Introduction
Esophageal cancer is one of the intractable
gastrointestinal cancers. Its incidence and mortality are close,
with recent statistics reporting 509,000 deaths out of 572,000
cases (1). Esophageal squamous cell
carcinoma (ESCC) is the major pathological histology throughout
Asia, whereas adenocarcinoma is major in Europe and the United
States (2). Owing to advances in
medical technology, cases of esophageal cancer when diagnosed early
can be resolved using endoscopy. Therefore, the survival rate of
patients with an early diagnosis of esophageal cancer and their
quality of life, with esophageal preservation, has been greatly
improved. In contrast, the overall survival rate of ESCC has been
reported to be at most 20–30%, with room for improvement (3). When detected early, the 5-year survival
rate of patients with ESCC is 80–90% (4,5);
however, early-stage esophageal cancer is less likely to show
clinical symptoms, and a lack of reliable noninvasive screening
methods has hindered its detection. Nevertheless, when ESCC is
diagnosed relatively early, the prognosis is clearly good, and the
method used for early diagnosis before the onset of symptoms is
important. Therefore, to improve the prognosis of ESCC, an
intractable cancer, it is necessary to search for a biomarker that
can be detected even earlier (6,7).
MicroRNA (miR), a non-coding RNA, is a functional
RNA that is eventually edited to 19–22 bases. This small RNA was
discovered in Caenorhabditis elegans in 1993 (8), and its name was proposed in 2001
(9). Many miRs are involved in the
onset and progression of various chronic diseases and malignant
tumors. In particular, miRs associated with malignant tumors are
classified into oncogenic miR (oncomiR) (10), a positively regulated miR; tumor
suppressor miR, a negatively regulated miR. In addition, miRs are
relatively stable in body fluids such as the blood by forming
complexes with proteins and being included in microparticles such
as exosomes. The usefulness of these miRs as biomarkers has been
previously reported, and we have also reported the usefulness of
miR-1246 as a biomarker in ESCC (11). miR-1246 is a biomarker not only in
esophageal cancer but also in gastrointestinal cancers such as
stomach and pancreatic cancers (12). Furthermore, in our previous report,
miR-106b expression was significantly lower in patient sera than in
that of healthy volunteers. miR-106b clusters together with miR-93
and miR-25, and has a tumor suppressor function in colorectal and
ovarian cancers (13–15). In addition, its expression is reduced
in the serum of gastric cancer patients (16–21).
The findings of our previous studies and other
previous reports on other organ cancers suggest the usefulness of
combination of miR-1246 and miR-106b as biomarkers. Here, we
investigated the expression levels of miR-1246 and miR-106b in the
serum of patients with ESCC and their clinicopathological
significance.
Materials and methods
Ethical approval
Written informed consent was obtained from all
patients, and the study was approved by the Ethics Committee of
Graduate School of Medicine, Chiba University (no. 889), and Chiba
Cancer Center (no. H29-0005) and performed in compliance with the
Declaration of Helsinki.
Sample collection
Between April 2013 and April 2016, venous blood
samples were collected from 55 patients with ESCC and 39 healthy
individuals at the Graduate School of Medicine, Chiba University in
Chiba, Japan. Between April 2016 and April 2019, venous blood
samples were collected from 101 patients with ESCC and 34 healthy
individuals at the Chiba Cancer Center in Chiba, Japan. These
samples were collected before any therapies, including endoscopic
resection, surgery, chemotherapy, or radiotherapy. Venous blood
samples were centrifuged at 1,500 × g for 5 min at 4°C to obtain
serum. The serum samples were then stored at −80°C until further
processing.
RNA extraction
Total RNA was extracted from 200 µl of serum using a
miRNeasy Serum/Plasma Kit (Qiagen) according to the manufacturer's
instructions. This kit contained the Caenorhabditis elegans
cel-miR-39, which was used as a spike-in control.
Reverse transcription
Total RNA was reverse transcribed to cDNA using a
miScript II RT Kit (Qiagen). In each reaction, 50 ng (12 µl) of
template RNA was combined with a master mix containing 4 µl 5×
miScript HiSpec Buffer, 2 µl 10× miScript Nucleics Mix, and 2 µl
miScript Reverse Transcriptase Mix and incubated for 60 min at
37°C. The reactions were then incubated for 5 min at 95°C to
inactivate the miScript Reverse Transcriptase Mix and placed on
ice.
miRNA microarray
Total RNA (100 ng) was labeled and hybridized
following the Human microRNA Microarray Kit protocol and using the
Human miRNA Microarray Kit (Release 16.0; Agilent Technologies).
Hybridization signals were detected with a DNA microarray scanner
G2505C (Agilent Technologies), and the scanned images were analyzed
using the Agilent Function Extraction Software program (v10.7.3.1).
Normalization was performed using the Agilent GeneSpring GX
software program version 11.0.2 (per chip: Normalization of control
genes; per gene: None). The Agilent Human miRNA Microarray (Design
ID: 031181) contained 1205 human miRs in total with 144 human viral
miRs without control probes. The miR expression profile of the
serum samples from patients with ESCC was examined using a
microarray analysis of three patients and four healthy individuals.
miR-16 was used as an endogenous control.
Reverse transcription-quantiative PCR
(RT-qPCR)
Quantitative RT-qPCR was performed using the
miScript SYBR®-Green PCR Kit (Qiagen) in a 7300
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The expression levels of miR-1246 (assay ID: 462575) and
miR-106b (assay ID: 000442) were analyzed using TaqMan quantitative
real-time PCR (TaqMan MicroRNA assays; Applied Biosystems; Thermo
Fisher Scientific, Inc.) and normalized to the levels of cel-miR-39
(assay ID: 001093). The parameters of the PCR reaction were as
follows: 95°C for 15 min followed by 40 cycles of 94°C for 15 sec,
55°C for 30 sec, and 70°C for 34 sec. All reactions were performed
in duplicate. Relative expression was calculated using comparative
cycle threshold (Ct) values. Relative miR-1246 or miR-106b
expression was calculated using the 2−ΔCt method, where
ΔCt=Ct (miR-1246 or miR-106b)-Ct (cel-miR-39) (22).
