Introduction
Ovarian cancer is the deadliest gynecological cancer
in developed countries and, in Japan, the fifth leading cause of
cancer death in women (1), with
4,745 deaths reported in 2017 (Cancer Registry and Statistics.
Cancer Information Service, National Cancer Center, Japan.
http://ganjoho.jp/reg_stat/statistics/dl/index.html).
Ovarian cancer is typically diagnosed at an advanced stage, after
peritoneal dissemination and massive ascites have developed
(2,3). Furthermore, no improvement in the
5-year survival rate of ovarian cancer patients is evident, despite
aggressive treatments involving surgery combined with platinum- and
taxane-based chemotherapy (2–5). The
most reliable prognostic factor predicting patient outcome is the
TNM stage. Whereas stage I ovarian cancer has a good prognosis, the
prognosis of stage IV is thought to be poor. According to this
finding, to identify prognostic factors predicting the outcomes of
patients with ovarian cancer, the objective of the study should be
limited to patients with intermediate stages (i.e. stages II and
III). Although pelvic examination, transvaginal ultrasonography,
and serum CA125 measurement are used as routine diagnostic
modalities, they fail to predict the prognosis of ovarian cancer
patients (6). Therefore, new
approaches to identify ovarian cancer markers are urgently needed
(7,8). At the molecular level, several genes
and pathways have been identified that may be closely associated
with the pathogenesis of ovarian cancer (9,10);
however, effective molecular markers predicting patient prognosis
have not been established (7,8). As
potential candidates, microRNAs (miRNAs) regulate cancer-related
gene expression and have been implicated in the etiology of ovarian
cancer (9,10). Recent studies have shown that
dysregulation of miRNA is closely associated with patient prognosis
in various cancers including ovarian cancer, melanoma and oral
cancer (9–16). In addition, expression of miRNA might
be useful as a diagnostic marker for cancer (9–16).
A recent study showed that cancer-associated
fibroblasts (CAFs), which are involved in the dynamic interaction
between cancer cells and the unique tumor microenvironment,
influence tumor progression as well as other genetic and epigenetic
events that markedly affect disease outcome and treatment response
1 (17–20). Based on this theory, both cancer
cells and the surrounding stromal cells play major roles in cancer
progression (17–20).
We used high-throughput genome-wide miRNA analysis
to evaluate the expression of specific miRNAs in cancer glands and
stromal cells, obtained using a crypt isolation method that
separates cancer glands from stromal cells, in ovarian high-grade
serous carcinoma (HGSC), a common histological and lethal ovarian
cancer variant.
Materials and methods
Patients
Thirty patients with HGSC of the ovary diagnosed at
Iwate Medical University were enrolled. These patients were divided
into cohort 1 (14 cases), whose samples were subjected to
comprehensive microarray analyses, and cohort 2 (16 cases in
addition to the 14 cases in cohort 1), whose samples were used to
verify the results from cohort 1 by RT-qPCR. The HGSC samples were
obtained from primary surgery in patients who had not received
chemotherapy. Two expert pathologists determined the histological
diagnoses, according to the General Rules for Ovarian Cancer of the
Japan Gynecological Cancer Group (21), using hematoxylin and eosin staining
to identify representative tumor areas, from which cores for
microarray analysis were obtained. The TNM classification of the
Union for International Cancer Control was used for disease staging
(22). Cases of low-grade serous
carcinoma were excluded from this study. The clinicopathological
variables examined, summarized in Table
I, included age, tumor size, FIGO stage, disease-free survival,
and overall survival. All patients provided written informed
consent, and the study was approved by the Iwate Medical University
Institutional Review Board (approval no. MH2018-528).
 | Table I.Clinicopathological findings of the
ovarian high-grade serous carcinoma we examined. |
Table I.
Clinicopathological findings of the
ovarian high-grade serous carcinoma we examined.
| Variable | Cohort
1a | Cohort
2b |
|---|
| Total | 14 | 30 |
| Median age (range),
years | 46.5 (31–79) | 56 (31–79) |
| Tumor size, median
(range), mm | 95 (28–176) | 89.5 (20–200) |
| FIGO stage (%) |
| II | 2
(14.3) | 8
