In 2010, it has been reported that pancreatic ductal
adenocarcinoma (PDAC) is a gastrointestinal malignancy with a
5-year survival rate <5% in the United States, giving patients
with pancreatic cancer the poorest prognosis among patients with
malignant cancer types (1). PDAC is
characterized by rapid progression and metastasis (2), and poses a great threat to human
health. At present, TNM staging (3)
is regarded as a convincing cancer staging system to guide clinical
treatment and offer a method for predicting the prognosis of
patients with cancer (4). However,
the clinical disadvantages of TNM staging are increasingly obvious
when further investigations are performed (5). Certain clinicopathological features,
such as TNM stage, lymph node involvement and differentiation, have
been demonstrated to be independent prognostic factors in patients
with pancreatic cancer (6).
Nonetheless, the majority of these prognostic markers are verified
after surgery. Therefore, it is crucial to identify useful
predictive factors for PDAC before surgery. With the development of
human gene sequencing technology, gene-based biomarkers have been
markedly improved, which provides an opportunity to investigate
effective biomarkers for guiding diagnosis, treatment and
evaluation of pancreatic cancer prognosis (7).
Emerging evidence has elucidated that local immune
suppression of the tumor microenvironment (TME) exhibits a strong
association with cancer growth, metastasis and even tumor immune
escape (8–10). Immune cells do not merely act as
tumor killers, but can also function as an activator of tumors.
They are capable of disturbing molecular signals and exert vital
roles in cancer biology, including those associated with tumor
growth, invasion and metastasis (11). However, certain types of cancer cells
are able to avoid being detected and escape immune attack in order
to promote tumor growth (12). At
present, although targeted therapies and immunotherapies are
effective for numerous solid malignancies, such as metastatic
melanoma and lung cancer, few clinical benefits can be observed in
pancreatic cancer (13,14). Therefore, the latent prognostic value
of immune genes can be illustrated through further investigation of
the association between immune genes and survival. In addition,
this may aid the identification of novel biomarkers. Ongoing
studies that intend to target the stroma and the immune
microenvironment either alone or in combination are expected to
present more sustained treatment outcomes (15,16). A
detailed description of the TME integrating tumor and host-related
factors may result in the verification of novel biomarkers for a
more targeted method for immunotherapy, and other combination
therapies to fix the immunosuppressive mechanisms in the
microenvironment of pancreatic cancer.
Immune scores and analyses derived from the
Estimation of Stromal and Immune Cells in Malignant Tumor Tissues
Using Expression Data (ESTIMATE) algorithm can contribute to the
quantification of the immune and stromal components in a tumor
(17), and the Cell Type
Identification by Estimating Relative Subset of Known RNA
Transcripts (CIBERSORT) algorithm is an approach for describing
immune cell components of tissues from their immune gene expression
profiles (18). Both can be used to
predict the immune microenvironment of pancreatic cancer.
Considering the association between immunity and cancer, the
present study was designed to identify immune-gene biomarkers for
the prediction of the prognosis of PDAC, and to examine the
differences in the immune microenvironment between high- and
low-risk patients with pancreatic cancer.
Gene expression profiles for use in the present
study were downloaded from four databases: Genotype-Tissue
Expression (19) via UCSC Xena
(GTEx; http://toil.xenahubs.net/download/gtex_RSEM_gene_fpkm.gz),
The Cancer Genome Atlas (20) (TCGA;
http://portal.gdc.cancer.gov/repository),
International Cancer Genome Consortium (21) (ICGC; http://dcc.icgc.org/releases/current/Projects/PACA-AU)
and Gene Expression Omnibus (22)
(GEO; http://www.ncbi.nlm.nih.gov/geo). The
fragments per kilobase of transcript per million mapped reads
(FPKM) data of normal pancreatic tissues were downloaded from the
GTEx-Pancreas database (Full metadata), and the RNA-sequencing FPKM
data of pancreatic tumor samples were obtained from the publicly
available TCGA-PAAD database. Additionally, normalized
high-throughput sequencing and microarray data of mRNAs (21,22) were
downloaded from the ICGC-PACA-AU database and GEO (GSE62452). Cases
with complete clinical information in the pancreatic cancer cohort
were included. Furthermore, patients with an overall survival ≤60
days were excluded, as these patients may have succumbed to factors
not associated with the tumor, such as hemorrhage and severe
infection (23). As samples from
GTEx obtained from normal pancreatic tissue samples, there is no
disease-related information. Eventually, a total of 459 samples
obtained from GTEx (n=156), TCGA (Table
SI; n=158), ICGC (Table SII;
n=79) and GEO (Table SIII; n=66)
databases were extracted for further analysis.
In order to screen IRGs with the most predictive
efficacy in univariate Cox regression analysis (survival package,
version 2.41–3; http://cran.r-project.org/web/packages/survival/index.html),
a least absolute shrinkage and selection operator (LASSO) analysis
(glmnet package, version 3.0–1; http://cran.r-project.org/web/packages/glmnet/index.html)
was adopted to deal with over-fitting of the model. Subsequently, a
prognostic model was constructed using the IRGs obtained from the
multivariate regression analysis. The risk score (RS) of each
patient was calculated by multiplying the expression level of each
IRG with its corresponding regression coefficients. The following
computational algorithm was used for this analysis (25–27):
RS=βgene(1) × exprgene(1) + βgene(2) × exprgene(2) + … + βgene(n) ×
exprgene(n). ‘β’ is the regression coefficient generated from
univariate Cox analysis of IRGs, and ‘exprgene’ refers to the
expression of IRGs in the sample. The optimal cut-off point of risk
value that was most relevant to survival was determined using the
surv_cutpoint function in the survminer package (version 0.4.3;
http://www.sthda.com/english/wiki/survminer-r-package-survival-data-analysis-and-visualization),
which was the standard to separate PDAC samples into high-and
low-RS subgroups.
CIBERSORT is a bioinformatics method used to
evaluate immune cell composition by transformation of standardized
gene expression (29). Based on the
CIBERSORT package (version 1.03; http://cibersort.stanford.edu/), the mRNA expression
matrix of patients from TCGA, corrected by the zoom function in the
limma package, was transformed into 22 subtypes of immune cells,
which quantify the cellular composition of the immune response
(18). After removing samples with
P>0.05 from the CIBERSORT analysis results, a Mann-Whitney U
test was performed to compare the differences in immune cell
subtypes between high- and low-RS groups. The association between
immune cells and modeling IGRs was also analyzed.
The Shapiro-Wilk normality test was used to measure
the normality of the variables between two groups. The statistical
significance of the discrepancy between normally distributed
variables was calculated via unpaired Student's t-test and the
association was estimated by Pearson's correlation coefficient.
Survival rates were measured by the Kaplan-Meier (KM) method, and
the significance of disparity between survival curves was assessed
via the log-rank test. Survival predictive accuracy of prognostic
models was assessed based on a time-dependent receiver operating
characteristic curve (ROC) analysis. Univariate ANOVA was used to
calculate whether the clinical outcome (alive, dead with tumor and
dead tumor free) was significantly affected by different grades of
clinical parameters. Mean and standard deviation reflected the
central trend and discrete trend of age in each clinical outcome.
All statistical analyses were performed using R software (version
3.6.1; http://www.r-project.org/). P<0.01
was used as the threshold of genes considered with prognostic value
calculated by univariate Cox analysis. P<0.05 was considered to
indicate a statistically significant difference.
A total of 554 IRGs were identified to be
differentially expressed between normal pancreatic tissues and
pancreatic tumors (Fig. S1A and B).
