Introduction
Combining chemotherapy drugs to treat cancer is a
common approach used in an attempt to enhance treatment strategies,
and to counteract the development of resistant cancer cells. For
example, utilising drugs such as folinic acid, 5-fluorouracil
(5-FU) and oxaliplatin (OXP) in combination that makes up the
FOLFOX chemotherapy regimen has brought good efficacy in approaches
to treat some colon cancers (1).
Similarly, in breast cancer, a combination of cyclophosphamide,
5-FU, epirubicin or methotrexate are often used together. However,
the use of particular chemotherapy drugs in certain cancer are
often selected based upon historical success of the single agent,
and rationales for chemotherapy combinations are often nebulous and
ill-defined (2).
Generally, drugs from different chemotherapeutic
classes are partnered with one another because they may exhibit
different mechanisms of action, have minimally overlapping spectra
of toxicity and target discrete phases of the cell cycle (3). This is thought to increase efficacy of
the treatments and reduces the probability of the tumour acquiring
resistance. However, despite cancer treatment algorithms and a
drive towards personalised medicine combination strategies can
still lack a rigorous scientific rationale, and few have been
assessed to establish optimal dose, schedule and delivery. Instead,
a ‘hit and hope’ approach can be adopted, which does not
necessarily lead to the most effective treatment for individual
patients. Indeed, the vagaries of combination regimen when
considered with the intrinsic diversity of tumours and
sensitivities to differing treatments, means a more rational plan
for the combination of chemotherapies is needed.
Identifying the drug combinations suitable for
certain diseases requires a more careful understanding of the
mechanisms of action for each drug involved in a regimen. It is
only by understanding profoundly the effects of each that the best
partners can be identified. However, and more bafflingly, it is
also not enough to understand the effect of a drug as these
typically are assessed in single-agent setting. Once drugs are
combined, the profile and complexity of the interactions can alter
the expected outcome. For example the use of monoclonal antibodies
to neutralise cytotoxic T-lymphocyte associated antigen 4 (CTLA-4)
can induce anticancer activity; however, this can cause a
compensatory increase in the programmed-death 1 (PD-1) receptor,
which can ultimately suppress the overall anticancer action. For
this reason, the sequential use of CTLA-4 and PD-1 antagonists
could overcome the resistance by using the former drug alone
(4).
In addition to simply killing, chemotherapeutic
drugs can interfere with intracellular processes rendering cells
more susceptible to other treatments, be they chemo-, immuno- or
radiotherapies (5–7). The effects on cell signalling can occur
even when the chemotherapy is used at doses that do not have a
great effect on cell number (8,9).
Attempts are being undertaken to assess the impact of the genetic
landscape on drug efficacy (10).
Knowledge of the signalling modulation and transcriptional changes
instigated by each chemotherapy will allow for a more holistic
approach to combination selection. Current thinking goes even
further than this with ‘precision’ chemotherapy approaches being
investigated, where even knowledge of the tumour's transcriptional
background being taken into consideration when designing treatment
regimens (8,11,12).
Chemotherapy drugs can be divided into a number of
different classes, including: Alkylating agents, such as
oxaliplatin (OXP), that work by covalently cross-linking
deoxyribonucleic acid (DNA) strands via their alkyl group;
antimetabolites, such as gemcitabine (GEM), that block DNA
replication; mitosis dysregulators, such as docetaxel (DOC), that
stop cancer cells completing mitosis by interfering with proper
microtubule function and others, such as the iMiDs, which target
cereblon and have anti-proliferative, anti-inflammatory and
anti-angiogenic properties (13,14), and
the artemisinins, which are anti-malarial drugs that have been
reported to not only retard tumour cell growth in vitro but
also be excellent combinatorial partners for other drugs (15,16).
Artemisinins initiate apoptosis in tumour cells through
iron-catalyzed lysosomal activation and reactive oxygen species
production (17,18).
In this study the colorectal tumour cell line,
HCT116, was treated with equi-active concentrations on OXP, GEM,
DOC, the iMiD lenalidomide (LEN) and a semi-synthetic derivative of
artemisinin, artesunate (ARS). A brief summary of each drug is
shown in Table I. The effect of each
drug on gene transcription was then measured by microarray and this
was used to predict whether particular drugs would be exhibit
synergistic or antagonistic characteristics when combined with one
another to inhibit tumour cell growth.
 | Table I.List of drugs used for microarray
experiments. |
Table I.
List of drugs used for microarray
experiments.
| Name | Class | Target |
|---|
| Artesunate | Anti-malarial | Lysosomal Iron |
| Camptothecin | Topoisomerase
Inhibitor | Topoisomerase |
| Docetaxel | Mitosis
inhibitor | Microtubules |
| Gemcitabine | Antimetabolite | DNA |
| Lenalidomide | Immune
modulatory | Cereblon |
| Naltrexone | Other | Not known |
| Oxaliplatin | Alkylating
agent | DNA |
Materials and methods
Tumour cell lines
The human colorectal cancer cell line HCT116 (Public
Health England) was grown in complete DMEM medium (Sigma-Aldrich,
Dorset, UK) supplemented with 10% foetal bovine serum (FBS)
(Invitrogen, Paisley, UK), 2 mM L-glutamine and 1%
penicillin/streptomycin (both Sigma). Authentication of this cell
line was performed by the service provider using the AmpFISTR
Identifier Plus PCR amplification kit looking for the presence of
<10 known loci for each cell line. Cells were incubated in a
humidified atmosphere with 5% CO2 in air at 37°C. When
approximately 75% confluency was reached, cells were harvested with
trypsin (Sigma-Aldrich) prior to washing and reseeding at a lower
cell density. Only cells with a passage number <15 were used in
experiments.
Drugs
GEM, OXP, CPM, camptothecin (all from
Sigma-Aldrich), artesunate (ARS) (St. George's Hospital Pharmacy)
and LEN (Celgene Corp.) were reconstituted in phosphate-buffered
saline (PBS) (Sigma), or in the case of LEN and DOC, dimethyl
sulfoxide (DMSO) (Sigma-Aldrich), to create a top stock solution of
100 mM, which and stored at −80°C long-term and −20°C short-term
(up to one month). Where necessary, all drugs were diluted in PBS.
