Methylation of cytosines, a common epigenetic
modification in eukaryotic cells, serves an important role in a
variety of genetic processes, including gene stability and
expression, chromosome accessibility and inactivation, and
nucleosome positioning (1).
5-methylcytosine (5mC) is produced by DNA methyltransferase
activity and is located in CG dinucleotides in DNA (2). Ten-eleven translocation (TET) family
proteins participate in oxidation reactions of 5mC to
5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine
(5hmC, 5fC, 5caC), which further decrease DNA methylation patterns
(2).
Post-translational modifications (PTMs) of proteins
facilitate immediate responses of cells to intracellular or
extracellular environmental stimuli by modifying functions of
targeted proteins (3). PTMs are
involved in various pathological processes such as proliferation,
apoptosis and migration in tumors (3). O-linked β-N-acetylglucosaminylation
(O-GlcNAcylation) is an atypical, dynamic and reversible PTM
consisting of addition of N-acetyl-D-glucosamine, a unique
non-elongated monosaccharide on proteins (4). Unlike classical glycosylation present
in the endoplasmic reticulum and Golgi apparatus, O-GlcNAcylation
takes place in the cytoplasm, nucleus and mitochondria and is
implicated in a wide range of effects on cellular function and
signaling in metabolic diseases and cancer (5). Compared to complex glycosyltransferase
and glycosidase system of classical glycosylation, O-GlcNAcylation
is only regulated by two enzymes: The glycosyltransferase OGT
(O-GlcNAc transferase) and the glycoside hydrolase OGA
(O-GlcNAcase) (3). Numerous recent
studies indicate a close connection between OGT and TET (5–7). OGT can
catalyze TET to form O-GlcNAcylated TET and can also interact with
TET to form a complex with the ability to further modify chromatin
which participated in regulating embryonic development and cancer
progression (6,7). The present review focuses on the
functional roles of TET family proteins and O-GlcNAcylation in
cancer progression, with focus on the connection between TET
proteins and OGT to clarify the effects of these proteins on cancer
development.
Epigenetic modifications, which include DNA
methylation and histone modifications can alter gene expression but
cannot change the primary sequence of DNA (8) Epigenetic modifications has been proven
to be widely involved in tumor development (9–11). DNA
hypermethylation is observed in myelodysplastic syndromes (MDS),
acute myeloid leukemia (AML), colorectal cancer, hepatocellular
carcinoma and ovarian cancer (12–14). TET
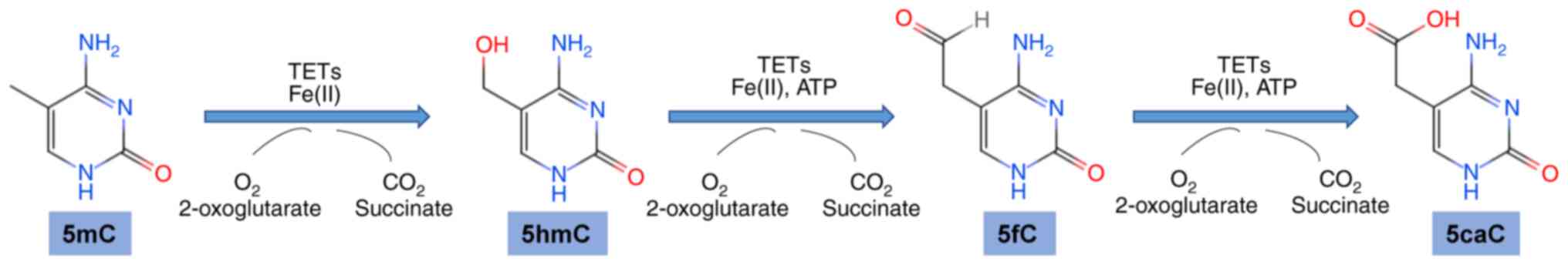
family proteins function as DNA hydroxymethylases in vertebrates
and can catalyze conversion of 5mC to 5hmC, and subsequently to 5fC
and 5caC (15) (Fig. 1). These three versions of oxidized
methylcytosines are all associated with DNA demethylation (16,17).