Statistical analysis
Data was conformed to a normal distribution using
Shapiro-Wilk's test. An unpaired Student's t-test was performed to
compare differences in age. A Mann-Whitney U test was performed to
compare differences in miR-1246 expression levels between patients
with cancer and healthy individuals. Spearman's rank correlation
coefficient was used to assess correlations between the miR-1246
expression levels in the three body fluids. The χ2 test
or Fisher's exact probability test was used to evaluate
correlations between serum and miR expression levels and
clinicopathological tumor factors. Receiver operating
characteristic (ROC) curves and area under the curve (AUC) were
used to assess the sensitivity and specificity of serum miR levels
in detecting ESCC. All tests were two-sided, and the significance
level was set at a P-value <0.05. The survival period of the
patients was defined as the duration from the time of surgery to
either death or the last follow-up day, and the survival rate was
calculated using the Kaplan-Meier method. Comparisons of two groups
as univariate analyses were performed using the log-rank test. JMP
14 (SAS Institute, Inc.) software was used for all statistical
analyses.
Results
MiRs with large fluctuations in
expression levels
A comprehensive expression analysis using microarray
identified six miRs that were significantly upregulated and five
that were significantly downregulated in ESCC serum samples
compared with controls (P<0.05; Table
I). miR-1246 expression levels were most significantly
upregulated in the serum of patients with ESCC, and miR-106b was
the second most downregulated miR. The fourth most downregulated
miR-93 forms a cluster with miR-106b, and thus miR-106b was
considered important. Therefore, miR-1246 and miR-106b were
selected as candidates for further analysis.
 | Table I.Differentially expressed miRNAs in
patients with esophageal squamous cell carcinoma. |
Table I.
Differentially expressed miRNAs in
patients with esophageal squamous cell carcinoma.
| A, Upregulated
miRNAs |
|---|
|
|---|
| miRNA | P-value |
|---|
| hsa-miR-1246 | 0.008 |
| hsa-miR-3202 | 0.032 |
| hsa-miR-23a | 0.038 |
| hsa-miR-718 | 0.042 |
| hsa-miR-3610 | 0.043 |
| hsa-miR-4271 | 0.044 |
|
| B, Downregulated
miRNAs |
|
| miRNA | P-value |
|
| hsa-miR-144 | 0.011 |
|
hsa-miR-106b-5p | 0.034 |
| hsa-miR-486-5p | 0.039 |
| hsa-miR-93 | 0.049 |
| hsa-miR-451 | 0.050 |
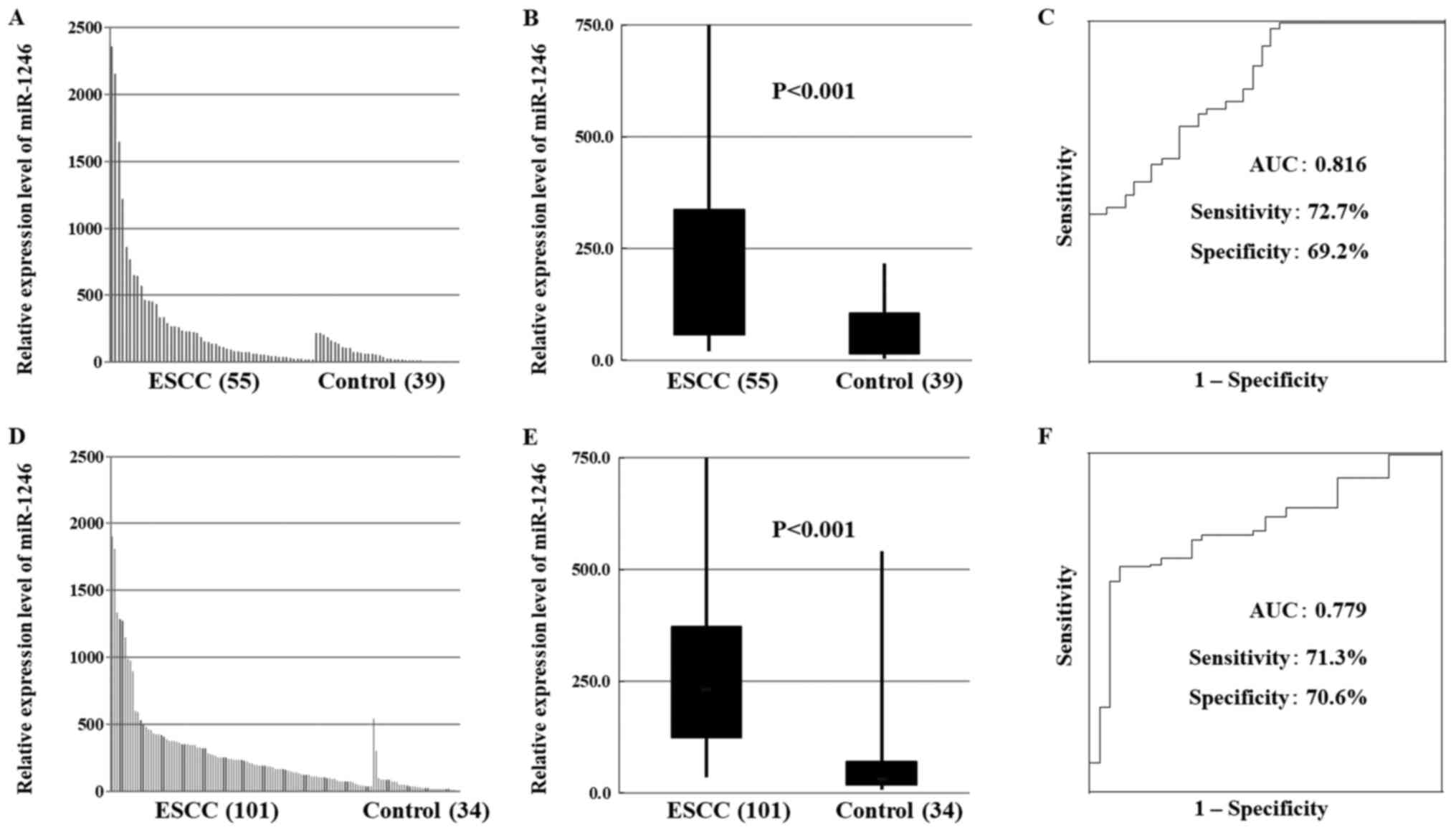
miR-1246 and miR-106b expression in
patients with ESCC
In total, 156 participants were recruited, 55 in the
test cohort and 101 in the validation cohort. In both the test and
validation cohorts, miR-1246 expression was significantly higher in
patients with ESCC than that in healthy controls (Fig. 1A, B, D and E). In contrast, the
expression of miR-106b was significantly lower in patients with
ESCC than that in healthy controls (Fig.
