Introduction
An understanding of the mechanisms by which gut
microbes affect tumor development and progression in the intestine
may provide a novel paradigm for colorectal cancer (CRC) therapy
(1). As one of the leading causes of
death among patients with cancer worldwide in recent years with an
age-standardized mortality rate of 8.9 per 100,000 patients, CRC is
a heterogeneous disease characterized by diversions of multiple
molecular pathways throughout its evolutionary process, such as
reduction in the apoptotic rate and development of metastasis
(2,3). Numerous contributing factors to CRC,
including genes and microbiota, have been identified (4–6).
Certain fecal genes have been demonstrated to serve
important molecular roles in cancer biology and molecular medicine
(7–9). Improving clinical outcomes depends on
identifying and understanding these genes and applying this
knowledge to CRC detection and chemotherapy (4,10,11). By
stimulating CRC cells, which exhibit high chemokine expression, the
gut microbiota recruit beneficial T cells into colonic tumor
lesions (12). Biomaterials in the
stool, including nucleic acids or gut microbiota, are associated
with the risk of CRC (1,13–15).
Furthermore, gut dysbiosis is associated with the development and
progression of CRC through DNA damage, activation of oncogenic
signaling pathways, production of tumor-promoting metabolites and
suppression of antitumor immunity (1).
Gut microbes affect gene expression in colonic cells
and may alter the progression of CRC (16–18).
Short-chain fatty acids (SCFAs), which are derived from microbial
metabolism in the gut, serve multiple roles in host homeostasis
(19). Certain reports have
indicated that decreased SCFA production is associated with
increased CRC risk and may have applications in CRC therapy
(20). Butyrate is a type of SCFA
with various molecular effects on intestinal cells, including
anticancer formation and cell immunity (21–23).
SCFA transporters and receptors act as molecular links between the
gut and microbes (24). For
instance, butyrate was demonstrated to alleviate gut inflammation
by coupling with a cell-surface G-protein-coupled receptor 43
(GPR43) (25,26) and regulate intestinal tight junction
proteins through a transporter, solute carrier family 5 member 8
(SLC5A8) (27). Furthermore,
butyrate-producing Butyricicoccus pullicaecorum (B.
pullicaecorum) has been revealed to prevent necrotic enteritis
and reduce pathogen abundance in the cecum and ileum (28,29).
Therefore, this next-generation probiotic has been reported to be
safe in a human intervention trial (28). The present study observed a trend
towards decreasing the abundance of B. pullicaecorum in the
stool of patients with late-stage CRC (28).
An understanding of the role of B.
pullicaecorum or its metabolites in CRC is crucial prior to
considering B. pullicaecorum a probiotic. However, the
anti-CRC effects of B. pullicaecorum or its metabolites,
particularly butyrate, on butyrate transporters and receptors have
not been directly confirmed. The aims of the present study were to
examine the effects of providing a supernatant of B.
pullicaecorum culture to investigate the growth of CRC cells
and evaluate 1,2-dimethylhydrazine (DMH)/dextran sulfate sodium
(DSS)-induced colonic tumors of mice with B. pullicaecorum
administration.
Materials and methods
Gut bacterium, mice and CRC
induction
The gut bacterium, B. pullicaecorum
(BCRC-81109), was purchased from the Bioresource Collection and
Research Center and was grown in a modified peptone yeast extract
broth (PY-X broth: 0.5 g peptone from meat; 0.5 g trypticase
peptone; 1.0 g yeast extract; 0.1 mg resazurin; 50 mg
L-cysteine-HCI.H2O; 0.5 g D-glucose; 10 mg
CaCl2.2H2O; 20 mg
MgS04.7H2O; 40 mg
K2HP04; 40 mg KH2P04;
0.4 g NaHC03; 0.8 mg NaCl; 0.5% xylan in 100 ml
distilled water; pH 7.0; DSMZ GmbH) for 3 days under anaerobic
conditions at 37°C. The conditioned medium from B.
pullicaecorum cultures was harvested by centrifugation (1,000 ×
g, 15 min) at room temperature to remove the bacteria and
sterilized by filtration with a 0.22 µm syringe filter (Thermo
Fisher Scientific, Inc.).
BALB/cByJNarl male mice (age, 4–6 weeks; weight
range, 20–25 g) were provided by the National Laboratory Animal
Center, National Applied Research Laboratories, Taipei, Taiwan. All
animal experiments complied with the Animal Research: Reporting of
In Vivo Experiments guidelines (30). All protocols were approved by the
Institutional Animal Care and Use Committees at Cathay General
Hospital, Taipei, Taiwan and followed the ‘3Rs’ (Reduction,
Refinement and Replacement) (31).
All efforts were made to minimize the number of animals and their
suffering. The body weight of mice was monitored every week and
mice were euthanized when they exhibited weakness and rapid weight
loss (15–20% of their body weight) within a few days (32). A total of 3–5 were housed per plastic
cage under pathogen-free conditions (humidity, 50±10%; light/dark
cycle, 12 h; temperature, 23±2°C) in an individually ventilated
cage rack system (Tecniplast S.p.A).
To evaluate the effect of B. pullicaecorum
(3.125×107 colony-forming units in 100 µl) on CRC
formation, CRC was induced with DMH (40 mg/kg; cat. no. D0741; TCI
America; Tokyo Chemical Industry)/DSS (2%; cat. no. D5144; TCI
America; Tokyo Chemical Industry), as reported by De Robertis et
al (33) and as described in the
legend of Fig. 1A. A total of 17
mice were used and randomly divided into the following groups to
study the induction of CRC by DMH and the in vivo effect of
B. pullicaecorum: i) Control group (CG; n=5), no DMH
treatment or B. pullicaecorum administration; ii) DMH mice
(n=6), administered DMH by intraperitoneal injection and DSS in
drinking water and no B. pullicaecorum administration; and
iii) DMH/B. pullicaecorum (BP) mice (n=6), administered DMH,
DSS, and B. pullicaecorum by oral gavage. Anuses of mice
were imaged on days 29, 32 and 36 following initial B.
pullicaecorum administration. Mice were sacrificed 8–9 weeks
later with CO2 euthanasia in a cage. The CO2
flow rate was set to displace 30% of the cage volume/minute. The
following parameters were used to confirm death: i) immobility; and
ii) lack of spontaneous breathing for >2 min. During the
experiments, the clinical characteristics of mice, including anal
bleeding, body weight and serum carcinoembryonic antigen (CEA)
levels were recorded.