TET proteins have a common cysteine-rich dioxygenase
region and C-terminal region which binds to ferrous iron and
α-ketoglutarate and catalyzes an oxidation reaction which involves
hydroxylation of 5mC to 5hmC and further to 5fC and 5caC (12). The three TET proteins (TET1, TET2,
TET3) have differing N-terminal regions (18). TET1 and TET3 have a CXXC-type zinc
finger domain (19). TET2 has no
CXXC DNA-binding domain, but can interact with a CXXC domain
protein, inhibitor of disheveled Dvl and Axin complex (IDAX)
(18).
Studies on aberrant expression of the three types of
TET proteins in various types of cancer are summarized in Table I.
O-GlcNAcylation is a reversible PTM that typically
targets proteins in the cytoplasm, cell nuclei (34), or mitochondria (35). It can regulate cellular processes at
various levels, such as transcription, translation, signal
transduction or cell metabolism (36). In general, proteins modified by
O-GlcNAcylation are phosphoproteins or parts of macromolecular
complexes (phosphoglycerate kinase 1), transcription complexes
(p53, c-myc), or nucleopores (transmembrane nucleoporin Pom121,
nucleoporin 155) (37).
O-GlcNAcylation has also been reported for numerous functional
proteins, including epigenetic regulation factors including the TET
proteins, the SIN3 transcription regulator family member A-histone
deacetylases and the Polycomb group proteins that regulate DNA
methylation, chromatin accessibility and chromatin modification
(38).
O-GlcNAcylation often affects subcellular
localization, stability and function of target proteins (7,39), and
in some cases helps modulate protein phosphorylation status,
protein stability, enzymatic activity, protein aggregation and
interactions with other proteins or DNA (5,36,40).
O-GlcNAc activity requires two enzymes: OGT and OGA (41). OGT catalyzes attachment of GlcNAc to
serine (Ser), threonine (Thr) and cysteine residues in proteins
(40,42).
OGT activity is highly sensitive to the uridine
diphosphate GlcNAc level, and is altered by variations of glucose,
glutamate or free fatty acid levels in cells (43). OGT activity is associated with
epithelial-mesenchymal transition (EMT), p53, Wnt and TGF-β
signaling pathways, inflammatory responses and apoptosis in
cervical cancer cells (44).
Knockdown of OGT in colon cells results in upregulation and altered
glycosylation of E-cadherin, an important factor in EMT progression
and may disrupt biosynthesis of glycosphingolipids
(lactosylceramide, gangliosides and globosides), with consequent
reduction of gangliosides (ganglioside 3 and ganglioside 2) but
increase of globosides (globoside 3 and globoside 4) (45). Chronic lymphocytic leukemia (CLL)
cells demonstrate high expression of O-GlcNAcylated proteins,
including p53, c-Myc, and Akt and enhanced protein glycosylation
alters intracellular signaling processes (p53 and PI3K/AKT/mTOR
signaling pathways) in these cells (46). O-GlcNAcylation increases downstream
signaling of toll-like receptors following cytokine stimulation in
CLL cells (46). On the other hand,
high baseline O-GlcNAc levels inhibit responses to such
stimulation, resulting in increased resistance to TLR agonists,
chemotherapeutic agents, B cell receptor crosslinking and mitogens
(46). Hart et al (36) reported that increased O-GlcNAcylation
of Thr58 on c-Myc inhibited c-Myc activity and reduced
transformation of non-Hodgkin's lymphoma cells.
OGA has O-GlcNAc hydrolase and associated enzymatic
activity of lysine acetyltransferase (47–49) and
can therefore hydrolyze O-GlcNAc residues from attached proteins
(41). Inhibition of OGA expression
in rats and mice resulted in increased O-GlcNAcylation of all
tissues (50). OGA shows high mRNA
expression in lung, colon and breast cancers (49). In colon cell lines, O-GlcNAcylation
level was increased by inhibition of OGA but decreased by
inhibition of OGT (51). OGA serves
an essential role in differentiation of ESCs (52,53).