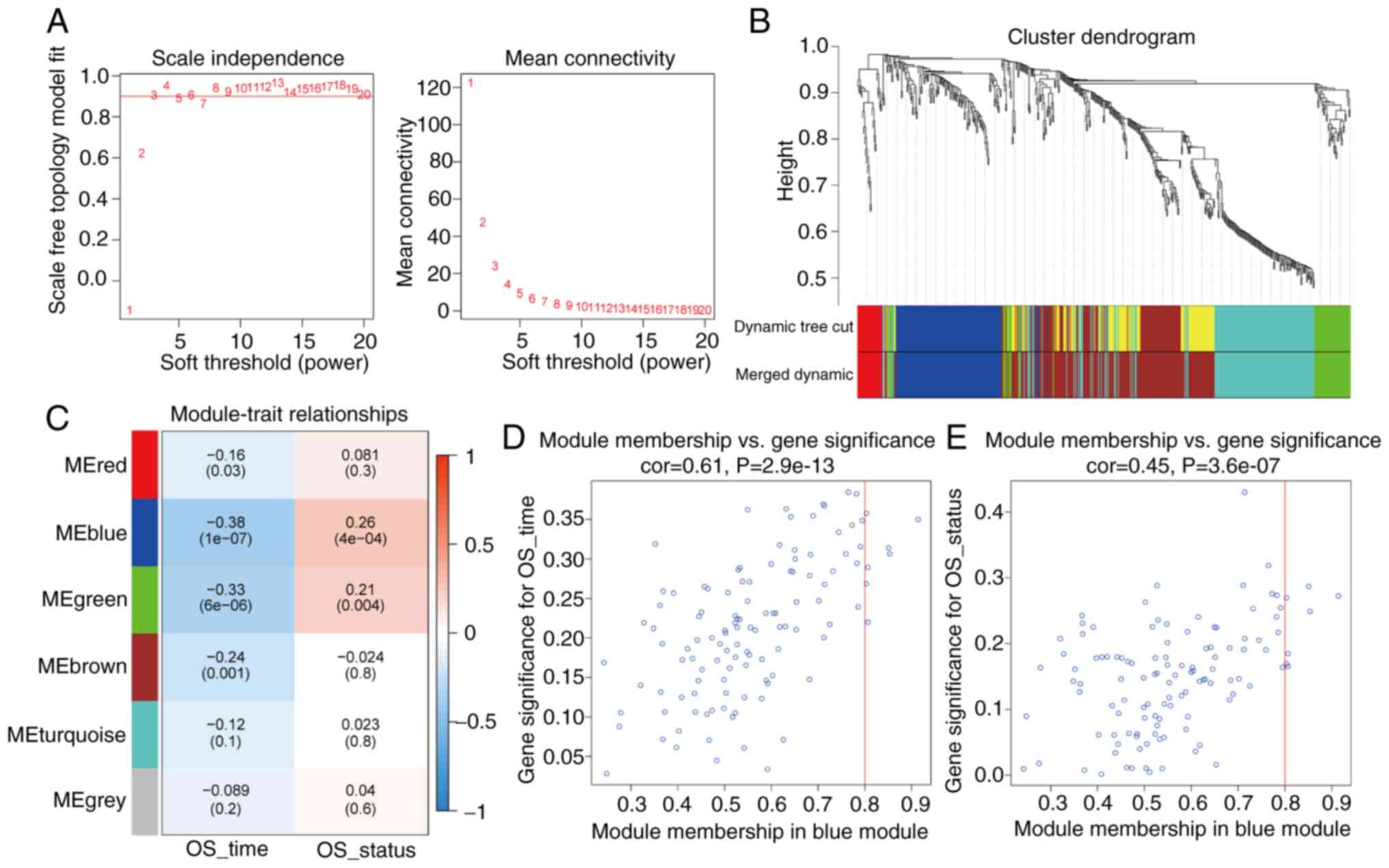
In the WGCNA of TCGA cohort, two important parameters,
R2 (R2>0.9) and average connectivity were
fully considered, hence β=4 was selected as the soft-thresholding
power (Fig. 1A). According to the
weighted adjacency matrix constructed by the soft-thresholding
power, the differential immune genes in the cohort from TCGA were
classified into six diverse gene modules (Fig. 1B). The present study assessed the
association between the module genes and characteristics including
the survival time and status of patients with pancreatic cancer in
the cohort from TCGA. Based on the correlation coefficient and the
P-value, it was revealed that the blue module had the highest
module significance (OS_time: cor=−0.38, P=1×10−7;
OS_status: cor=0.26, P=4×10−4; Fig. 1C) and gene significance (OS_time:
cor=0.61, P=2.9e-13; OS_status: cor=0.45, P=3.6e-7; Fig. 1D and E). Therefore, the blue module,
which contained 117 IRGs, was selected for further analysis.
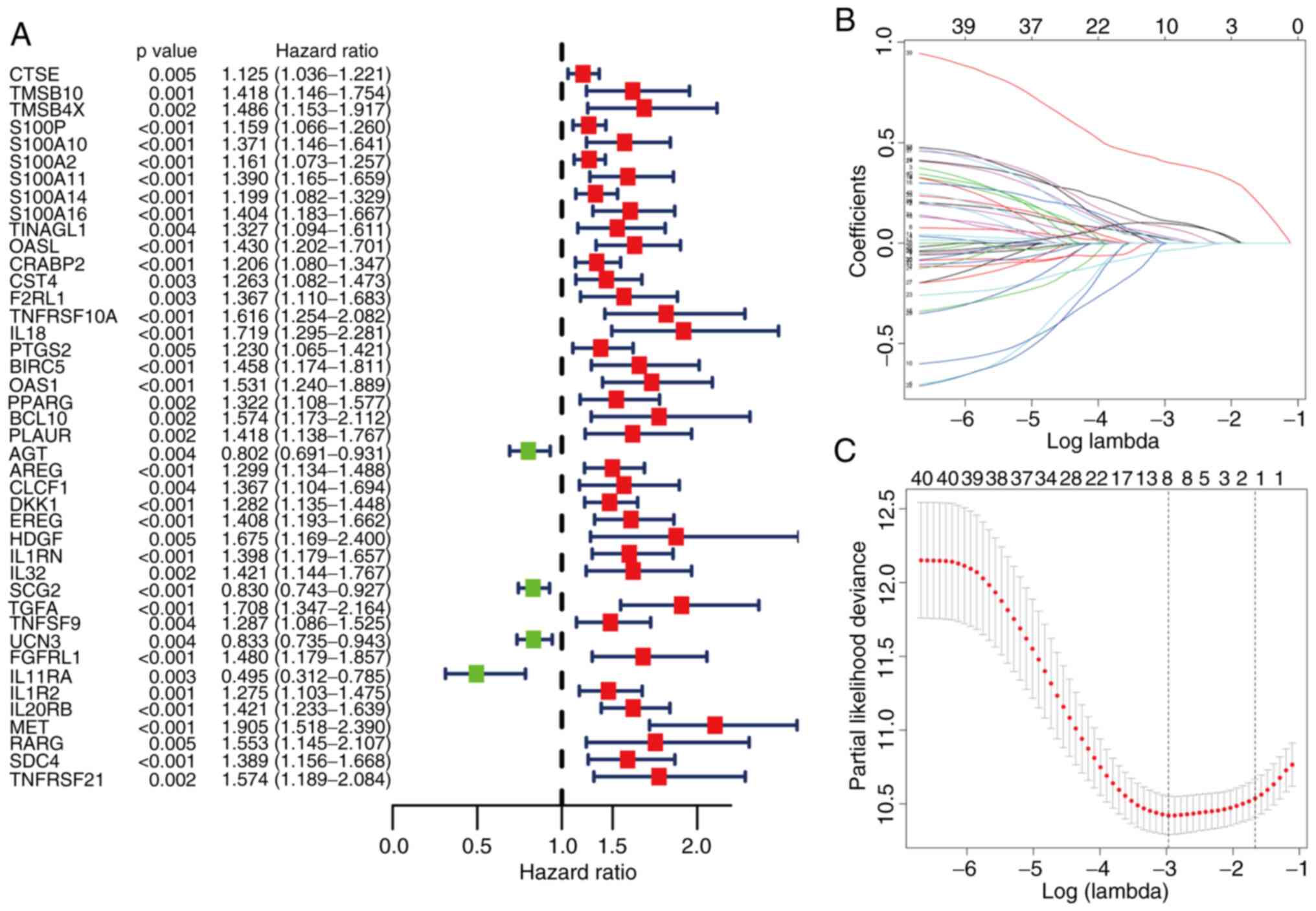
Based on univariate Cox regression analysis of TCGA
cohort, 42 genes with significant prognostic value were further
extracted from the blue module (P<0.01; Fig. 2A). LASSO regression analysis was
applied to avoid over-simulation of the model by adjusting the
complexity of the classifier, and IRGs that correlated highly with
one another were deleted (Fig. 2B and
C). In the case of dimensionality reduction of the preliminary