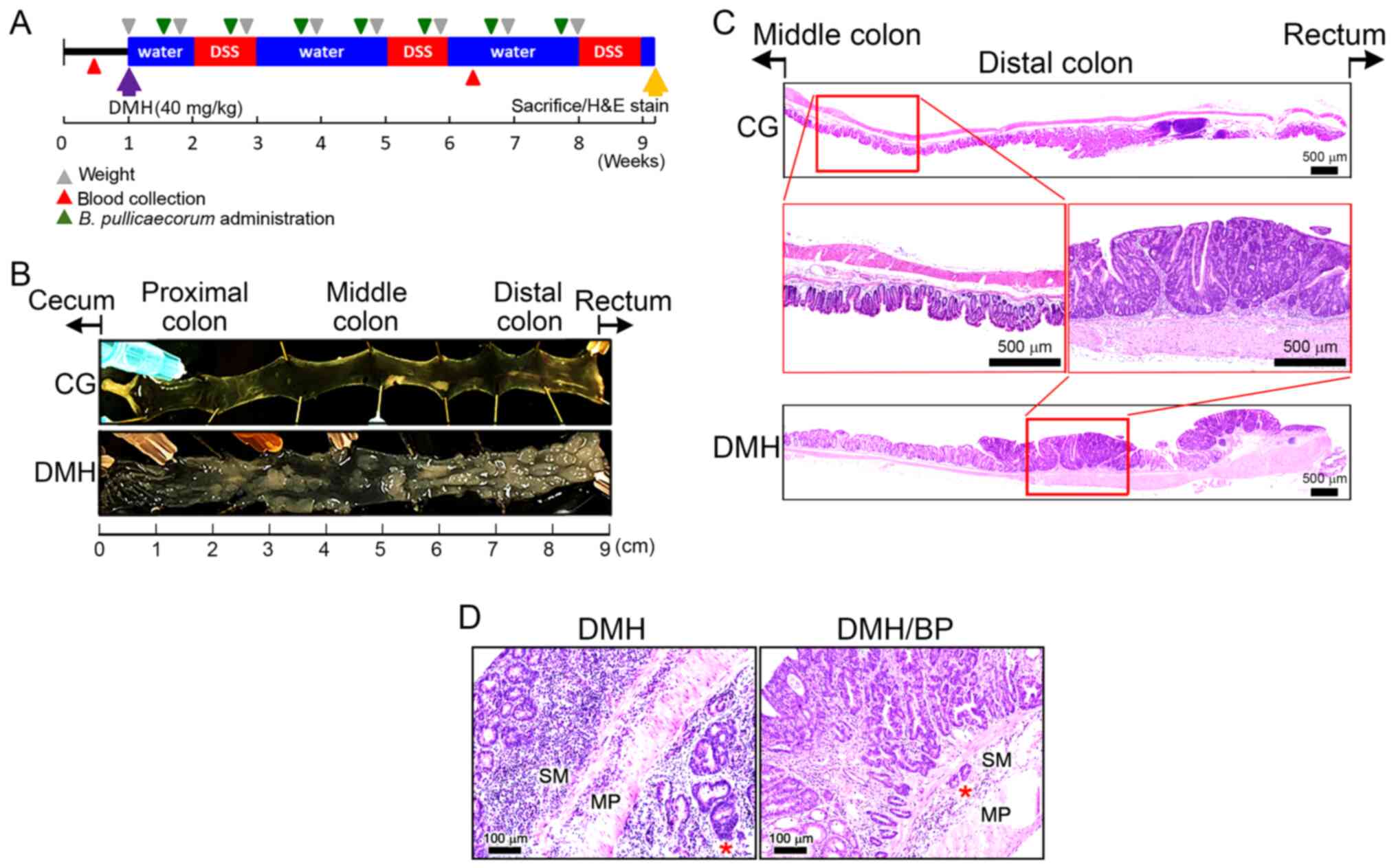 | Figure 1.Reduction in DMH/DSS-induced CRC
formation following B. pullicaecorum administration. (A)
Timing of DMH and DSS induction of CRC and B. pullicaecorum
administration. Purple arrow, subcutaneous injection of DMH 40
mg/kg body weight. Yellow arrow, sacrifice and colon sampling of
H&E staining; blue box, water drinking; red box, DSS drinking;
gray triangle, recording body weight; red triangle, blood
collection; green triangle, B. pullicaecorum administration.
(B) Inner layer of the colon of mice. (C) Histopathological
examination of the colon of mice. Red square, higher magnification.
Scale bar, 500 µm. (D) The effect of B. pullicaecorum
administration on DMH-induced CRC. Scale bar, 100 µm. DMH,
1,2-dimethylhydrazine; DSS, dextran sulfate sodium; CRC, colorectal
cancer; CG, control group; BP, B. pullicaecorum; H&E,
hematoxylin and eosin; SM, submucosa; MP, muscularis propria; *,
the maximal depth of tumor invasion. |
CRC cell lines, cell culture and cell
counts
SW480 colon cancer cell line (ATCC CRL-1459; AJCC
stage II) and SW620 lymph node metastatic derivative cell line
(ATCC CRL-1831; AJCC stage III) from the same patient were
purchased from American Type Culture Collection. The two cell lines
were expanded in complete medium [Leibovitz's L-15 medium (Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(NQBB International Biological Co., Ltd.)] under 100% atmospheric
air (without CO2) in a 37°C humidified incubator. To
evaluate the effects of B. pullicaecorum on CRC cell
proliferation, the numbers of SW480 and SW620 cells were
respectively counted at days 1 and 5. Briefly, cells incubated with
or without 10% conditioned medium from B. pullicaecorum
cultures in complete medium were imaged under an Olympus BX41 light
microscope (Olympus Corporation) and analyzed using ImageJ software
(version 1.52; National Institutes of Health) (34).
Reverse transcription PCR
Total RNA from SW480 and SW620 cells, which were
untreated or treated with 5 mM sodium butyrate (NaB) for 48 h as
reported in our previous study (21)
was independently extracted using a RNeasy Mini kit (Qiagen, Inc.),
according to the manufacturer's protocol. Total RNA (1 µM) was
reverse-transcribed to single-stranded cDNA using a high-capacity
cDNA Reverse Transcription kit (cat. no. 4368813; Thermo Fisher
Scientific, Inc.), as previously described (35). To quantify the expression of GPR43,
the reaction mixture containing the cDNA sample, QuantiTect
SYBR-Green PCR Master mix (Qiagen GmbH), QuantiTect Primer assay
(cat. no. Hs_FFAR2_1_SG; Qiagen GmbH) and RNase-free water was
amplified using the following cycling program: 10 min at 95°C,
followed by 40 cycles at 95°C for 15 sec and at 60°C for 1 min. To
quantify the expression of SLC5A8, the reaction mixture containing
forward (5′-TCTTCCTCCCGGTGTTCTAC-3′) and reverse primers
(5′-GAACACATTTGTTAAATCGAAGTTCT-3′), no. 48 universal probe (Roche
Diagnostics GmbH), LightCycler TaqMan Master (Roche Diagnostics
GmbH) and RNase-free water was amplified by a program (10 min at
95°C, proceeding with 60 cycles at 95°C for 10 sec and at 60°C for
20 sec). The expression of GAPDH (no. 60 universal probe (Roche
Diagnostics GmbH); forward primer, 5′-CTCTGCTCCTCCTGTTCGAC-3′;
reverse primer, 5′-ACGACCAAATCCGTTGACTC-3′) was used as an internal
control for calibration. For control purposes, the human reference
cDNA (Clontech Laboratories) was used as a positive control to
estimate the relative expression levels in the cells and the
non-template reaction was a negative control. All RT-PCR reactions
were run in a LightCycler 96 (Roche Diagnostics GmbH) and the data
were analyzed using the 2−ΔΔCq method (36).
Histological and immunohistochemical
staining of mouse tissues
Colon tissues were obtained immediately following
sacrifice and death and embedded in paraffin. Hematoxylin and eosin
(H&E) were added to the paraffin section (thickness, 3–5 µm) to
identify non-CRC and CRC areas using the method reported by Fischer
et al (37).
Immunohistochemistry (IHC) was performed as described by Huang
et al (9). Briefly, colon
tissue sections were hybridized with anti-GPR43 (1:100 in blocking
solution; cat. no. BS-13536R) and anti-SLC5A8 (1:100 in blocking
solution; cat. no. BS-6106R) polyclonal antibodies obtained from
Bioss Antibodies. GPR43 and SLC5A8 were visualized using
3,3¢-diaminobenzidine (Vector Laboratories, Inc.) as the substrate.
A pathologist viewed and categorized the H&E and IHC-stained
sections.
Measurement of serum CEA levels
Blood samples (100 µl) were collected from a tail
incision in each mouse. Sample was centrifuged at 1,000 × g for 15
min at room temperature to isolate the serum. Serum CEA levels were
measured using a commercial ELISA kit (cat. no. E-EL-M0232;
Elabscience), according to the manufacturer's protocol.
Statistical analysis
One-way ANOVA with Fisher's least significant
difference (LSD) post-hoc test was performed to identify
significant differences between groups. Differences in gene
expression were compared using Student's t-test. Statistical
analyses were performed using the SPSS statistics software (version
22.0; IBM Corp.). Data for cell numbers, body weight and serum CEA
levels are presented for a >3 mice or ≥3 experiments with
similar results. Data are presented as mean ± SEM. P<0.05 was
considered to indicate a statistically significant difference.