(26.7) |
|
III | 12 (85.7) | 22 (73.3) |
| Recurrence (%) |
|
Present | 7
(50.0) | 16 (53.3) |
|
Absent | 7
(50.0) | 14 (46.7) |
| Survival (%) |
|
Dead | 2
(14.3) | 6
(20.0) |
|
Alive | 12 (85.7) | 24 (80.0) |
| Disease free
survival, median (range), days | 637
(381–1,098) | 592 (25–2,166) |
| Overall survival,
median (range), days | 1,035
(400–3,343) | 1,084
(259–3,343) |
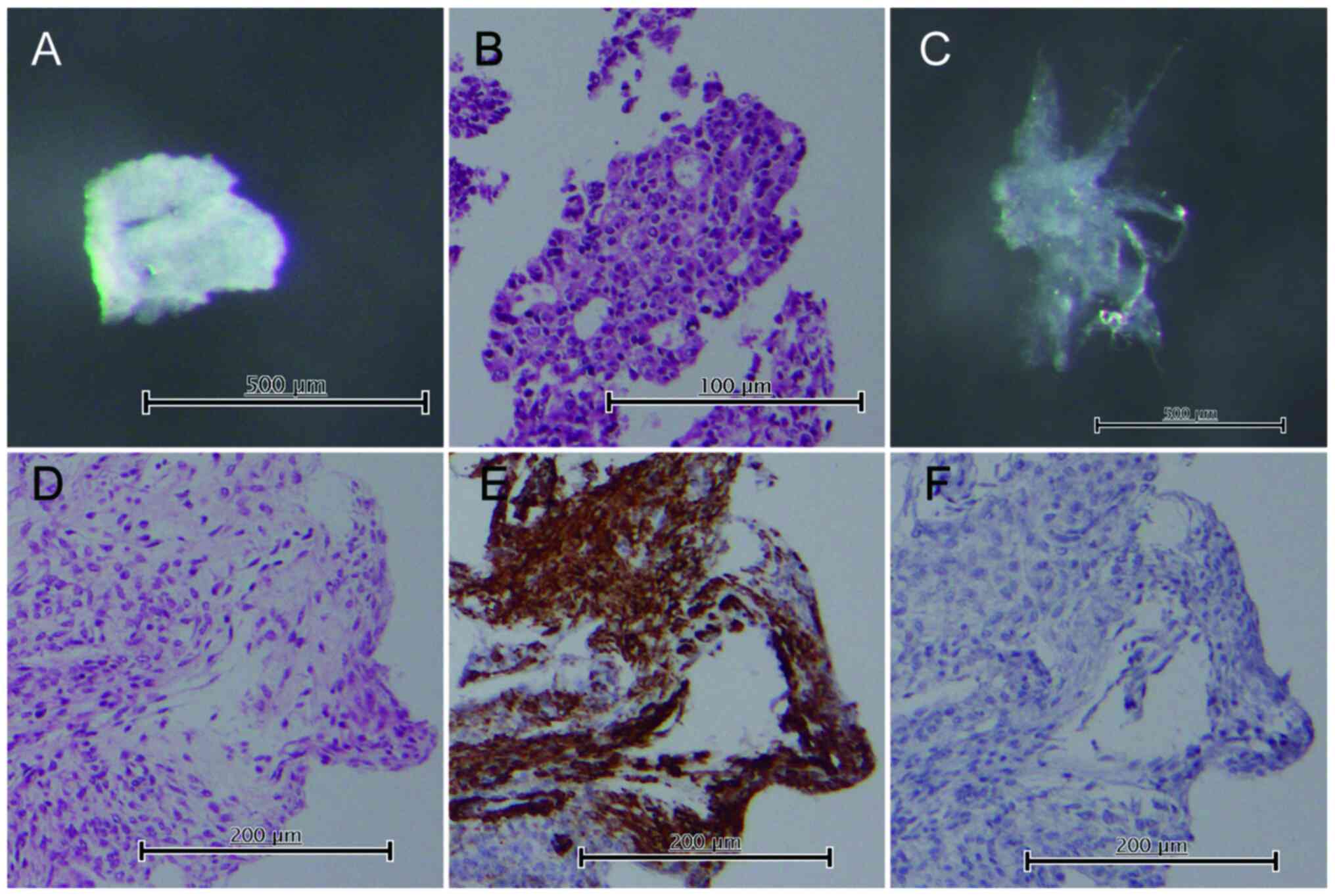
Gland isolation
Crypts were isolated from tumor and normal tissues,
as described previously, to obtain pure tumor glands separately
from the surrounding stromal tissues (23–25).
Tumor glands were isolated from the solid tumor region involved in
the invasion front, and this involvement was confirmed using tissue
sections prepared for the pathological diagnosis. Gland cells were
obtained from the tumor tissues after careful separation of the
stromal cells (i.e. CAFs) adjacent to the glands, performed under a
dissecting microscope. Cells from normal fallopian tube tissue and
normal fibroblasts within the Fallopian tubes were also obtained as
controls. Paraffin-embedded tissue sections of the isolated samples
were routinely processed to confirm the histology. Immunostaining
using antibodies against smooth muscle actin (clone 1A4; Dako) and
desmin (clone D33; Dako) was performed in the stromal cells to
confirm the exclusive presence of fibroblasts, determined according
to negative staining of smooth muscle actin and positive staining
of desmin. However, we could not exclude the possibility of stromal
cell contamination with other non-epithelial cells, such as
inflammatory and vessel cells. Representative images are shown in
Fig. 1.
RNA extraction
miRNAs in isolated tumor glands and the
corresponding stromal cells were extracted using the mirVana™ miRNA
Isolation kit (Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. The quantity and quality of the
obtained RNA were evaluated using the DU730 spectrophotometer
(Beckman Coulter) and the integrity by gel electrophoresis.
miRNA microarray analysis
For microarray analysis, 200 ng RNA was
polyadenylated and labelled using a FlashTag™ Biotin HSR RNA
Labelling kit and then treated with DNA ligase. The labeled RNA was
hybridized to GeneChip miRNA 4.0 microarrays (Thermo Fisher
Scientific) at 48°C for 16 h, followed by washing and staining
using a streptavidin-PE solution. The stained arrays were scanned
using a GeneChip™ Scanner 3000 7G System (Thermo Fisher
Scientific). The Affymetrix miRNA 4.0 microarray contains 6,631
probes on the array, including 2,570 mature miRNA probes. Detailed
methods have been described previously (26).
Verification of miRNA expression by
RT-qPCR in cohort 2 samples
RT-qPCR was used to confirm miRNA expression levels
in HGSC (isolated cancer glands and surrounding stromal cells) and
normal tissue samples using the Applied Biosystem (ABI) Detection
System (Step One Plus) (27). First,
cDNA was reverse-transcribed from 10 ng total RNA using the TaqMan
MicroRNA Reverse Transcription Kit (cat. no. 4366596; ABI). Then,
PCR was performed using TaqMan Universal Master Mix II, no UNG
(cat. no.: 4440040), and the following TaqMan assays:
Hsa-miR-188-5p (assay ID: 002320), hsa-miR-214-3p (assay ID:
002306), hsa-miR-505-5p (assay ID: 002087), hsa-miR-4455 (assay ID:
463355_mat), hsa-miR-6753-3p (assay ID: 466443_mat) and
hsa-miR-6877-3p (assay ID: 467048_mat). In addition, the expression
levels of hsa-miR-101-5p (assay ID: 002143), hsa-miR-320c (assay
ID: 241053_mat), hsa-miR-320d (assay ID: 241066_mat), hsa-miR-320e
(assay ID: 243005_mat), hsa-miR-378f (assay ID: 462794_mat),
hsa-miR-455-3p (assay ID: 002244), hsa-miR-4429 (assay ID:
464083_mat) in the isolated stromal cells were examined by the same
method. All TaqMan miRNA assays were obtained from ABI. The primer
sequences are provided in Table SI.
The 2−ΔΔCq method was used to determine the relative
expression levels, with hsa-let-7a as an internal control that was
determined based on pre-analytical experiments in which expression
levels of RNU6B, let-7a and miRNA-21 were examined in all samples
including isolated cancer glands and isolated stromal tissue
samples. The expression levels of let-7a were constant and stable
in both isolated cancer gland and isolated stromal tissue
samples.
Statistical analysis
Differences in miRNA expression levels were analyzed
using the TAC4.0 (Thermo Fisher Scientific Inc.) and JMP pro 13.0
software package for Windows (SAS Institute Inc.).