results using the LASSO method, the genes and their coefficients
involved in the model were determined by multivariate Cox
regression analysis. The coefficients of four IRGs are presented in
Table SIV. Ultimately, four optimal
IRGs [2′-5′-oligoadenylate synthetase 1 (OAS1), MET proto-oncogene,
receptor tyrosine kinase (MET), interleukin 1 receptor type 2
(IL1R2) and interleukin 20 receptor subunit β (IL20RB)] were
included in the prognostic model to calculate the RS, and the
formula was as follows: RS=0.221350 × OAS1 + 0.515099 × MET +
0.179351 × IL1R2 + 0.141478 × IL20RB. The cut-off value was
determined to be 4.15 using the surv_cutpoint function in the
survminer R package, and the patients were divided into high- and
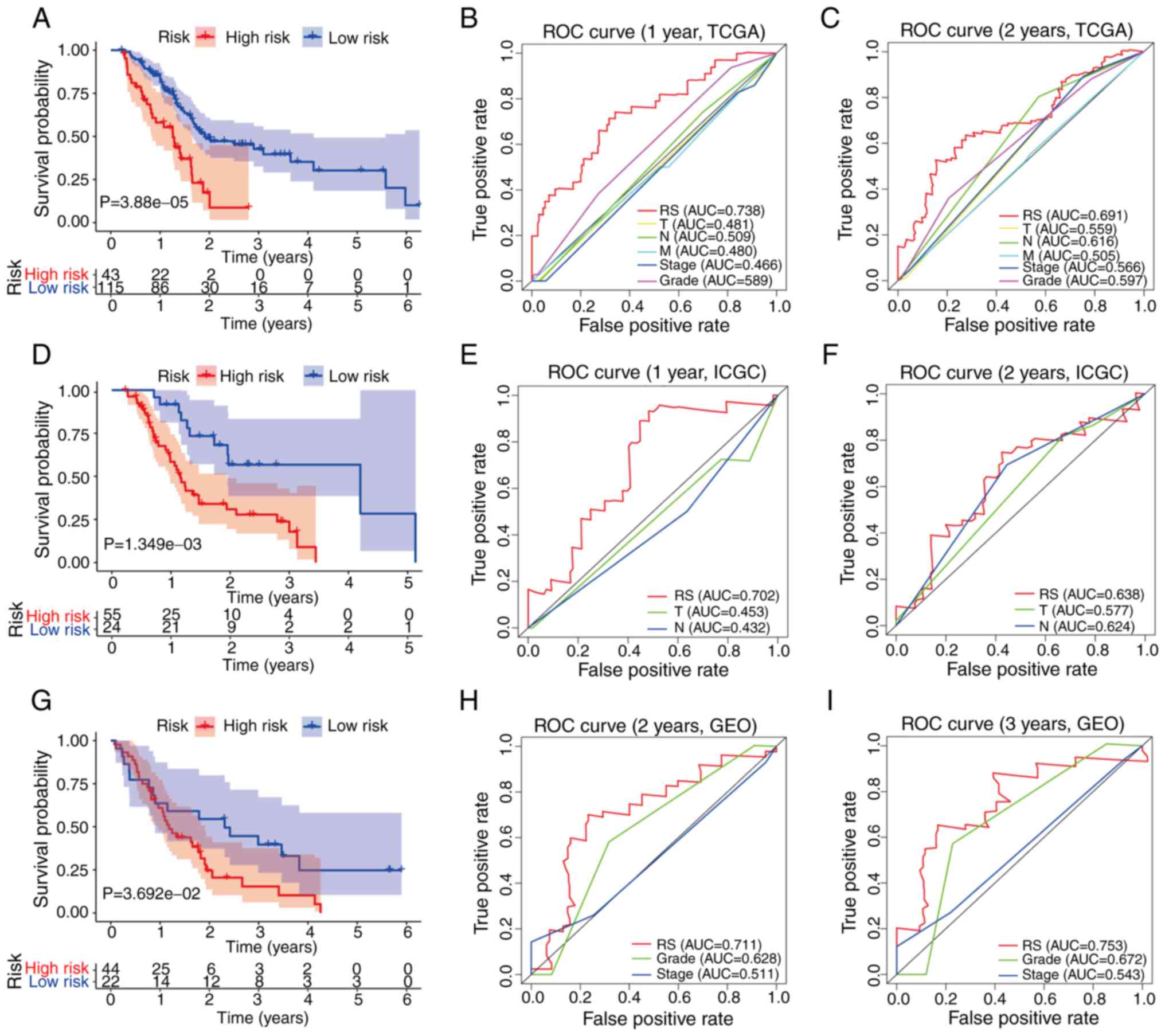
low-RS groups. In the TCGA training set, the KM survival curve
analysis demonstrated that the low-RS group had significantly
improved survival compared with the high-RS group (P=3.88e-05;
Fig. 3A), and the ROC curve revealed
that the model had a good predictive function for the prognosis of
patients with pancreatic cancer (ROC 1-year=0.738; Fig. 3B; ROC 2-years =0.691; Fig. 3C). Additionally, it was revealed that
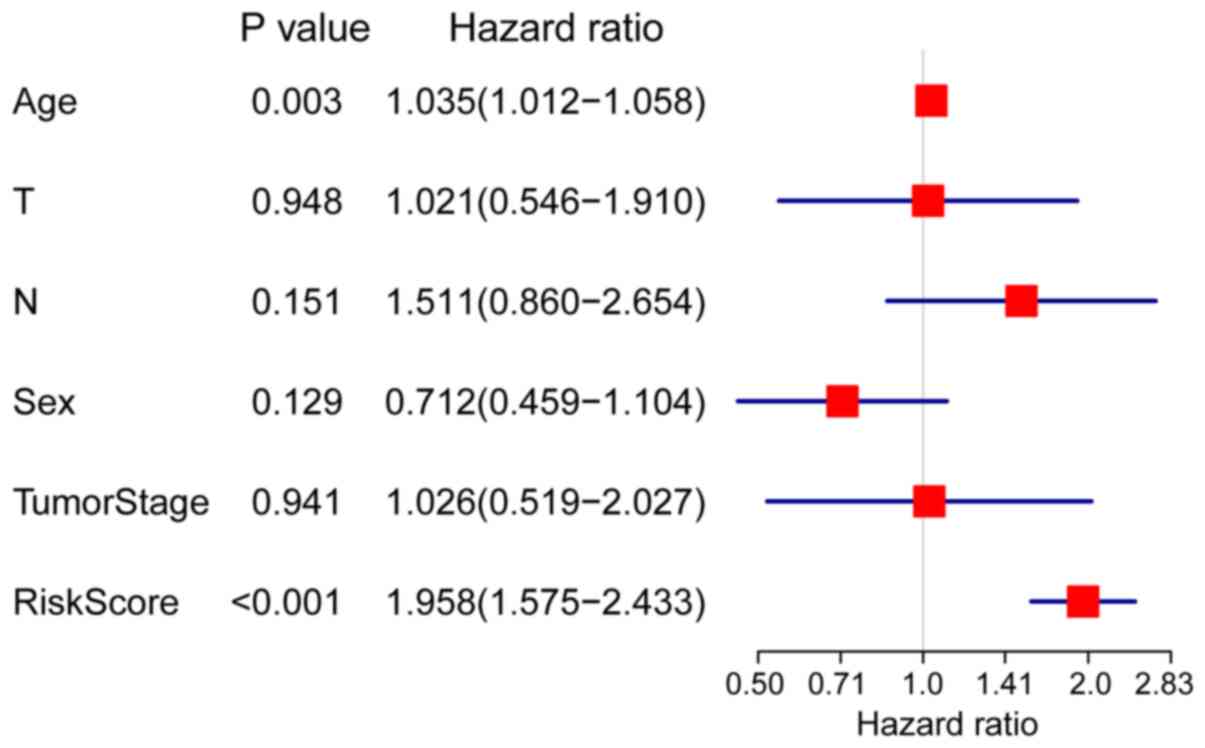
the RS could be used as an independent predictor [hazard ratio
(HR), 1.958; P<0.001; Fig. 4] for
the prognosis of patients with pancreatic cancer in the training
cohort by multivariate regression analysis including RS, sex, age,
TNM stage and tumor stage.
