Introduction
Gastric cancer (GC) is the fifth most common cancer
type and ranks as the second leading cause of cancer-associated
death worldwide (1). Due to the lack
of distinct symptoms at the early stage, the majority of patients
are diagnosed in advanced stages and lose the opportunity of
radical surgery, and chemotherapy-based treatment remains the main
strategy. However, no remarkable advances were achieved in the past
decades (2). With the developments
in molecular biology, certain molecular targets were discovered,
which have been successfully used in the treatment of tumors
(3,4). However, the prospect of targeted
therapy for GC remains uncertain and the underlying obstacle may be
the lack of effective molecular targets. Therefore, it is urgent to
explore novel mechanisms of anti-GC drugs and provide new targets
in order to improve the prognosis of GC.
Proton pump inhibitors (PPIs) such as benzimidazole
derivatives, are safely used to treat a wide range of
gastrointestinal disorders such as peptic ulcer, gastritis and
reflux esophagitis (5). Depending on
their structure and chemical properties, PPIs may have different
mechanisms of action. Recently, PPIs have been repurposed,
including their application to decrease cisplatin-induced
nephrotoxicity, target viral replication and inhibit the
thioesterase activity of human fatty acid synthase (6–8). In
addition, PPIs have demonstrated antitumor activity in a variety of
tumor types and the antitumor mechanism may be associated with
apoptosis, autophagy and the acidic microenvironment (9). Esomeprazole (ESO), which is well known
as a powerful stomach acid inhibitor, has been recently
investigated regarding its growth inhibition, drug synergy and drug
resistance reversal functions in cancer cells (10,11). Due
to limited research, the anti-tumor effect and mechanisms of action
of ESO in GC cells have remained elusive.
Long non-coding RNAs (lncRNAs) are defined as
autonomously transcribed non-coding RNAs longer than 200
nucleotides with no coding function. lncRNAs have various roles,
such as remodeling of chromatin and genome architecture, RNA
stabilization and transcriptional regulation (12). Circular RNAs (circRNAs) are a new
type of endogenous noncoding RNA with a closed circular structure,
which regulate linear RNA transcription, downstream gene expression
and protein production (13).
MicroRNAs (miRNAs or miRs) either inhibit mRNA translation or
trigger mRNA degradation by binding to complementary sequences in
the 3′-untranslated regions of their target mRNAs (14). lncRNAs and circRNAs may act as
competing endogenous RNAs (ceRNAs) to control miRNA translation
(15–17). lncRNAs, together with circRNAs,
miRNAs, mRNAs and their interactions, provide insight into the
molecular pathogenesis of GC and a novel direction for therapeutic
approaches for this disease (17).
Therefore, it is worthwhile to explore the underlying molecular
mechanisms and targets (among lncRNAs, circRNAs, miRNAs and mRNAs)
of ESO as a promising antitumor agent in GC cells.
The present study investigated the effects of ESO on
the proliferation, metastasis, apoptosis and chemosensitivity in
AGS cells. The differential expression profiles were determined
using chip analysis and RNA-sequencing. Furthermore, an integrative
network analysis among lncRNAs, circRNAs, miRNAs and mRNAs, was
performed using bioinformatics methods.
Materials and methods
Cell line and cell culture
The human gastric cancer cell line AGS was provided
by the Shanghai Cell Bank of the Chinese Academy of Sciences. Cells
were cultured in F-12K medium (Hyclone; Cytiva) supplemented with
10% FBS (Hangzhou Sijiqing Biological Engineering Materials, Co.,
Ltd.) and antibiotics (100 U/ml penicillin and 100 µg/ml
streptomycin) in humidified air with 5% CO2 at 37°C.
Proliferation assays
Cell proliferation assays were performed with MTT
(Sigma-Aldrich; Merck KGaA). AGS cells were seeded into 96-well
plates in 100 µl F-12K medium containing 10% FBS at a density of
1×104 cells per well and incubated overnight for cell
attachment. Next, the cells were treated with ESO (AstraZeneca),
adriamycin (ADM; Pfizer, Inc.) and cisplatin (DDP; Qilu
Pharmaceutical Co., Ltd.), for 24 or 48 h. MTT solution (20 µl per
well) was then added and the plate was incubated at 37°C for an
additional 4 h. Next, the medium was discarded and 150 µl DMSO was
added to each well, followed by incubation for 10 min until the
formazan crystals that had formed were completely dissolved. The
absorbance was measured at 490 nm using a microplate reader (BioTek
Instruments, Inc.).
Transwell assays
The assays were performed in Transwell insert
chambers (pore size, 8 µm; Corning, Inc.). Approximately
1×105 cells treated with ESO (0, 10, 20 and 40 µg/ml)
alone, or combined with ADM (0.2 µg/ml) or DDP (20 µg/ml), were
seeded into the upper chamber in serum-free medium in triplicate
with or without Matrigel (BD Biosciences) for the invasion and
migration assay, respectively. A total of 600 µl F-12K medium with
10% FBS was added to the lower chamber. After incubation with the
above drugs for 12 h, the upper chambers were fixed with 4%
paraformaldehyde for 30 min at 37°C, and then stained with 0.1%
crystal violet for 30 min at 37°C. The migrating and invading cells
were counted in at least 6 visual fields per membrane under a light
microscope (Olympus Corp.).
Flow cytometric analysis
In brief, 2×105 cells in 500 µl
serum-free medium were seeded into 24-well plates and treated with
ESO (10 µg/ml) alone or in the presence of ADM (0.05 µg/ml) or DDP
(2.5 µg/ml), for apoptosis analysis, while cells were treated with
ESO (40 µg/ml) alone or in the presence of ADM (0.2 µg/ml) or DDP
(20 µg/ml), for cell cycle analysis. After incubation at 37°C for
12–48 h, the cells were collected for apoptosis and cell cycle
analyses using an Annexin V-FITC/PI apoptosis assay kit (BD
Biosciences). The cells were stained with 400 µl 1X binding buffer
and 5 µl Annexin V-FITC for 15 min, followed by incubation with 5
µl PI for 5 min at room temperature in the dark for the apoptosis
assay. For cell cycle analysis, cells were fixed with ice-cold
ethanol at 4°C overnight and suspended in ice-cold PBS containing
50 µg/ml PI at room temperature for 30 min. The cells were
immediately analyzed on an Accuri C6 flow cytometer (BD
Biosciences).
Microarray assay
The microarray Agilent Human lncRNA V6 (Agilent
Technologies, Inc.) was used. Total RNA extracted from the control
and experimental group (0 and 40 µg/ml ESO treatment, respectively)
was quantified with the NanoDrop ND-2000 (Thermo Fisher Scientific,
Inc.) and the RNA integrity was assessed using an Agilent
Bioanalyzer 2100 (Agilent Technologies, Inc.). Sample labeling,
microarray hybridization and washing were performed based on the
manufacturer's standard protocols. In brief, total RNA was
transcribed into double-strand complementary (c)DNA, then
reverse-transcribed into cRNA and labeled with cyanine-3-cytidine
triphosphate. The labeled cRNAs were hybridized onto the
microarray. After washing, the arrays were scanned with an Agilent
Scanner G2505C (Agilent Technologies, Inc.).
Feature Extraction software (version 10.7.1.1;
Agilent Technologies, Inc.) was used to analyze array images to
obtain raw data. GeneSpring (version 13.1; Agilent Technologies,
Inc.) was employed to normalize the raw data with the quantile
algorithm. Differentially expressed genes (DEGs) or transcripts
were identified through fold-change and P-value as calculated with
Student's t-test. The threshold set for up- and downregulated genes
or transcripts was fold change >2.0 and P<0.05.