Results
Reduction in DMH/DSS-induced CRC
formation following B. pullicaecorum administration
Fig. 1A presented the
timing of DMH and DSS induction of CRC and B. pullicaecorum
administration. Numerous irregular and different sized crypt foci
were observed in the inner layer of the colon of the DMH group,
particularly in the distal colon and near the rectum (Fig. 1B). The histopathological examination
demonstrated no foci in the colons of CG mice and the intestinal
tract had an intact mucosal epithelium without any significant
dysplastic changes (Fig. 1C, top
panel). Conversely, aberrant crypt foci and multiple exophytic
tumors in the mucosal layer were observed in the intestinal tract
of DMH mice (Fig. 1C, bottom
panel).
Visualization at higher magnification detected no
specific histological changes in the intestinal mucosa, submucosa
or muscular layers of CG mice (Fig.
1C, left middle panel). Furthermore, the neoplastic glands of
DMH mice exhibited a complex or fused glandular pattern and notable
nuclear atypia (Fig. 1C, right
middle panel).
Fig. 1D demonstrated
the regressive effect of B. pullicaecorum administration on
DMH-induced CRC. In DMH/BP mice, tumors appeared to be downstaged
histopathologically. Invasive carcinoma was limited to the
submucosa with narrow maximal tumor invasion depth and fewer
inflammatory cells infiltrates were observed in the periglandular
area. The colonic tumors of DMH mice exhibited an early stage of
the carcinogenesis process, which ranged from in situ to
minimally invasive adenocarcinoma.
Conditional induction of SLC5A8 and
GPR43 following B. pullicaecorum administration
Levels of SCFA transporter (SLC5A8) and receptor
(GPR43) were examined by IHC staining of the colon of mice. As
shown in Fig. 2, SLC5A8 and GPR43
were detected mainly in the superficial portion of the crypt glands
in CG mice, while faint and randomly distributed reactivity was
observed in DMH mice. Notably, transporters and receptors were
diffusely and moderately to strongly expressed in neoplastic
epithelial cells in DMH/BP mice. In addition to the epithelial cell
populations, patchy areas with variable cytoplasmic staining for
SLC5A8 and GPR43 were observed in the underlying muscular
layer.
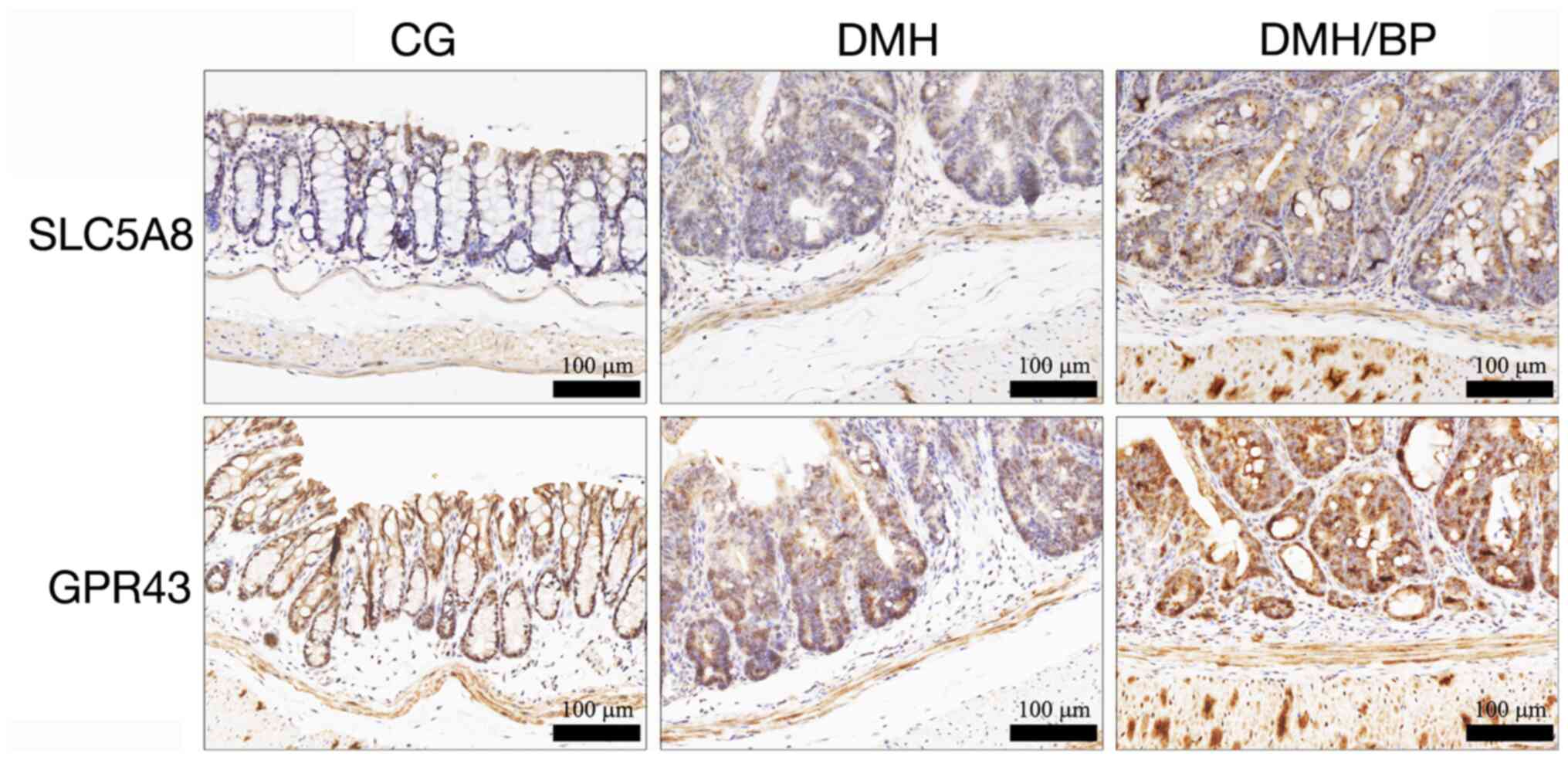 | Figure 2.IHC staining of SLC5A8 and GPR43 in
the colons of mice. SLC5A8 and GPR43 were examined by IHC staining
of the colon of mice. CG: Control group, no DMH/DSS treatment or
B. pullicaecorum administration. Scale bar, 100 µm. SLC5A8,
solute carrier family 5 member 8; GPR43, G-protein-coupled receptor
43; IHC, immunohistochemistry; CG, control group; DMH,
1,2-dimethylhydrazine; DSS, dextran sulfate sodium; BP, B.