In order to confirm the robustness of the prognostic
classifier used in the present study, the duplicate formula for RS
and cut-off value were used for the analysis of the ICGC and GEO
validation sets. In each cohort, the 79 patients in the ICGC cohort
and 66 patients in the GSE62452 dataset were divided into high- and
low-RS groups, respectively. The KM survival curves (P=0.03692;
Fig. 3D and G) and ROC curves
(Fig. 3E, F, H and I) validated the
reliability of the prognostic model used in the present study.
Among them, although RS was not effective in predicting the 1-year
survival rate (P>0.05; data not shown) of patients with
pancreatic cancer in the GSE62452 dataset, it exerted a preferable
effect on calculating the 3-year survival rate (Fig. 3G and I). Furthermore, to emphasize
the superiority of the RS in predicting overall survival of
patients with PDAC, time-dependent ROC curve analysis was used to
compare the predictive effect of RS with classic disease
classification parameters, and this revealed that RS was the best
index in three independent databases (Fig. 3). The RS distribution, survival
status and risk gene expression in the training cohort and
validation cohort are presented in Fig.
S2A-I.
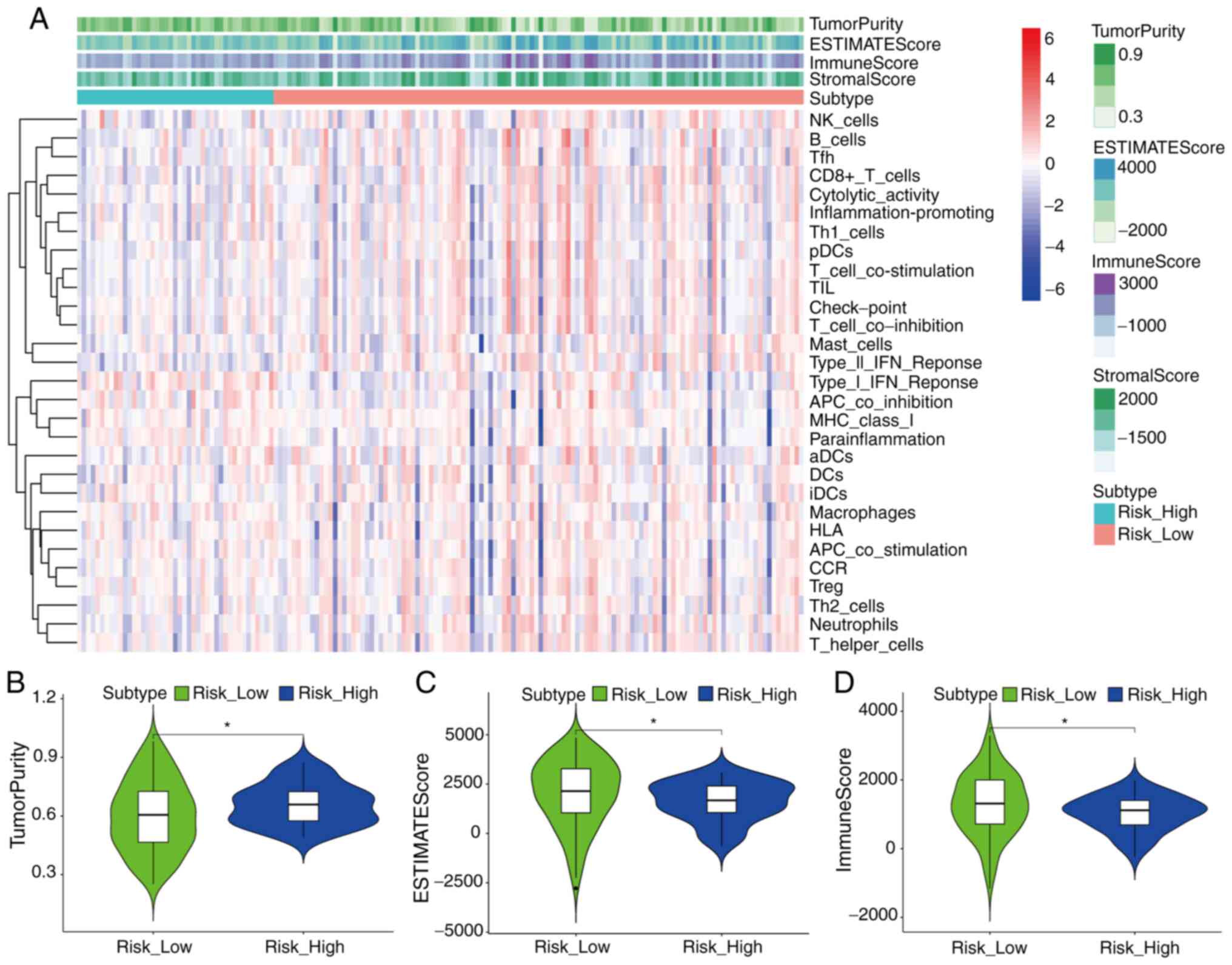
The analysis results of the ESTIMATE algorithm
revealed the difference of immune status between high- and low-RS
groups in the cohort from TCGA (Fig.
5A-D). ssGSEA analysis revealed that the high-RS group had
lower enrichment levels in multiple positively regulated immunity
gene sets (‘Cytolytic activity’ and ‘Inflammation-promoting’) than
the low-RS group (Fig. 5A), which
suggested that various immunity-associated biological processes and
mechanisms were activated in both the aforementioned two groups,
and increased immune activity may help patients with pancreatic
cancer achieve improved outcomes. Notably, the high-RS group had a
higher level of tumor purity and lower immune score than the low-RS
group (P<0.05; Fig. 5B and D).
The high-RS group also had a lower ESTIMATE score which reflected
the comprehensive level of tumor purity, immune score and stromal
score (P<0.05; Fig. 5C).