When adding drug to cell culture, volume and DMSO concentration
were maintained for each condition, DMSO concentration was always
<0.01%. Cells were allowed to adhere overnight before drugs were
added at equi-active concentrations based on our previous work
(5,15). Final concentrations were: ARS (1 µM),
DOC (10 nM), GEM (100 nM), LEN (1 µM), NAL (1 µM) and OXP (500 nM).
Tumour cells were then cultured for a further 48 h, at which time,
cell numbers were assessed by MTT. Additionally, the effect of
treatment on the gene expression was assessed by isolating RNA from
cells cultured with the drugs for 4 h. These samples were processed
for subsequent microarray analyses.
Illumina microarrays
RNA was isolated from control or HCT116 cells
treated with ARS, DOC, GEM, LEN, NAL or OXP using the Qiagen
mini-kit according to the instructions of the manufacturer. The
concentration and quality of the resultant RNA was determined using
NanoDrop and 2100 Bioanalyzer (Agilent Technologies). The RNA was
found to be extremely pure with OD260/280 ratios
measured by nanodrop >1.90 and electropherograms confirmed
intact RNA. Microarrays were performed by Dr Jayne Dennis at the
St. George's, University of London Biomics Centre. Biotinylated
cRNA was generated from 100 ng total RNA using the Illumina
TotalPrep RNA Amplification Kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to manufacturer's instructions. Equal
amounts (750 ng) of cRNA were hybridised to the Illumina human
HT12-v3 arrays for 18 h. These arrays consisted of more than 48,000
probes covering RefSeq and UniGene annotated genes, and
acquisitions were subsequently processed according to
manufacturer's instructions before scanning on an Illumina
BeadArray Reader. The image data were processed using default
values in GenomeStudio v2009.1 with imputation of missing data,
before loading onto GeneSpring v9.0 for data normalisation and
filtering. Gene-ontology (GO)-enrichment analysis was also
performed (19–21). Gene activity values represent the
mean of three separate experiments that were used as scientific
replicates i.e. each experiment represented a single well on the
microarray chip.
Methylthiazoletetrazolium (MTT)
assays
To study the effect of drugs on the number of viable
cells, cells growing exponentially were added to 96-well plates at
a density of 3×104 cells/ml in 180 µl complete medium.
After the cells had been allowed to adhere overnight, 20 µl of drug
stock solutions were added to the wells to the appropriate final
concentration. The number of viable cells was measured at 48 h
using a standard MTT-based assay without modifications. Briefly,
MTT (Sigma-Aldrich) was added to each well to give a working
concentration of 0.4 mg/ml, and plates returned to 37°C for a
further h. After this time, the medium was aspirated off, 200 µl
DMSO added to each well and plates agitated gently for one minute
before measuring optical density at 550 nm using a microplate
reader (Dynex-MRX II; Dynex Technologies Ltd.). Each sample was run
in triplicate and experiment was completed three times.
Statistical analysis of correlation
and gene ontology analysis
R2 values and Spearman's Rank Correlation
Coefficients were obtained using Microsoft Excel 2019 v1808. Gene
ontology (GO) analysis was performed using Gene Ontology Resource
(http://geneontology.org/).
Results
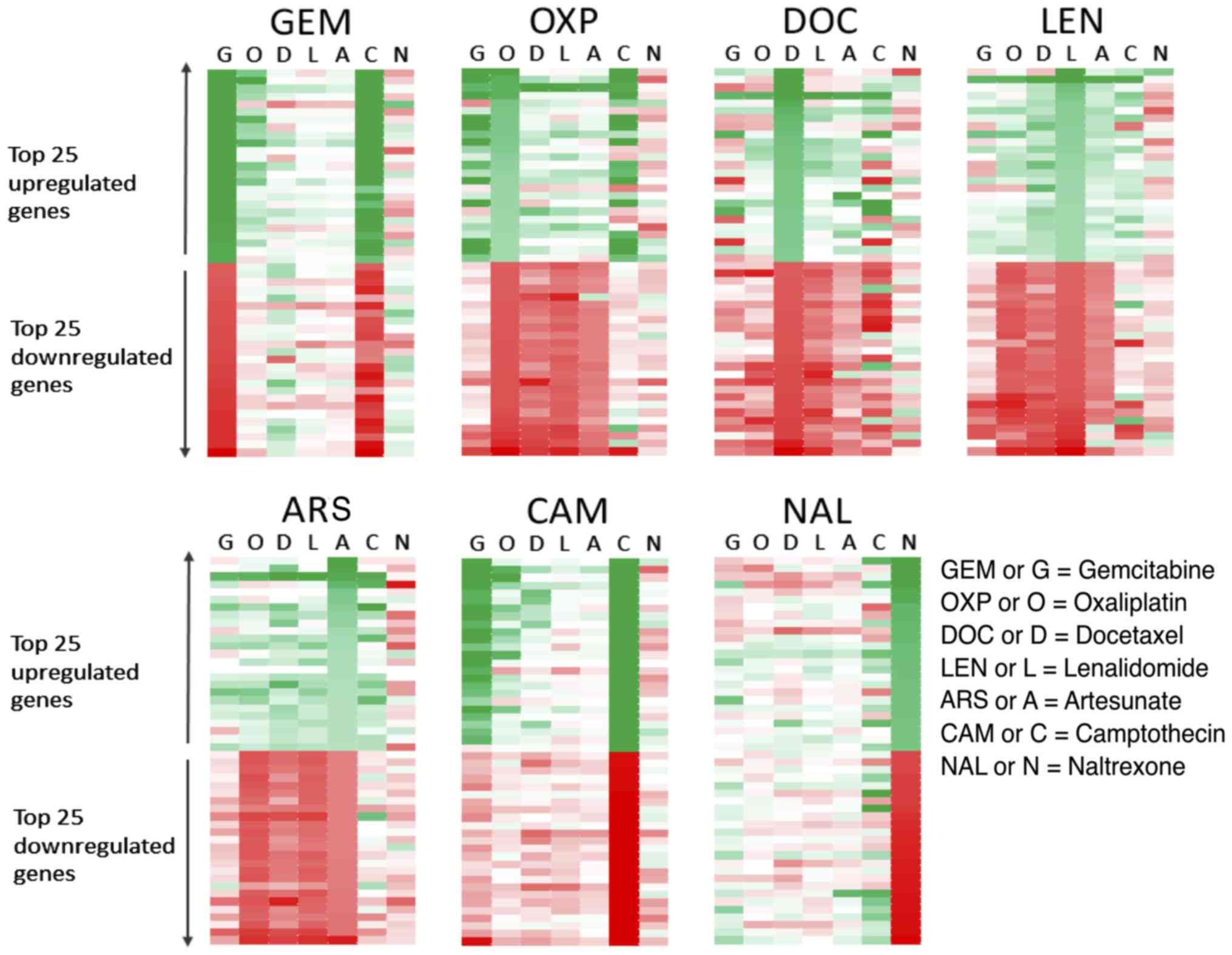
Each drug produced a specific
signature of gene activation/inhibition in HCT116 cells
HCT116 cells treated with equi-active concentrations
of various drugs were assessed by microarray. The 25 most
upregulated and 25 most downregulated genes were then listed for
each treatment and the change in those genes in response to the
other drugs was then evaluated (Fig.