2A, B, D and E).
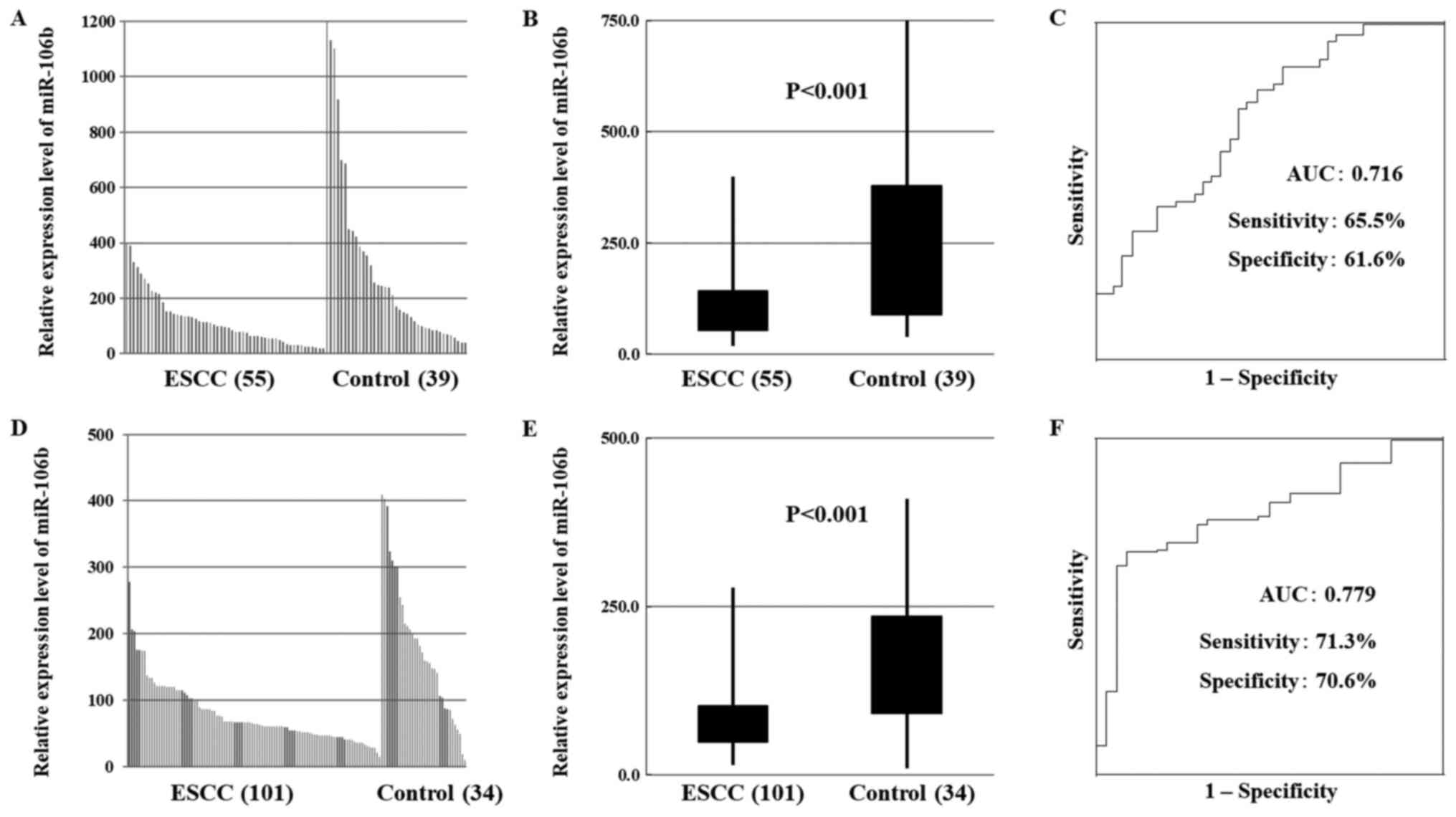 | Figure 2.Serum miR-106b expression in patients
with ESCC. (A) In the test cohort, miR-106b expression was
decreased in the serum of patients with ESCC compared with healthy
controls. (B) In the test cohort, miR-106b expression was
statistically significantly decreased in patients with ESCC
compared with healthy controls (P<0.001). (C) In the test
cohort, receiver operating characteristic curve analysis revealed
the sensitivity of miR-106b as a diagnostic indicator of ESCC. (D)
In the validation cohort, miR-106b expression was decreased in the
serum of patients with ESCC compared with healthy controls. (E) In
the validation cohort, miR-106b expression was statistically
significantly decreased ESCC patients than in healthy controls. (F)
In the validation cohort, receiver operating characteristic curve
analysis revealed the sensitivity of miR-106b as a diagnostic
indicator of ESCC. miR, microRNA; ESCC, esophageal squamous cell
carcinoma; AUC, area under the curve; miR, microRNA; ESCC,
esophageal squamous cell carcinoma; AUC, area under the curve. |
Diagnostic ability of serum levels of
miR-1246 and miR-106b
ROC curve analysis revealed the sensitivity of
miR-1246 as a diagnostic indicator of ESCC (Fig. 1C and F). The AUC was 0.816
(sensitivity 72.7%, specificity 69.2%) and 0.779 (sensitivity
71.3%, specificity 70.6%) in the test and validation cohorts,
respectively. In addition, ROC curve analysis revealed the
sensitivity of miR-106b levels as a diagnostic indicator of ESCC
(Fig. 2C and F). The AUC was 0.716
(sensitivity 65.5%, specificity 61.6%) and 0.815 (sensitivity
74.3%, specificity 73.5%) in the test and validation cohorts,
respectively.
Diagnostic ability of
miR-1246/miR-106b ratio
We examined the diagnostic ability of the ratio of
miR-1246 whose expression was elevated and miR-106b whose
expression was reduced in patient sera. We found that the ratio of
miR-1246/miR-106b had indeed, a better AUC of 0.901 (sensitivity
80.0%, specificity 80.0%) and 0.903 (sensitivity 82.1%, specificity
82.3%) in the test and validation cohorts, respectively (Fig. 3A-F).