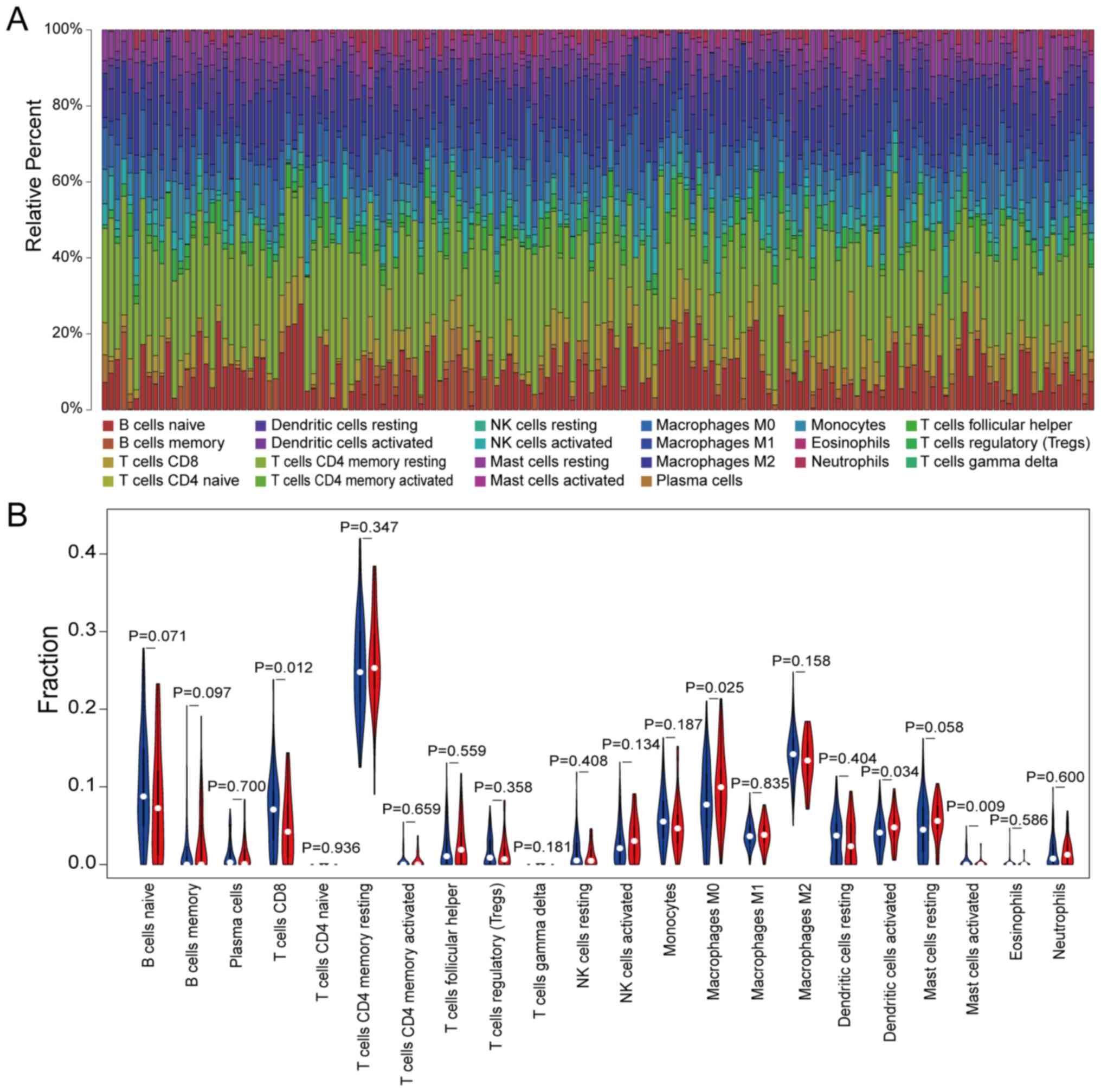
The CIBERSORT algorithm was used to evaluate the
composition of 22 immune cell types in TCGA cohort. It was revealed
that the resting CD4 memory cells occupied the highest proportion
among all cells, while activated CD4 memory cells accounted for a
small proportion (Fig. 6A). This
suggested that the absence of immune cell components with immune
activation functions in the immune microenvironment may serve an
important role in accelerating the development of pancreatic
cancer. By comparing the composition of immune cells in the high-RS
group and the low-RS group of TCGA cohort, it was revealed that the
high-RS group had a lower infiltration of CD8 T cells and a higher
composition of M0 macrophages (Fig.
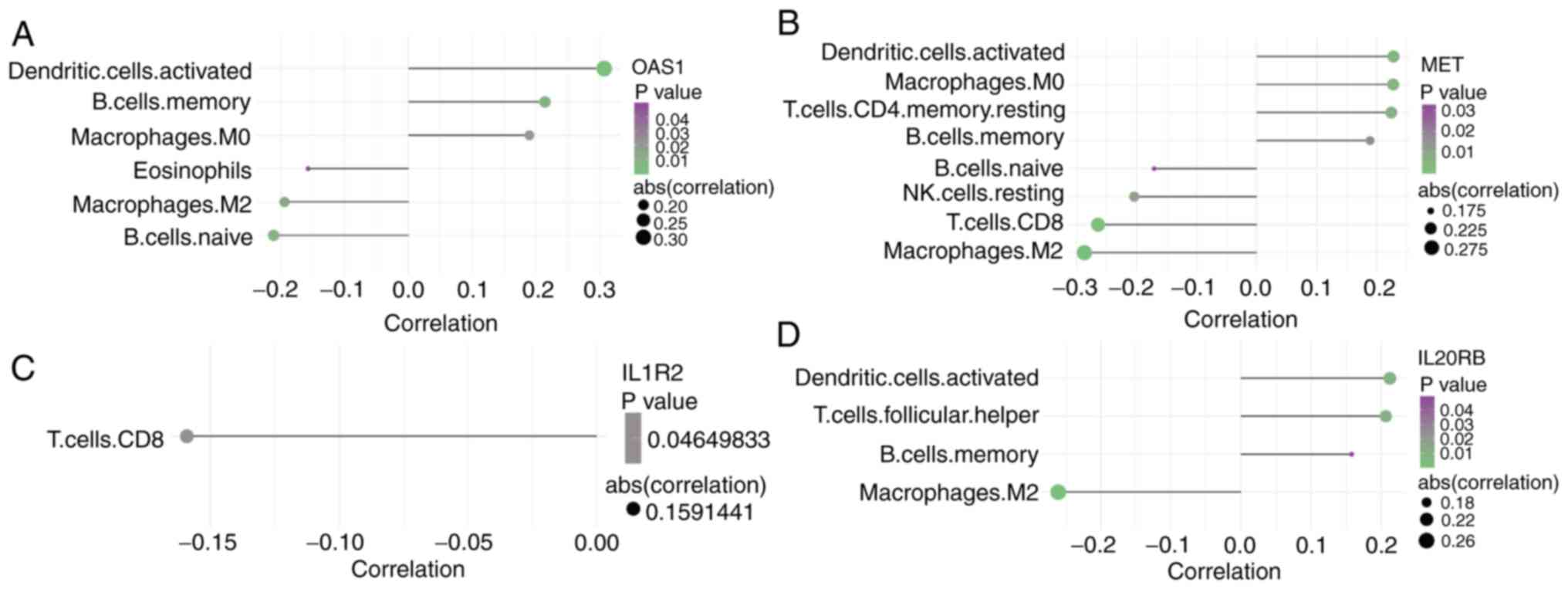
6B). Finally, the present study analyzed the correlation
between the gene expression of four marker genes in the TCGA cohort
and the composition of 22 immune cells. The results revealed that
the expression levels of OAS1, MET and IL20RB genes were negatively
correlated with the composition of M2macrophages (Fig. 7A, B and D; P<0.01). With the
increase of CD8 T cells, the gene expression levels of MET and
IL1R2 were decreased (Fig. 7B and C;
P<0.01), which suggested that CD8 T cells may repress the
activation of the oncogenes observed in pancreatic cancer.