Expression levels of miRNAs were analyzed using
Mann-Whitney U test. We determined the cutoff expression levels
using receiver operating characteristic (ROC) analysis. We
calculated disease-free survival (without recurrence including
metachronous metastasis) from the date of surgery to the
development of recurrence (including metachronous metastasis) or
the last follow-up, and overall survival from the date of surgery
to death or the last follow-up using Kaplan-Meier analysis. A log
rank test was conducted after Kaplan-Meier analysis to determine
significance. Potential factors associated with survival were
identified by univariate and multivariate analyses using Cox
proportional hazards regression models conducted using JMP 13.0
software. P<0.01 and P<0.05 were considered significant in
array analysis and non-array analysis, respectively.
Results
Association of clinicopathological
findings between cohort 1 and 2
There was no significant difference in the
clinicopathological variables between cohorts 1 and 2.
miRNA expression profiling in ovarian
HGSC
To identify potential miRNA biomarkers of ovarian
HGSC, we performed global miRNA expression profiling in the 14
HGSCs and normal tissues samples, as well as in the corresponding
surrounding stromal and normal fibrous tissues. Using the criteria
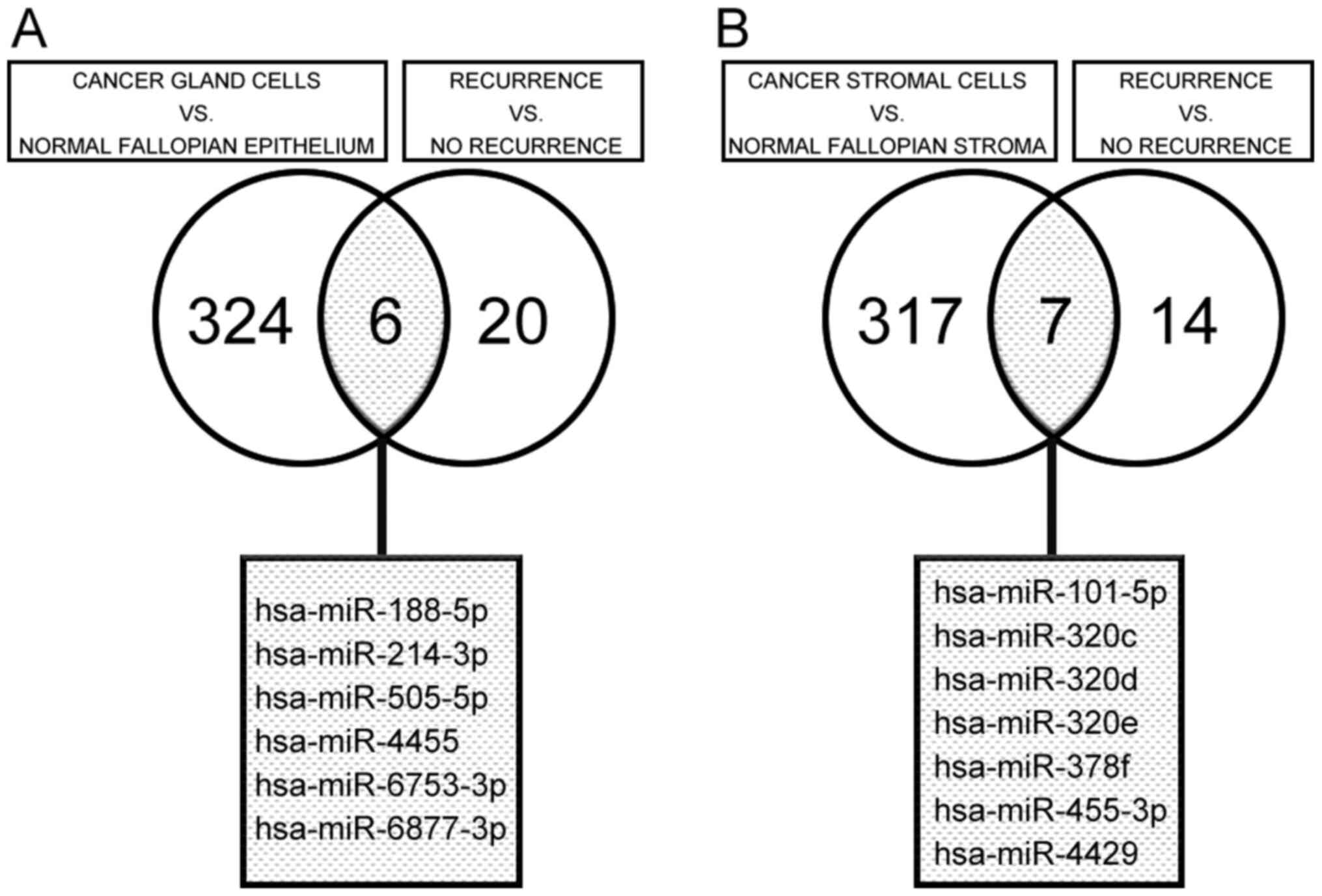
of a fold change in expression <-2 or >2 and P<0.01, we
performed two comparisons among the isolated cancer gland samples.
First, we compared expression levels between isolated cancer glands
and normal crypts and identified 330 differentially expressed
miRNAs (115 downregulated and 215 upregulated). Second, we compared
miRNA expression in isolated cancer glands between HGSC cases with
recurrence (including metachronous metastasis) and those without
recurrence (including metachronous metastasis) and identified 26
differentially expressed miRNAs (18 downregulated and 8
upregulated). Of these differentially expressed miRNAs, six
(hsa-miR-188-5p, hsa-miR-214-3p, hsa-miR-505-5p, hsa-miR-4455,
hsa-miR-6753-3p, and hsa-miR-6877-3p) were identified as
differentially expressed (two significantly downregulated and four
significantly upregulated) in both comparisons in isolated cancer
glands, based on a Venn diagram (Fig.
2A).
Using the same differential expression criteria
(fold change <-2 or >2 and P<0.01), we performed two
comparisons in the isolated stromal cell samples, similar to those
made in the isolated cancer glands. We identified 324
differentially expressed miRNAs (102 significantly downregulated
and 222 significantly upregulated) between cancer and normal
stromal cells and 21 differentially expressed miRNAs (13
significantly downregulated and 8 significantly upregulated) in
cancer stromal cells between HGSC cases with recurrence (including
metachronous metastasis) and those without recurrence. Of these
differentially expressed miRNAs, seven (hsa-miR-101-5p,
hsa-miR-320c, hsa-miR-320d, hsa-miR-320e, hsa-miR-378f,
hsa-miR-455-3p and hsa-miR-4429) were identified as differentially
expressed (six significantly downregulated and one significantly
upregulated) in both sets of comparisons in stromal cells, based on
a Venn diagram (Fig. 2B).