Blocking of O-GlcNAc cycling in mice by OGA knockdown resulted in
anatomical defects and notable changes in expression of
pluripotency markers such as Nanog, Sox2 and Orthodenticle homeobox
2 (53). OGA knockdown in mouse
hematopoietic stem cells reduced progenitor pools, reduced cell
stemness of cells, altered transcription of several crucial genes
such as hypoxia inducible factor-1α and cyclin dependent kinase
inhibitor 1C and increased apoptotic cell number in bone marrow
(54).
TET proteins mediate DNA demethylation, while OGT
mediates protein O-GlcNAcylation (39,55).
These two enzymatic activities may seem to be independent of each
other. However, several recent studies have revealed the physical
and functional interactions between TETs and OGT.
Although TET proteins and OGT have been hot topics
of research in recent years, very limited knowledge of TET function
and TET O-GlcNAcylation in cancer development and progression
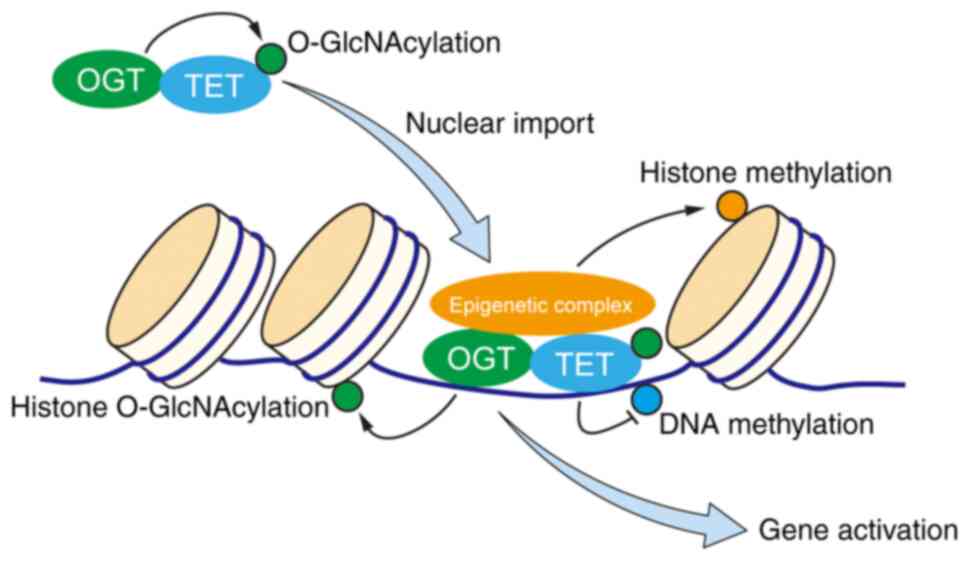
exists. TET/OGT complex contributes to certain epigenetic
modifications, such as DNA demethylation, histone O-GlcNAcylation,
histone methylation associated with positive regulation of gene
expression, hence providing a direct link between epigenetics and
cellular metabolism (62). Hsu et
al (64) reported that TET1
demonstrated reduced expression in prostate and breast cancers, and
suppresses cancer cell invasion by promoting expression of tissue
inhibitors of metalloproteinases. In a study of cervical cancer
cells, Guan et al (65)
observed that nuclear localization and O-GlcNAcylation of TET3 were
modulated by glucose metabolism, and that gene expression was
regulated through TET/OGT-mediated epigenetic changes in response
to nutrient availability. The role of O-GlcNAcylated TET proteins
in cancer progression is an exciting topic for future study. Based
on current finding in the field, a working model of the role of
TET/OGT complex in regulation of target proteins during cancer
development may be proposed (Fig.
2).