RNA-sequencing
The preparation of whole-transcriptome libraries and
deep sequencing were performed by Illumina analysis (Shanghai OE
Biotech Co., Ltd.). For primary analysis, the length distribution
of the RNA sequences in the reference genome (ftp://ftp.ncbi.nlm.nih.gov/genomes) was
determined. The known miRNAs were identified by alignment against
the miRBase v.21 database (http://www.mirbase.org/) and patterns in different
samples were analyzed. Unannotated small RNAs were analyzed by the
software miRDeep2 (version 0.0.8; GitHub, Inc.) to predict novel
miRNAs.
Functional enrichment analysis
Functional enrichment analysis was performed with
the Database for Annotation, Visualization and Integrated Discovery
(DAVID; http://david.ncifcrf.gov/) to
determine the roles of differentially expressed RNAs. Gene Ontology
(GO) analysis was performed to obtain significantly enriched terms
in order to deduce important biological functions involving
multiple RNAs. Furthermore, Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analysis was used to identify the likely functions
and pathways associated with DEGs.
Construction of a ceRNA regulatory
network
The overlapped regions of the miRNA sequence binding
sites both on lncRNAs/circRNAs and mRNAs were searched to predict
lncRNA/circRNA-miRNA-mRNA interactions with the software miRanda
v3.3a (http://www.microrna.org). The ceRNA
networks were performed using Cytoscape (version 3.6.1; http://cytoscape.org/).
Statistical analysis
Values are expressed as the mean ± standard
deviation from at least three independent determinations. One-way
ANOVA followed by Bonferroni's post hoc test was used for multiple
comparisons. Comparisons between two groups were performed with
two-tailed Student's t-tests. All statistical analyses were
performed using SPSS v21.0 (IBM Corp.). The plotting of all
statistical graphs was performed with GraphPad Prism 8.0 (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
ESO inhibits proliferation and
metastasis, and increases chemosensitivity in AGS cells
MTT assays were performed to clarify whether ESO
exerted cytotoxic effects on AGS cells. The results indicated that
ADM, DDP and ESO significantly inhibited cell proliferation
(Fig. 1A-D). Furthermore, ESO
enhanced the susceptibility of AGS cells to the cytotoxic effects
of ADM and DDP (Fig. 1E and F). The
additional cytotoxic effect of ESO when combined with ADM and DDP
was dose- and time-dependent. To further determine the effects of
ESO, Transwell assays were performed (Fig. 1G and H). As expected, the migration
and invasion abilities of AGS cells were markedly suppressed in a
dose-dependent manner when the cells were treated with ESO, either
alone or in combination with ADM or DDP. In addition, the
combination of the above three drugs displayed a lower metastasis
potential compared with that caused by the combination of two
drugs. These results suggested that ESO suppressed proliferation
and metastasis, and enhanced chemosensitivity of AGS cells.
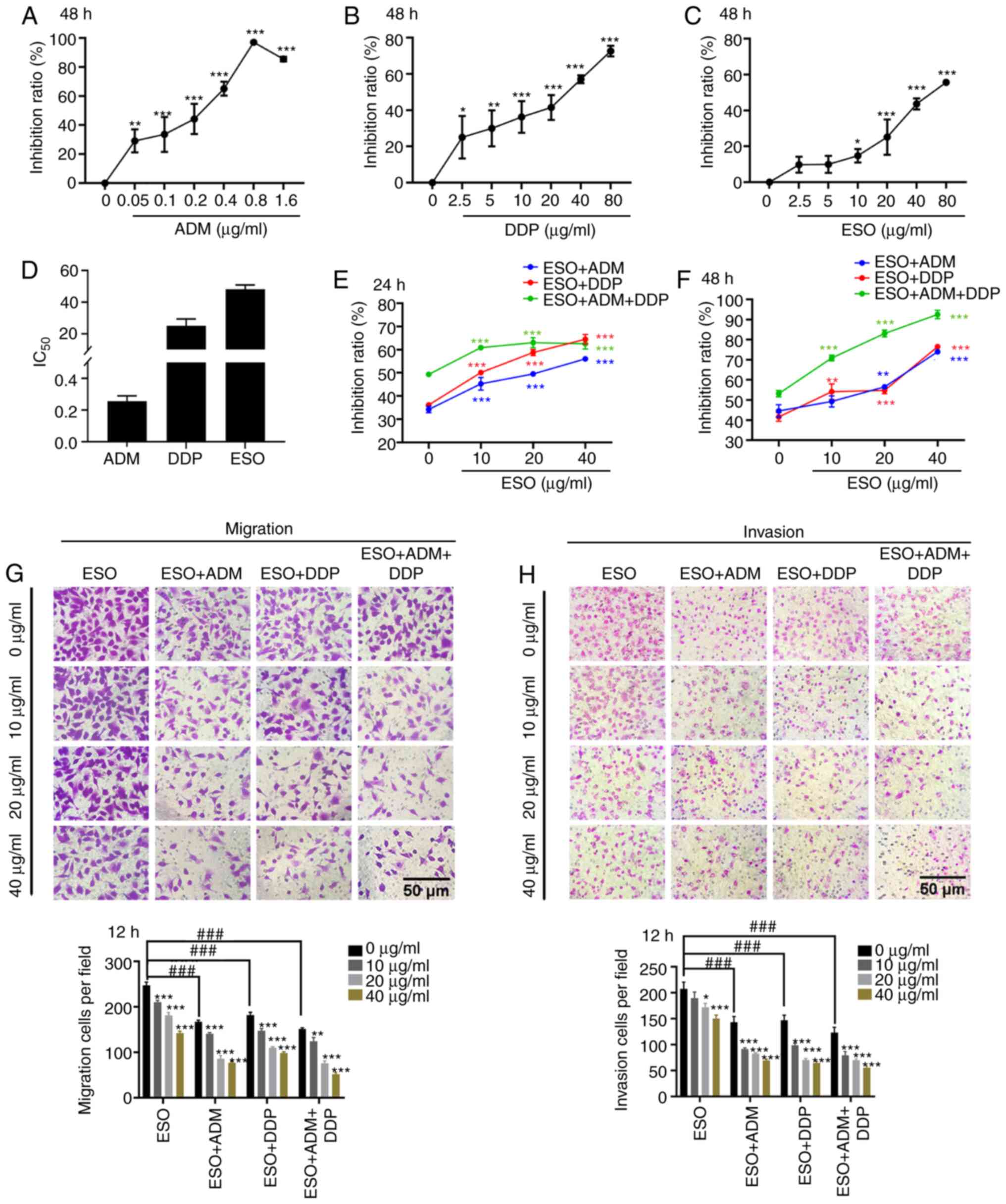 | Figure 1.ESO inhibits proliferation and
metastasis, and increases chemosensitivity in AGS cells. (A-C) MTT
assays demonstrated that (A) ADM, (B) DDP and (C) ESO inhibited
cell proliferation. (D) The IC50 of ADM, DDP and ESO in
AGS cells. (E and F) ESO significantly increased chemosensitivity
in AGS cells in a dose- and time-dependent manner (E) 24 h and (F)
48 h. (G and H) Transwell assays verified that ESO inhibited the
(G) migration and (H) invasion of AGS cells, alone or in
combination with 0.2 µg/ml ADM or 20 µg/ml DDP (scale bars, 50 µm).