1). Genes with a ≥3-fold mRNA expression increase after
treatment are shown in green, graded to white which indicates no
change. Genes with a ≥3-fold mRNA decrease after treatment
are shown in red, again graded to white which indicates no change.
The left-hand panel shows the genes that were most strongly
upregulated or downregulated in response to GEM and how those same
genes were affected by the other treatments. OXP and CAM appeared
to upregulate some of the same genes as GEM, as shown by the number
of genes highlighted in green in the second lane (O) and sixth lane
(C) in the top half of the panel. Genes that were strongly
upregulated by GEM were not greatly affected by DOC, LEN and ARS,
indicated by the pale grading of the genes in these lanes.
Conversely, genes that were downregulated by GEM, shown in the
bottom half of the left panel and coloured red were upregulated by
DOC and consequently appear green. This discordance between GEM and
DOC holds true when the analysis is performed on the genes most
affected by DOC treatment (3rd panel on top row), where many of the
genes that are most upregulated by treatment with DOC are
downregulated by GEM. By analysing the data in this way it is
possible to infer which treatments are more similar or more
different with regards to their effect on gene
transcription. The drugs DOC, LEN and ART seemed to produce a
similar pattern of gene changes in HCT116 cells, as did GEM, CAM
and OXP. The gene changes induced by naltrexone treatment did not
appear to share many similarities with the other treatments.
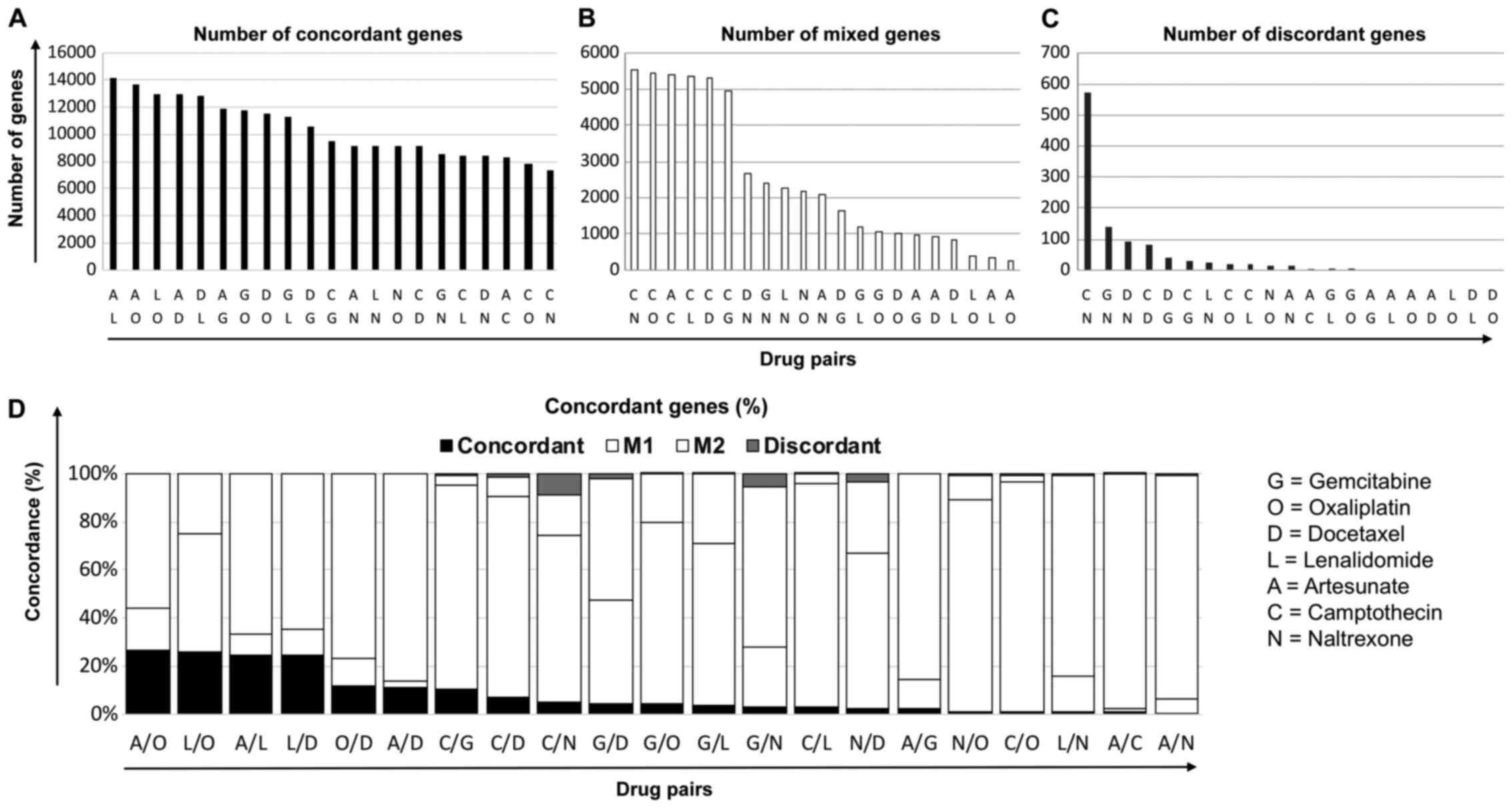
The pattern of affected genes was
similar between drugs with disparate mechanisms
To understand whether similarities in genetic
profiles of different drugs could determine the combination
benefit, HCT116 cells were cultured with equi-active concentrations
of each of the drug, and RNA extracted after 4 h for gene
microarray analyses. Changes in gene expression relative to the
untreated control were then sorted for each drug, and the
similarities in their patterns determined. Gene expression was
termed ‘altered by treatment’ if the expression had changed ±25% or
more from control and otherwise regarded as unchanged. Gene lists
were then judged against each other to determine how changes in the
expression of the individual genes compared and contrasted
following a particular treatment.