The TET proteins (TET1, TET2, TET3) catalyze
conversion of 5mC to 5hmC. O-GlcNAcylation is a reversible PTM and
it can O-GlcNAcylate TETs (59). OGT
and O-GlcNAcylation have been clearly demonstrated to serve an
important role in tumorigenesis and metastasis (66). TET proteins can recruit the OGT to
chromatin, which promotes post-transcriptional modifications of
histones and facilitates gene expression (40). It was reported that TET2 mediates OGT
modification on H2B Ser112 and is associated with highly
transcribed genes (62). In
addition, TET/OGT complex can serve as the scaffold for epigenetic
complexes (7,63).
However, the present review had some limitations,
such as the research of TET/OGT complex mainly focused on the
function during embryonic development (6,7,55). The role of TETs and their
O-GlcNAcylation in cancer development is largely unknown (60). The essential characteristic of cancer
is uncontrolled cell proliferation resulting from accumulated
alterations of cell metabolism and signaling pathways (67). One trait of cancer initiation is the
dynamics of O-GlcNAcylation are highly sensitive to availability of
nutrients and oxygen, determined by the cellular microenvironment
(68). Aberrant glucose metabolism
in cancer cells may alter O-GlcNAcylation of TET proteins and
therefore affect their stability; conversely, TET loss-of function
in cancer may influence the nuclear and/or cytoplasmic distribution
of OGT, which in turn may affect the stability of tumor suppressors
and oncogenes such as p53 (69), MYC
(70), and β-catenin (71). The dysregulated expression and
loss-of-function mutation of TET family proteins participated in
the progress of a variety of cancers especially hematopoietic
malignancies (29). Hence, it is
logical to raise the question about whether TET/OGT is involved in
cancer development and how they get involved. TETs can be
post-translationally modified by the nutrient-sensing enzyme OGT,
also suggesting a connection between metabolism and the epigenome
(6,62). In addition to suggesting a broader
role for the TET/OGT complex, the present review provides
information about the interaction between OGT and TET proteins,
which may provide new insights into the development of cancer.
TET proteins can interact with and undergo
O-GlcNAcylation by OGT and O-GlcNAcylation can alter properties of
TET enzymes (62). TET/OGT complexes
are primarily targeted to promoter regions through interaction of
TET with DNA, and TET-linked OGT can O-GlcNAcylate a wide variety
of proteins (58). Relationships
between OGT and TETs during cancer pathological processes remain to
be elucidated. Identification of modified proteins present upstream
and downstream of TET/OGT complex will be useful in this regard.
The functions of TETs and their O-GlcNAcylation in cancer
development is an important topic for future studies.
Not applicable.
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81470294, 81770123
and 32071274), National Science and Technology Major Project of
China (grant no. 2018ZX10302205), 13115 Key Projects of Scientific
and Technical Innovation of Shaanxi Province (grant no.
2010ZDKG-53), Natural Science Foundation of Shaanxi Province (grant
no. 2018JM3014), Youth Innovation Team of Shaanxi Universities, and
Hundred-Talent Program of Shaanxi Province.
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
XL, XCP, HJL, and BXL designed the study and
co-wrote the manuscript. YW, FG, and YY were involved in study
conception and design, and revised the manuscript for important
intellectual content. All authors have read and approved the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Schubeler D: Function and information
content of DNA methylation. Nature. 517:321–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scott-Browne JP, Lio CJ and Rao A: TET
proteins in natural and induced differentiation. Curr Opin Genet
Dev. 46:202–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang X and Qian K: Protein
O-GlcNAcylation: Emerging mechanisms and functions. Nat Rev Mol
Cell Biol. 18:452–465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wells L, Vosseller K and Hart GW:
Glycosylation of nucleocytoplasmic proteins: Signal transduction
and O-GlcNAc. Science. 291:2376–2378. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hart GW, Housley MP and Slawson C: Cycling
of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins.
