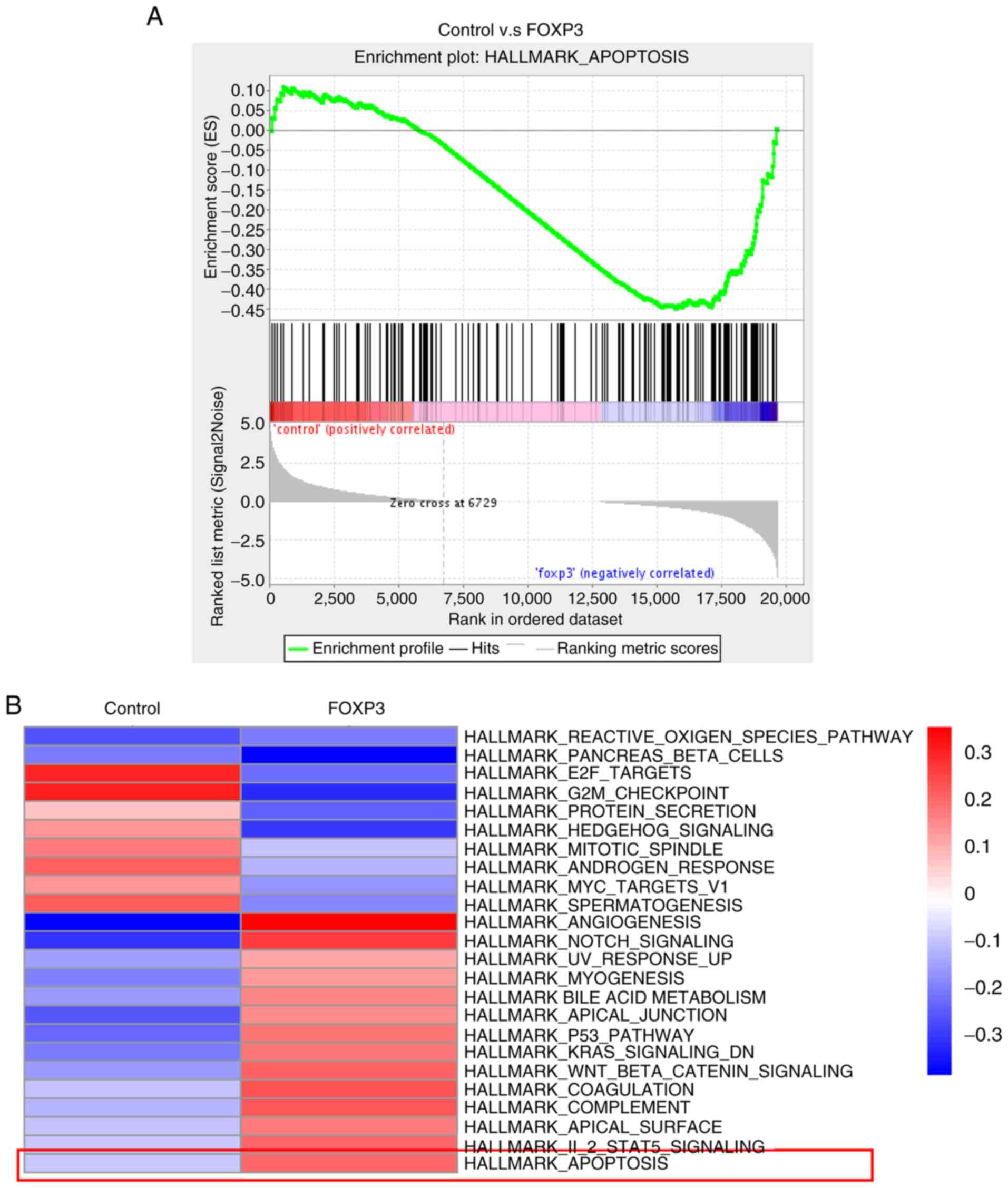
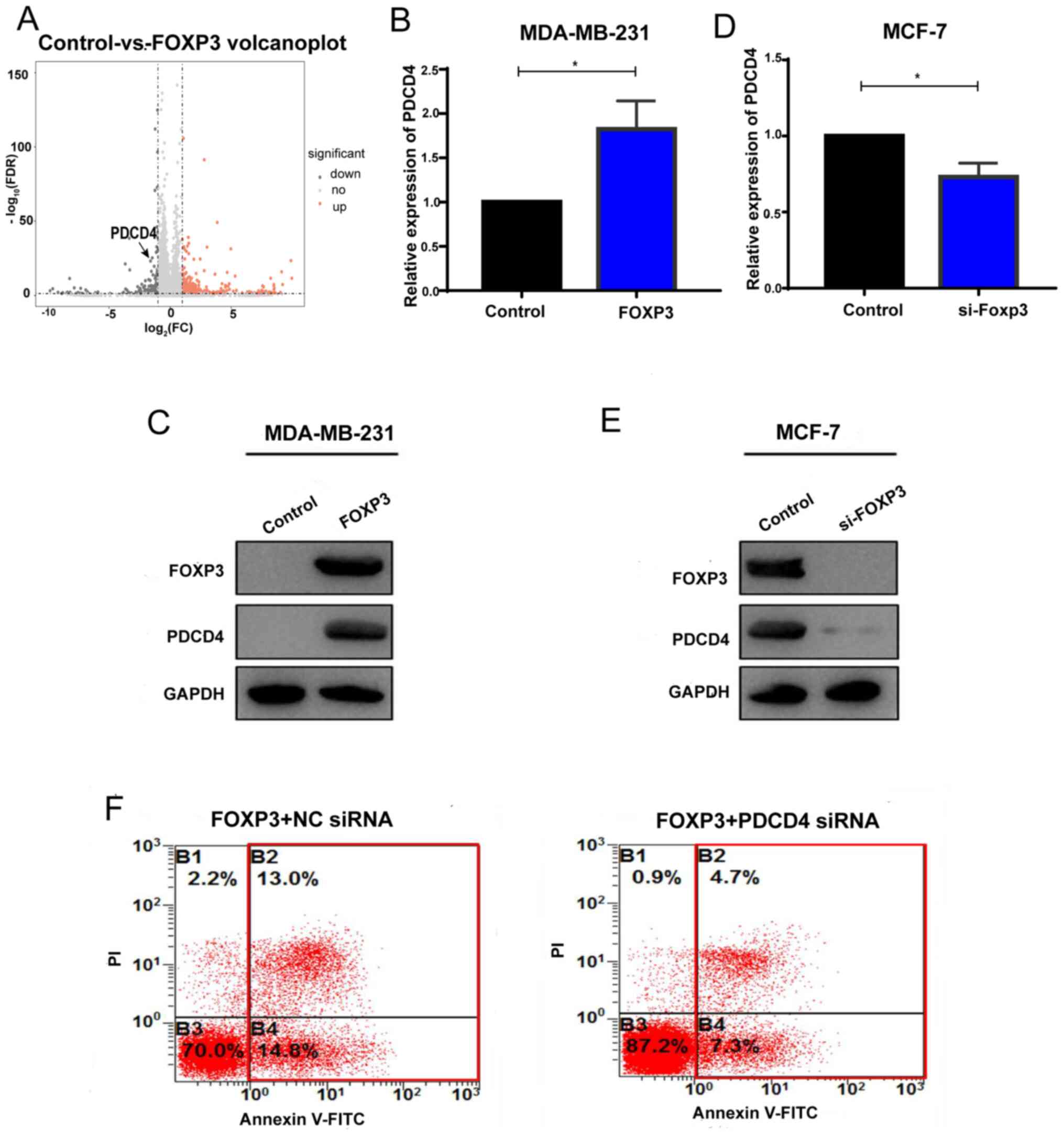
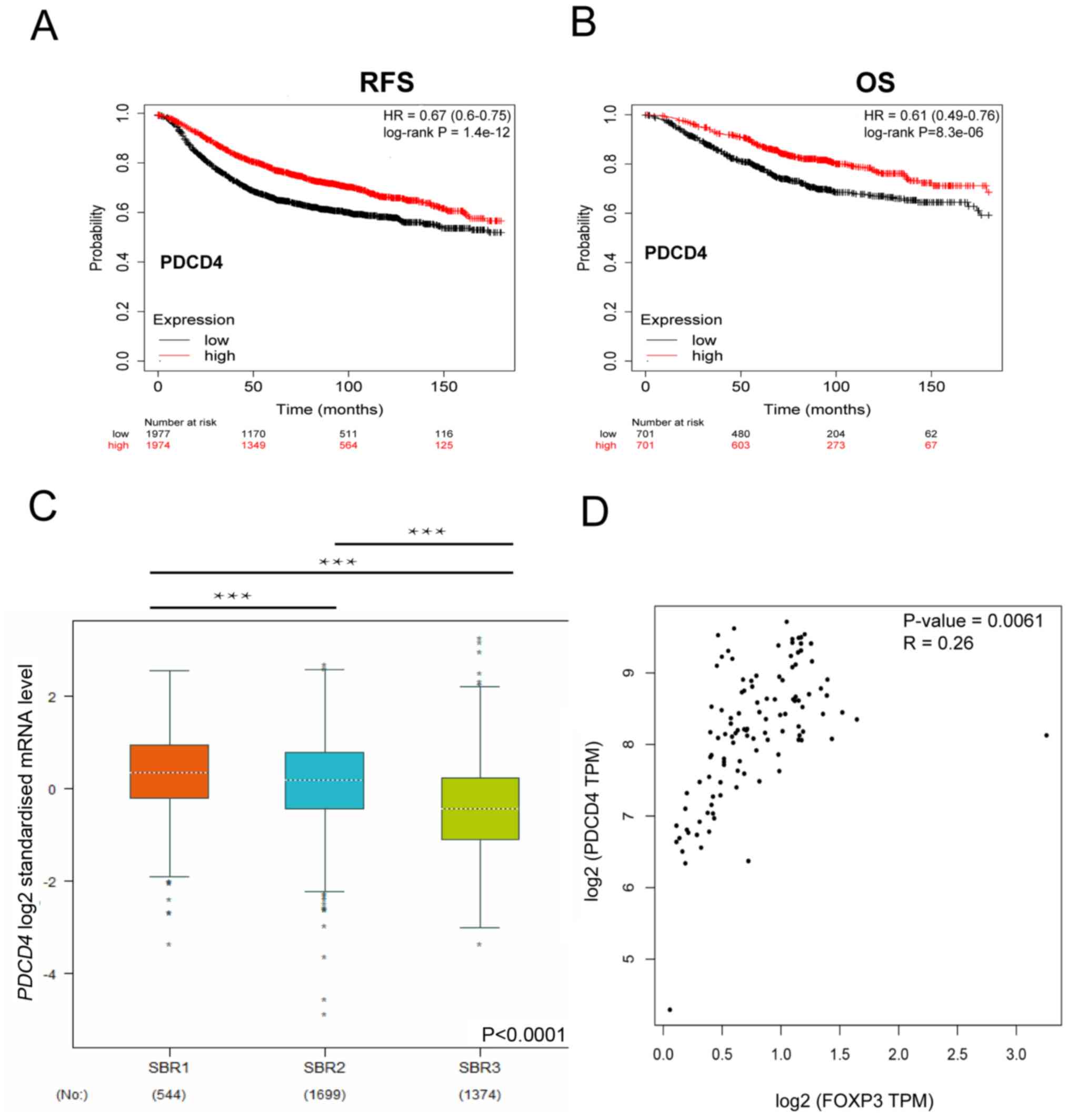
|
1
|
Deng C, Zhang Q, Jia M, Zhao J, Sun X,
Gong T and Zhang Z: Tumors and their microenvironment
dual-targeting chemotherapy with local immune adjuvant therapy for
effective antitumor immunity against breast cancer. Adv Sci
(Weinh). 6:18018682019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu
ZY, Shi W, Jiang J, Yao PP and Zhu HP: Risk factors and preventions
of breast cancer. Int J Biol Sci. 13:1387–1397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pranavathiyani G, Thanmalagan RR,
Leimarembi Devi N and Venkatesan A: Integrated transcriptome
interactome study of oncogenes and tumor suppressor genes in breast
cancer. Genes Dis. 6:78–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brunkow ME, Jeffery EW, Hjerrild KA,
Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF
and Ramsdell F: Disruption of a new forkhead/winged-helix protein,
scurfin, results in the fatal lymphoproliferative disorder of the
scurfy mouse. Nat Genet. 27:68–73. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rudensky AY: Regulatory T cells and Foxp3.