The effect of the treatment on each gene in the
microarray was assessed and changes that were in agreement in terms
of the direction of change, irrespective of magnitude, were defined
as being concordant. For example, expression of the gene, aurora
kinase A, was increased following treatment with GEM and CAM,
decreased after DOC and NAL and unchanged in response to OXP, LEN
and ART. Consequently, this gene was called concordant between GEM
and CAM, discordant between GEM and DOC or NAL and mixed between
GEM and the other treatment pairs. By using this method of
analysis, pairs of drugs could be assessed for the extent of the
concordance in genes. Results showed that ART and LEN had the
greatest number of genes (14,186) that were altered in a similar
manner following treatment (Fig.
2A). Conversely, CAM and NAL had the fewest number of
concordant genes (7343) and the most mixed (Fig. 2B) and discordant genes (Fig. 2C). As a larger proportion of
concordant genes could be those unchanged after treatment, these
were excluded in subsequent analysis where the percentage of
concordant, discordant or mixed genes for each treatment pair is
shown (Fig. 2D).
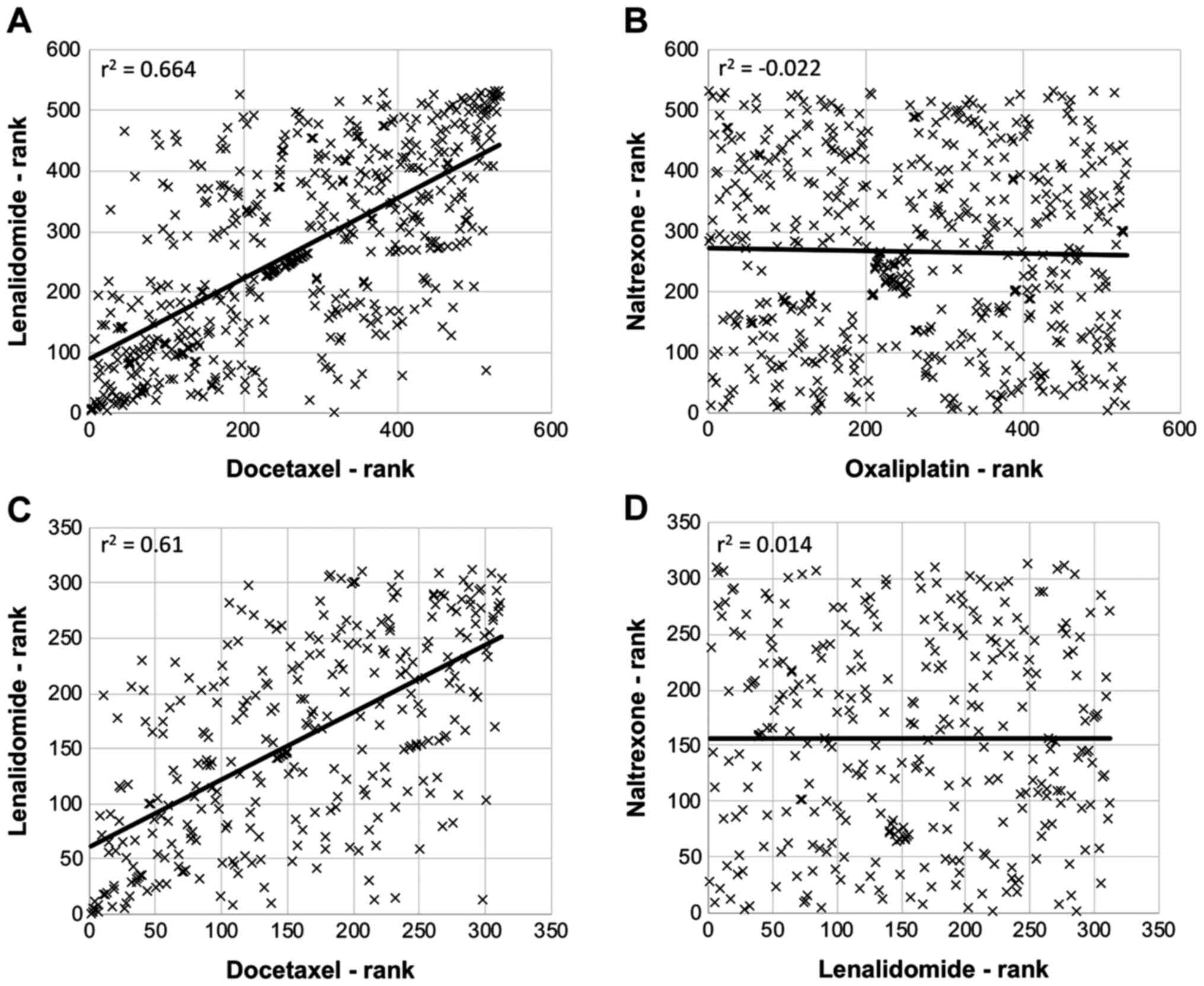
There were good correlations in the
changes to certain gene families following treatment
The effect of each of the drugs on genes named on
the Qiagen ‘apoptosis gene list’ was assessed. The changes were
then ranked in order of the magnitude of change for each gene, and
these ranked-lists compared with each other. For example, the
r2-value was 0.664 for lenalidomide vs. docetaxel, which
suggested a good positive correlation in apoptosis-genes affected
by the drugs (Fig. 3A). Conversely,
there was no correlation between the apoptosis-related genes
affected by naltrexone and oxaliplatin (r2=−0.022;
Fig. 3B). A similar analysis using
lists of genes involved in the cell cycle showed similarities and
differences between the drugs; with lenalidomide and docetaxel
being the drug pair displaying the highest correlation
(r2=0.610; Fig. 3C) and
naltrexone and lenalidomide displaying the least correlation
(r2=0.014; Fig. 3D). In
addition to any effects on apoptosis or cell cycle genes, gene
ontology enrichment analysis revealed which cellular processes were
impacted by each treatment (Table
II). Genes related to the DNA damage response, cellular stress,
apoptosis and cell cycle were overrepresented in the 100 most
affected genes after culturing cells with the DNA damaging
chemotherapies such as CAM, GEM and OXP. Each of these drugs seemed
to be involved in the activation or repression of numerous cellular
processes. In contrast, DOC showed only enrichment in ‘cell
division’ and LEN in no processes at all. That is not to say that
LEN did not affect any cellular process, just that the drug did not
affect any particular process disproportionately.
 | Table II.The one hundred most affected genes
(by fold change) from microarray data for each drug were used as
gene sets for analysis using the GO Resource Enrichment Analysis
Tool. This analysis determines which GO terms are over- or
under-represented using annotations for these gene sets. The table
shows significant shared GO Biological Process terms used to
describe each set of genes. These data reveal which cellular
mechanisms may be hit by each drug. |
Table II.