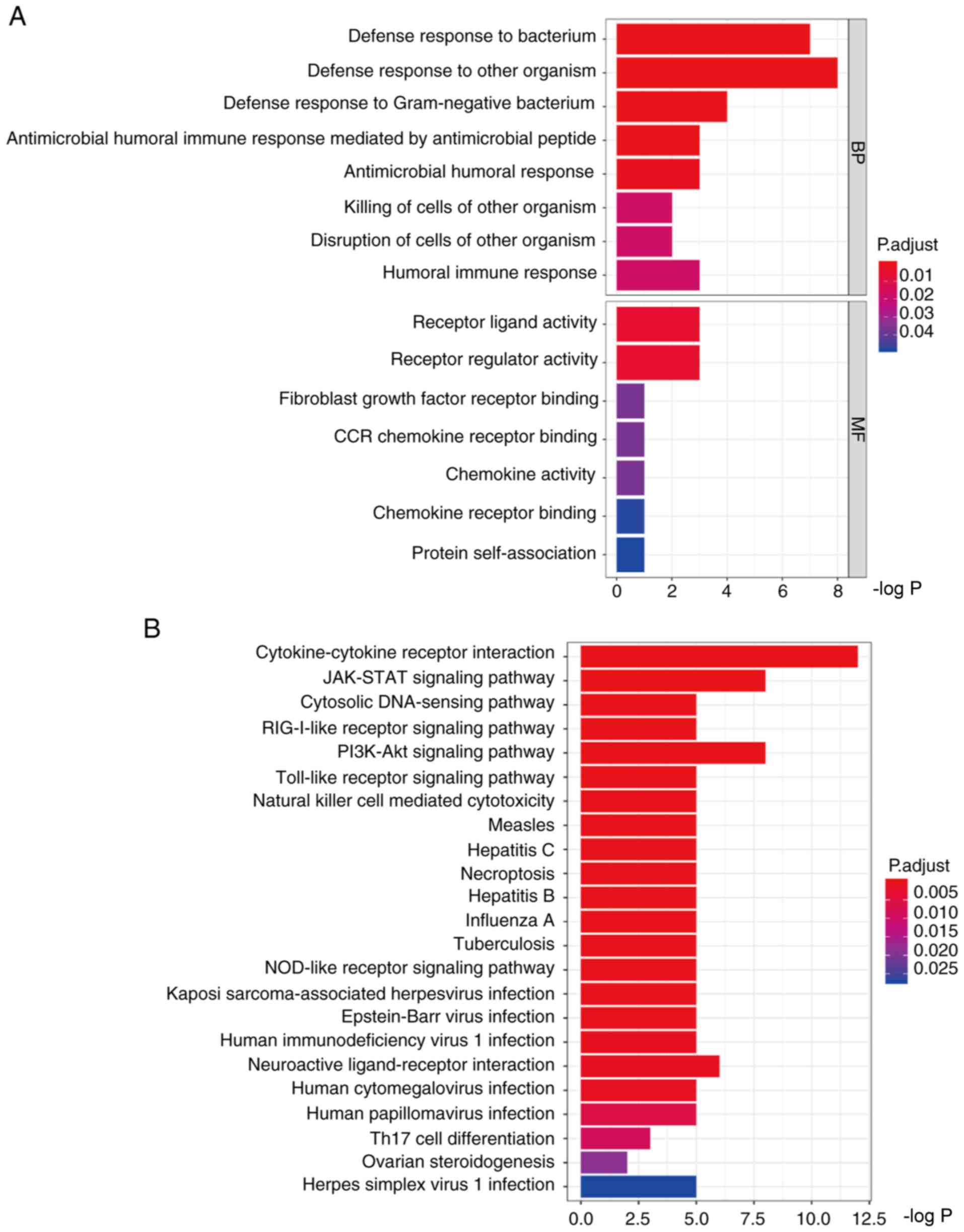
The results revealed that the upregulated genes were
associated with 8 biological processes and 7 molecular functions,
including ‘defense response to bacterium’, ‘defense response to
other organisms’ and ‘receptor ligand activity’(Fig. 8A). Through pathway analyses, the
present study revealed that upregulated genes were associated with
‘cytokine-cytokine receptor interaction’ and ‘JAK-STAT signaling
pathway’ (Fig. 8B).
As a highly malignant tumor, over half of all PDAC
cases are diagnosed at an advanced stage for which mortality rates
closely parallel incidence rates. PDAC is also the fourth leading
cause for cancer-associated mortality worldwide (30,31).
With characteristics of insidious onset and early metastasis, ~80%
of patients are diagnosed at a late stage of disease (32). At present, tumor immunotherapy
results in marked effects in cancer treatment (33). The impact of the immune
microenvironment on tumor cells has been demonstrated in numerous
studies (34,35). Immunotherapy has emerged as an option
for pancreatic cancer (36).
Wartenberg et al (37)
suggested that immunophenotypic classification is associated with
tumor characteristics leading to pancreatic cancer with
prognostic/predictive significance. Considering the significance of
the immune microenvironment in cancer progression, finding
immunity-associated biomarkers to predict the prognosis of patients
with PDAC is necessary, and may also serve an important role in
immunotherapy (38).
The present study revealed that a module including
117 IRGs was associated with pancreatic cancer prognosis using
WGCNA. In the univariate Cox regression and LASSO analyses, four
IRGs were included in the classifier (OAS1, MET, IL1R2 and IL20RB).
The model could effectively predict the prognosis of patients with
PDAC in the TCGA cohort [1-year area under the curve (AUC)=0.738;
2-years AUC=0.691]. In addition, the performance of the model was
assessed using patients in the ICGC cohort (1-year AUC=0.702;
2-years AUC=0.638) and GSE62452 (2-years AUC=0.711; 3-years
AUC=0.753). Using multivariate regression analyses, the RS of the
model was demonstrated to be an independent factor for predicting
the prognosis of patients with pancreatic cancer in the TCGA cohort
(HR, 1.958; P<0.001).
The discovery of the following four genes provides
guidance for searching for targeted genes for immunotherapy of
pancreatic cancer. It has been reported that immune response by
four IRGs (OAS1, MET, IL1R2 and IL20RB) is associated with the
prognosis of pancreatic cancer (39–42). The
OAS system is an antiviral signaling pathway induced by interferon
and OAS genes that are described as interferon-stimulated genes
(43,44). There are three types of OAS proteins
in humans, OAS1, OAS2 and OAS3. OAS1 has been demonstrated to be
associated with pancreatic cancer and prostate cancer (41,45).
Oncolytic virus therapy is a promising treatment option for
pancreatic cancer; however, high expression levels of OAS in cell
lines are associated with resistance to this therapy (46). Therefore, OAS1 could be a potential
immunotherapy target in pancreatic cancer and a prognostic
biomarker to identify suitability of patients with pancreatic
cancer to receive immunotherapy (45,47). The
MET receptor, with a hepatocyte growth factor receptor ligand, is
upregulated in several malignancies, including breast, lung and
pancreatic cancer (48,49). Activation of several intracellular
signaling pathways that is mediated by MET has led to the emergence
of diverse cellular hallmarks of cancer, including cell
proliferation, survival, invasion, migration, metastasis and
inhibition of apoptosis (50–52). MET
may be involved in the malignant process of pancreatic cancer, and
can serve as a biomarker for assessing the prognosis of patients
with pancreatic cancer (53). IL1R2,
as an endogenous inhibitor, can be highly expressed in the
follicular helper T cells of mice, and the specific marker
molecules, such as IL1R2, on the surface of tumor infiltrating Treg
cells in colorectal cancer and non-small cell lung cancer have also
been demonstrated to be upregulated (54,55).
Furthermore, another study on breast cancer revealed that IL1R2 is
more abundant in tumor-infiltrating T cells compared with Treg
cells or peripheral blood T cells in normal breast tissues
(56). In conclusion, IL1R2 is
regarded as a negative regulatory factor that can be used as a
potential target for immunotherapy. IL20R, a heterodimeric
receptor, is composed of two chains, interleukin-20 receptor
subunit α and IL20RB (57). IL20RB
may affect the IL20 signaling pathway. Several studies have
demonstrated that IL20RB serves a significant role in a number of
different types of cancer, including breast cancer (58), lung cancer (59), nasopharyngeal carcinoma (60) and pancreatic cancer (40). Overall, the results of the present
study may provide potential mRNA targets for immunotherapy and
prognostic evaluation to help improve the clinical outcomes of
pancreatic cancer.
The CIBERSORT algorithm is able to assess 22 types
of immune cells based on the gene expression data with high
sensitivity and specificity (18).
Several studies have suggested that the relative levels of various
immune cells, analyzed by the CIBERSORT algorithm, could estimate
the immune cell composition in tumors (69–71). In
the present study, the difference in immune infiltration of
pancreatic cancer cells in TCGA cohorts mainly consisted of
macrophage M0 and T cells CD8, as determined by the CIBERSORT
algorithm. In the immune infiltration analysis, the proportion of
CD8 T cells in the high-RS group was relatively lower compared with
the low-RS group. Additionally, the proportion of M0 macrophages in
the high-RS group was relatively higher than low-RS group. This
indicated that an imbalance in the CD8 T cells and M0 macrophage
ratio may lead to a lower survival rate in the high-RS group.
Tahkola et al (72) revealed
that the 5-year survival rates of the CD8 T cells low-, medium- and
high-infiltration groups in pancreatic cancer were 4.2, 13.4 and
31.5%, respectively. Another study demonstrated that an imbalance
in the proportion of immune cells was closely associated with low
survival rate and poor clinical outcomes in patients with cancer
(73). However, the specific effect
of these differentially expressed chemotactic factors on immune
infiltration of pancreatic cancer requires further
investigation.
To summarize, in the present study, an IRG prognosis
model of pancreatic cancer was constructed, and the effectiveness
of the model was verified in the independent validation set. The
difference in overall survival between high- and low-RS groups
based on this model may be caused by the difference of immune
infiltration and TME. The findings of the present study added
certain guidance values to the analysis of pancreatic cancer
pathogenesis and provide a reference for clinical treatment. There
were some limitations to the present study, and thus, the validity
of the conclusions should be confirmed by an investigation
involving a larger number of cases, and further animal studies or
cellular experiments are required to test the prognostic accuracy
of the signatures.