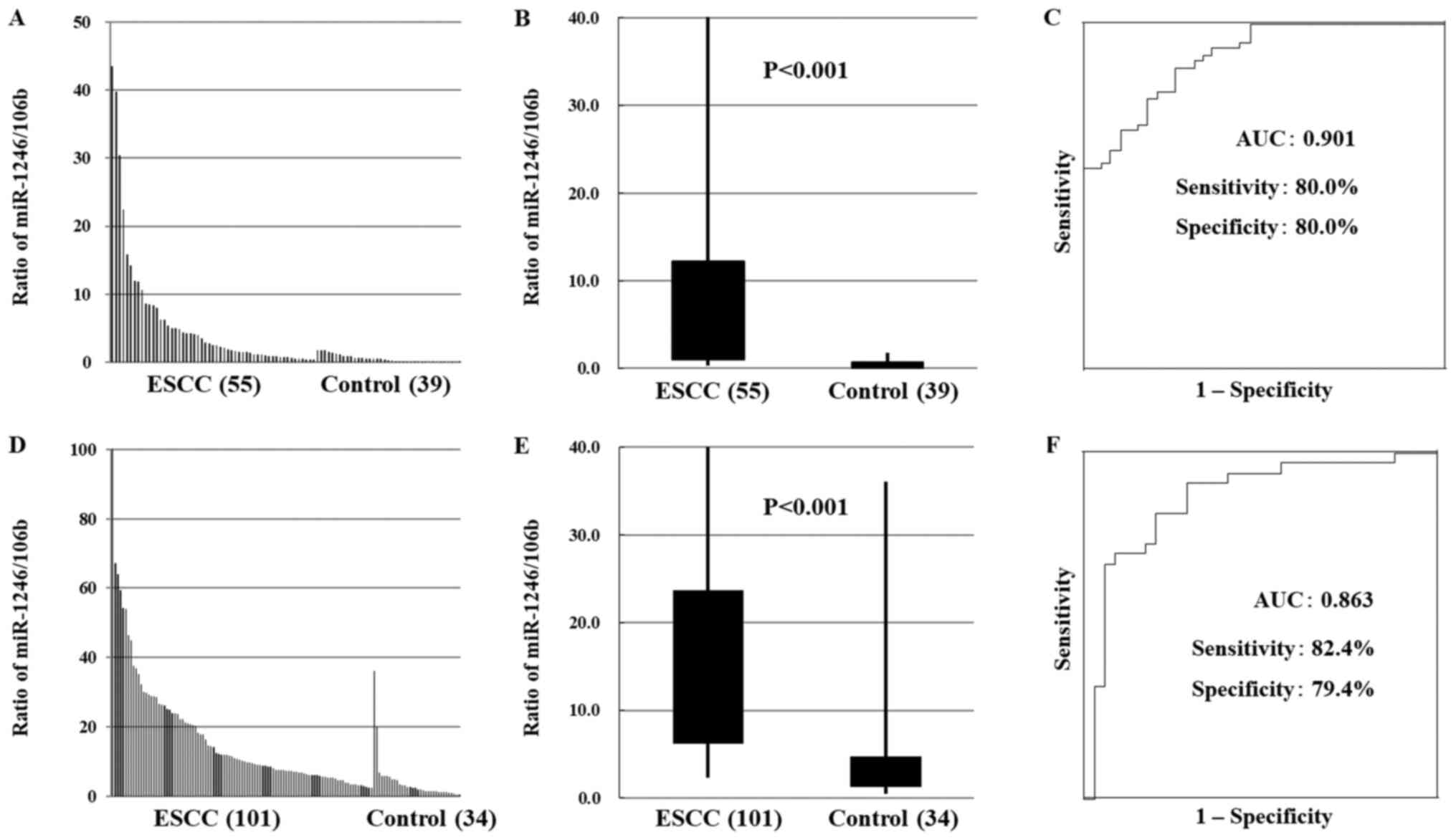 | Figure 3.Serum miR-1246/106b ratio in patients
with ESCC. (A) In the test cohort, miR-1246/106b ratio was higher
in the serum of ESCC patients than in that of healthy controls. (B)
In the test cohort, miR-1246/106b ratio was statistically
significantly higher in ESCC patients than in healthy controls
(P<0.001). (C) The AUC value of the miR-1246/miR-106b ratio was
as high as 0.901 (sensitivity, 80.0%; specificity, 80.0%) in the
test cohort. (D) In the validation cohort, miR-1246/106b ratio was
higher in the serum of ESCC patients than in that of healthy
controls. (E) In the validation cohort, miR-1246/106b ratio was
statistically significantly higher in ESCC patients than in healthy
controls (P<0.001). (F) The AUC value of the miR-1246/miR-106b
ratio was as high as 0.903 (sensitivity 82.1%, specificity 82.3%)
in the validation cohort and 0.903 (sensitivity, 82.1%;
specificity, 82.3%) in the validation cohort. miR, micro RNA; ESCC,
esophageal squamous cell carcinoma; AUC, area under the curve. |
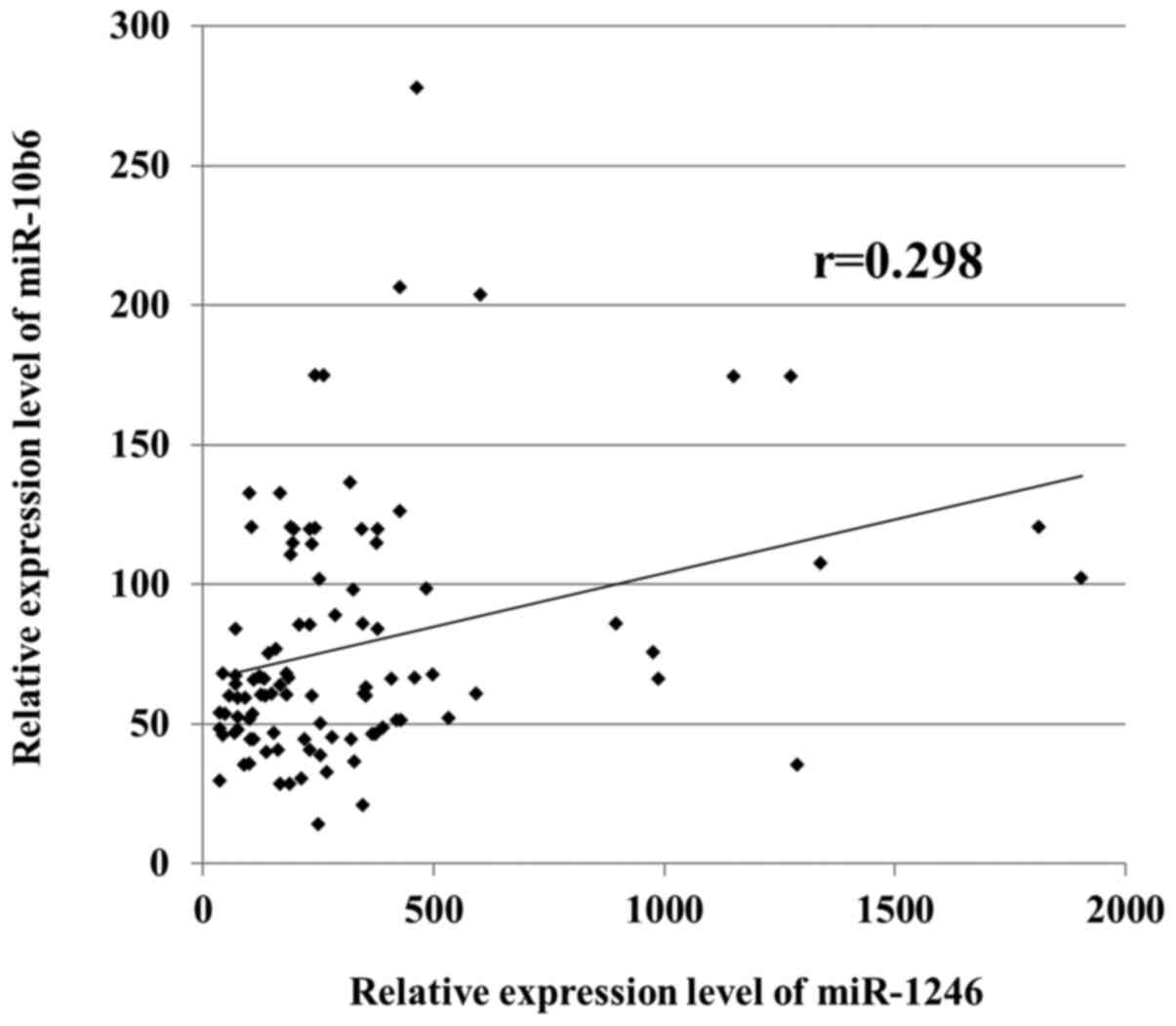
Relationship between miR-1246 and
miR-106b levels in ESCC
A correlation between the expression levels of
miR-1246 and miR-106b was evaluated for patients with ESCC in the
validation cohort. However, no significant correlation was observed
between the respective expression levels (Fig. 4).
Relationship between the miR-1246/106b
ratio and clinicopathological factors of ESCC
Statistical analysis was performed to determine the
relationships between miR-1246/miR-106b levels and
clinicopathological factors of ESCC (Table II). Patient samples were based on
the median miR-1246/106b expression levels to obtain high and low
groups. High serum miR-1246/106b expression showed a significant
association with tumor depth, positive lymph node metastasis,
stage, and survival of patients.
 | Table II.Correlation between the 1246/106b
ratio and the clinicopathological features of patients with
ESCC. |
Table II.
Correlation between the 1246/106b
ratio and the clinicopathological features of patients with
ESCC.
|
Characteristics | Total | High 1246/106b