Fold-changes in miRNA expression for each comparison
(e.g., recurrence vs. no recurrence; crypt vs. stroma) are
presented in Table SII.
Comparison of candidate miRNA
expression between cancer gland and stromal tissue samples in
cohorts 1 and 2
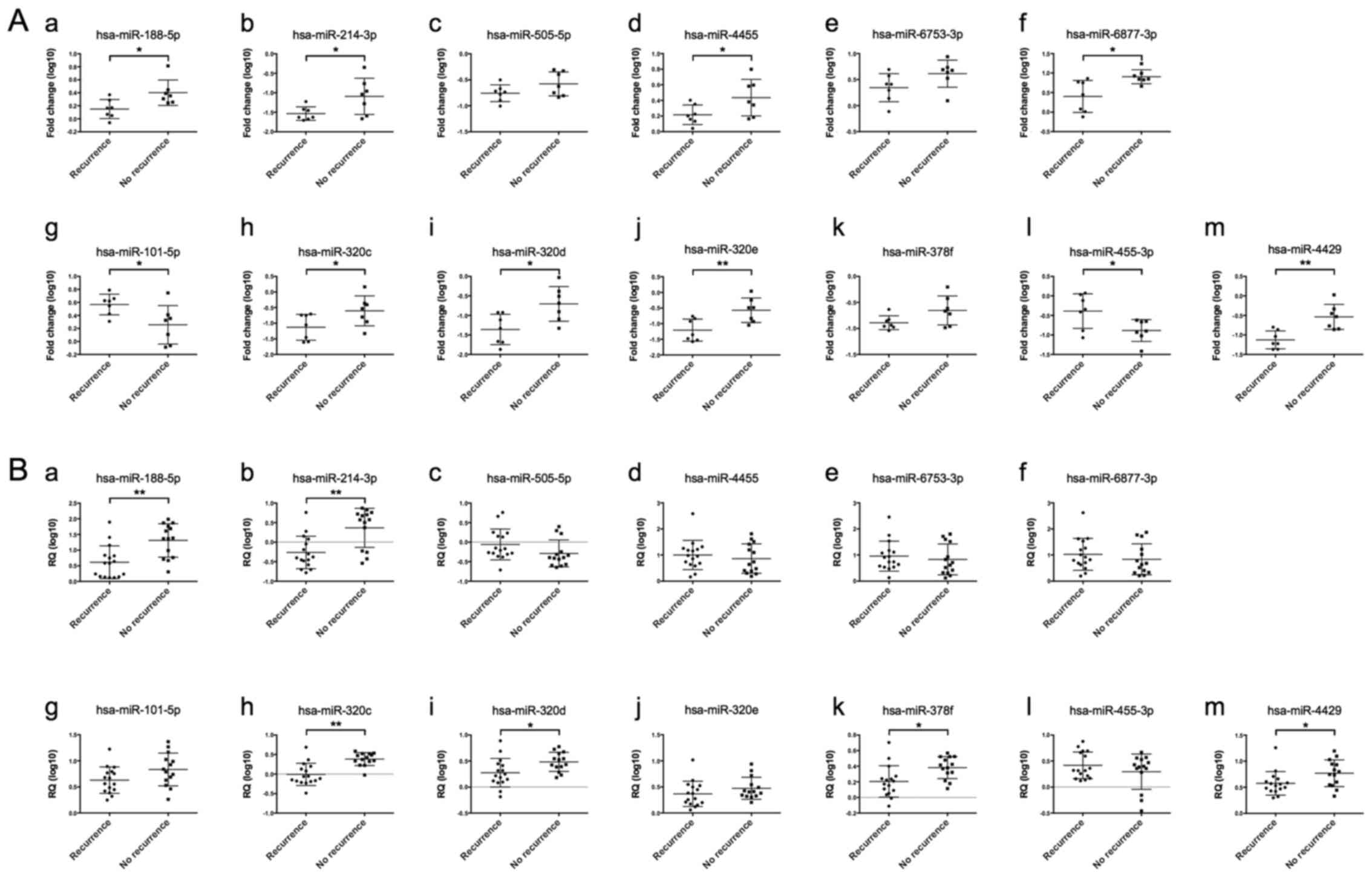
Among the six differentially expressed miRNAs
identified in isolated cancer glands from patients in cohort 1,
statistically significant differences in the expression level were
seen for four miRNAs (hsa-miR-188-5p, hsa-miR-214-3p, hsa-miR-4455
and hsa-miR-6877-3p) in comparisons of HGSCs with recurrence
(including metachronous metastasis) and without recurrence.
Meanwhile, for patients in cohort 2, two miRNAs (hsa-miR-188-5p and
hsa-miR-214-3p) had statistically different expression levels
between HGSCs with recurrence (including metachronous metastasis)
and without recurrence (including metachronous metastasis)
(Fig. 3Aa-f and Ba-f). Of the seven
differentially expressed miRNAs identified in isolated stromal
cells from cohort 1, differences were seen in the expression level
of five miRNAs including hsa-miR-101-5p, hsa-miR-320c,
hsa-miR-320d, hsa-miR-320e, hsa-miR-455-3p and hsa-miR-4429 between
HGSCs with recurrence (including metachronous metastasis) and those
without recurrence (cohort 1). In cohort 2, four miRNAs
(hsa-miR-320c, hsa-miR-320d, hsa-miR-378f, and hsa-miR-4429) had
statistically different expression levels between HGSCs with
recurrence (including metachronous metastasis) and those without
recurrence (Fig. 3Ag-m and
Bg-m).
Ability of miRNA expression to predict
patient survival
We determined the cutoff expression levels of
miRNA-214-3p and miRNA-320c that are potentially predictive of the
development of recurrence (metachronous metastasis) in HGSC, using
receiver operating characteristic (ROC) analysis (Fig. S1). In the present study, we did not
perform absolute quantification in order to determine standard
value for prediction of patient prognosis with HGSC. At each
expression level, the sensitivity and specificity for the outcome
under study (recurrence including metachronous metastasis) were
plotted to generate a ROC curve. If a ROC curve was generated from
pairs of weighted mean sensitivities and mean specificities, then
the weighted mean sensitivities and specificities for each miRNA
expression level were plotted to generate ROC curves, and the area
under the curve was used to determine the ability of the miRNA to
discriminate between the presence and absence of recurrence
(including metachronous metastasis). As a result, miRNA-214-3p and
miRNA-320c expression levels were identified as the best predictors
of recurrence (including metachronous metastasis) in HGSC among the
miRNAs examined (area under the curve: 0.81696 for isolated cancer
glands and 0.88393 for isolated stromal tissue). Consequently, less
than 0.278271 for miRNA-214-3p expression level (Fig. S1Ab) and less than 0.198724 for
miRNA-320c expression level (Fig.