Not applicable.
No funding was received.
All data generated or analyzed during this study are
included in this published article.
SY, JF, YZ,YX and FF contributed to the study
concept and design, and the acquisition, interpretation and
analysis of the data. SY, JF and FF drafted the manuscript. FF was
responsible for the integrity of the work as a whole. All authors
read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lian EY, Hyndman BD, Moodley S, Maritan SM
and Mulligan LM: RET isoforms contribute differentially to invasive
processes in pancreatic ductal adenocarcinoma. Oncogene. Sep
3–2020.(Epub ahead of print). View Article : Google Scholar
|
|
3
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bertero L, Massa F, Metovic J, Zanetti R,
Castellano I, Ricardi U, Papotti M and Cassoni P: Eighth edition of
the UICC Classification of Malignant Tumours: An overview of the
changes in the pathological TNM classification criteria-What has
changed and why? Virchows Archiv. 472:519–531. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galon J, Pagès F, Marincola FM, Thurin M,
Trinchieri G, Fox BA, Gajewski TF and Ascierto PA: The immune score
as a new possible approach for the classification of cancer. J
Transl Med. 10:12012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiang J, Liu L, Wang W, Xu H, Wu C, Xu J,
Liu C, Long J, Ni Q and Yu X: Metabolic tumor burden: A new
promising way to reach precise personalized therapy in PDAC. Cancer
Lett. 359:165–168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Jing R, Xu J, Liu K, Xue J, Wen Z
and Li M: Comparative analysis of oncogenes identified by
microarray and RNA-sequencing as biomarkers for clinical prognosis.
Biomark Med. 9:1067–1078. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fridman WH, Pagès F, Sautès-Fridman C and
Galon J: The immune contexture in human tumours: Impact on clinical
outcome. Nat Rev Cancer. 12:298–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moon YW, Hajjar J, Hwu P and Naing A:
Targeting the indoleamine 2,3-dioxygenase pathway in cancer. J
Immunother Cancer. 3:512015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harden JL and Egilmez NK: Indoleamine
2,3-dioxygenase and dendritic cell tolerogenicity. Immunol Invest.
41:738–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martinez-Bosch N, Vinaixa J and Navarro P:
Immune evasion in pancreatic cancer: From mechanisms to therapy.
Cancers (Basel). 10:62018. View Article : Google Scholar
|
|
13
|
Dreyer SB, Chang DK, Bailey P and Biankin
AV: Pancreatic cancer genomes: Implications for clinical management
and therapeutic development. Clin Cancer Res. 23:1638–1646. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Knudsen ES, Vail P, Balaji U, Ngo H,
Botros IW, Makarov V, Riaz N, Balachandran V, Leach S, Thompson DM,
et al: Stratification of pancreatic ductal adenocarcinoma:
Combinatorial genetic, stromal, and immunologic markers. Clin
Cancer Res. 23:4429–4440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Connor AA, Denroche RE, Jang GH, Timms L,
Kalimuthu SN, Selander I, McPherson T, Wilson GW, Chan-Seng-Yue MA,
Borozan I, et al: Association of distinct mutational signatures
with correlates of increased immune activity in pancreatic ductal
adenocarcinoma. JAMA Oncol. 3:774–783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sherman MH, Yu RT, Tseng TW, Sousa CM, Liu
S, Truitt ML, He N, Ding N, Liddle C, Atkins AR, et al: Stromal
cues regulate the pancreatic cancer epigenome and metabolome. Proc
Natl Acad Sci USA. 114:1129–1134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jia D, Li S, Li D, Xue H, Yang D and Liu
Y: Mining TCGA database for genes of prognostic value in
glioblastoma microenvironment. Aging (Albany NY). 10:592–605. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
GTEx Consortium: Human genomics. The
Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene
regulation in humans. Science. 348:648–660. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Research Network.
Electronic address: andrew_aguirre@dfci.harvard.edu; Cancer Genome
Atlas Research Network: Integrated genomic characterization of
pancreatic ductal adenocarcinoma. Cancer Cell. 32:185–203.e13.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bailey P, Chang DK, Nones K, Johns AL,
Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC,
et al: Genomic analyses identify molecular subtypes of pancreatic
cancer. Nature. 531:47–52. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang S, He P, Wang J, Schetter A, Tang W,
Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al: A
novel MIF signaling pathway drives the malignant character of
pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bhattacharya S, Andorf S, Gomes L, Dunn P,
Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J,
et al: ImmPort: Disseminating data to the public for the future of
immunology. Immunol Res. 58:234–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng W, Ren X, Zhang C, Cai J, Liu Y, Han
S and Wu A: Bioinformatic profiling identifies an immune-related
risk signature for glioblastoma. Neurology. 86:2226–2234. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ,
Yan W, Liu YW, Zhang W, Chen L and Jiang T: Prognostic value of a
nine-gene signature in glioma patients based on mRNA expression
profiling. CNS Neurosci Ther. 20:112–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei C, Liang Q, Li X, Li H, Liu Y, Huang
X, Chen X, Guo Y and Li J: Bioinformatics profiling utilized a nine
immune-related long noncoding RNA signature as a prognostic target
for pancreatic cancer. J Cell Biochem. 120:14916–14927. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barbie DA, Tamayo P, Boehm JS, Kim SY,
Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al:
Systematic RNA interference reveals that oncogenic KRAS-driven
cancers require TBK1. Nature. 462:108–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang C, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kota J, Hancock J, Kwon J and Korc M:
Pancreatic cancer: Stroma and its current and emerging targeted
therapies. Cancer Lett. 391:38–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dieterich LC and Bikfalvi A: The tumor
organismal environment: Role in tumor development and cancer
immunotherapy. Semin Cancer Biol. 65:197–206. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang X, Li Y, Li J, Li L, Zhu H, Chen H,
Kong R, Wang G, Wang Y, Hu J and Sun B: Cell-in-cell phenomenon and
its relationship with tumor microenvironment and tumor progression:
A review. Front Cell Dev Biol. 7:3112019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Lazarus J, Steele NG, Yan W, Lee
HJ, Nwosu ZC, Halbrook CJ, Menjivar RE, Kemp SB, Sirihorachai VR,
et al: Regulatory T cell depletion alters the tumor
microenvironment and accelerates pancreatic carcinogenesis. Cancer
Discov. 10:422–439. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sohal DPS, Kennedy EB, Khorana A, Copur
MS, Crane CH, Garrido-Laguna I, Krishnamurthi S, Moravek C,
O'Reilly EM, Philip PA, et al: Metastatic pancreatic cancer: ASCO
clinical practice guideline update. J Clin Oncol. 36:2545–2556.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wartenberg M, Cibin S, Zlobec I, Vassella
E, Eppenberger-Castori S, Terracciano L, Eichmann MD, Worni M,
Gloor B, Perren A and Karamitopoulou E: Integrated genomic and
immunophenotypic classification of pancreatic cancer reveals three
distinct subtypes with prognostic/predictive significance. Clin
Cancer Res. 24:4444–4454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rückert F, Dawelbait G, Winter C, Hartmann
A, Denz A, Ammerpohl O, Schroeder M, Schackert HK, Sipos B, Klöppel
G, et al: Examination of apoptosis signaling in pancreatic cancer
by computational signal transduction analysis. PLoS One.
5:e122432010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Haider S, Wang J, Nagano A, Desai A,
Arumugam P, Dumartin L, Fitzgibbon J, Hagemann T, Marshall JF,
Kocher HM, et al: A multi-gene signature predicts outcome in
patients with pancreatic ductal adenocarcinoma. Genome Med.