Nature. 446:1017–1022. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vella P, Scelfo A, Jammula S, Chiacchiera
F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin
K and Pasini D: Tet proteins connect the O-linked
N-acetylglucosamine transferase Ogt to chromatin in embryonic stem
cells. Mol Cell. 49:645–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hrit J, Goodrich L, Li C, Wang BA, Nie J,
Cui X, Martin EA, Simental E, Fernandez J, Liu MY, et al: OGT binds
a conserved C-terminal domain of TET1 to regulate TET1 activity and
function in development. Elife. 7:e348702018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darılmaz Yüce G and Ortaç Ersoy E: Lung
cancer and epigenetic modifications. Tuberk Toraks. 64:163–170.
2016.(In Turkish). View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sasanakietkul T, Murtha TD, Javid M, Korah
R and Carling T: Epigenetic modifications in poorly differentiated
and anaplastic thyroid cancer. Mol Cell Endocrinol. 469:23–37.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alam R, Abdolmaleky HM and Zhou JR:
Microbiome, inflammation, epigenetic alterations, and mental
diseases. Am J Med Genet B Neuropsychiatr Genet. 174:651–660. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ciesielski P, Jozwiak P and Krzeslak A:
TET proteins and epigenetic modifications in cancers. Postepy Hig
Med Dosw (Online). 69:1371–1383. 2015.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li D and Zeng Z: Epigenetic regulation of
histone H3 in the process of hepatocellular tumorigenesis. Biosci
Rep. 39:BSR201918152019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Losi L, Lauriola A, Tazzioli E, Gozzi G,
Scurani L, D'Arca D and Benhattar J: Involvement of epigenetic
modification of TERT promoter in response to all-trans retinoic
acid in ovarian cancer cell lines. J Ovarian Res. 12:622019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L and
Rao A: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song J, Moscinski L, Zhang H, Zhang X and
Hussaini M: Does SF3B1/TET2 double mutation portend better or worse
prognosis Than Isolated SF3B1 or TET2 Mutation? Cancer Genomics
Proteomics. 16:91–98. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen L, Wu H, Diep D, Yamaguchi S,
D'Alessio AC, Fung HL, Zhang K and Zhang Y: Genome-wide analysis
reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics.
Cell. 153:692–706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ko M, An J, Bandukwala HS, Chavez L, Aijö
T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al:
Modulation of TET2 expression and 5-methylcytosine oxidation by the
CXXC domain protein IDAX. Nature. 497:122–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Good CR, Madzo J, Patel B, Maegawa S,
Engel N, Jelinek J and Issa JJ: A novel isoform of TET1 that lacks
a CXXC domain is overexpressed in cancer. Nucleic Acids Res.
45:8269–8281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh KP, Yabuuchi A, Rao S, Huang Y,
Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky
G, et al: Tet1 and Tet2 regulate 5-hydroxymethylcytosine production
and cell lineage specification in mouse embryonic stem cells. Cell
Stem Cell. 8:200–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dawlaty MM, Breiling A, Le T, Barrasa MI,
Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F and
Jaenisch R: Loss of Tet enzymes compromises proper differentiation
of embryonic stem cells. Dev Cell. 29:102–111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W,
Xie ZG, Shi L, He X, Jin SG, et al: The role of Tet3 DNA
dioxygenase in epigenetic reprogramming by oocytes. Nature.
477:606–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ito S, Shen L, Dai Q, Wu SC, Collins LB,
Swenberg JA, He C and Zhang Y: Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Science. 333:1300–1303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng J, Guo S, Chen S, Mastriano SJ, Liu
C, D'Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al: An
extensive network of TET2-targeting microRNAs regulates malignant
hematopoiesis. Cell Rep. 5:471–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu
J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al: Tumor development is
associated with decrease of TET gene expression and
5-methylcytosine hydroxylation. Oncogene. 32:663–669. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haferlach T, Nagata Y, Grossmann V, Okuno
Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T,
et al: Landscape of genetic lesions in 944 patients with
myelodysplastic syndromes. Leukemia. 28:241–247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fernandez-Mercado M, Yip BH, Pellagatti A,
Davies C, Larrayoz MJ, Kondo T, Pérez C, Killick S, McDonald EJ,
Odero MD, et al: Mutation patterns of 16 genes in primary and
secondary acute myeloid leukemia (AML) with normal cytogenetics.