S1Bb), respectively, determined by RT-qPCR were regarded as
presence of downregulation.
Association of clinicopathological
findings with candidate miRNAs identified in cancer gland and
stromal tissue samples
We examined associations of clinicopathological
findings including sex, age, tumor size and histological grade with
expression of miRNA-214-3p and miRNA-320c in samples from isolated
cancer glands and stromal tissues, respectively. In univariate
analysis of clinicopathological findings, we observed no
significant difference in the expression of miRNAs including
miRNA-214-3p and miRNA-320c.
Associations between patient survival
and the candidate miRNAs identified in the cancer gland and stromal
tissue samples
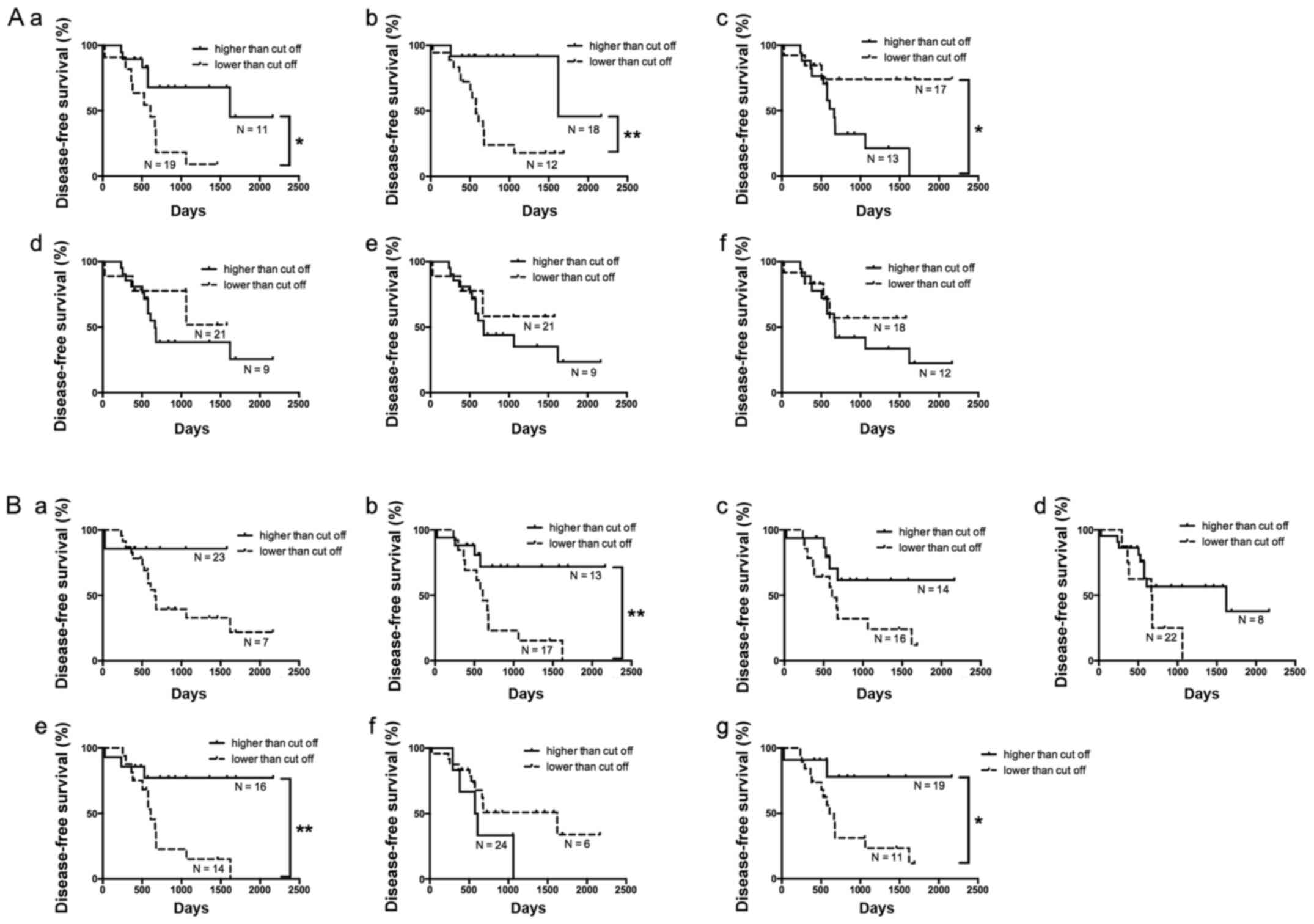
Of the 30 HGSC cases, 14 (46.7%) had no recurrence
(including metachronous metastasis). Disease-free survival (without
recurrence) was compared according to the expression of each miRNA
differentially expressed in the isolated cancer gland samples using
Kaplan-Meier analysis (Fig. 4A). The
presence of recurrence including metachronous metastasis was
associated with downregulation of hsa-miRNA-214-3p. Cox
proportional hazards models were used to determine any independent
associations of disease-free survival with the clinicopathological
findings and hsa-miRNA-214-3p expression. We initially performed
univariate analyses to determine the associations of the following
variables with the presence of recurrence including metachronous
metastasis: Age, tumor size, FIGO stage, and the six differentially
expressed miRNAs identified in the isolated cancer glands. The
results indicated that FIGO stage and expression of
hsa-miRNA-188-5p and hsa-miRNA-214-3p were associated with the
presence of recurrence including metachronous metastasis. The FIGO
stage and hsa-miRNA-214-3p expression retained significance in the
multivariate analysis. The summarized results are shown in Table II.
 | Table II.Univariate and multivariate analyses
of clinicopathologic findings and miRNA expression as predictors of
recurrence in cancer gland cells of ovarian high-grade serous
carcinoma using Cox proportional hazards model. |
Table II.
Univariate and multivariate analyses
of clinicopathologic findings and miRNA expression as predictors of
recurrence in cancer gland cells of ovarian high-grade serous
carcinoma using Cox proportional hazards model.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | 0.984 | 0.944–1.024 | 0.4320 |
|
|
|
| Tumor size
(mm) | 0.995 | 0.982–1.007 | 0.4157 |
|
|
|
| FIGO stage |
| III vs.