Immunol Rev. 241:260–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gregorczyk I and Maslanka T: Significant
expression of Foxp3 in murine extrathymic CD4+CD8+ double positive
T cells. Pol J Vet Sci. 20:815–817. 2017.PubMed/NCBI
|
|
11
|
Pierini A, Nishikii H, Baker J, Kimura T,
Kwon HS, Pan Y, Chen Y, Alvarez M, Strober W, Velardi A, et al:
Foxp3+ regulatory T cells maintain the bone marrow
microenvironment for B cell lymphopoiesis. Nat Commun. 8:150682017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Douglass S, Ali S, Meeson AP, Browell D
and Kirby JA: The role of FOXP3 in the development and metastatic
spread of breast cancer. Cancer Metastasis Rev. 31:843–854. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karanikas V, Speletas M, Zamanakou M,
Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI and
Germenis AE: Foxp3 expression in human cancer cells. J Transl Med.
6:192008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zuo T, Wang L, Morrison C, Chang X, Zhang
H, Li W, Liu Y, Wang Y, Liu X, Chan MW, et al: FOXP3 is an X-linked
breast cancer suppressor gene and an important repressor of the
HER-2/ErbB2 oncogene. Cell. 129:1275–1286. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Gao Y, Li J, Zhang K, Han J, Li W,
Hao Q, Zhang W, Wang S, Zeng C, et al: FOXP3 inhibits angiogenesis
by downregulating VEGF in breast cancer. Cell Death Dis. 9:7442018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang C, Xu Y, Hao Q, Wang S, Li H, Li J,
Gao Y, Li M, Li W, Xue X, et al: FOXP3 suppresses breast cancer
metastasis through downregulation of CD44. Int J Cancer.
137:1279–1290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan H, Xin S, Huang Y, Bao Y, Jiang H,
Zhou L, Ren X, Li L, Wang Q and Zhang J: Downregulation of PDCD4 by
miR-21 suppresses tumor transformation and proliferation in a nude
mouse renal cancer model. Oncol Lett. 14:3371–3378. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang HS, Matthews CP, Clair T, Wang Q,
Baker AR, Li CC, Tan TH and Colburn NH: Tumorigenesis suppressor
Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase
kinase 1 expression to suppress colon carcinoma cell invasion. Mol
Cell Biol. 26:1297–1306. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ghislain I, Zikos E, Coens C, Quinten C,
Balta V, Tryfonidis K, Piccart M, Zardavas D, Nagele E,
Bjelic-Radisic V, et al: Health-related quality of life in locally
advanced and metastatic breast cancer: Methodological and clinical
issues in randomised controlled trials. Lancet Oncol. 17:e294–e304.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Y, Wang Z, Hao Q, Li W, Xu Y, Zhang J,
Zhang W, Wang S, Liu S, Li M, et al: Loss of ERα induces
amoeboid-like migration of breast cancer cells by downregulating
vinculin. Nat Commun. 8:144832017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qin H, Yu H, Sheng J, Zhang D, Shen N, Liu
L, Tang Z and Chen X: PI3Kgamma inhibitor attenuates
immunosuppressive effect of poly(l-Glutamic Acid)-combretastatin A4
conjugate in metastatic breast cancer. Adv Sci (Weinh).