pullicaecorum. |
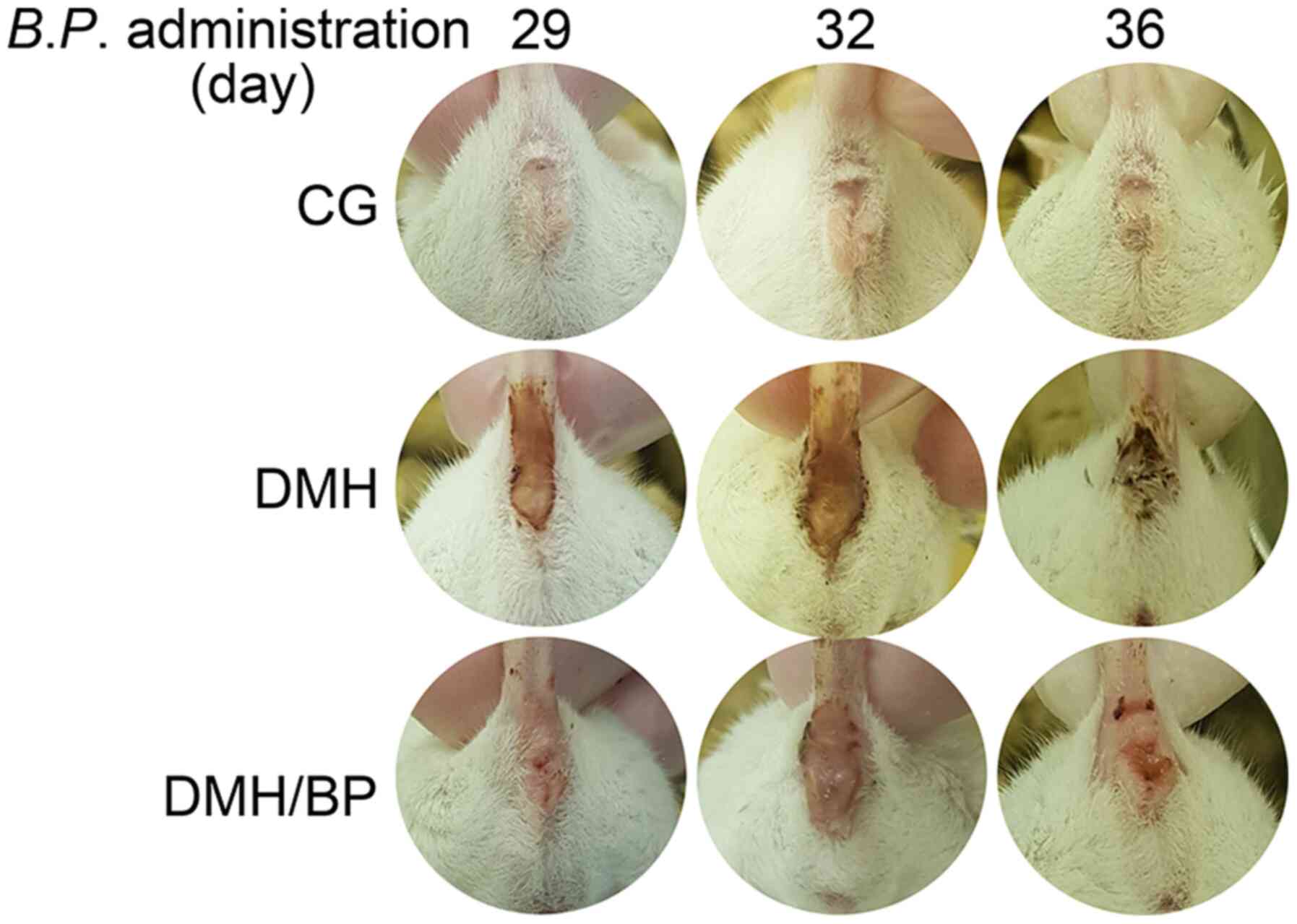
Characteristics of DMH-injected mice
administered B. pullicaecorum
Phenotypic abnormalities and clinical
characteristics of DMH and DMH/BP mice were analyzed. Compared with
CG mice, anal bleeding was worse in the DMH mice; however, this was
improved in DMH/BP mice (Fig. 3).
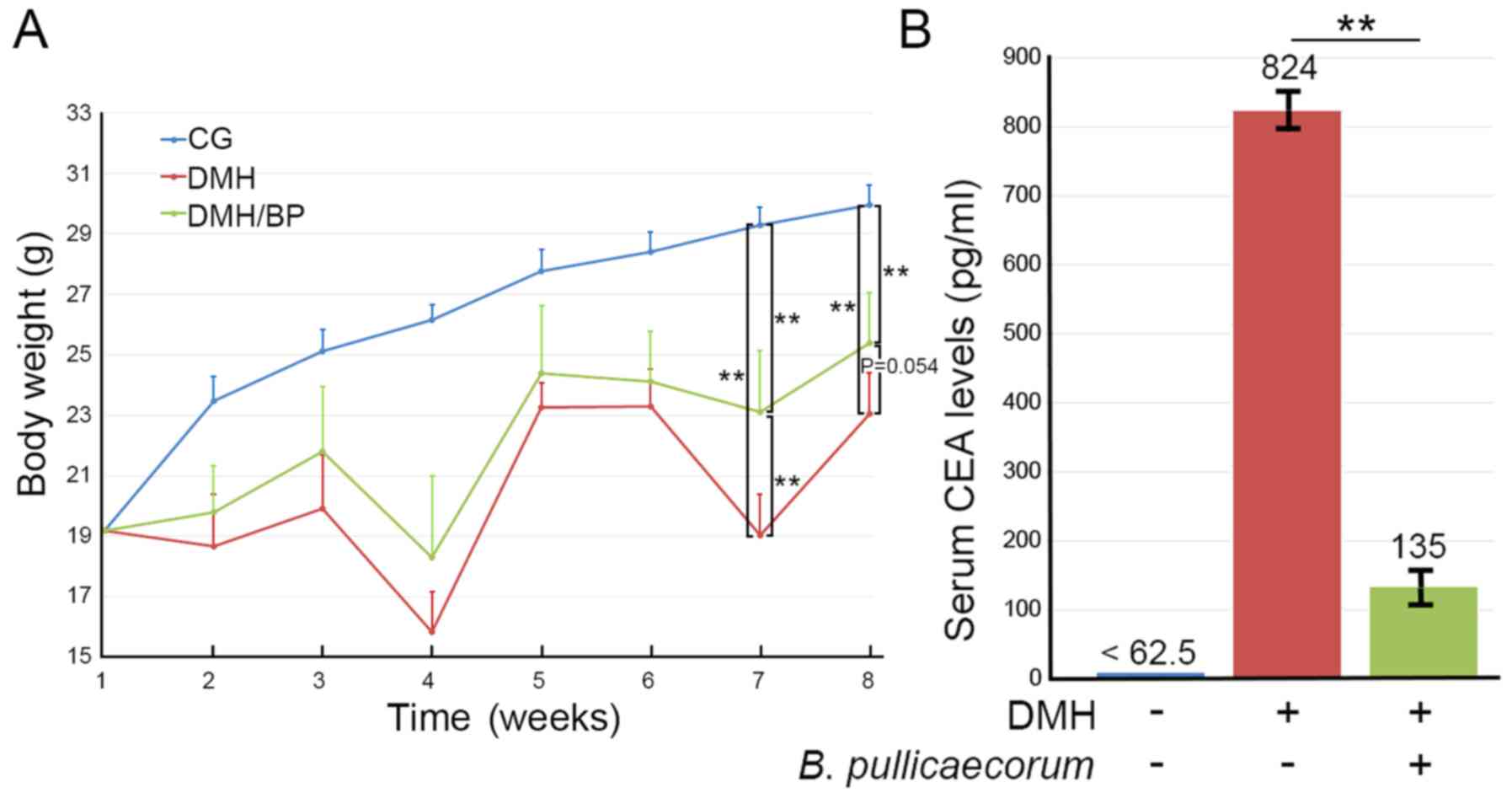
Body weights and serum CEA levels of mice were measured. As shown
in Fig. 4A, the mean body weight
increased significantly from 19.2 to 30.0 g in 8 weeks in CG mice
and no mice died during this time. Mice in the DMH/BP group
exhibited significant weight loss, which was previously reported by
Kim et al (38). The rate of
weight gain was slower in mice in the groups administered DMH (DMH
and DMH/BP mice) compared with CG mice. DMH mice (initial n=6, two
died during the experiment) exhibited the lowest weight. DMH/BP
mice (initial n=6, one died during the experiment) lost less weight
than DMH mice. Briefly, statistical significance was observed at
week 7 (P<0.01) and near significance was observed at week 8
(P=0.054) when comparing the body weights of DMH mice and DMH/BP
mice. Overall, three mice (two DMH mice and one DMH/BP mouse) died
during the experiments due to colon tumors. The rest of the mice
were euthanized at the humane endpoint. DMH/BP mice exhibited
longer survival rates (83.3±5.2%; Fig.
S1), reduced body weight loss and lower serum CEA levels
compared with DMH mice (Fig.
4B).