II | 8.455 | 1.680–153.834 | 0.0056 | 14.745 | 2.185–321.081 | 0.0029 |
| miRNA expression in
cancer gland cells |
|
hsa-miR-188-5p | 0.376 | 0.123–0.965 | 0.0416 | 4.462 | 0.572–35.610 | 0.1510 |
|
hsa-miR-214-3p | 0.257 | 0.069–0.754 | 0.0119 | 0.075 | 0.008–0.648 | 0.0187 |
|
hsa-miR-4455 | 1.257 | 0.439–3.479 | 0.6639 |
|
|
|
|
hsa-miR-505-5p | 2.527 | 0.663–9.278 | 0.1699 |
|
|
|
|
hsa-miR-6753-3p | 1.182 | 0.432–3.060 | 0.7368 |
|
|
|
|
hsa-miR-6877-3p | 1.394 | 0.537–3.461 | 0.4841 |
|
|
|
Using the same process, the association of the
expression of specific miRNAs with disease-free survival was
examined in isolated stromal tissue. According to Kaplan-Meier
analysis, downregulation of hsa-miRNA-320c was associated with the
patients who had developed recurrence including metachronous
metastasis (Fig. 4B). Then,
univariate analyses were conducted to determine whether age, tumor
size, FIGO stage, and the seven candidate miRNAs identified in
isolated stromal tissue were independent predictors of disease-free
survival in HGSC patients. According to the results, the FIGO stage
and hsa-miRNA-320c expression were associated with an increased
rate of recurrence including metachronous metastasis, and both
factors remained significant in the multivariate analysis. These
results are summarized in Table
III.
 | Table III.Univariate and multivariate analyses
of clinicopathologic findings and miRNA expression as predictors of
recurrence in cancer stroma of ovarian high-grade serous carcinoma
using Cox proportional hazards model. |
Table III.
Univariate and multivariate analyses
of clinicopathologic findings and miRNA expression as predictors of
recurrence in cancer stroma of ovarian high-grade serous carcinoma
using Cox proportional hazards model.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | 0.984 | 0.944–1.024 | 0.4320 |
|
|
|
| Tumor size
(mm) | 0.995 | 0.982–1.007 | 0.4157 |
|
|
|
| FIGO stage |
|
|
|
|
|
|
| III vs.
II | 8.455 | 1.680–153.834 | 0.0056 | 5.926 | 1.074–110.573 | 0.0438 |
| miRNA expression in
cancer stroma |
|
|
|
|
|
|
|
hsa-miR-101-5p | 0.272 | 0.038–1.728 | 0.1704 |
|
|
|
|
hsa-miR-320c | 0.086 | 0.012–0.502 | 0.0061 | 0.129 | 0.013–0.947 | 0.0398 |
|
hsa-miR-320d | 0.145 | 0.018–1.234 | 0.0767 |
|
|
|
|
hsa-miR-320e | 0.212 | 0.011–2.506 | 0.2327 |
|
|
|
|
hsa-miR-378f | 0.062 | 0.003–1.013 | 0.0511 |
|
|
|
|
hsa-miR-455-3p | 3.198 | 0.632–23.207 | 0.1707 |
|
|
|
|
hsa-miR-4429 | 0.090 | 0.006–1.072 | 0.0572 |
|
|
|
Discussion
The cancer microenvironment, comprising cancer cells
and adjacent stromal cells, was recently reported to have a key
role in human cancer pathogenesis (17,18).
However, despite advances in molecular cancer science, for ovarian
cancer the detailed molecular mechanisms associated with the cancer
microenvironment remain unclear in part because ovarian cancer is a
heterogeneous disease that is strongly influenced by genetic and
epigenetic alterations (28,29). Moreover, the tumor environment in
ovarian cancer involves complex interconnected signaling networks
(17,18). CAFs interact with several immune
cells, including tumor-associated macrophages, T cells, natural
killer cells, and cytokines, to promote the growth and metastasis
of ovarian cancer cells (30).
Therefore, understanding the pathogenesis and unique tumor
microenvironment, which may determine malignancy, of ovarian cancer
is crucial for developing more sensitive tools predicting patient
prognosis, in turn influencing the choice of effective treatment
options (11,12). We attempted to identify specific
miRNAs potentially predicting the prognosis of patients with HGSC,
a common variant of ovarian cancer, based on their expression
levels in isolated cancer glands and surrounding stromal cells.
Here, we used a crypt isolation method to isolate
both pure tumor glands as well as the surrounding stromal tissues.
Although this method can accurately isolate pure tumor glands based
on our extensive experience (23–25), it
is possible that our isolated stromal cells were contaminated with
cancer cells (23). To avoid such
contamination, we confirmed that the histological sections of the
isolated stromal tissues were devoid of cancer gland cells and
comprised primarily CAFs. We believe that the cancer glands and
stromal tissue were successfully separated in the present
study.
Recently, significantly lower miRNA-214-3p
expression was found in two esophageal squamous cancer cell lines
relative to that in normal esophageal epithelial cells (31,32).
Another study demonstrated that reduced miRNA-214-3p expression
inhibited chemoresistance in esophageal cancer cells by targeting
survivin (an inhibitor of apoptotic proteins) and CUG-binding
protein 1 (an RNA binding protein), which increases survivin
expression (32). Maternal embryonic
leucine zipper kinase (MELK), a member of the anti-apoptotic Bcl-2
family and essential for cancer growth, was identified as a target
gene of miRNA-214-3p; MELK was also identified as a target gene of
PRDI-BF1/Blimp-1, a transcriptional corepressor for specific
subsets of DNA-binding transcription factors, suggesting a possible
mechanism underlying the proliferation and resistance to apoptosis
of hepatocellular carcinoma cells (32). This was supported by the association
between MELK expression and gastric cancer progression detected in
clinical samples (33). These
findings suggest that decreased miRNA-214-3p expression affects the
progression of specific cancers by targeting MELK (31,32). A
recent study showed downregulated miRNA-214-3p expression and a
negative correlation between miRNA-214-3p and fibroblast growth
factor receptor 1 (FGFR1) expression in lung cancer patients
(34). A regulatory mechanism
involving miRNA-214-3p and FGFR1 was proposed, suggesting that
miRNA-214-3p is an important therapeutic target in lung cancers
with FGFR1 gene amplification (34). Thus, miRNA-214-3p expression may play
a crucial role in human cancers by targeting specific proteins such
as MELK and FGFR1 (31,34). In the current study, downregulation
of miRNA-214-3p was correlated with disease-free survival in
isolated cancer glands obtained from primary HGSC specimens. As
immunohistochemical expression of miRNA-214-3p target proteins,
including MELK and FGFR1, was not examined in this study, further
studies are needed to identify whether MELK and FGFR1 expression is
related to downregulation of miRNA-214-3p in HGSC. Finally, this
finding highlights a possible association of miRNA 214–3p
expression with cancer-stroma interactions. The role of miR-214-3p
in modulating expression of genes encoding proteins involved in
extracellular matrix (ECM) degradation, tumor invasion and
metastasis that is associated with the tumor microenvironment
comprising heterogeneous cancer cells and surrounding interstitial
cells has been the focus of several studies in the carcinogenesis
field (34). For bladder cancer,
down-regulation of miR-214-3p expression is associated with
prognosis and modulation of MMP-9 and NGAL (Neutrophil
gelatinase-associated lipocalin) gene expression and may reflect
formation of the tumor microenvironment (31,35,36). In
addition, downregulated hsa-miR-214-3p expression could in turn
alter the expression of genes involved in both epithelia
mesenchymal transition (EMT) and NGAL/MMP-9 pathways, suggesting
that downregulation of miR214-3p expression could facilitate EMT
via activation of NGAL/MMP-9 pathways (35). The present study is the first to
suggest that downregulation of miRNA-214-3p is correlated with
disease-free survival in HGSC.