The one hundred most affected genes
(by fold change) from microarray data for each drug were used as
gene sets for analysis using the GO Resource Enrichment Analysis
Tool. This analysis determines which GO terms are over- or
under-represented using annotations for these gene sets. The table
shows significant shared GO Biological Process terms used to
describe each set of genes. These data reveal which cellular
mechanisms may be hit by each drug.
| Drug | GO Terms |
|---|
| Artesunate | Lipid droplet
organization; intrinsic apoptotic signaling pathway in response to
endoplasmic reticulum stress; positive regulation of neuron
apoptotic process; cellular protein localization; negative
regulation of gene expression; regulation of cellular metabolic
process; regulation of primary metabolic process; regulation of
nitrogen compound metabolic process. |
| Camptothecin | Positive regulation
of extrinsic apoptotic signaling pathway via death domain
receptors; intrinsic apoptotic signaling pathway in response to
oxidative stress; DNA damage response, signal transduction by p53
class mediator resulting in cell cycle arrest; apoptotic
mitochondrial changes; cellular response to UV; positive regulation
of protein kinase B signalling; cellular response to starvation;
positive regulation of protein kinase activity; regulation of cell
population proliferation; regulation of response to stress. |
| Docetaxel | Cell division. |
| Gemcitabine | Mitotic chromosome
movement towards spindle pole; meiotic sister chromatid cohesion,
centromeric; spindle assembly involved in meiosis; mitotic
metaphase plate congression; mitotic spindle assembly checkpoint;
DNA damage response, signal transduction by p53 class mediator
resulting in cell cycle arrest; cellular response to amino acid
starvation; positive regulation of cyclin-dependent protein kinase
activity; regulation of cyclin-dependent protein serine/threonine
kinase activity; regulation of mitotic spindle organization;
centromere complex assembly; mitotic spindle organization;
regulation of cytokinesis; G2/M transition of mitotic cell cycle;
cellular response to UV; anaphase-promoting complex-dependent
catabolic process; cell division; intrinsic apoptotic signaling
pathway; regulation of G2/M transition of mitotic cell cycle;
positive regulation of cellular catabolic process; DNA conformation
change; positive regulation of cell population proliferation;
regulation of apoptotic process. |
| Lenalidomide | N/A |
| Oxaliplatin | Response to
corticosterone; DNA damage response, signal transduction by p53
class mediator resulting in cell cycle arrest; positive regulation
of cysteine-type endopeptidase activity involved in apoptotic
process; intrinsic apoptotic signaling pathway; transcription
initiation from RNA polymerase II promoter; cellular response to
extracellular stimulus; regulation of neuron death; response to
radiation; regulation of apoptotic signaling pathway; cell
development; regulation of gene expression. |
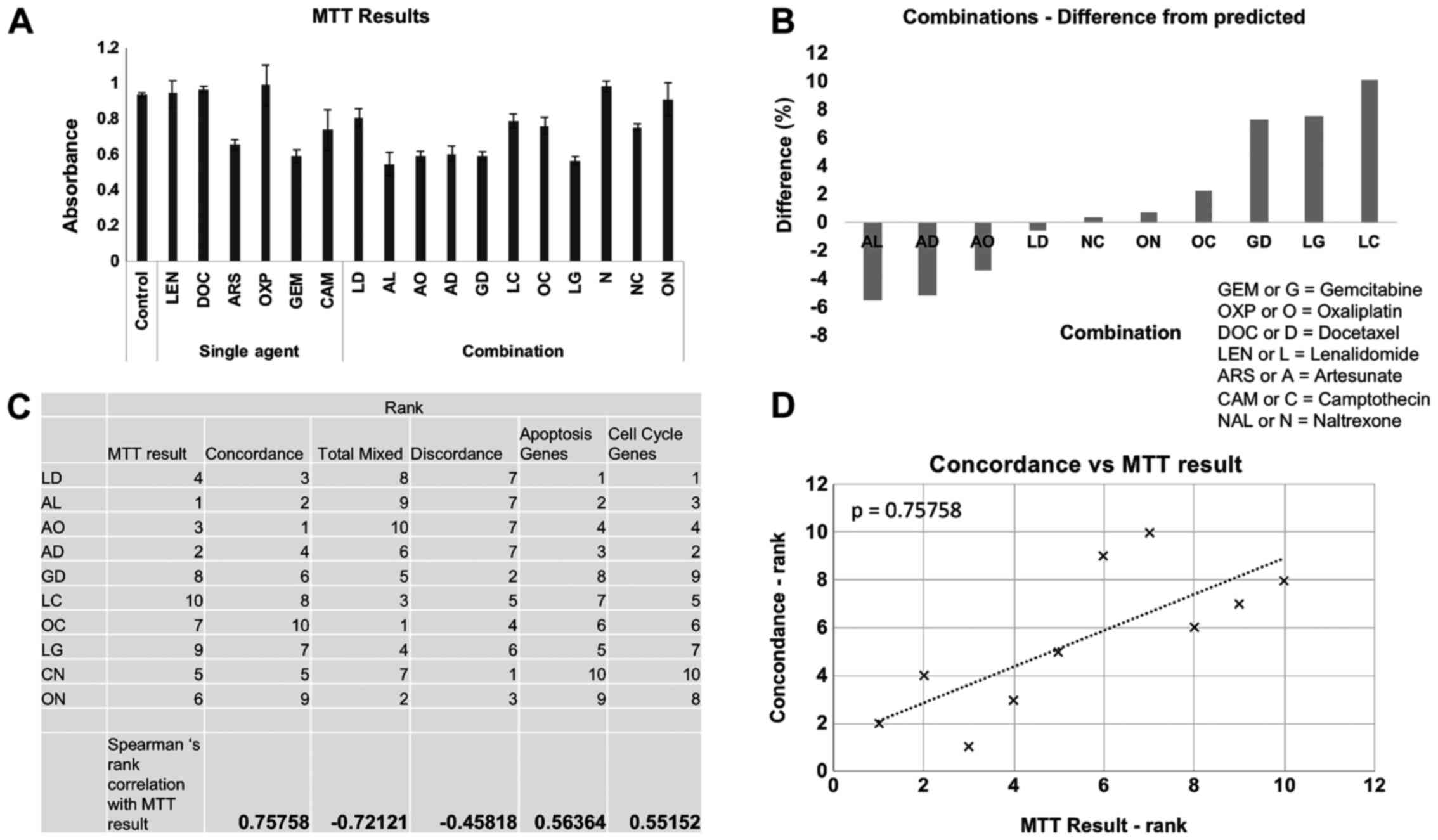
Combinational value correlated with
gene concordance
MTT analysis was performed on HCT116 cells that had
been cultured with the chemotherapeutic drugs either as single
agents or as selected combinations for 48 h, a representative
example of data is shown in Fig. 4A.