*P<0.05, **P<0.01 and ***P<0.001 vs. control;
###P<0.001 as indicated. ESO, esomeprazole; ADM,
adriamycin; DDP, cisplatin; IC50, half maximal
inhibitory concentration. |
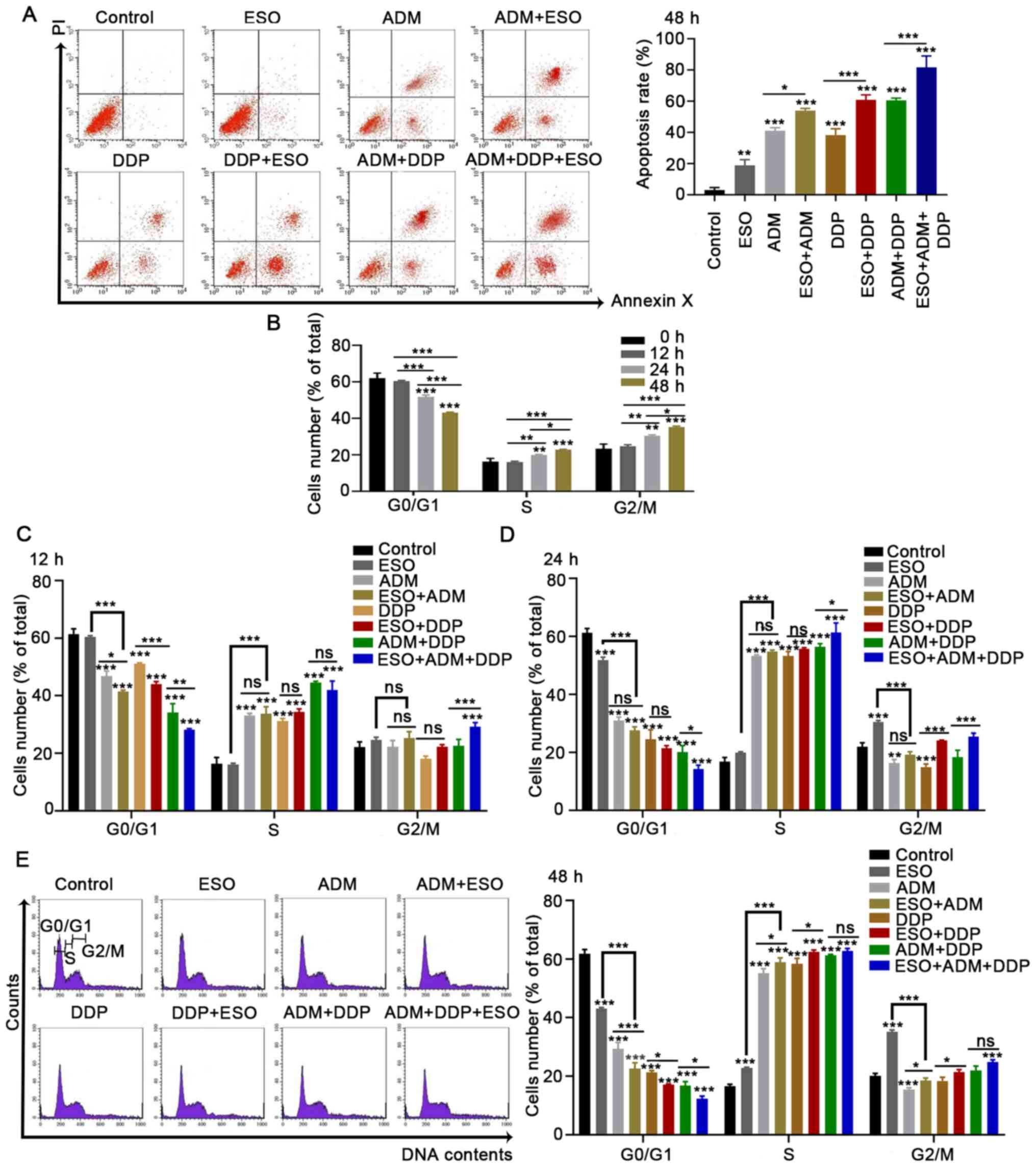
ESO induces AGS cell apoptosis via
causing cell cycle arrest at the S and G2/M phases
ESO induced AGS cell apoptosis, as demonstrated by
flow cytometry (Fig. 2A). The
apoptosis rates were significantly increased in AGS cells treated
with ESO, ADM or DDP. Furthermore, when treating the cells with a
combination of the above three drugs, the highest number of
apoptotic cells was observed. Cell cycle analysis suggested that
the proportion of cells in G0/G1 phase decreased significantly,
while the proportion of cells in the S and G2/M phases increased
after ESO treatment in a time-dependent manner (Fig. 2B). Different phenomena were observed
when ADM and DDP were added to the cells, either alone or in
combination with ESO. The numbers of cells arrested in S phase were
markedly increased, while those in G0/G1 phase were significantly
decreased (Fig. 2C-E). Taken
together, the present results suggested that ESO induced AGS cell
apoptosis, and caused cell cycle arrest in the S and G2/M phases in
a time-dependent manner. In combination with ADM and DDP, ESO
induced S-phase arrest in AGS cells.
Identification of differentially
expressed lncRNAs, circRNAs, miRNAs and mRNAs
To further analyze DEGs involved in the regulatory
effects of ESO on AGS cells, microarray analysis and RNA-sequencing
were performed. A total of 948 lncRNAs (487 upregulated and 461
downregulated), 114 circRNAs (32 upregulated and 82 downregulated),
1,197 mRNAs (498 upregulated and 699 downregulated) and 199 miRNAs
(71 upregulated and 128 downregulated) were identified to be
differentially expressed with a fold change >2.0 and P<0.05
(Fig. 3A-F). The top 10 DEGs were
distributed on multiple chromosomes (chrs), particularly chr10 and
chr19. The top 10 upregulated and downregulated lncRNAs, circRNAs,
miRNAs and mRNAs were identified from the microarrays (Tables SI–IV).
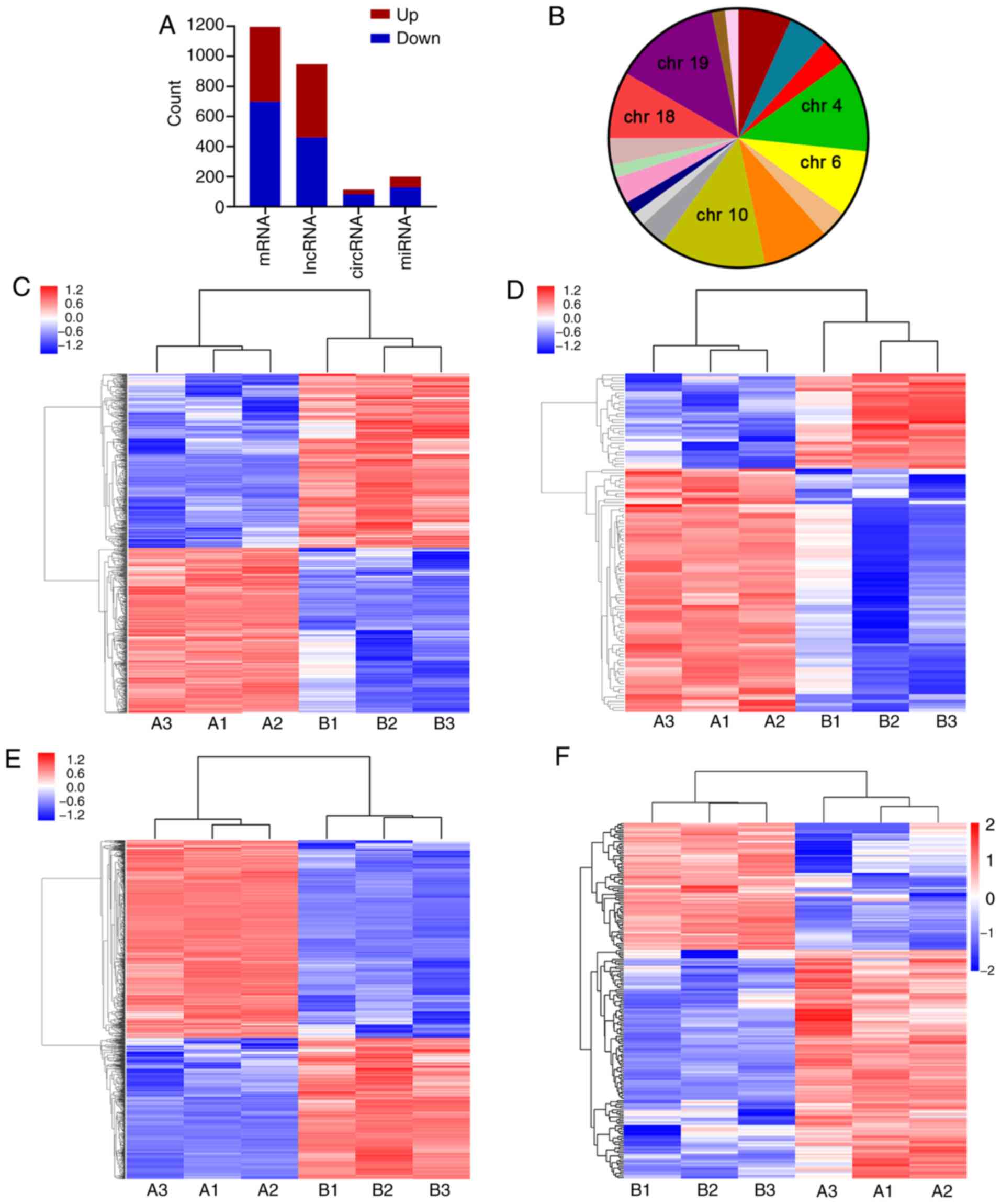 | Figure 3.Differentially expressed genes and
transcripts in AGS cells treated with ESO. (A) Statistical analysis
of the results of differentially expressed genes and transcripts.