ratio | Low 1246/106b
ratio | P-value |
|---|
| Total | 101 | 51 (50.5) | 50 (49.5) |
|
| Sex |
|
|
| 0.5564 |
|
Male | 85 | 44 (43.6) | 41 (40.6) |
|
|
Female | 16 | 7 (6.9) | 9 (8.9) |
|
| Age, years |
|
|
| 0.1707 |
|
<65 | 37 | 22 (21.8) | 15 (14.9) |
|
|
≥65 | 64 | 29 (28.7) | 35 (34.6) |
|
| Tumor depth |
|
|
| <0.001 |
|
T1-2 | 42 | 11 (10.9) | 31 (30.7) |
|
|
T3-4 | 59 | 40 (39.6) | 19 (18.8) |
|
| Lymph node
metastasis |
|
|
| <0.001 |
|
Negative | 97 | 18 (17.8) | 35 (34.6) |
|
|
Positive | 4 | 33(32.7) | 15 (14.9) |
|
| Stage |
|
|
| <0.001 |
|
I–II | 55 | 19 (18.8) | 36 (35.6) |
|
|
III–IV | 46 | 32 (31.7) | 14 (13.9) |
|
| Patient
survival |
|
|
| <0.005 |
|
Alive | 59 | 22 (21.8) | 37 (36.6) |
|
|
Dead | 42 | 29 (28.7) | 13 (12.9) |
|
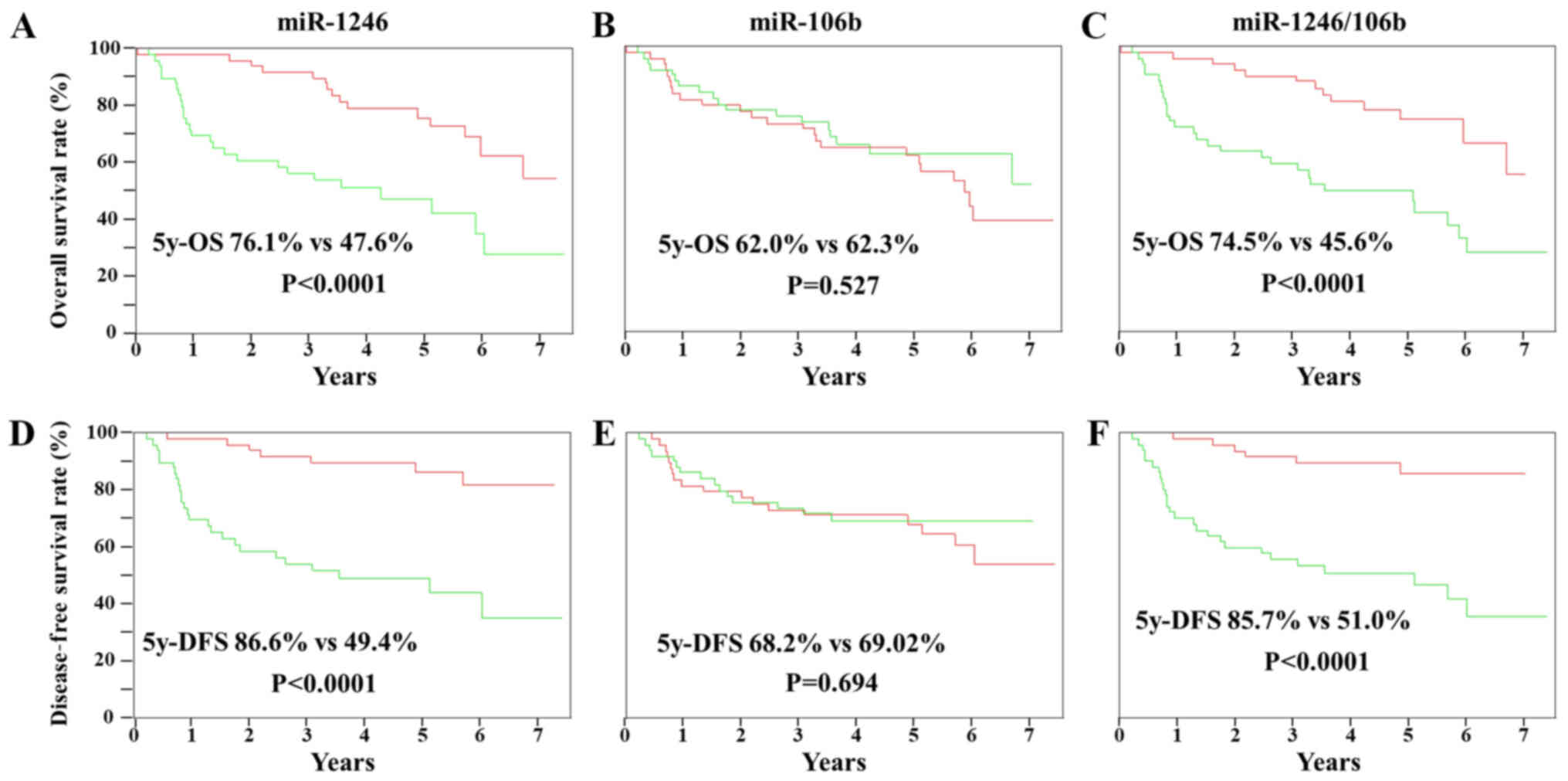
Association between miR-1246 and
miR-106b expression levels or miR-1246/106b ratio and ESCC
prognosis
Overall survival analysis was performed using the
Kaplan-Meier approach, with statistical analysis performed using
the log-rank test. Using the patient groups described above, the
prognostic values of miR-1246, miR-106b, and miR-1246/miR-106b
expression levels were evaluated (Fig.
5). The prognosis of the group with high serum miR-1246
expression was significantly worse than that of the group with low
serum miR-1246 expression (P<0.001). The 5-year overall survival
rates were 76.1 and 47.6% for the groups with high and low miR-1246
expression, respectively. The 5-year disease free survival rates
were 86.6 and 49.4% for the groups with high and low miR-1246
expression, respectively. In contrast, there was no association
between the serum levels of miR-106b expression and prognosis. In
addition, the miR-1246/miR-106b ratio showed an association with
prognosis as well as miR-1246. The 5-year overall survival rates
were 74.5 and 45.6% for the groups with high and low
miR-1246/miR-106b ratio, respectively. Furthermore, the 5-year
disease free survival rates were 85.7 and 51.0% for the groups with
high and low m iR-1246/miR-106b ratio, respectively.
Discussion
There have been numerous reports on the expression