The difference between the expected effect on cell number by
combining two drugs (generated by adding together of the results of
the effect on cell number of each single agent) was then compared
to the actual result of combining the two drugs together. Using
this analysis it was possible to find drug combinations that
performed better or worse than the predicted effect by single-agent
analysis (Fig. 4B). The combination
of artesunate and lenalidomide performed the best, exceeding the
predicted effect on cell number. The combinations of artesunate and
docetaxel and artesunate and oxaliplatin also performed better than
expected. The combination of lenalidomide and camptothecin
performed worst and had a less than predicted effected on cell
number suggesting antagonism between these two drugs at the
concentrations used here. The combinations were ranked and then
compared with the rankings of the drugs in terms of their
transcriptional concordance, discordance, apoptosis response and
cell cycle response (Fig. 4C). The
best correlation occurred when MTT data was compared with overall
gene concordance (Fig. 4D),
suggesting that this was the best predictive measure for
combinational efficiency.
Discussion
The current study compares the mRNA expression
profiles of HCT116 tumour cells after they have been cultured with
different chemotherapeutic agents. The tumour cells were cultured
with suboptimal concentrations of different classes of chemotherapy
and the response measured by whole-genome microarray. Despite the
different targets and mechanisms of action of the drugs, there were
some similarities in the pattern of gene activation/inhibition for
some of the drugs. Moreover, drugs that displayed general
concordance in gene signature were shown to exhibit synergistic
traits when combined with one another.
A number of drugs have multiple mechanisms of
action, and so deciding drug-partners in combination regimens
simply on the basis of the principal mechanism of action may
inadvertently miss opportunities. For example, we have shown
naltrexone, which is a proficient antagonist of opioid receptors
can also reduce the expression of the cyclin-dependent kinase
inhibitor p21waf1 (22).
This suggests it could be used in combination with other drugs that
modify cell cycle functions. Previous studies have investigated the
correlation between the mRNA expression patterns in tumour cells
and; growth inhibition by agents (23), drug resistance (24,25) and
prognosis (26), but these studies
have usually focussed on untreated cells, and not attempted to
predict combination value by comparing the mRNA expression profiles
of tumour cells after culture with different classes of drug. In
the current study, we specifically examined the gene signatures in
one colon cancer cell line after treatment with a range of
chemotherapy drugs, and analysed these profiles for patterns that
could identify similarities in MOAs. Data pertaining to certain
genes from cells treated with gemcitabine has been published in our
previous papers (5,7). Data pertaining to naltrexone has been
published previously (30).
For some time now, the identification of genetic
patterns in patients with certain types of cancer has been used as
a prognostic indicator. Indeed, in some cases, establishing the
genetic fingerprint of the a cancer can guide the type of treatment
(27,28). The current study differs from this
more general approach by first identifying the genes that are
changed following treatment. It is hoped that by matching these
signatures, certain drugs that have similar genetic profiles may be
supportive of one another. A similar study examining the RNAi
signatures of mammalian cell death genes highlighted the importance
of mechanism in drug combination and revealed, for example, that
the mapping of 17AAG and taxol in the same region in the principal
component analysis space, supported the prediction that 17AAG would
reinforce a taxol-like action (29).
Our previous work has shown that the same agent can
have diverse effects on genes when used at different concentrations
(30). At high concentrations, a
number of drugs will cause a large amount of cell death and have a
catastrophic effect on gene translation, so for this reason, we
cultured cells with drugs at sub-optimal concentrations.
Additionally, microarrays were performed on RNA extracted from
cells after 4 h of culture with drug to ensure that the
secondary/tertiary effects of the drugs and non-specific effects on
gene expression were minimised and so the primary fingerprint of
effect could be determined for each drug. Artesunate, camptothecin,
docetaxel, gemcitabine, lenalidomide, naltrexone and oxaliplatin
each initiated a particular gene response when cultured with HCT116
cells at suboptimal concentrations. However, despite the varying
molecular targets and mechanisms of action for the drugs, there
were similarities between the patterns of genes hits by certain
pairs of drugs. For example, there was a remarkable concordance in
the mRNA expression of tumour cells cultured with artesunate and
lenalidomide. This despite the disparate nature of the mechanisms
of action and molecular targets of these drugs, artesunate
potentially working through iron-catalysed alkylating cytotoxic
free radicals in tumours and lenalidomide targeting cereblon
(31). Conversely, drugs such as
gemcitabine and docetaxel, and, naltrexone and camptothecin, had
very different microarray responses, in many cases genes that were
upregulated by one drug would be downregulated by the other. This
kind of reciprocal response may be expected for drugs that, for
example, target different points on the cell cycle, and although GO
analysis (Table II) of the 50 most
upregulated and the 50 most downregulated genes suggested that
gemcitabine, although multifaceted, affects G2/M
transition, while docetaxel affects cell division only, there were
cell cycle protein-specific changes that highlighted this
difference. For example, gemcitabine upregulated cyclin E and
marginally downregulated cyclin A, while for docetaxel the opposite
was true, cyclin E being reduced, and cyclin A being
upregulated.