(B) Chromosome distribution of the top 10 differentially expressed
lncRNAs, circRNAs and mRNAs. Hierarchical clustering analysis of
(C) lncRNAs, (D) circRNAs, (E) mRNAs and (F) miRNAs. Groups: A,
control group; B, experimental group (40 µg/ml ESO). ESO,
esomeprazole; chr, chromosome; lncRNA, long non-coding RNA;
circRNA, circular RNA; miRNA, microRNA. |
GO and KEGG pathway analyses
The results of the functional enrichment analysis
suggested that upregulated lncRNAs were mainly involved in
intrinsic apoptotic signaling pathway in response to endoplasmic
reticulum stress (GO:0070059) in the category biological process
(BP) (Fig. S1A). DNA replication,
nucleosome and protein heterodimerization activity were the most
meaningful downregulated terms in BP, cellular component (CC) and
molecular function (MF), respectively (Fig. S1B). From the KEGG pathway analysis,
EGFR tyrosine kinase inhibitor resistance [path: Homo
sapiens (hsa)01521] and miRNAs in cancer were most
significantly enriched by upregulated lncRNAs (Fig. S1C). However, cell cycle and DNA
replication were the top pathways enriched by downregulated lncRNAs
(Fig. S1D). As expected, regulation
of transcription, DNA-templated and protein binding were the top
terms associated with miRNAs in the categories BP and MF,
respectively (Fig. 4). And hydrogen;
potassium-exchanging ATPase complex (H+,
K+-ATPase), positive regulation of sodium;
potassium-exchanging ATPase (Na+, K+-ATPase)
activity and regulation of ATPase activity were also noted from GO
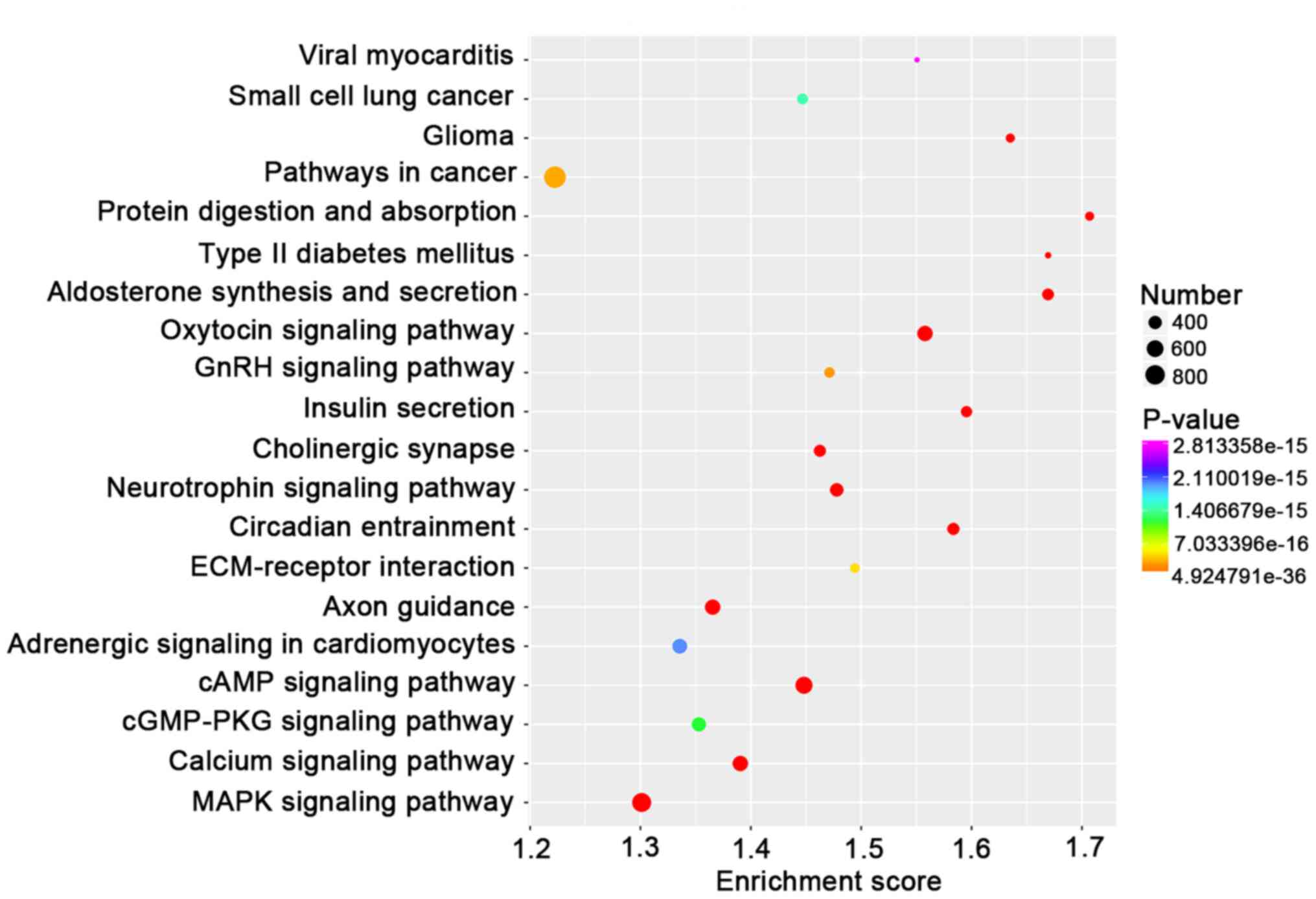
analysis (Table SV). Furthermore,
KEGG analysis indicated that target genes of miRNAs were
significantly enriched in the oxytocin signaling pathway and a
larger number of target genes were associated with pathways in
cancer (Fig. 5). Of note, the cell
cycle and DNA replication were also highlighted in the
lncRNA/circRNA-miRNA-mRNA ceRNA networks (Fig. 6). These results suggested that ESO
exerted its anti-GC effect via those co-expression networks,
impacting both lncRNAs and circRNAs associated with DNA replication
and the cell cycle.