of miRs being associated with cancer and its significance (23,24).
Many studies have also been reported on the expression of miRs in
body fluids including serum (25).
The mechanism by which miRs are stably present in the serum is
thought to be related to protection from RNase degradation by being
sequestered in exosomes or forming a protein complex with argonaute
2 protein (26–28). In this study, we performed a
comprehensive analysis of serum levels of miRs in patients with
ESCC that differ from healthy subjects using microarrays. We
observed that the expression of multiple miRs was significantly
increased or attenuated. Similar observations have been previously
reported using serum from patients with other cancer types
(29,30). Here, we focused on miR-1246, a miR
with increased expression, and miR-106b, a miR with attenuated
expression.
miR-1246 was shown to have elevated serum levels in
patients with ESCC in our previous study. Since then, other
researchers have reported increased serum levels of this miR in
other carcinomas (31,32). In a systematic review of
gastrointestinal cancer, it was reported that the expression of
miR-1246 in serum is more useful as a biomarker than that of
miR-21, which is an oncomiR (12).
Moshiri et al demonstrated the utility of plasma and serum
miR biomarkers in hepatocellular carcinoma using combinations of
multiple miRs including miR-1246. However, their analysis included
a single cohort, and it is considered that validation using other
cohorts is required (33).
Therefore, here we used different cohorts to increase the
reliability of the findings. The function of miR-1246 in cancer has
been reported so far in studies using cancer tissues and cell
lines. In a report using serum from 11 patients with lung cancer,
miR-1246 was the circulating miR with the highest expression
compared with healthy subjects. In addition, in studies using A549,
a lung cancer cell line, miR-1246 regulates the Wnt/β-catenin
pathway by targeting GSK-3β/β-catenin, thereby regulating cell
migration ability and metastatic potential (34). In addition, miR-1246 is implicated in
curcumin's anticancer effect and attenuated radiation sensitivity.