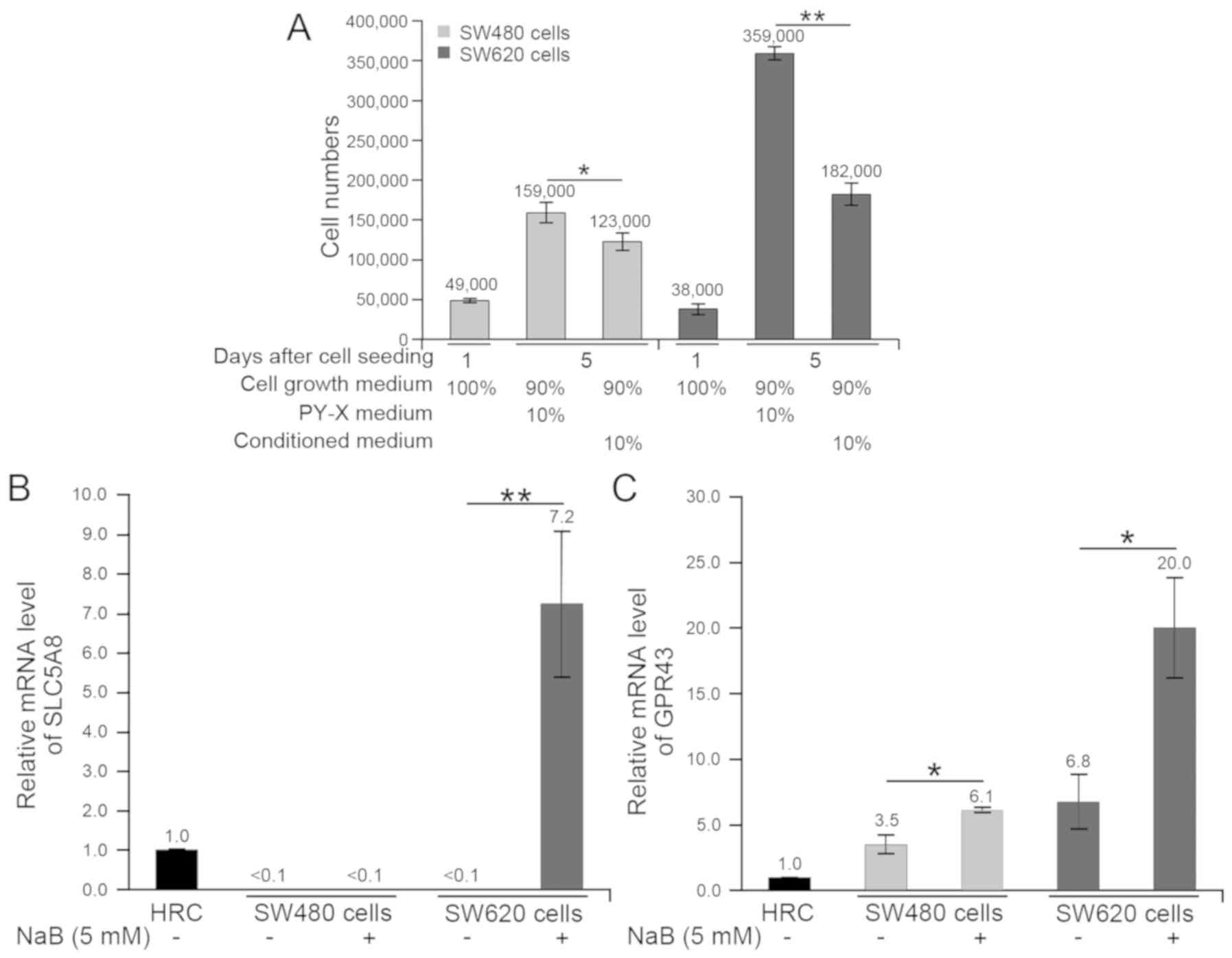
Effects of butyrate on CRC cells
Gas chromatography-mass spectrometry analysis
demonstrated that 3.05 mM butyrate was the predominant metabolite
in the medium following cultivation of B. pullicaecorum
(Table SI). The changes in SW480
and SW620 cells following treatment with the butyrate-containing
supernatant from B. pullicaecorum cultures were evaluated
using ImageJ software (Fig. 5A). On
day 5, SW480 and SW620 cell numbers were lower in cells cultured in
90% cell growth medium + 10% filter-sterilized conditioned medium
compared with cells cultured with the control medium (90% cell
growth medium + 10% PY-X broth). However, the growth of SW620 cells
was more sensitive than SW480 cells to cultivation with 10%
filter-sterilized conditioned medium. In the cultures of SW480 and
SW620 cells with 10% supernatant from B. pullicaecorum,
SW620 cell growth was reduced to 50.7% (from 359,000 cells to
182,000 cells) and SW480 cells to 77.4% (from 159,000 cells to
123,000 cells). This inhibitory effect on the CRC cell growth
following butyrate treatment was also reported in our previous
study (21). SLC5A8 and GPR43
expressions in butyrate-treated CRC cells were quantified (Fig. 5B and C for SLC5A8 and GPR43,
respectively). In comparison with untreated cells, the NaB-treated
SW480 cells only presented higher GPR43 (P<0.05), but the
NaB-treated SW620 cells expressed the most SLC5A8 (P<0.01) and
GPR43 (P<0.05).
Discussion
A greater understanding of gut microbes may lead to
different use of medicine in CRC. Gut microbe-based therapeutics
are actively used for cancer treatment or prevention (39). Numerous studies have reported on the
use of microbial markers, including microbes and metabolites, to
target CRC (40,41). Wang et al (18) reported that a target microbe affected
CRC or reduced the occurrence in an animal model. The present study
demonstrated that B. pullicaecorum administration following
DMH/DSS-induced tumorigenesis led to CRC regression in mice. Gao
et al (41) revealed that gut
microbe-mediated CRC suppression may occur through the production
of histamine, a specific metabolite of Lactobacillus
reuteri.
In a previous study, Butyricicoccus spp. was
observed in the stools of patients with CRC (21). The species of Butyricicoccus, B.
pullicaecorum, was originally isolated from the cecal content
of a broiler chicken and is an anaerobic and butyrate-producing
bacterium that may protect chickens from harmful microorganisms and
necrotic enteritis (29,42). Patients with infectious
gastroenteritis, including inflammatory bowel disease (IBD),
exhibit fewer B. pullicaecorum in their stools (43,44).
Butyrate has been reported to be involved in CRC cell function,
including the regulation of gene expression (45,46),
cell signaling (47) and repression
of cell growth (48).
The present study demonstrated that SLC5A8 and GPR43
were upregulated in neoplastic epithelial cells in the underlying
muscular layer of the colon following B. pullicaecorum
administration. SLC5A8 and GPR43 interact with butyrate in the
lumen of the colon (49,50). Their upregulation may be associated
with the production of butyrate, a major metabolite of B.
pullicaecorum. Furthermore, SLC5A8 and GPR43 are known to serve
as tumor suppressors (51,52). As reviewed by Zaiss et al
(53), mice lacking SLC5A8 develop
CRC. Additionally, previous studies reported that GPR43 activation
prevented colon inflammation and carcinogenesis (52,53). The
high signal intensities of SLC5A8 and GPR43 staining observed in
the current study indicated that B. pullicaecorum
administration may be associated with CRC prognosis. For instance,
the present and other previous studies have observed that butyrate
reduced histone deacetylase (HDAC) activity and acted as an HDAC
inhibitor (21,54). SLC5A8-dependent inhibition of HDAC
activity can be caused by the production of butyrate (55). These results indicated that the
reduction of HDAC activity caused by butyrate may be due to
SLC5A8-dependent inhibition. This hypothesis is supported by our
previous studies concerning in vivo animal and in
vitro cell models.