Sub-pathway analysis in the lncRNA
co-expression network
To investigate the antitumor mechanism of ESO in GC,
four representative sub-pathways were selected through KEGG pathway
analysis, including the EGFR tyrosine kinase inhibitor resistance
pathway, FOXO signaling pathway, p53 signaling pathway and platinum
drug resistance pathway. There were complex associations between
mRNAs and multiple lncRNAs. The results indicated that upregulated
vascular endothelial growth factor A (VEGFA), transforming growth
factor α (TGFA), EGFR, FOXO3 and son of sevenless homolog (SOS)1,
and downregulated fibroblast growth factor receptor 2 (FGFR2) and
platelet-derived growth factor receptor α (PDGFRA), were associated
with the EGFR tyrosine kinase inhibitors resistance signaling
pathway (Fig. S2). Furthermore,
certain genes, such as cyclin B1 (CCNB1), cyclin B2 (CCNB2) and
S-phase kinase-associated protein 2 (SKP2) were downregulated,
while other genes (FOXO3, EGFR, phosphatidylinositol 3-kinase,
catalytic, 110-KD, α (PIK3CA) and SOS1) were upregulated in the
FOXO signaling pathway (Fig. S3).
Furthermore, the results demonstrated that lncRNAs regulated the
development of GC after ESO treatment by targeting
phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1),
cyclin-dependent kinase inhibitor 1A (CDKN1A), CCNB1 and CCNB2 in
the p53 signaling pathway (Fig.
S4). In addition, upregulation of CDKN1A, PMAIP1,
mitogen-activated protein kinase kinase kinase-5 (MAP3K5), PIK3CA
and downregulation of baculoviral inhibitor of apoptosis repeat
containing 5 (BIRC5) were closely associated with the platinum drug
resistance signaling pathway (Fig.
S5).
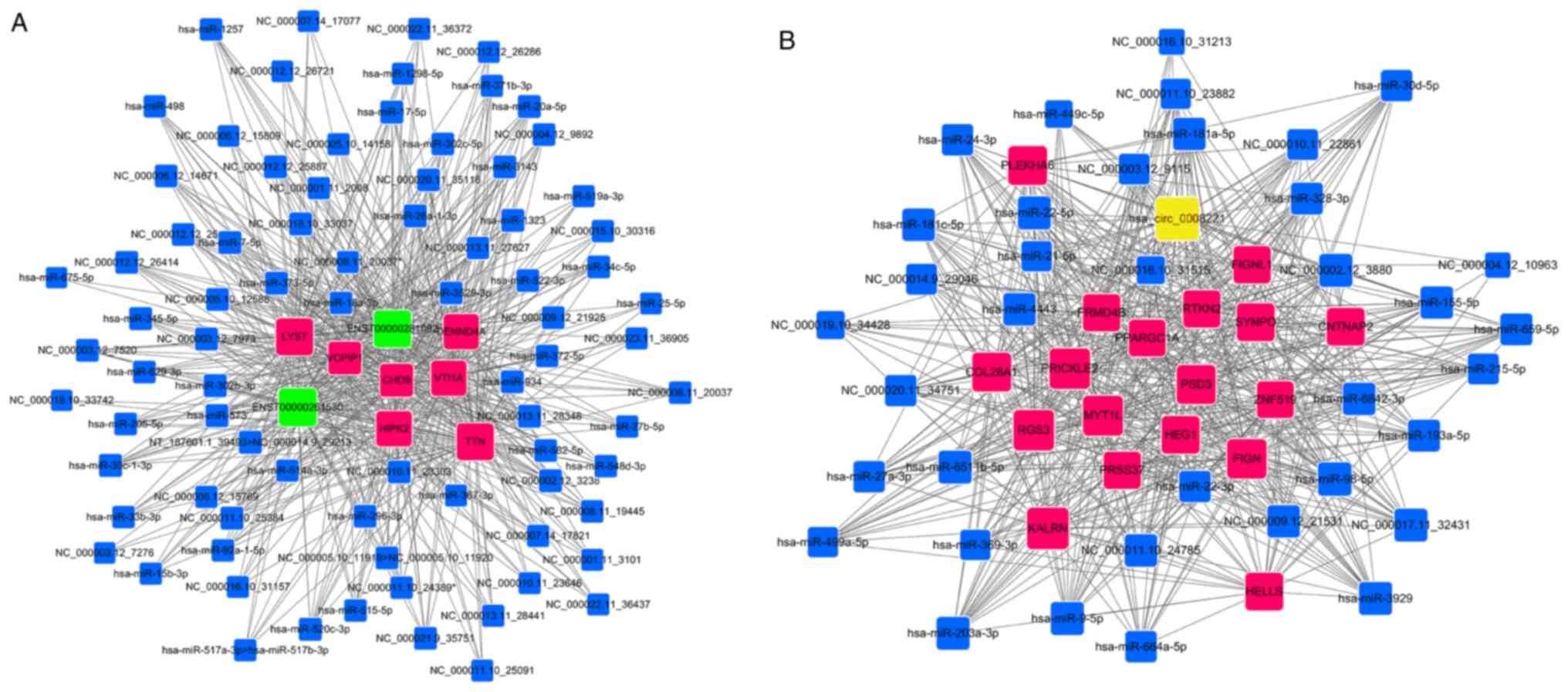
Prediction of
lncRNA/circRNA-miRNA-mRNA ceRNA network
In the present study, a total of 132,195
lncRNA-miRNA-mRNA ceRNA networks and 15,410 circRNA-miRNA-mRNA
ceRNA networks were created using the differentially expressed
lncRNAs (n=944), circRNAs (n=114), miRNAs (n=199) and mRNAs
(n=939). The top lncRNAs and circRNAs regulating multiple miRNAs
were highlighted, such as ENST00000261530, ENST00000281092 and
hsa_circ_0008221, which may be defined as key nodes of the ceRNA
network. There was a sub-network containing 2 lncRNAs, 81 miRNAs
and 7 mRNAs (Fig. 7A). In addition,
a ceRNA network consisted of 1 circRNA, 37 miRNAs and 18 mRNAs, as
presented in Fig. 7B. Integrating of
the lncRNA/circRNA-mRNA interactions indicated that
microtubule-associated protein 2 (MAP2), homeodomain-interacting
protein kinase 2 (HIPK2) and ankyrin 2 (ANK2) were targeted by both
lncRNAs and circRNAs (Figs. S6 and
S7). The list of data (Tables SVI and SVII) suggested that lncRNAs (NR_033268,
ENST00000261530 and ENST00000281092) and circRNAs (hsa_circ_0076332
and hsa_circ_0059713) were the ceRNAs of hsa-miR-372-5p targeting
MAP2, HIPK2 and ANK2. The above results revealed that the antitumor
mechanism of ESO in GC may be mediated by lncRNA/circRNA-miRNA-mRNA
ceRNA networks.
Discussion
It is well known that PPIs inhibit H+
transport and disrupt the acidic microenvironment on tumors by
inhibiting the activity of V-ATPase (a vacuolar proton pump), which
is the pivotal basis for the anti-cancer mechanism of PPIs
(9,11,18). In
addition, growing evidence revealed that PPIs have numerous novel
mechanisms responsible for their antitumor effects, including
influencing intracellular signal transduction, chromatin
remodeling, phosphorylation, autophagy and stress response
(19–21). Subsequently, it was indicated that
the mechanisms of PPIs to inhibit cell growth were closely
associated with miRNAs. Omeprazole inhibited cell proliferation and
induced cell cycle arrest through upregulating miR-203a-3p
expression in Barrett's esophagus cells (22). Unexpectedly, ESO not only impacted
the survival, metastatic potential and chemotherapy resistance of
esophageal cancer cells, but also affected the expression of
resistance-associated miRNAs (10).
Furthermore, high doses of PPIs regulated the pathways associated
with tumor malignancy and the microenvironment via inhibiting the
release of exosomes, which contain miRNAs (23). The present data suggested that ESO
also affected the proliferation, metastasis, chemosensitivity and
apoptosis of GC cells through regulating lncRNA/circRNA-miRNA-mRNA
ceRNA networks.