6:1052014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang D, Wu Q, Yuan Z, Xu J, Zhang H, Jin
Z, Zhang Q, Xu M, Wang Z, Dai Z, et al: Identification of key
pathways and genes changes in pancreatic cancer cells (BXPC-3)
after cross-talk with primary pancreatic stellate cells using
bioinformatics analysis. Neoplasma. 66:681–693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qian X, Chen Z, Chen SS, Liu LM and Zhang
AQ: Integrated analyses identify immune-related signature
associated with Qingyihuaji formula for treatment of pancreatic
ductal adenocarcinoma using network pharmacology and weighted gene
co-expression network. J Immunol Res. 2020:75036052020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lundberg M, Krogvold L, Kuric E,
Dahl-Jorgensen K and Skog O: Expression of interferon-stimulated
genes in insulitic pancreatic islets of patients recently diagnosed
with type 1 diabetes. Diabetes. 65:3104–3110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Carey CM, Govande AA, Cooper JM, Hartley
MK, Kranzusch PJ and Elde NC: Recurrent loss-of-function mutations
reveal costs to OAS1 antiviral activity in primates. Cell Host
Microbe. 25:336–343.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mandal S, Abebe F and Chaudhary J: 2′-5′
oligoadenylate synthetase 1 polymorphism is associated with
prostate cancer. Cancer. 117:5509–5518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Moerdyk-Schauwecker M, Shah NR, Murphy AM,
Hastie E, Mukherjee P and Grdzelishvili VZ: Resistance of
pancreatic cancer cells to oncolytic vesicular stomatitis virus:
Role of type I interferon signaling. Virology. 436:221–234. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ito M, Shichijo S, Tsuda N, Ochi M,
Harashima N, Saito N and Itoh K: Molecular basis of T cell-mediated
recognition of pancreatic cancer cells. Cancer Res. 61:2038–2046.
2001.PubMed/NCBI
|
|
48
|
Escorcia FE, Houghton JL, Abdel-Atti D,
Pereira PR, Cho A, Gutsche NT, Baidoo KE and Lewis J: ImmunoPET
predicts response to Met-targeted radioligand therapy in models of
pancreatic cancer resistant to met kinase inhibitors. Theranostics.
10:151–165. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan
C, Wu Y, Li X, Li X, Li G, et al: Function of the c-Met receptor
tyrosine kinase in carcinogenesis and associated therapeutic
opportunities. Mol Cancer. 17:452018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yamaoka T, Kusumoto S, Ando K, Ohba M and
Ohmori T: Receptor tyrosine kinase-targeted cancer therapy. Int J
Mol Sci. 19:34912018. View Article : Google Scholar
|
|
51
|
Giovannetti E, van der Borden CL, Frampton
AE, Ali A, Firuzi O and Peters GJ: Never let it go: Stopping key
mechanisms underlying metastasis to fight pancreatic cancer. Semin
Cancer Biol. 44:43–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Moosavi F, Giovannetti E, Saso L and
Firuzi O: HGF/MET pathway aberrations as diagnostic, prognostic,
and predictive biomarkers in human cancers. Crit Rev Clin Lab Sci.
56:533–566. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hu CY, Xu XM, Hong B, Wu ZG, Qian Y, Weng
TH, Liu YZ, Tang TM, Wang MH and Yao HP: Aberrant RON and MET
co-overexpression as novel prognostic biomarkers of shortened
patient survival and therapeutic targets of tyrosine kinase
inhibitors in pancreatic cancer. Front Oncol. 9:13772019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
De Simone M, Arrigoni A, Rossetti G,
Gruarin P, Ranzani V, Politano C, Bonnal RJP, Provasi E, Sarnicola
ML, Panzeri I, et al: Transcriptional landscape of human tissue
lymphocytes unveils uniqueness of tumor-infiltrating T regulatory
cells. Immunity. 45:1135–1147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ritvo PG, Churlaud G, Quiniou V, Florez L,
Brimaud F, Fourcade G, Mariotti-Ferrandiz E and Klatzmann D:
Tfr cells lack IL-2Rα but express decoy IL-1R2 and
IL-1Ra and suppress the IL-1-dependent activation of Tfh
cells. Sci Immunol. 2:eaan03682017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Plitas G, Konopacki C, Wu K, Bos PD,
Morrow M, Putintseva EV, Chudakov DM and Rudensky AY: Regulatory T
cells exhibit distinct features in human breast cancer. Immunity.
45:1122–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Madouri F, Barada O, Kervoaze G, Trottein
F, Pichavant M and Gosset P: Production of interleukin-20 cytokines
limits bacterial clearance and lung inflammation during infection
by Streptococcus pneumoniae. EBioMedicine. 37:417–427. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Omarini C, Bettelli S, Caprera C,
Manfredini S, Caggia F, Guaitoli G, Moscetti L, Toss A, Cortesi L,
Kaleci S, et al: Clinical and molecular predictors of long-term
response in HER2 positive metastatic breast cancer patients. Cancer
Biol Ther. 19:879–886. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Baird AM, Gray SG and O'Byrne KJ: IL-20 is
epigenetically regulated in NSCLC and down regulates the expression
of VEGF. Eur J Cancer. 47:1908–1918. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC,
Li J, Zhang YQ, Shi JW, Lin XL, Yang S, et al: miR-9 modulates the
expression of interferon-regulated genes and MHC class I molecules
in human nasopharyngeal carcinoma cells. Biochem Biophys Res
Commun. 431:610–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Manzo T, Prentice BM, Anderson KG, Raman
A, Schalck A, Codreanu GS, Nava Lauson CB, Tiberti S, Raimondi A,
Jones MA, et al: Accumulation of long-chain fatty acids in the
tumor microenvironment drives dysfunction in intrapancreatic CD8+ T
cells. J Exp Med. 217:e201919202020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hessmann E, Buchholz SM, Demir IE, Singh
SK, Gress TM, Ellenrieder V and Neesse A: Microenvironmental
determinants of pancreatic cancer. Physiol Rev. 100:1707–1751.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Junttila MR and de Sauvage FJ: Influence
of tumour micro-environment heterogeneity on therapeutic response.
Nature. 501:346–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Petitprez F, Vano YA, Becht E, Giraldo NA,
de Reyniès A, Sautès-Fridman C and Fridman WH: Transcriptomic
analysis of the tumor microenvironment to guide prognosis and
immunotherapies. Cancer Immunol Immunother. 67:981–988. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mahajan UM, Langhoff E, Goni E, Costello
E, Greenhalf W, Halloran C, Ormanns S, Kruger S, Boeck S, Ribback
S, et al: Immune cell and stromal signature associated with
progression-free survival of patients with resected pancreatic
ductal adenocarcinoma. Gastroenterology. 155:1625–1639.e2. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Efstathiou JA, Mouw KW, Gibb EA, Liu Y, Wu
CL, Drumm MR, da Costa JB, du Plessis M, Wang NQ, Davicioni E, et
al: Impact of immune and stromal infiltration on outcomes following
bladder-sparing trimodality therapy for muscle-invasive bladder
cancer. Eur Urol. 76:59–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jusakul A, Cutcutache I, Yong CH, Lim JQ,
Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et
al: Whole-genome and epigenomic landscapes of etiologically
distinct subtypes of cholangiocarcinoma. Cancer Discov.
7:1116–1135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rohr-Udilova N, Klinglmüller F,
Schulte-Hermann R, Stift J, Herac M, Salzmann M, Finotello F,
Timelthaler G, Oberhuber G, Pinter M, et al: Deviations of the
immune cell landscape between healthy liver and hepatocellular
carcinoma. Sci Rep. 8:62202018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Xiong Y, Liu L, Xia Y, Qi Y, Chen Y, Chen
L, Zhang P, Kong Y, Qu Y, Wang Z, et al: Tumor infiltrating masT
cells determine oncogenic HIF-2alpha-conferred immune evasion in
clear cell renal cell carcinoma. Cancer Immunol Immunother.
68:731–741. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tahkola K, Mecklin JP, Wirta EV, Ahtiainen
M, Helminen O, Böhm J and Kellokumpu I: High immune cell score
predicts improved survival in pancreatic cancer. Virchows Archiv.
472:653–665. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bense RD, Sotiriou C, Piccart-Gebhart MJ,
Haanen JBAG, van Vugt MATM, de Vries EGE, Schröder CP and Fehrmann
RSN: Relevance of tumor-infiltrating immune cell composition and
functionality for disease outcome in breast cancer. J Natl Cancer
Inst. 109:djw1922017. View Article : Google Scholar
|