It has been stated that miR-1246 directly targets p53 and inhibits
p53 translation (35). In uterine
cervical cancer, miR-1246 has been reported to inhibit
thrombospondin-2, which regulates cell adhesion and migration via
hydrolysis of the extracellular matrix and a signaling pathway
(36,37). From the above reports, it is presumed
that miR-1246 functions as an oncomiR, and its expression may be
increased in the serum of ESCC patients.
miR-106b-5p has been identified as an oncogenic miR
in hepatocellular carcinoma, laryngeal carcinoma, breast carcinoma,
and glioma (38). miR-106b is
present in the intron of the MCM7 gene and forms a cluster with
miR-25 and miR-93 (39,40). Reports on the function of miR-106b
are controversial, wherein few report its tumor suppressive
function in colorectal and ovarian cancers while others report its
action as an oncomir in colorectal and other cancers. Functionally
silencing ectopic or endogenous miR-106b-5p expression inhibited or
promoted colorectal cancer (CRC) cell invasion and metastasis in
vitro and in vivo. The long non-coding RNA MALAT1
mediates SLAIN2-related MT motility and regulates CRC progression
by regulating miR-106b-5p expression and functioning as a competing
endogenous RNA in vitro and in vivo (21). However, in our study, the expression
of miR-93 in the same cluster as miR-106b was attenuated. In this
regard, expression of the host of miR-25, miR-93, and miR-106b
clusters, and MCM7 is attenuated in patients with ESCC, resulting
in attenuated expression of these miRs that are located in the MCM7
intron. However, RT-qPCR showed that MCM7 expression increased in
ESCC tissues compared with healthy tissues (Fig. S1A). Furthermore, no significant
change in tissue expression of any miRs in the miR-25-93 cluster
present in the intron of MCM7 was observed (supplementary Fig. S1B-D). There are many reports on
increased and decreased miR serum levels, in addition to our report
on miRs with changes in expression levels in the serum identified
by comprehensive analysis using microarray. However, no paper
mentions the cause of the attenuation. We suggest that the cause of
attenuated miR-106b expression in ESCC patients may be that
miR-106b target genes are increased, thereby activating miR-106b
consumption in tissues. The gene expression profile in tissues was
compared using microarrays between the high and low miR-106b
expression groups. We observed that the expression levels of target
genes (programmed cell death 1 ligand 2, hematological and
neurological expressed 1, BTG family member 3, etc.) of miR-106b
were increased in the low miR-106b expression group (data not
shown). In other words, it was suggested that low expression of
miR-106b in tumors might have increased expression of miR-106b
target genes including oncogenes. Alternatively, irrespective of
expression in cancer tissue, in patients in the process of
developing ESCC, miR-106b expression is attenuated by some
mechanism, and thus miR-106b serum levels may be reduced.
Nevertheless, further research is required to uncover the mechanism
of the reduced expression of miRs in the serum of cancer
patients.
Our previous paper reported that the expression of
miR-1246 in the serum of patients with ESCC was an independent
prognostic factor (11). However,
the AUC on the ROC curve was 0.754, and both sensitivity and
specificity were ~70%. Therefore, we decided to focus on miRs,
which decreases in serum of patients with ESCC. However, as
expected, miR-106b had a significant decrease in its expression in
the serum, but the AUC was ~0.7 and it was inferior to that of
miR-1246. Therefore, we expected that improvement could be obtained
by combining miR (miR-1246) that significantly increases and miR
(miR-106b) that significantly decreases. Unfortunately, no
significant association was found between each expression. However,
as a result, it was clarified that the ratio of miR-1246/miR-106b
was the most useful with an AUC of ~0.9. On the other hands,
regarding the correlation with pathological factors, we showed that
the ratio of miR-1246/miR-106b was associated with tumor invasion
depth, lymph node metastasis, progression, life and death.
Actually, similar results were already obtained in a single report
of miR-1246 (11). In other words,
it may be sufficient to investigate miR-1246 alone for prognosis
and correlation with histopathological factors. However, again,
when focusing on the usefulness as a biomarker, the sensitivity and
specificity of miR-1246 alone was ~70%, but by using the ratio of
miR-1246 and miR-106b was ~80%. In other words, we believe that the
miR1-1246/miR-106b ratio is useful for distinguishing between ESCC
patients and healthy subjects.
However, although this combination improved the
diagnostic ability in the present study, a clear improvement in the
accuracy as a prognostic marker was not confirmed.
Our study has several limitations. We used sera
obtained at multiple facilities, but it is a retrospective study,
and we considered it necessary to verify the findings of this study
by a prospective study that includes other facilities. The
reliability of miR whose expression is reduced in serum was
clarified using multiple samples, but the underlying mechanism
remains unclear, which should be addressed in future studies.
Furthermore, in ESCC, early detection is directly associated with a
good prognosis, but the present findings did not show its
usefulness in early diagnosis.
In this study, we reported the usefulness of serum
levels of miR-1246 and miR-106b in ESCC patients. In addition, it
has been clarified that the diagnostic ability can further be
improved by using the ratio of a miR having higher expression to a
miR having lower expression. We suggest that such a combination of
miRs in the serum may be used clinically as a screening marker in
the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Japan Society for the
Promotion of Science KAKENHI for scientific research (grant no.
17K10616) and Cancer Research Funds for Patients and Family (grant
no. H30-11).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
IH planned, analyzed and conducted all experiments
and wrote the manuscript. FI, YI, HG and NK performed RT-qPCR and
analyzed resulting data. FS, YN NT and HM contributed to study
conception and design, acquisition of data and the analysis and
interpretation of data. FI and YI acquired, analyzed and
interpreted the data, drafted the manuscript and revised it
critically for important intellectual content. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by Ethical Committees
of the Graduate School of Medicine, Chiba University (approval no.
889), and the Chiba Cancer Center (grant no. H29-0005). All
procedures were performed in accordance with the ethical standards
of the responsible committee on human experimentation and with the
Helsinki Declaration of 1964 and its later amendments. Informed
consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from the
patients for the publication of the current study and accompanying
clinicopathological data.
Authors' information
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ESCC
|
esophageal squamous cell carcinoma
|
|
miR
|
microRNA
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
|
ICC
|
intraclass correlation coefficient
|
|
CRC
|
colorectal cancer
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brown LM, Devesa SS and Chow WH: Incidence
of adenocarcinoma of the esophagus among white Americans by sex,
stage, and age. J Natl Cancer Inst. 100:1184–1187. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Law S and Wong J: The current management
of esophageal cancer. Adv Surg. 41:93–119. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Headrick JR, Nichols FC III, Miller DL,
Allen MS, Trastek VF, Deschamps C, Schleck CD, Thompson AM and
Pairolero PC: High-grade esophageal dysplasia: Long-term survival
and quality of life after esophagectomy. Ann Thorac Surg.