In cytotoxicity assays, ESO inhibited cell
proliferation and enhanced the susceptibility of the cells to ADM
and DDP in a dose- and time-dependent manner. Furthermore, it
significantly inhibited the migration and invasion of AGS cells in
a dose-dependent manner. In addition, the inhibitory effect of ESO
was more obvious when combined with ADM and DDP. The present
results were consistent with those of previous studies (10,23).
Thus, it was hypothesized that ESO suppressed the proliferation and
metastatic potential, and increased the chemosensitivity of AGS
cells in a dose-dependent manner.
Induction of apoptosis and cell cycle arrest are
currently considered to be important mechanisms underlying the
anticancer effects of potential drugs (22,24). The
present results suggested that ESO significantly induced AGS cell
apoptosis, which was enhanced after combination with ADM and DDP.
Of note, ESO inhibited the progression into S and G2/M phase in a
time-dependent manner, while ADM and DDP inhibited DNA synthesis by
causing S-phase arrest in AGS cells. Omeprazole was reported to
dose-dependently inhibit the growth of Barrett's esophagus and
induce cell cycle arrest in G0/G1 phase (22). Pantoprazole treatment caused cell
cycle arrest in G0/G1 phase to induce apoptosis in glioma cells
(24). The different of structures
of PPIs, cell lines and experimental conditions cannot be ruled
out. After treatment with ESO, ADM and DDP, cells were distinctly
accumulated in the S-phase. This phenomenon may be associated with
the fact that PPIs reverse the pH gradient and assist the
chemotherapeutic drugs to enter the cells, thus affecting the cell
cycle (9,18). The present results suggested a role
of PPIs in promoting apoptosis of GC cells, suggesting further
exploration of the anti-tumor mechanisms of ESO in preclinical
studies.
Accumulating evidence indicated that dysregulation
of lncRNAs, circRNAs, miRNAs and mRNAs contributes to the
development and progression of GC (17,25).
Based on lncRNA microarray, it was detected that the top 10
lncRNAs, circRNAs and mRNAs were mostly concentrated on chr10 and
chr19. Cytochrome P450 family 1 subfamily A member 1 (CYP1A1),
which is closely associated with the metabolism of PPIs (26), was upregulated 5-fold. At present,
multiple chemotherapeutic drugs require metabolic activation by
CYP1A1 to exert their cytostatic action (27). As a classic target gene of the Aryl
hydrocarbon receptor pathway, upregulated CYP1A1 may mediate the
growth and apoptosis in GC (28). It
was hypothesized that CYP1A1 may be an important target of PPIs in
GC to have antitumor effects. Among the genes detected on chr10,
the downregulation of aldo-keto reductase family 1 member C1
significantly reversed oxaliplatin resistance in GC and ATP binding
cassette subfamily C member 2–24C>T polymorphism was associated
with the response to platinum/5-fluorouracil-based neoadjuvant
chemotherapy in advanced GC (29,30).
Furthermore, the present results revealed that deleted in malignant
brain tumors 1 (DMBT1) was upregulated after ESO treatment. Paresi
et al (31) revealed a
potential link between benzimidazole compounds (the same effects as
those of PPIs) and DMBT1 for H. pylori eradication and
mucosal protection. Whether DMBT1 is a direct target of PPIs and
has a specific regulatory mechanism remains to be further
confirmed. Wang et al (32)
suggested that chromosomal instability was associated with the
aggressiveness of peritoneal metastasis in GC, such as chr19 gain.
Therefore, it was speculated that chr10 and chr19 may be closely
linked to the chemotherapy response and aggressiveness in GC,
respectively. Thus, studies should further explore the associated
genes on these chromosomes to identify relevant targets for the
treatment of GC.
Apoptotic signaling pathway and DNA replication were
pivotal terms accumulated by the up- and downregulated lncRNAs,
respectively. In the category CC, altered lncRNAs were enriched in
cytosol, extracellular exosome and nucleosome. KEGG pathway
analysis indicated that EGFR tyrosine kinase inhibitor resistance
and microRNAs in cancer were enriched by the upregulated lncRNAs.
Cell cycle and DNA replication were significant among the
downregulated lncRNAs, which were both enriched in the
lncRNA/circRNA-miRNA-mRNA ceRNA networks. Exosomes are small
vesicles containing multiple miRNAs and proteins. High doses of
PPIs were observed to suppress the malignant features of GC via
inhibiting the release of exosomes (23). Exosomes and miRNAs may be promising
for the detection and acquisition of ideal biomarkers, and may
provide a basis for novel therapeutic strategies in GC. In
addition, it was inferred that ESO mainly suppressed GC progression
by disrupting DNA synthesis and cell cycle progression, and
promoting the apoptotic signaling pathway.
In terms of the classical tumor signaling pathways
reported, such as the EGFR tyrosine kinase inhibitor resistance
pathway and the FOXO signaling pathway, it was indicated that EGFR,
FOXO3 and SOS1 were all upregulated after treatment with ESO. Of
note, both FOXO3 and EGFR were targets of ENST00000613376, and
lnc-endogenous retrovirus FRD 1-1:1(ERVFRD-1-1:1) was able to
regulate FOXO3 and SOS1 simultaneously. FOXO3 overexpression and
acidic stress have important roles in inducing apoptosis and
autophagy in AGS cells via the PI3K/AKT signaling pathway (33). Knockdown of EGFR-antisense 1
inhibited cell proliferation by suppressing the EGFR-dependent
PI3K/AKT pathway in GC (34).
However, panaxydol exposure activated EGFR in MCF-7 cells, which
triggered endoplasmic reticulum stress and induced cell apoptosis
(35). As a regulator downstream of
EGFR, FOXO3 was phosphorylated and degraded in colon cancer
following EGFR activation (36). The
SOS family mediates multiple signaling cascade connections. Growth
factor receptor-bound protein 2 (GRB2) knockdown led to decreased
phosphorylation of EGFR, and phosphorylated EGFR and the GRB2/SOS1
complex mediated resistance to osimertinib in acquired
afatinib-resistant non-small cell lung cancer with sustained KRAS
activation (37). Whether the
upregulation of EGFR is related to the upregulation of SOS1 remains
elusive. In addition, whether there is an antitumor effect by
targeting ENST00000613376 or lnc-ERVFRD-1-1:1 to activate the
EGFR/FOXO3/SOS1 signaling pathway remains to be explored.
The p53 pathway is associated with proliferation,
apoptosis and cell cycle changes in cancer cells, which are
regulated by multiple genes (38,39).
Platinum drug resistance remains is an intractable challenge in
anticancer treatment. Cell cycle arrest was indicated to be
predominantly mediated by transcriptionally increased expression of
growth arrest and DNA-damage-inducible 45 α (GADD45A) and CDKN1A,
and by decreased SKP2 levels (40).
MAPK1/3, BIRC5 and SKP2 have important roles in the apoptosis of
osteosarcoma cells (40). It was
proven that downregulation of BIRC5 (41), CCNB1 (38) and G2 and S phase-expressed-1 (GTSE-1)
(42) has key roles in tumor
inhibition. PMAIP1 belongs to the pro-apoptotic BH3-only family. It
was reported that upregulated PMAIP1 has a crucial role in inducing
apoptosis in bladder cancer (43).
These results were consistent with those of the present study.
By integrating the lncRNA/circRNA-miRNA-mRNA
co-expression networks, the complex ceRNA networks were
constructed. In addition, there were two signaling pathways:
NR_033268, ENST00000261530, ENST00000281092/hsa_circ_0076332,
hsa_circ_0059713-hsa-miR-372-5p-MAP2, HIPK2 and ANK2. MAP2 is a
novel prognostic marker in gemcitabine-resistant pancreatic cancer
(44). HIPK family members are
potent oncogenes and drive epithelial-to-mesenchymal transition.