This kind of inverse response on gene transcription
suggests that the drugs, when combined, would neutralise each
other, abrogating the response of both in terms of gene changes and
cytotoxicity/cytostasis. What may help to indicate an efficacious
combination is by examining the effects of the drugs on genes
regulating key processes such as apoptosis and the cell cycle. By
focusing on apoptosis-associated genes, it was revealed that
docetaxel and lenalidomide had the most similar response i.e. both
drugs showed a similar pattern of response in genes associated with
apoptosis. This despite the fact that docetaxel is generally viewed
as a potent cytotoxic agent, whilst lenalidomide is generally
thought of as non-toxic. Parallel to this, docetaxel and
lenalidomide produced a similar response in cell cycle-associated
genes in HCT116 cells again signifying that although docetaxel and
lenalidomide have different potencies and mechanisms of action, the
end result of their use in HCT116 tumour cells is transcriptionally
similar.
Cells contain numerous sensors that perceive damage
and then relay this to machinery involved with cellular repair,
death or senescence (32). The
present study shows that even when using vastly differing
chemotherapies with diverse molecular targets and MOAs, there can
be an overlap in the mRNA-transcripts that are affected. Moreover,
correlation between chemotherapy and gene response cannot
necessarily be predicted by the similarity of drugs. For example,
gemcitabine and oxaliplatin both block DNA replication by inserting
into DNA strands; however, in terms of the general mRNA response,
only 4.2% of affected genes were ‘concordant’. Conversely,
artesunate and oxaliplatin supposedly have totally different MOAs
and yet 26.5% of genes were impacted by the individual drugs in the
same way.
Upon determining the gene response to the various
chemotherapies, we next wanted to know whether these could be
predictive of the combination effect when using the drugs
concomitantly. Intuitively, combining drugs with discordant gene
signatures would lead to a situation where the effects imparted by
the individual drugs would negate the actions of the other, thereby
neutralising the response. However, combining drugs that were not
discordant could potentially be beneficial as they could, for
example, better activate desired gene responses in a cooperative
manner (concordance) or activate more than one desired response
(mixed). Indeed, using MTT analysis it was found that additivity
between drugs correlated most strongly to overall gene concordance
between drugs. That is to say, drugs with similarities in their
transcriptional responses combined better to reduce cell number,
compared to those with transcriptional effects that were less
similar.
The fundamental rationale for combination therapy in
an oncology setting is to optimise overall action by using drugs
that work together to target tumour masses that contain cells that
are often heterogeneous (33). This
heterogeneity means that the cell types can be diverse enough to
prevent drugs with focussed MOAs lacking the activity to elicit a
therapeutic effect. Therefore, mixing drugs to allow a more diverse
range of action could be beneficial. Similarly, it could be argued
that the over-reliance of a drug concentrating on one specific
target could allow resistance to the drug to develop more readily
(34). However, even drugs with very
similar MOAs can have very different responses, such as those
reported with platinum derivatives in cisplatin-resistant cell
lines (35). Despite this, most
clinical data and scientific dogma would suggest that combining
drugs with identical MOAs would not be best practise and this idea
is still maintained in the present study, which suggests not
combining similar drugs but combining very different drugs that
have a similarity of response at the transcriptional level. Further
investigation of this effect using more cell lines and different
chemotherapies is needed to determine whether this theory holds
true.
Due to the nature of the study the combination that
performed best in terms of reduction of cell number may not have
been the combination that reduced cell number to the largest
extent, as combinations were ranked in terms of performance
compared to predicted performance, not which combination reduced
cell number the most. This study is also limited by the fact that
only one concentration of each of the drugs was used. We can say
with some certainty that titration of the drugs to different
concentrations will produce a different pattern of transcriptional
response and it may also be that changing the concentrations will
also influence whether the drug combinations retain their
beneficial or detrimental effects on tumour cell number. A further
confounding factor not even touched upon in this study is the
importance of drug sequence when using more than one agent
(22,36). For these reasons, these data can only
be used as a guide as to which drugs may combine well with one
another and it is essential that further studies include an
investigation of drug dose.
The present study suggests that an important factor
in the strategic combination of chemotherapies may be that there is
some correlation in the transcriptional response induced by the
chemotherapeutics that are being combined. That is not to say that
the drugs should not have different targets and mechanisms but the
present data suggest that the outcome of this should be a similar
pattern of gene expression to allow the best chance for
combinational synergy.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Institute for
Cancer Vaccines and Immunotherapy.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request. Microarray data is available in the GEO respository
accession number GSE122985.
Authors' contributions
AMG and WML designed the current study, acquired and
analysed the data and wrote the manuscript. JLD performed the
experiments and acquisition of data. AGD and JC designed the
current study, interpreted data and wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
AMG is presently funded by Celgene Corporation. WML
is presently funded by LDN Pharma Limited. The remaining authors
declare that they have no competing interests.
References
|
1
|
O'Connell MJ: Current status of adjuvant
therapy for colorectal cancer. Oncology (Williston Park).
18:751–755; discussion 755–758. 2004.PubMed/NCBI
|
|
2
|
Almendro V, Ametller E, García-Recio S,
Collazo O, Casas I, Augé JM, Maurel J and Gascón P: The role of
MMP7 and its cross-talk with the FAS/FASL system during the
acquisition of chemoresistance to oxaliplatin. PLoS One.
4:e47282009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yardley DA: Drug resistance and the role
of combination chemotherapy in improving patient outcomes. Int J
Breast Cancer. 2013:1374142013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buchbinder EI and Desai A: CTLA-4 and PD-1
pathways: Similarities, differences, and implications of their
inhibition. Am J Clin Oncol. 39:98–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gravett AM, Dalgleish AG and Copier J: In
vitro culture with gemcitabine augments death receptor and NKG2D
ligand expression on tumour cells. Sci Rep. 9:15442019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scott KA, Dalgleish AG and Liu WM: The
combination of cannabidiol and Δ9-tetrahydrocannabinol enhances the
anticancer effects of radiation in an orthotopic murine glioma
model. Mol Cancer Ther. 13:2955–2967. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gravett AM, Trautwein N, Stevanović S,
Dalgleish AG and Copier J: Gemcitabine alters the proteasome
composition and immunopeptidome of tumour cells. Oncoimmunology.