73:1697–1703. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Research Network:
Analysis Working Group: Asan University; BC Cancer Agency; Brigham
and Women's Hospital; Broad Institute; Brown University; Case
Western Reserve University; Dana-Farber Cancer Institute; Duke
University; Greater Poland Cancer Centre, et al, . Integrated
genomic characterization of oesophageal carcinoma. Nature.
541:169–175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee RC, Feinbaum RL and Ambros V: The C.
Elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ruvkun G: Molecular biology. Glimpses of a
tiny RNA world. Science. 294:797–799. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et
al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei C, Li Y, Huang K, Li G and He M:
Exosomal miR-1246 in body fluids is a potential biomarker for
gastrointestinal cancer. Biomark Med. 12:1185–1196. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni S, Weng W, Xu M, Wang Q, Tan C, Sun H,
Wang L, Huang D, Du X and Sheng W: miR-106b-5p inhibits the
invasion and metastasis of colorectal cancer by targeting CTSA.
OncoTargets Ther. 11:3835–3845. 2018. View Article : Google Scholar
|
|
14
|
Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L
and Jia L: miR-106b and miR-93 regulate cell progression by
suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell
Death Dis. 8:e27962017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
Inhibition of ovarian epithelial carcinoma tumorigenesis and
progression by microRNA 106b mediated through the RhoC pathway.
PLoS One. 10:e01257142015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen G, Jia H, Tai Q, Li Y and Chen D:
miR-106b downregulates adenomatous polyposis coli and promotes cell
proliferation in human hepatocellular carcinoma. Carcinogenesis.
34:211–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang A, Hao J, Wang K, Huang Q, Yu K,
Kang C, Wang G, Jia Z, Han L and Pu P: Down-regulation of miR-106b
suppresses the growth of human glioma cells. J Neurooncol.
112:179–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Wang K, Gao W, Zhang C, Huang F, Wen
S and Wang B: MicroRNA-106b regulates the tumor suppressor RUNX3 in
laryngeal carcinoma cells. FEBS Lett. 587:3166–3174. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu F, Gong J, Huang W, Wang Z, Wang M,
Yang J, Wu C, Wu Z and Han B: MicroRNA-106b-5p boosts glioma
tumorigensis by targeting multiple tumor suppressor genes.
Oncogene. 33:4813–4822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prasad R and Katiyar SK: Down-regulation
of miRNA-106b inhibits growth of melanoma cells by promoting
G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1
protein. Oncotarget. 5:10636–10649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhuang M, Zhao S, Jiang Z, Wang S, Sun P,
Quan J, Yan D and Wang X: MALAT1 sponges miR-106b-5p to promote the
invasion and metastasis of colorectal cancer via SLAIN2 enhanced
microtubules mobility. EBioMedicine. 41:286–298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hoshino I and Matsubara H: MicroRNAs in
cancer diagnosis and therapy: From bench to bedside. Surg Today.
43:467–478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hermeking H: MicroRNAs in the p53 network:
Micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Khoury S and Tran N: Circulating
microRNAs: Potential biomarkers for common malignancies. Biomark
Med. 9:131–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kumar S, Sharawat SK, Ali A, Gaur V, Malik
PS, Kumar S, Mohan A and Guleria R: Identification of
differentially expressed circulating serum microRNA for the
diagnosis and prognosis of Indian non-small cell lung cancer
patients. Curr Probl Cancer. 44:1005402020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshida K, Yokoi A, Kagawa T, Oda S,
Hattori S, Tamauchi S, Ikeda Y, Yoshikawa N, Nishino K, Utsumi F,
et al: Unique miRNA profiling of squamous cell carcinoma arising
from ovarian mature teratoma: Comprehensive miRNA sequence analysis
of its molecular background. Carcinogenesis. 40:1435–1444.
2019.PubMed/NCBI
|
|
31
|
Chuma M, Toyoda H, Matsuzaki J, Saito Y,
Kumada T, Tada T, Kaneoka Y, Maeda A, Yokoo H, Ogawa K, et al:
Circulating microRNA-1246 as a possible biomarker for early tumor
recurrence of hepatocellular carcinoma. Hepatol Res. 49:810–822.
2019.PubMed/NCBI
|
|
32
|
Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang
J, Yang X and Liu Z: Exosomal miR-1246 in serum as a potential
biomarker for early diagnosis of gastric cancer. Int J Clin Oncol.
25:89–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moshiri F, Salvi A, Gramantieri L,
Sangiovanni A, Guerriero P, De Petro G, Bassi C, Lupini L, Sattari
A, Cheung D, et al: Circulating miR-106b-3p, miR-101-3p and
miR-1246 as diagnostic biomarkers of hepatocellular carcinoma.
Oncotarget. 9:15350–15364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang F, Xiong H, Duan L, Li Q, Li X and
Zhou Y: miR-1246 promotes metastasis and Invasion of A549 cells by
targeting GSK-3β-mediated Wnt/β-catenin pathway. Cancer Res Treat.
51:1420–1429. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu R, Li H, Wu S, Qu J, Yuan H, Zhou Y and
Lu Q: MicroRNA-1246 regulates the radio-sensitizing effect of
curcumin in bladder cancer cells via activating P53. Int Urol
Nephrol. 51:1771–1779. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Yao D, Zhao S, He C, Ding N, Li L
and Long F: miR-1246 promotes SiHa cervical cancer cell
proliferation, invasion, and migration through suppression of its
target gene thrombospondin 2. Arch Gynecol Obstet. 290:725–732.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Du P, Lai YH, Yao DS, Chen JY and Ding N:
Downregulation of microRNA-1246 inhibits tumor growth and promotes
apoptosis of cervical cancer cells by targeting thrombospondin-2.
Oncol Lett. 18:2491–2499. 2019.PubMed/NCBI
|
|
38
|
Cai K, Wang Y and Bao X: miR-106b promotes
cell proliferation via targeting RB in laryngeal carcinoma. J Exp
Clin Cancer Res. 30:732011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mehlich D, Garbicz F and Włodarski PK: The
emerging roles of the polycistronic miR-106b-25 cluster in cancer -
A comprehensive review. Biomed Pharmacother. 107:1183–1195. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tamilzhalagan S, Rathinam D and Ganesan K:
Amplified 7q21-22 gene MCM7 and its intronic miR-25 suppress COL1A2
associated genes to sustain intestinal gastric cancer features. Mol
Carcinog. 56:1590–1602. 2017. View Article : Google Scholar : PubMed/NCBI
|