The results of the animal experiments indicated that
B. pullicaecorum exhibited potential anti-CRC activity by
increasing the expression of SLC5A8 and GPR43 in an animal model of
DMH/DSS tumorigenesis. In the in vitro cell model,
conditioned medium from cultured B. pullicaecorum affected
the growth of SW480 and SW620 cells. There was a minor difference
between the two cell lines: The SW480 and SW620 cells were derived
from primary tumor and lymph-node metastasis, respectively, from
the same patient. SW620 cells exhibit a short doubling time (~25 h)
and grow faster than SW480 cells (50 h doubling time) under regular
incubation conditions (56,57). Growth reduction was found in SW480
cells and SW620 cells, indicating that a metabolite of B.
pullicaecorum, butyrate, served a potential role in the
inhibition of CRC cell growth. Notably, the growth inhibition
effect was more apparent for cells from an advanced-stage of CRC,
such as in SW620 cells. The present study further revealed that
butyrate upregulated the expression of SLC5A8 and GPR43 in CRC
cells. Additionally, SLC5A8 has been reported to mediate the
concentrative entry of butyrate from the lumen into colonocytes
(58). Moreover, advanced CRC cells,
including SW620 cells, have been demonstrated to express very low
GPR43 (59,60). Therefore, the changes in the
expression of SLC5A8 and GPR43 in CRC cells needs to be
elucidated.
The histopathological and other results of the
present study demonstrated that DMH/BP mice exhibited decreased
colon tumor progression. These findings indicated positive effects
of B. pullicaecorum in CRC-bearing mice. CRC-bearing mice
that were not treated with B. pullicaecorum exhibited
advanced CRC tumors. Additionally, the butyrate-induced upregulated
expression of SLC5A8 and GPR43 may alter the progression of CRC
cells in vitro or in vivo.
The importance of probiotics in the prevention and
treatment of CRC has been studied (61) and there is increasing evidence of the
clinical significance of butyrate-producing gut microbes (44,62,63). For
instance, another butyrate-producing probiotic in the same family
of B. pullicaecorum, Clostridium butyricum, has been
demonstrated to prevent tumor development in the intestinal barrier
(64,65). Other families (e.g.
Lachnospiraceae and Ruminococcaceae) also include a
number of butyrate-producing microbes (63). Therefore, one limitation of the
present study was that studying B. pullicaecorum alone
cannot fully reflect all the butyrate-producing probiotics in
CRC.
B. pullicaecorum is considered to be a
potential next-generation probiotic that is safe following oral
administration (28). The microbial
metabolite SCFAs, including butyrate, are involved in the
regulation of intestinal homeostasis and IBD pathogenesis (66). Dysbiosis leading to reduction in SCFA
levels is associated with numerous human diseases, such as stroke
and non-small-cell lung cancer (62,63,67).
Additionally, SCFAs are associated with certain physiological
processes, including immune function (68), anti-inflammatory effects (69) and glucose homeostasis (70–72).
SCFAs are considered to have potential as therapeutic agents
against gastrointestinal cancers (73). Wang et al (18) reported that the four-carbon molecule
butyrate may lower the risk of CRC in the absence of dysbiosis in
the gut.
Animals are regarded as independent entities that
coexist with microbiota in a symbiotic relationship (74,75).
Understanding the interactions between the colonic tract and gut
microbiota may lead to improved opportunities in the treatment of
CRC (75). Accordingly, further
studies are required to explore the clinical application of B.
pullicaecorum and other butyrate-producing microbes or their
major metabolite, butyrate, for patients with CRC. In conclusion,
the present study demonstrated that the administration of B.
pullicaecorum or its metabolite(s) improved the clinical
outcome of CRC in a mouse model. These results indicated that B.
pullicaecorum was a probiotic with anti-CRC potential (21,76).
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Shih-Hsuan Chou
(a metabolomics application scientist at Biotools Co., New Taipei,
Taiwan) for performing the gas chromatography-mass spectrometry
analysis, as well as Dr Chun-Chao Chang (Division of
Gastroenterology and Hepatology, Department of Internal Medicine,
Taipei Medical University Hospital, Taipei, Taiwan) and Dr
Chi-Cheng Huang (Department of Surgery, Taipei-Veterans General
Hospital, Taipei, Taiwan) for revising the manuscript.
Funding
The present study was supported by funding from the
Cathay General Hospital, Taiwan, China (grant nos. CGH-MR-A106013
to Dr CMP and 2019 to Dr CJH) and the Tri-Service General Hospital,
Taiwan, China (grant no. TSGH-C108-178 to Dr JMH).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SCC, MHS and CJH designed and conducted the
experiments. SCC and MHS conceived the present study. CYL and CJH
performed the pathological analyses. CMP and JMH contributed to the
data interpretation. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
B. pullicaecorum
|
Butyricicoccus
pullicaecorum
|
|
DMH
|
1,2-dimethylhydrazine
|
|
SCFA
|
short-chain fatty acids
|
|
CEA
|
carcinoembryonic antigen
|
References
|
1
|
Mima K, Ogino S, Nakagawa S, Sawayama H,
Kinoshita K, Krashima R, Ishimoto T, Imai K, Iwatsuki M, Hashimoto
D, et al: The role of intestinal bacteria in the development and
progression of gastrointestinal tract neoplasms. Surg Oncol.
26:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zarkavelis G, Boussios S, Papadaki A,
Katsanos KH, Christodoulou DK and Pentheroudakis G: Current and
future biomarkers in colorectal cancer. Ann Gastroenterol.
30:613–621. 2017.PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gonzalez-Vallinas M, Vargas T,
Moreno-Rubio J, Molina S, Herranz J, Cejas P, Burgos E, Aguayo C,
Custodio A, Reglero G, et al: Clinical relevance of the
differential expression of the glycosyltransferase gene GCNT3 in
colon cancer. Eur J Cancer. 51:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takakura Y, Ikeda S, Imaoka Y, Urushihara
T and Itamoto T: An elevated preoperative serum carbohydrate
antigen 19-9 level is a significant predictor for peritoneal
dissemination and poor survival in colorectal cancer. Colorectal
Dis. 17:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tilg H, Adolph TE, Gerner RR and Moschen
AR: The intestinal microbiota in colorectal cancer. Cancer Cell.
33:954–964. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kisiel JB, Klepp P, Allawi HT, Taylor WR,
Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann
S, et al: Analysis of DNA methylation at specific loci in stool
samples detects colorectal cancer and high-grade dysplasia in
patients with inflammatory bowel disease. Clin Gastroenterol
Hepatol. 17:914–921.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang CJ, Yang SH, Lee CL, Cheng YC, Tai
SY and Chien CC: Ribosomal protein S27-like in colorectal cancer: A
candidate for predicting prognoses. PLoS One. 8:e670432013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang CJ, Lee CL, Yang SH, Chien CC, Huang
CC, Yang RN and Chang CC: Upregulation of the growth
arrest-specific-2 in recurrent colorectal cancers, and its
susceptibility to chemotherapy in a model cell system. Biochim
Biophys Acta. 1862:1345–1353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kimura Y, Sumiyoshi M and Baba K:
Antitumor activities of synthetic and natural stilbenes through
antiangiogenic action. Cancer Sci. 99:2083–2096. 2008.PubMed/NCBI
|
|
11
|
Watine JC and Bunting PS: Mass colorectal
cancer screening: Methodological quality of practice guidelines is
not related to their content validity. Clin Biochem. 41:459–466.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cremonesi E, Governa V, Garzon JFG, Mele
V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D,
Däster SR, et al: Gut microbiota modulate T cell trafficking into
human colorectal cancer. Gut. 67:1984–1994. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ahmed FE: miRNA as markers for the
diagnostic screening of colon cancer. Expert Rev Anticancer Ther.
14:463–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nishioka Y, Ueki T, Hokazono K, Nagayoshi
K and Tanaka M: Comparative detection of aberrantly methylated DNA
in preoperative and postoperative stool from patients with
colorectal cancers. Int J Biol Markers. 30:e81–e87. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pittman ME: Fecal microbiota and screening
for colorectal cancer. Clin Chem. 64:1273–1274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bishop KS, Xu H and Marlow G: Epigenetic
regulation of gene expression induced by butyrate in colorectal
cancer: Involvement of MicroRNA. Genet Epigenet.