6:19003272019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao Y, Li X, Shu Z, Zhang K, Xue X, Li W,
Hao Q, Wang Z, Zhang W, Wang S, et al: Nuclear galectin-1-FOXP3
interaction dampens the tumor-suppressive properties of FOXP3 in
breast cancer. Cell Death Dis. 9:4162018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tian T, Wang M, Zheng Y, Yang T, Zhu W, Li
H, Lin S, Liu K, Xu P, Deng Y, et al: Association of two FOXP3
polymorphisms with breast cancer susceptibility in Chinese Han
women. Cancer Manag Res. 10:867–872. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Recouvreux MS, Grasso EN, Echeverria PC,
Rocha-Viegas L, Castilla LH, Schere-Levy C, Tocci JM, Kordon EC and
Rubinstein N: RUNX1 and FOXP3 interplay regulates expression of
breast cancer related genes. Oncotarget. 7:6552–6565. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Katoh H, Zheng P and Liu Y: Signalling
through FOXP3 as an X-linked tumor suppressor. Int J Biochem Cell
Biol. 42:1784–1787. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lopes JE, Torgerson TR, Schubert LA,
Anover SD, Ocheltree EL, Ochs HD and Ziegler SF: Analysis of FOXP3
reveals multiple domains required for its function as a
transcriptional repressor. J Immunol. 177:3133–3142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zuo T, Liu R, Zhang H, Chang X and Liu Y,
Wang L, Zheng P and Liu Y: FOXP3 is a novel transcriptional
repressor for the breast cancer oncogene SKP2. J Clin Invest.
117:3765–3773. 2007.PubMed/NCBI
|
|
34
|
Douglass S, Meeson AP, Overbeck-Zubrzycka
D, Brain JG, Bennett MR, Lamb CA, Lennard TW, Browell D, Ali S and
Kirby JA: Breast cancer metastasis: Demonstration that FOXP3
regulates CXCR4 expression and the response to CXCL12. J Pathol.
234:74–85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abraha AM and Ketema EB: Apoptotic
pathways as a therapeutic target for colorectal cancer treatment.
World J Gastrointest Oncol. 8:583–591. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hipfner DR and Cohen SM: Connecting
proliferation and apoptosis in development and disease. Nat Rev Mol
Cell Biol. 5:805–815. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gudipaty SA, Conner CM, Rosenblatt J and
Montell DJ: Unconventional ways to live and die: Cell death and
survival in development, homeostasis, and disease. Annu Rev Cell
Dev Biol. 34:311–332. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Modelska A, Turro E, Russell R, Beaton J,
Sbarrato T, Spriggs K, Miller J, Graf S, Provenzano E, Blows F, et
al: The malignant phenotype in breast cancer is driven by
eIF4A1-mediated changes in the translational landscape. Cell Death
Dis. 6:e16032015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goggin MM, Nelsen CJ, Kimball SR,
Jefferson LS, Morley SJ and Albrecht JH: Rapamycin-sensitive
induction of eukaryotic initiation factor 4F in regenerating mouse
liver. Hepatology. 40:537–544. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Song X, Zhang X, Wang X, Zhu F, Guo C,
Wang Q, Shi Y, Wang J, Chen Y and Zhang L: Tumor suppressor gene
PDCD4 negatively regulates autophagy by inhibiting the expression
of autophagy-related gene ATG5. Autophagy. 9:743–755. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Li Y, Wan L, Liu Y, Sun Y, Liu Y,
Shi Y, Zhang L, Zhou H, Wang J, et al: Downregulation of PDCD4
induced by progesterone is mediated by the PI3K/AKT signaling
pathway in human endometrial cancer cells. Oncol Rep. 42:849–856.
2019.PubMed/NCBI
|
|
42
|
Liu R, Liu C, Chen D, Yang WH, Liu X, Liu
CG, Dugas CM, Tang F, Zheng P, Liu Y and Wang L: FOXP3 controls an
miR-146/NF-KB negative feedback loop that inhibits apoptosis in
breast cancer cells. Cancer Res. 75:1703–1713. 2015. View Article : Google Scholar : PubMed/NCBI
|