Despite reports that the miRNA-320 family is
involved in several different human malignancies, its role in
ovarian cancer is not fully understood (37,38). In
colorectal cancer, SOX4, FOXM1, and FOXQ1 were
identified as novel targets of the miRNA-320 family (37), and Rac1 was found to be a
direct target of miRNA-320a (39).
Sun et al showed that miRNA-320a inhibits colorectal cancer
cell growth by targeting the β-catenin signaling pathway (38). Interestingly, Zhang et al
demonstrated that miRNA-320d was downregulated in stem cells
derived from HT29 colon cancer cells expressing CD133 compared with
those without CD133 expression, suggesting that miRNA-320 might be
involved in stem cell differentiation (40). In another study, abnormal expression
of miRNA-320 was examined in several human malignant tumors and
found to be downregulated in malignant cholangiocarcinoma, in which
this miRNA negatively regulated the expression of anti-apoptotic
Mcl-1 and Bcl-2 (41). Furthermore,
in prostate cancer, upregulation of miRNA-320 inhibited the
Wnt/β-catenin pathway and decreased CD44 expression, a marker of
cancer stem cells, in tumor-initiating cells (42). However, little has been reported on
the expression of the miRNA-320 family in CAFs, especially those in
ovarian cancer. In the present study, downregulation of miRNA-320c
in isolated stromal cells was correlated with disease-free survival
in patients with HGSC. As a potential mechanism, decreased
miRNA-320 expression in CAFs surrounding the tumor leads to
upregulation of several miRNA-320 target genes that possibly induce
tumor progression and drug resistance (42). According to this finding, we propose
that the miRNA-320 family serves as potential therapeutic targets
for the future management of HGSC.
This study has several limitations. First, the
sample was relatively small since very few HGSC cases do not
receive chemotherapy in routine clinical practice. In addition,
determining miRNA expression profiles in CAFs could aid the
diagnosis and treatment of HGSC. Second, in situ
hybridization may be required to identify the expression of
specific miRNAs in target cells. However, we did not perform in
situ hybridization because it is a very difficult procedure in
our laboratory. Alternatively, we examined whether stromal tissue
is composed of pure CAFs in examined isolated stromal tissue, as a
result, the histological finding of isolated stromal tissue was
considered to be a CAF.
In conclusion, we examined miRNA expression by
high-throughput genome-wide screening in isolated cancer glands and
the surrounding stromal tissue separately. We found that
downregulation of miRNA-214-3p in isolated cancer glands and
downregulation of miRNA-320c in stromal tissue each correlated with
a lack of recurrence (disease-free survival). This information can
be valuable to increase the number of prognostic markers and
treatment options for HGSC. Additional studies will be needed to
verify the results seen for the present study.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. E. Sugawara and
Ms. C. Ishikawa (Department of Molecular Diagnostic Pathology,
School of Medicine, Iwate Medical University) for their technical
assistance.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CS constructed the figures and tables, and performed
the statistical analyses. MO generated the figures and tables, and
performed the statistical analyses. TN, HI and TB provided the
clinical data, and examined the association between molecular
findings and clinical features. HS assisted with the molecular
techniques. TS contributed to the preparation of the manuscript,
including all aspects of the data collection and analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent, and
the study was approved by the Iwate Medical University
Institutional Review Board (approval no. MH2018-528).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lisio MA, Fu L, Goyeneche A, Gao ZH and
Telleria C: High-grade serous ovarian cancer: Basic sciences,
clinical and therapeutic standpoints. Int J Mol Sci. 20:9522019.
View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Falzone L, Salomone S and Libra M:
Evolution of cancer pharmacological treatments at the turn of the
third millennium. Front Pharmacol. 9:13002018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chandra A, Pius C, Nabeel M, Nair M,
Vishwanatha JK, Ahmad S and Basha R: Ovarian cancer: Current status
and strategies for improving therapeutic outcomes. Cancer Med.
8:7018–7031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mathieu KB, Bedi DG, Thrower SL, Qayyum A
and Bast RC Jr: Screening for ovarian cancer: Imaging challenges
and opportunities for improvement. Ultrasound Obstet Gynecol.
51:293–303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mills GB, Bast RC Jr and Srivastava S:
Future for ovarian cancer screening: Novel markers from emerging
technologies of transcriptional profiling and proteomics. J Natl
Cancer Inst. 93:1437–1439. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bandera CA, Ye B and Mok SC: New
technologies for the identification of markers for early detection
of ovarian cancer. Curr Opin Obstet Gynecol. 15:51–55. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Liu T, Zhang Z, Payne SH, Zhang
B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, et al:
Integrated proteogenomic characterization of human high-grade
serous ovarian cancer. Cell. 166:755–765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Davidson B: Biomarkers of drug resistance
in ovarian cancer-an update. Expert Rev Mol Diagn. 19:469–476.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Elias KM, Guo J and Bast RC Jr: Early
detection of ovarian cancer. Hematol Oncol Clin North Am.