9:1179237X177299002017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang
X, Jia W, Cai S and Zhao L: Structural segregation of gut
microbiota between colorectal cancer patients and healthy
volunteers. ISME J. 6:320–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Huang D, Chen KY, Cui M, Wang W,
Huang X, Awadellah A, Li Q, Friedman A, Xin WW, et al: Fucosylation
deficiency in mice leads to colitis and adenocarcinoma.
Gastroenterology. 152:193–205.e10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Puertollano E, Kolida S and Yaqoob P:
Biological significance of short-chain fatty acid metabolism by the
intestinal microbiome. Curr Opin Clin Nutr Metab Care. 17:139–144.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang G, Yu Y, Wang YZ, Wang JJ, Guan R,
Sun Y, Shi F, Gao J and Fu XL: Role of SCFAs in gut microbiome and
glycolysis for colorectal cancer therapy. J Cell Physiol.
234:17023–17049. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang CC, Shen MH, Chen SK, Yang SH, Liu
CY, Guo JW, Chang KW and Huang CJ: Gut butyrate-producing organisms
correlate to placenta specific 8 protein: Importance to colorectal
cancer progression. J Adv Res. 22:7–20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li JY, Yu M, Pal S, Tyagi AM, Dar H, Adams
J, Weitzmann MN, Jones RM and Pacifici R: Parathyroid
hormone-dependent bone formation requires butyrate production by
intestinal microbiota. J Clin Invest. 130:1767–1781. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosser EC, Piper CJM, Matei DE, Blair PA,
Rendeiro AF, Orford M, Alber DG, Krausgruber T, Catalan D, Klein N,
et al: Microbiota-derived metabolites suppress arthritis by
amplifying aryl-hydrocarbon receptor activation in regulatory B
cells. Cell Metab. 31:837–851.e10. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ganapathy V, Thangaraju M, Prasad PD,
Martin PM and Singh N: Transporters and receptors for short-chain
fatty acids as the molecular link between colonic bacteria and the
host. Curr Opin Pharmacol. 13:869–874. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bartoszek A, Moo EV, Binienda A, Fabisiak
A, Krajewska JB, Mosińska P, Niewinna K, Tarasiuk A, Martemyanov K,
Salaga M and Fichna J: Free fatty acid receptors as new potential
therapeutic target in inflammatory bowel diseases. Pharmacol Res.
152:1046042020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ,
Yao S, Xiao Y, Huang X, Eaves-Pyles TD, Golovko G, et al: GPR43
mediates microbiota metabolite SCFA regulation of antimicrobial
peptide expression in intestinal epithelial cells via activation of
mTOR and STAT3. Mucosal Immunol. 11:752–762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen J, Xuan YH, Luo MX, Ni XG, Ling LQ,
Hu SJ, Chen JQ, Xu JY, Jiang LY, Si WZ, et al: Kaempferol
alleviates acute alcoholic liver injury in mice by regulating
intestinal tight junction proteins and butyrate receptors and
transporters. Toxicology. 429:1523382020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boesmans L, Valles-Colomer M, Wang J,
Eeckhaut V, Falony G, Ducatelle R, Van Immerseel F, Raes J and
Verbeke K: Butyrate producers as potential next-generation
probiotics: Safety assessment of the administration of
Butyricicoccus pullicaecorum to healthy volunteers.
mSystems. 3:e00094–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eeckhaut V, Wang J, Van Parys A,
Haesebrouck F, Joossens M, Falony G, Raes J, Ducatelle R and Van
Immerseel F: The Probiotic Butyricicoccus pullicaecorum
reduces feed conversion and protects from potentially harmful
intestinal microorganisms and necrotic enteritis in broilers. Front
Microbiol. 7:14162016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McGrath JC, Drummond GB, McLachlan EM,
Kilkenny C and Wainwright CL: Guidelines for reporting experiments
involving animals: The ARRIVE guidelines. Br J Pharmacol.
160:1573–1576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hampshire VA and Gilbert SH: Refinement,
reduction, and replacement (3R) strategies in preclinical testing
of medical devices. Toxicol Pathol. 47:329–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vidali S, Aminzadeh-Gohari S, Feichtinger
RG, Vatrinet R, Koller A, Locker F, Rutherford T, O'Donnell M,
Stöger-Kleiber A, Lambert B, et al: The ketogenic diet is not
feasible as a therapy in a CD-1 nu/nu mouse model of renal cell
carcinoma with features of Stauffer's syndrome. Oncotarget.
8:57201–57215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Robertis M, Massi E, Poeta ML, Carotti
S, Morini S, Cecchetelli L, Signori E and Fazio VM: The AOM/DSS
murine model for the study of colon carcinogenesis: From pathways
to diagnosis and therapy studies. J Carcinog. 10:92011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rueden CT, Schindelin J, Hiner MC, DeZonia
BE, Walter AE, Arena ET and Eliceiri KW: ImageJ2: ImageJ for the
next generation of scientific image data. BMC Bioinformatics.
18:5292017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hung CS, Wang YC, Guo JW, Yang RN, Lee CL,
Shen MH, Huang CC, Huang CJ, Yang JY and Liu CY: Expression pattern
of placenta specific 8 and keratin 20 in different types of
gastrointestinal cancer. Mol Med Rep. 21:659–666. 2020.PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Protoc. 2008:pdb.prot4986. 2008.
|
|
38
|
Kim JJ, Shajib MS, Manocha MM and Khan WI:
Investigating intestinal inflammation in DSS-induced model of IBD.
J Vis Exp. 60:36782012.
|
|
39
|
Vargason AM and Anselmo AC: Clinical
translation of microbe-based therapies: Current clinical landscape
and preclinical outlook. Bioeng Transl Med. 3:124–137. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shah MS, DeSantis T, Yamal JM, Weir T,
Ryan EP, Cope JL and Hollister EB: Re-purposing 16S rRNA gene
sequence data from within case paired tumor biopsy and
tumor-adjacent biopsy or fecal samples to identify microbial
markers for colorectal cancer. PLoS One. 13:e02070022018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao C, Ganesh BP, Shi Z, Shah RR, Fultz R,
Major A, Venable S, Lugo M, Hoch K, Chen X, et al: Gut
microbe-mediated suppression of inflammation-associated colon
carcinogenesis by luminal histamine production. Am J Pathol.
187:2323–2336. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Eeckhaut V, Van Immerseel F, Teirlynck E,
Pasmans F, Fievez V, Snauwaert C, Haesebrouck F, Ducatelle R, Louis
P and Vandamme P: Butyricicoccus pullicaecorum gen. nov.,
sp. nov., an anaerobic, butyrate-producing bacterium isolated from
the caecal content of a broiler chicken. Int J Syst Evol Microbiol.
58:2799–2802. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sitkin S and Pokrotnieks J: Clinical
potential of anti-inflammatory effects of faecalibacterium
prausnitzii and butyrate in inflammatory bowel disease. Inflamm
Bowel Dis. 25:e40–e41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Eeckhaut V, Machiels K, Perrier C, Romero
C, Maes S, Flahou B, Steppe M, Haesebrouck F, Sas B, Ducatelle R,
et al: Butyricicoccus pullicaecorum in inflammatory bowel
disease. Gut. 62:1745–1752. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kurata N, Tokashiki N, Fukushima K,
Hasuoka N, Kitagawa K, Mashimo M, Regan JW, Murayama T and Fujino
H: Short chain fatty acid butyrate uptake reduces expressions of
prostanoid EP4 receptors and their mediation of cyclooxygenase-2
induction in HCA-7 human colon cancer cells. Eur J Pharmacol.