7:e14381072018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nars MS and Kaneno R: Immunomodulatory
effects of low dose chemotherapy and perspectives of its
combination with immunotherapy. Int J Cancer. 132:2471–2478. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsimberidou AM, Eggermont AM and Schilsky
RL: Precision cancer medicine: The future is now, only better. Am
Soc Clin Oncol Educ Book. 2014:61–69. 2014. View Article : Google Scholar
|
|
10
|
Hu HM, Zhao X, Kaushik S, Robillard L,
Barthelet A, Lin KK, Shah KN, Simmons AD, Raponi M, Harding TC and
Bandyopadhyay S: A quantitative chemotherapy genetic interaction
map reveals factors associated with PARP inhibitor resistance. Cell
Rep. 23:918–929. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McKenna MT, Weis JA, Brock A, Quaranta V
and Yankeelov TE: Precision medicine with imprecise therapy:
Computational modeling for chemotherapy in breast cancer. Transl
Oncol. 11:732–742. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng X and Nakamura Y: Cancer precision
medicine: From cancer screening to drug selection and personalized
immunotherapy. Trends Pharmacol Sci. 38:15–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan B and Lentzsch S: The application and
biology of immunomodulatory drugs (IMiDs) in cancer. Pharmacol
Ther. 136:56–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hideshima T, Chauhan D, Shima Y, Raje N,
Davies FE, Tai YT, Treon SP, Lin B, Schlossman RL, Richardson P, et
al: Thalidomide and its analogs overcome drug resistance of human
multiple myeloma cells to conventional therapy. Blood.
96:2943–2950. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu WM, Gravett AM and Dalgleish AG: The
antimalarial agent artesunate possesses anticancer properties that
can be enhanced by combination strategies. Int J Cancer.
128:1471–1480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gravett AM, Liu WM, Krishna S, Chan WC,
Haynes RK, Wilson NL and Dalgleish AG: In vitro study of the
anti-cancer effects of artemisone alone or in combination with
other chemotherapeutic agents. Cancer Chemother Pharmacol.
67:569–577. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hamacher-Brady A, Stein HA, Turschner S,
Toegel I, Mora R, Jennewein N, Efferth T, Eils R and Brady NR:
Artesunate activates mitochondrial apoptosis in breast cancer cells
via iron-catalyzed lysosomal reactive oxygen species production. J
Biol Chem. 286:6587–6601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang ND, Tan SH, Ng S, Shi Y, Zhou J, Tan
KS, Wong WS and Shen HM: Artesunate induces cell death in human
cancer cells via enhancing lysosomal function and lysosomal
degradation of ferritin. J Biol Chem. 289:33425–33441. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
The Gene Ontology Consortium: The gene
ontology resource: 20 Years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mi H, Huang X, Muruganujan A, Tang H,
Mills C, Kang D and Thomas PD: PANTHER version 11: Expanded
annotation data from gene ontology and reactome pathways, and data
analysis tool enhancements. Nucleic Acids Res. 45:D183–D189. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scott KA, Dalgleish AG and Liu WM:
Anticancer effects of phytocannabinoids used with chemotherapy in
leukaemia cells can be improved by altering the sequence of their
administration. Int J Oncol. 51:369–377. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wallqvist A, Rabow AA, Shoemaker RH,
Sausville EA and Covell DG: Establishing connections between
microarray expression data and chemotherapeutic cancer
pharmacology. Mol Cancer Ther. 1:311–320. 2002.PubMed/NCBI
|
|
24
|
Tseng AH, Chung FH, Lee HC, Wu LC, Chen CH
and Su LJ: Microarray analysis and establishment of drug screening
platform using 5-fluorouracil resistance HCT116 colon cancer cells.
Genomic Med Biomarkers Health Sci. 4:21–27. 2012. View Article : Google Scholar
|
|
25
|
Lønning PE, Sørlie T, Perou CM, Brown PO,
Botstein D and Børresen-Dale AL: Microarrays in primary breast
cancer-lessons from chemotherapy studies. Endocr Relat Cancer.
8:259–263. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kao KJ, Chang KM, Hsu HC and Huang AT:
Correlation of microarray-based breast cancer molecular subtypes
and clinical outcomes: Implications for treatment optimization. BMC
Cancer. 11:1432011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kamel HF and Al-Amodi HS: Exploitation of
gene expression and cancer biomarkers in paving the path to era of
personalized medicine. Genomics Proteomics Bioinformatics.
15:220–235. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang TH and Chao A: Microarray analysis of
gene expression of cancer to guide the use of chemotherapeutics.
Taiwan J Obstet Gynecol. 46:222–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pritchard JR, Bruno PM, Gilbert LA, Capron
KL, Lauffenburger DA and Hemann MT: Defining principles of
combination drug mechanisms of action. Proc Natl Acad Sci USA.
110:E170–E179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu WM, Scott KA, Dennis JL, Kaminska E,
Levett AJ and Dalgleish AG: Naltrexone at low doses upregulates a
unique gene expression not seen with normal doses: Implications for
its use in cancer therapy. Int J Oncol. 49:793–802. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fink EC and Ebert BL: The novel mechanism
of lenalidomide activity. Blood. 126:2366–2369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jackson SP and Bartek J: The DNA-damage
response in human biology and disease. Nature. 461:1071–1078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu WM: Enhancing the cytotoxic activity
of novel targeted therapies-is there a role for a combinatorial
approach? Curr Clin Pharmacol. 3:108–117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Housman G, Byler S, Heerboth S, Lapinska
K, Longacre M, Snyder N and Sarkar S: Drug resistance in cancer: An
overview. Cancers (Basel). 6:1769–1792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rixe O, Ortuzar W, Alvarez M, Parker R,
Reed E, Paull K and Fojo T: Oxaliplatin, tetraplatin, cisplatin,
and carboplatin: Spectrum of activity in drug-resistant cell lines
and in the cell lines of the national cancer institute's anticancer
drug screen panel. Biochem Pharmacol. 52:1855–1865. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shah MA and Schwartz GK: The relevance of
drug sequence in combination chemotherapy. Drug Resist Updat.
3:335–356. 2000. View Article : Google Scholar : PubMed/NCBI
|