32:903–914. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Falzone L, Romano GL, Salemi R, Bucolo C,
Tomasello B, Lupo G, Anfuso CD, Spandidos DA, Libra M and Candido
S: Prognostic significance of deregulated microRNAs in uveal
melanomas. Mol Med Rep. 19:2599–2610. 2019.PubMed/NCBI
|
|
14
|
Staicu CE, Predescu DV, Rusu CM, Radu BM,
Cretoiu D, Suciu N, Crețoiu SM and Voinea SC: Role of microRNAs as
clinical cancer biomarkers for ovarian cancer: A short overview.
Cells. 9:1692020. View Article : Google Scholar
|
|
15
|
Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka
Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda
T, et al: Integrated extracellular microRNA profiling for ovarian
cancer screening. Nat Commun. 9:43192018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Falzone L, Lupo G, La Rosa GRM, Crimi S,
Anfuso CD, Salemi R, Rapisarda E, Libra M and Candido S:
Identification of novel MicroRNAs and their diagnostic and
prognostic significance in oral cancer. Cancers (Basel).
11:6102019. View Article : Google Scholar
|
|
17
|
Cirri P and Chiarugi P: Cancer associated
fibroblasts: The dark side of the coin. Am J Cancer Res. 1:482–497.
2011.PubMed/NCBI
|
|
18
|
Cirri P and Chiarugi P:
Cancer-associated-fibroblasts and tumour cells: A diabolic liaison
driving cancer progression. Cancer Metastasis Rev. 31:195–208.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dasari S, Fang Y and Mitra AK: Cancer
associated fibroblasts: Naughty neighbors that drive ovarian cancer
progression. Cancers (Basel). 10:4062018. View Article : Google Scholar
|
|
20
|
Sun W and Fu S: Role of cancer-associated
fibroblasts in tumor structure, composition and the
microenvironment in ovarian cancer. Oncol Lett. 18:2173–2178.
2019.PubMed/NCBI
|
|
21
|
Japanese Society of Obstetrics and
Gynecology, the Japanese Society of Pathology, . The General Rules
for Clinical and Pathological Management of Ovarian Tumors Part 1:
Histological Classification and Color Atlas of Ovarian Tumors.
(2nd). Kanehara. 1–41. 2009.
|
|
22
|
Prat J and FIGO Committee on Gynecologic
Oncology, . Staging classification for cancer of the ovary,
fallopian tube, and peritoneum. Int J Gynaecol Obstet. 124:1–5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sato A, Fujita Y, Otsuka K, Sasaki A,
Suzuki H, Matsumoto T and Sugai T: Differential expression of
microRNAs in colorectal cancer: Different patterns between isolated
cancer gland and stromal cells. Pathol Int. 70:21–30. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Habano W, Sugai T, Nakamura S and Yoshida
T: A novel method for gene analysis of colorectal carcinomas using
a crypt isolation technique. Lab Invest. 74:933–940.
1996.PubMed/NCBI
|
|
25
|
Sugai T, Habano W, Nakamura SI, Uesugi N,
Sasou S and Itoh C: A unique method for mutation analysis of tumor
suppressor genes in colorectal carcinomas using a crypt isolation
technique. Arch Pathol Lab Med. 124:382–386. 2000.PubMed/NCBI
|
|
26
|
Miles GD, Seiler M, Rodriguez L, Rajagopal
G and Bhanot G: Identifying microRNA/mRNA dysregulations in ovarian
cancer. BMC Res Notes. 5:1642012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shiomi E, Sugai T, Ishida K, Osakabe M,
Tsuyukubo T, Kato Y, Takata R and Obara W: Analysis of expression
patterns of microRNAs that are closely associated with renal
carcinogenesis. Front Oncol. 9:4312019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter
ME: MicroRNAs as mediators and communicators between cancer cells
and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kossaï M, Leary A, Scoazec JY and Genestie
C: Ovarian cancer: A heterogeneous disease. Pathobiology. 85:41–49.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Comerford SA, Clouthier DE, Hinnant EA and
Hammer RE: Induction of hepatocyte proliferation and death by
modulation of T-Antigen expression. Oncogene. 22:2515–2530. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Li Y, Chen Y, Xie Q, Dong N, Gao Y,
Deng H, Lu C and Wang S: MicroRNA-214-3p inhibits proliferation and
cell cycle progression by targeting MELK in hepatocellular
carcinoma and correlates cancer prognosis. Cancer Cell Int.
17:1022017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phatak P, Byrnes KA, Mansour D, Liu L, Cao
S, Li R, Rao JN, Turner DJ, Wang JY and Donahue JM: Overexpression
of miR-214-3p in esophageal squamous cancer cells enhances
sensitivity to cisplatin by targeting survivin directly and
indirectly through CUG-BP1. Oncogene. 35:2087–2097. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Du T, Qu Y, Li J, Li H, Su L, Zhou Q, Yan
M, Li C, Zhu Z and Liu B: Maternal embryonic leucine zipper kinase
enhances gastric cancer progression via the FAK/Paxillin pathway.
Mol Cancer. 13:1002014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Li Z, Yuan H, Ji W, Wang K, Lu T,
Yu Y, Zeng Q, Li F, Xia W and Lu S: Reciprocal regulatory mechanism
between miR-214-3p and FGFR1 in FGFR1-amplified lung cancer.
Oncogenesis. 8:502019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Xu Y, Wang J and Ying H:
Circulating miR-214-3p predicts nasopharyngeal carcinoma recurrence
or metastasis. Clin Chim Acta. 503:54–60. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vishnubalaji R, Hamam R, Yue S, Al-Obeed
O, Kassem M, Liu FF, Aldahmash A and Alajez NM: MicroRNA 320
suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1.
Oncotarget. 7:35789–35802. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun JY, Huang Y, Li JP, Zhang X, Wang L,
Meng YL, Yan B, Bian YQ, Zhao J, et al: MicroRNA-320a suppresses
human colon cancer cell proliferation by directly targeting
β-catenin. Biochem Biophys Res Commun. 420:787–792. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao H, Dong T, Zhou H, Wang L, Huang A,
Feng B, Quan Y, Jin R, Zhang W, Sun J, et al: MiR-320a suppresses
colorectal cancer progression by targeting Rac1. Carcinogenesis.
35:886–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y
and Zhang F: MicroRNA expression profile of colon cancer stem-like
cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun.
404:273–278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu
Q, Li L, Huang DD, Ding J, Shen F, et al: The role of microRNA
expression pattern in human intrahepatic cholangiocarcinoma. J
Hepatol. 50:358–369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
downregulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|