853:308–315. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kazemi Sefat NA, Mohammadi MM, Hadjati J,
Talebi S, Ajami M and Daneshvar H: Sodium butyrate as a histone
deacetylase inhibitor affects toll-like receptor 4 expression in
colorectal cancer cell lines. Immunol Invest. 48:759–769. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Luo S, Li Z, Mao L, Chen S and Sun S:
Sodium butyrate induces autophagy in colorectal cancer cells
through LKB1/AMPK signaling. J Physiol Biochem. 75:53–63. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou Q, Li G, Zuo S, Zhu W and Yuan X: RNA
sequencing analysis of molecular basis of sodium butyrate-induced
growth inhibition on colorectal cancer cell lines. Biomed Res Int.
2019:14278712019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pirozzi C, Francisco V, Guida FD, Gómez R,
Lago F, Pino J, Meli R and Gualillo O: Butyrate modulates
inflammation in chondrocytes via GPR43 receptor. Cell Physiol
Biochem. 51:228–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gopal E, Miyauchi S, Martin PM, Ananth S,
Roon P, Smith SB and Ganapathy V: Transport of nicotinate and
structurally related compounds by human SMCT1 (SLC5A8) and its
relevance to drug transport in the mammalian intestinal tract.
Pharm Res. 24:575–584. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cimadamore A, Santoni M, Massari F,
Gasparrini S, Cheng L, Lopez-Beltran A, Montironi R and Scarpelli
M: Microbiome and cancers, with focus on genitourinary tumors.
Front Oncol. 9:1782019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gurav A, Sivaprakasam S, Bhutia YD,
Boettger T, Singh N and Ganapathy V: Slc5a8, a Na+-coupled
high-affinity transporter for short-chain fatty acids, is a
conditional tumour suppressor in colon that protects against
colitis and colon cancer under low-fibre dietary conditions.
Biochem J. 469:267–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zaiss MM, Jones RM, Schett G and Pacifici
R: The gut-bone axis: How bacterial metabolites bridge the
distance. J Clin Invest. 129:3018–3028. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen JS, Faller DV and Spanjaard RA:
Short-chain fatty acid inhibitors of histone deacetylases:
Promising anticancer therapeutics? Curr Cancer Drug Targets.
3:219–236. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Singh N, Thangaraju M, Prasad PD, Martin
PM, Lambert NA, Boettger T, Offermanns S and Ganapathy V: Blockade
of dendritic cell development by bacterial fermentation products
butyrate and propionate through a transporter (Slc5a8)-dependent
inhibition of histone deacetylases. J Biol Chem. 285:27601–27608.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Turowski GA, Rashid Z, Hong F, Madri JA
and Basson MD: Glutamine modulates phenotype and stimulates
proliferation in human colon cancer cell lines. Cancer Res.
54:5974–5980. 1994.PubMed/NCBI
|
|
57
|
Leibovitz A, Stinson JC, McCombs WB III,
McCoy CE, Mazur KC and Mabry ND: Classification of human colorectal
adenocarcinoma cell lines. Cancer Res. 36:4562–4569.
1976.PubMed/NCBI
|
|
58
|
Gupta N, Martin PM, Prasad PD and
Ganapathy V: SLC5A8 (SMCT1)-mediated transport of butyrate forms
the basis for the tumor suppressive function of the transporter.
Life Sci. 78:2419–2425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tang Y, Chen Y, Jiang H, Robbins GT and
Nie D: G-protein-coupled receptor for short-chain fatty acids
suppresses colon cancer. Int J Cancer. 128:847–856. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Thangaraju M, Cresci GA, Liu K, Ananth S,
Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ,
Lambert NA, et al: GPR109A is a G-protein-coupled receptor for the
bacterial fermentation product butyrate and functions as a tumor
suppressor in colon. Cancer Res. 69:2826–2832. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Eslami M, Yousefi B, Kokhaei P, Hemati M,
Nejad ZR, Arabkari V and Namdar A: Importance of probiotics in the
prevention and treatment of colorectal cancer. J Cell Physiol.
234:17127–17143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gui Q, Li H, Wang A, Zhao X, Tan Z, Chen
L, Xu K and Xiao C: The association between gut butyrate-producing
bacteria and non-small-cell lung cancer. J Clin Lab Anal.
34:e233182020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zeng X, Gao X, Peng Y, Wu Q, Zhu J, Tan C,
Xia G, You C, Xu R, Pan S, et al: Higher risk of stroke is
correlated with increased opportunistic pathogen load and reduced
levels of butyrate-producing bacteria in the gut. Front Cell Infect
Microbiol. 9:42019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen D, Jin D, Huang S, Wu J, Xu M, Liu T,
Dong W, Liu X, Wang S, Zhong W, et al: Clostridium
butyricum, a butyrate-producing probiotic, inhibits intestinal
tumor development through modulating Wnt signaling and gut
microbiota. Cancer Lett. 469:456–467. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hagihara M, Kuroki Y, Ariyoshi T, Higashi
S, Fukuda K, Yamashita R, Matsumoto A, Mori T, Mimura K, Yamaguchi
N, et al: Clostridium butyricum modulates the microbiome to
protect intestinal barrier function in mice with antibiotic-induced
dysbiosis. iScience. 23:1007722020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sun M, Wu W, Liu Z and Cong Y: Microbiota
metabolite short chain fatty acids, GPCR, and inflammatory bowel
diseases. J Gastroenterol. 52:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Alvarez-Mercado AI, Navarro-Oliveros M,
Robles-Sánchez C, Plaza-Díaz J, Sáez-Lara MJ, Muñoz-Quezada S,
Fontana L and Abadía-Molina F: Microbial population changes and
their relationship with human health and disease. Microorganisms.
7:682019. View Article : Google Scholar
|
|
68
|
Roduit C, Frei R, Ferstl R, Loeliger S,
Westermann P, Rhyner C, Schiavi E, Barcik W, Rodriguez-Perez N,
Wawrzyniak M, et al: High levels of butyrate and propionate in
early life are associated with protection against atopy. Allergy.
74:799–809. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li M, van Esch BCAM, Henricks PAJ,
Folkerts G and Garssen J: The anti-inflammatory effects of short
chain fatty acids on lipopolysaccharide- or tumor necrosis factor
α-stimulated endothelial cells via activation of GPR41/43 and
inhibition of HDACs. Front Pharmacol. 9:5332018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Scorletti E, Afolabi PR, Miles EA, Smith
DE, Almehmadi A, Alshathry A, Moyses HE, Clough GF, Wright M, Patel
J, et al: Design and rationale of the INSYTE study: A randomised,
placebo controlled study to test the efficacy of a synbiotic on
liver fat, disease biomarkers and intestinal microbiota in
non-alcoholic fatty liver disease. Contemp Clin Trials. 71:113–123.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lau WL and Vaziri ND: Gut microbial
short-chain fatty acids and the risk of diabetes. Nat Rev Nephrol.
15:389–390. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cornejo-Pareja I, Muñoz-Garach A,
Clemente-Postigo M and Tinahones FJ: Importance of gut microbiota
in obesity. Eur J Clin Nutr. 72 (Suppl 1):S26–S37. 2019. View Article : Google Scholar
|
|
73
|
Gill PA, van Zelm MC, Muir JG and Gibson
PR: Review article: Short chain fatty acids as potential
therapeutic agents in human gastrointestinal and inflammatory
disorders. Aliment Pharmacol Ther. 48:15–34. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Esser D, Lange J, Marinos G, Sieber M,
Best L, Prasse D, Bathia J, Rühlemann MC, Boersch K, Jaspers C and
Sommer F: Functions of the microbiota for the physiology of animal
metaorganisms. J Innate Immun. 11:393–404. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Koliarakis I, Messaritakis I, Nikolouzakis
TK, Hamilos G, Souglakos J and Tsiaoussis J: Oral bacteria and
intestinal dysbiosis in colorectal cancer. Int J Mol Sci.
20:41462019. View Article : Google Scholar
|
|
76
|
Chen J and Vitetta L:
Inflammation-modulating effect of butyrate in the prevention of
colon cancer by dietary fiber. Clin Colorectal Cancer.
17:e541–e544. 2018. View Article : Google Scholar : PubMed/NCBI
|