Overexpression of HIPK promoted excessive cell proliferation and
invasion (45). Silencing of ANK2
restrained the migration and invasive potential of pancreatic
carcinoma (46). Furthermore,
miR-647 inhibited proliferation and metastasis in GC by
downregulating ANK2 (47).
Consequently, MAP2, HIPK2 and ANK2 may be important targets of ESO,
exerting an anti-tumor effect in AGS cells. It was hypothesized
that lncRNAs and circRNAs regulate mRNA expression and degradation
through co-competition of miRNAs, and further affect the occurrence
and development of tumors.
However, certain limitations of the present study
should be considered. First, as activated prodrugs in an acidic
environment, PPIs inhibit the activity of H+ and
K+-ATPase and induce cancer cell death by affecting pH
homeostasis (48). However, various
anti-cancer targets and mechanisms of PPIs also have been realized
under neutral pH conditions (11,19–24). The
objective of the present study was to explore the anti-GC effect of
ESO repurposed as a chemical anti-cancer agent under normal culture
conditions and to explore the underlying mechanisms. Whether PPIs
have a similar impact on GC and associated mechanisms in a low-pH
environment remains to be determined. The present data may provide
a basis for additional experiments under different pH conditions.
Furthermore, while the powerful anticancer effects of ESO have been
demonstrated in mouse models of melanoma, the potency of ESO
requires to be confirmed in GC in in vivo experiments
(49). Finally, the potential
therapeutic targets and signaling pathways determined in the
present study require further verification. Therefore, validation
experiments will be performed in the future in order to strengthen
the support for the application of ESO as a clinical treatment.
In conclusion, the present results confirmed that
ESO inhibits the proliferation, migration and invasion of AGS
cells, while strongly enhancing the cells' chemosensitivity and
inducing apoptosis by causing cell cycle arrest at the S and G2/M
phases. Furthermore, the profiles of RNAs regulated by ESO,
including lncRNAs, circRNAs, miRNAs and mRNAs, were determined for
the first time, to the best of our knowledge. HIPK2, MAP2 and ANK2
may be valuable targets for diagnosis and treatment of GC.
Furthermore, the EGFR tyrosine kinase inhibitor resistance pathway,
FOXO signaling pathway, p53 signaling pathway and platinum drug
resistance pathway may be closely associated with the antitumor
effect of ESO in AGS cells, although its specific mechanism
requires to be further verified in detail. These novel results
enhance the current understanding of the complex molecular
mechanisms of the effect of ESO on GC cell growth and provide
prospective targets for gene therapy.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Henan Medical Science
Foundation (grant no. 201701015 to ZZ and grant no. 2018020239 to
NB) and the Henan Scientific and Technological Foundation (grant
no. 172102310076 to ZZ).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ and NB conceived and designed the study. QX and
XJ wrote the manuscript and were responsible for cytological data
analysis. QW performed the cytological experiments. QX, LS and XX
were responsible for data analysis and interpretation of chip
detection and RNA-sequencing data. LS and XX revised the initial
manuscript critically for important intellectual content. ZM and HZ
performed the acquisition and collation of data, contributed to the
statistical analysis and plotted all statistical graphs and tables.
ZZ, NB, LS and XX revised the manuscript and approved the final
version of the manuscript submitted for publication. All authors
read and approved the manuscript and agreed to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work were appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ilson DH: Advances in the treatment of
gastric cancer. Curr Opin Gastroenterol. 33:473–476. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giustini NP, Jeong AR, Buturla J and
Bazhenova L: Advances in treatment of locally advanced or
metastatic Non-Small cell lung cancer: Targeted Therapy. Clin Chest
Med. 41:223–235. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song YH, He L, Wang YL, Wu Q and Huang WZ:
Molecularly targeted therapy and immunotherapy for hormone
Receptor-positive/human epidermal growth factor receptor 2-negative
advanced breast cancer (Review). Oncol Rep. 44:3–13.
2020.PubMed/NCBI
|
|
5
|
Kinoshita Y, Ishimura N and Ishihara S:
Advantages and disadvantages of Long-term proton pump inhibitor
use. J Neurogastroenterol Motil. 24:182–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ikemura K, Hiramatsu S and Okuda M: Drug
repositioning of proton pump inhibitors for enhanced efficacy and
safety of cancer chemotherapy. Front Pharmacol. 8:9112017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Watanabe SM, Ehrlich LS, Strickland M, Li
X, Soloveva V, Goff AJ, Stauft CB, Bhaduri-McIntosh S, Tjandra N
and Carter C: Selective targeting of virus replication by proton
pump inhibitors. Sci Rep. 10:40032020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fako VE, Wu X, Pflug B, Liu JY and Zhang
JT: Repositioning proton pump inhibitors as anticancer drugs by
targeting the thioesterase domain of human fatty acid synthase. J
Med Chem. 58:778–784. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu ZN, Tian B and Guo XL: Repositioning of
proton pump inhibitors in cancer therapy. Cancer Chemother
Pharmacol. 80:925–937. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lindner K, Borchardt C, Schöpp M, Bürgers
A, Stock C, Hussey DJ, Haier J and Hummel R: Proton pump inhibitors
(PPIs) impact on tumour cell survival, metastatic potential and
chemotherapy resistance, and affect expression of
resistance-relevant miRNAs in esophageal cancer. J Exp Clin Cancer
Res. 33:732014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He J, Shi XY, Li ZM, Pan XH, Li ZL, Chen
Y, Yan SJ and Xiao L: Proton pump inhibitors can reverse the YAP
mediated paclitaxel resistance in epithelial ovarian cancer. BMC
Mol Cell Biol. 20:492019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic Non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Ge YZ, Xu L and Jia R: Circular RNA
ITCH: A novel tumor suppressor in multiple cancers. Life Sci.
254:1171762020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hausser J and Zavolan M: Identification
and consequences of miRNA-target Interactions-beyond repression of
gene expression. Nat Rev Genet. 15:599–612. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu J, Zhang X, Gao W, Hu H, Wang X and
Hao D: lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar
intervertebral disc degeneration. Mol Med Report. 20:3160–3174.
2019.
|
|
17
|
Li J, Wang X, Lu W, Xiao Y, Yu Y, Wang X,
Xu C and Shen B: Comprehensive analysis of differentially expressed
Non-coding RNAs and mRNAs in gastric cancer cells under hypoxic
conditions. Am J Transl Res. 10:1022–1035. 2018.PubMed/NCBI
|
|
18
|
Iessi E, Logozzi M, Mizzoni D, Di Raimo R,
Supuran CT and Fais S: Rethinking the combination of proton
exchanger inhibitors in cancer therapy. Metabolites. 8:22018.
View Article : Google Scholar
|
|
19
|
Zhang B, Ling T, Zhaxi P, Cao Y, Qian L,
Zhao D, Kang W, Zhang W, Wang L, Xu G and Zou X: Proton pump
inhibitor pantoprazole inhibits gastric cancer metastasis via
suppression of telomerase reverse transcriptase gene expression.
Cancer Lett. 452:23–30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marino ML, Fais S, Djavaheri-Mergny M,
Villa A, Meschini S, Lozupone F, Venturi G, Della Mina P, Pattingre
S, Rivoltini L, et al: Proton pump inhibition induces autophagy as
a survival mechanism following oxidative stress in human melanoma
cells. Cell Death Dis. 1:e872010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao Y, Chen M, Tang D, Yan H, Ding X, Zhou
F, Zhang M, Xu GF, Zhang W, Zhang S, et al: The proton pump
inhibitor pantoprazole disrupts protein degradation systems and
sensitizes cancer cells to death under various stresses. Cell Death
Dis. 9:6042018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hou Y, Hu Q, Huang J and Xiong H:
Omeprazole inhibits cell proliferation and induces G0/G1 cell cycle
arrest through Up-regulating miR-203a-3p expression in barrett's
esophagus cells. Front Pharmacol. 8:9682017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guan XW, Zhao F, Wang JY, Wang HY, Ge SH,
Wang X, Zhang L, Liu R, Ba Y, Li HL, et al: Tumor microenvironment
interruption: A novel Anti-cancer mechanism of Proton-pump
inhibitor in gastric cancer by suppressing the release of
microRNA-carrying exosomes. Am J Cancer Res. 7:1913–1925.
2017.PubMed/NCBI
|
|
24
|
Geeviman K, Babu D and Prakash Babu P:
Pantoprazole induces mitochondrial apoptosis and attenuates NF-κB
signaling in glioma cells. Cell Mol Neurobiol. 38:1491–1504. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Han T, Li J, Cai H, Xu J, Chen L
and Zhan X: Comprehensive analysis of the regulatory network of
differentially expressed mRNAs, lncRNAs and circRNAs in gastric
cancer. Biomed Pharmacother. 122:1096862020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu D, Qiu T, Zhang Q, Kang H, Yuan S, Zhu
L and Zhu R: Systematic toxicity mechanism analysis of proton pump
inhibitors: An in silico study. Chem Res Toxicol. 28:419–430. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mescher M and Haarmann-Stemmann T:
Modulation of CYP1A1 metabolism: From adverse health effects to
chemoprevention and therapeutic options. Pharmacol Ther. 187:71–87.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yin XF, Chen J, Mao W, Wang YH and Chen
MH: A selective aryl hydrocarbon receptor modulator
3,3′-Diindolylmethane inhibits gastric cancer cell growth. J Exp
Clin Cancer Res. 31:462012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen CC, Chu CB, Liu KJ, Huang CY, Chang
JY, Pan WY, Chen HH, Cheng YH, Lee KD, Chen MF, et al: Gene
expression profiling for analysis acquired oxaliplatin resistant
factors in human gastric carcinoma TSGH-S3 cells: The role of IL-6
signaling and Nrf2/AKR1C axis identification. Biochem Pharmacol.
86:872–887. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Xing X, Shan F, Li S, Li Z, Xiao A,
Xing Z, Xue K, Li Z, Hu Y, et al: ABCC2-24C>T polymorphism is
associated with the response to platinum/5-Fu-based neoadjuvant
chemotherapy and better clinical outcomes in advanced gastric
cancer patients. Oncotarget. 7:55449–55457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paresi CJ, Liu Q and Li YM: Benzimidazole
covalent probes and the gastric H(+)/K(+)-ATPase as a model system
for protein labeling in a Copper-free setting. Mol Biosyst.
12:1772–1780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang R, Song S, Harada K, Ghazanfari
Amlashi F, Badgwell B, Pizzi MP, Xu Y, Zhao W, Dong X, Jin J, et
al: Multiplex profiling of peritoneal metastases from gastric
adenocarcinoma identified novel targets and molecular subtypes that
predict treatment response. Gut. 69:18–31. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Y, Qi W, Sun L, Lv J, Qiu W and Liu S:
FOXO3 Inhibits human gastric adenocarcinoma (AGS) cell growth by
promoting autophagy in an acidic microenvironment. Cell Physiol
Biochem. 49:335–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu J, Qian Y, Peng L, Ma L, Qiu T, Liu Y,
Li X and Chen X: Long noncoding RNA EGFR-AS1 promotes cell
proliferation by increasing EGFR mRNA stability in gastric cancer.
Cell Physiol Biochem. 49:322–334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim HS, Lim JM, Kim JY, Kim Y, Park S and
Sohn J: Panaxydol, a component of Panax ginseng, induces apoptosis
in cancer cells through EGFR activation and ER stress and inhibits
tumor growth in mouse models. Int J Cancer. 138:1432–1441. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qi W, Weber CR, Wasland K, Roy H, Wali R,
Joshi S and Savkovic SD: Tumor suppressor FOXO3 mediates signals
from the EGF receptor to regulate proliferation of colonic cells.
Am J Physiol Gastrointest Liver Physiol. 300:G264–G272. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakatani K, Yamaoka T, Ohba M, Fujita KI,
Arata S, Kusumoto S, Taki-Takemoto I, Kamei D, Iwai S, Tsurutani J
and Ohmori T: KRAS and amplifications mediate resistance to
rociletinib and osimertinib in acquired afatinib-resistant NSCLC
harboring exon 19 deletion/T790M in EGFR. Mol Cancer Ther.
18:112–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang H, Zhang X, Li X, Meng WB, Bai ZT,
Rui SZ, Wang ZF, Zhou WC and Jin XD: Effect of CCNB1 silencing on
cell cycle, senescence, and apoptosis through the p53 signaling
pathway in pancreatic cancer. J Cell Physiol. 234:619–631. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
40
|
Kleinsimon S, Longmuss E, Rolff J, Jäger
S, Eggert A, Delebinski C and Seifert G: GADD45A and CDKN1A are
involved in apoptosis and cell cycle modulatory effects of viscumTT
with further inactivation of the STAT3 pathway. Sci Rep.
8:57502018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shang X, Liu G, Zhang Y, Tang P, Zhang H,
Jiang H and Yu Z: Downregulation of BIRC5 inhibits the migration
and invasion of esophageal cancer cells by interacting with the
PI3K/Akt signaling pathway. Oncol Lett. 16:3373–3379.
2018.PubMed/NCBI
|
|
42
|
Guo L, Zhang S, Zhang B, Chen W, Li X,
Zhang W, Zhou C, Zhang J, Ren N and Ye Q: Silencing GTSE-1
expression inhibits proliferation and invasion of hepatocellular
carcinoma cells. Cell Biol Toxicol. 32:263–274. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cui J, Sun W, Hao X, Wei M, Su X, Zhang Y,
Su L and Liu X: EHMT2 inhibitor BIX-01294 induces apoptosis through
PMAIP1-USP9X-MCL1 axis in human bladder cancer cells. Cancer Cell
Int. 15:42015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Le Large TYS, El Hassouni B, Funel N, Kok
B, Piersma SR, Pham TV, Olive KP, Kazemier G, van Laarhoven HWM,
Jimenez CR, et al: Proteomic analysis of gemcitabine-resistant
pancreatic cancer cells reveals that microtubule-associated protein
2 upregulation associates with taxane treatment. Ther Adv Med
Oncol. 11:17588359198412332019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Blaquiere JA, Wong KKL, Kinsey SD, Wu J
and Verheyen EM: Homeodomain-interacting protein kinase promotes
tumorigenesis and metastatic cell behavior. Dis Model Mech.
11:dmm0311462018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen Y, Löhr M and Jesnowski R: Inhibition
of ankyrin-B expression reduces growth and invasion of human
pancreatic ductal adenocarcinoma. Pancreatology. 10:586–596. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cao W, Wei W, Zhan Z, Xie D, Xie Y and
Xiao Q: Role of miR-647 in human gastric cancer suppression. Oncol
Rep. 37:1401–1411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mullin JM, Gabello M, Murray LJ, Farrell
CP, Bellows J, Wolov KR, Kearney KR, Rudolph D and Thornton JJ:
Proton pump inhibitors: Actions and reactions. Drug Discov Today.
14:647–660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
De Milito A, Canese R, Marino ML, Fais S,
Venturi G, Rodolfo M, Borghi M, Villa A, Della Mina P, Lozupone F,
et al: pH-dependent antitumor activity of proton pump inhibitors
against human melanoma is mediated by inhibition of tumor acidity.
Int J Cancer. 127:207–219. 2010. View Article : Google Scholar : PubMed/NCBI
|