PLoS One. 7:e423342012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shih AH, Abdel-Wahab O, Patel JP and
Levine RL: The role of mutations in epigenetic regulators in
myeloid malignancies. Nat Rev Cancer. 12:599–612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li R, Zhou Y, Cao Z, Liu L, Wang J, Chen
Z, Xing W, Chen S, Bai J, Yuan W, et al: TET2 loss dysregulates the
behavior of bone marrow mesenchymal stromal cells and accelerates
Tet2−/−Driven myeloid malignancy progression. Stem Cell
Reports. 10:166–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Cai X, Cai CL, Wang J, Zhang W,
Petersen BE, Yang FC and Xu M: Deletion of Tet2 in mice leads to
dysregulated hematopoietic stem cells and subsequent development of
myeloid malignancies. Blood. 118:4509–4518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Ozark PA, Smith ER, Zhao Z,
Marshall SA, Rendleman EJ, Piunti A, Ryan C, Whelan AL, Helmin KA,
et al: TET2 coactivates gene expression through demethylation of
enhancers. Sci Adv. 4:eaau69862018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan W, Zhu S, Qu K, Meeth K, Cheng J, He
K, Ma H, Liao Y, Wen X, Roden C, et al: The DNA Methylcytosine
Dioxygenase Tet2 sustains immunosuppressive function of
Tumor-infiltrating myeloid cells to promote melanoma progression.
Immunity. 47:284–297.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Itoh H, Kadomatsu T, Tanoue H, Yugami M,
Miyata K, Endo M, Morinaga J, Kobayashi E, Miyamoto T, Kurahashi R,
et al: TET2-dependent IL-6 induction mediated by the tumor
microenvironment promotes tumor metastasis in osteosarcoma.
Oncogene. 37:2903–2920. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Levine ZG and Walker S: The Biochemistry
of O-GlcNAc Transferase: Which functions make it essential in
mammalian cells? Annu Rev Biochem. 85:631–657. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma J, Banerjee P, Whelan SA, Liu T, Wei
AC, Ramirez-Correa G, McComb ME, Costello CE, O'Rourke B, Murphy A
and Hart GW: Comparative proteomics reveals dysregulated
mitochondrial O-GlcNAcylation in diabetic hearts. J Proteome Res.
15:2254–2264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hart GW, Slawson C, Ramirez-Correa G and
Lagerlof O: Cross talk between O-GlcNAcylation and phosphorylation:
Roles in signaling, transcription, and chronic disease. Annu Rev
Biochem. 80:825–858. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Love DC and Hanover JA: The hexosamine
signaling pathway: Deciphering the ‘O-GlcNAc code’. Sci STKE.
2005:re132005.PubMed/NCBI
|
|
38
|
Gambetta MC and Muller J: A critical
perspective of the diverse roles of O-GlcNAc transferase in
chromatin. Chromosoma. 124:429–442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bond MR and Hanover JA: O-GlcNAc cycling:
A link between metabolism and chronic disease. Annu Rev Nutr.
33:205–229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hanover JA, Krause MW and Love DC:
Bittersweet memories: Linking metabolism to epigenetics through
O-GlcNAcylation. Nat Rev Mol Cell Biol. 13:312–321. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mulloy B, Dell A, Stanley P and James HP:
Structural analysis of glycans. In: Essentials of Glycobiology 3rd.
Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, et al: Cold
Spring Harbor; NY: pp. 639–652. 2015, PubMed/NCBI
|
|
42
|
Maynard JC, Burlingame AL and
Medzihradszky KF: Cysteine S-linked N-acetylglucosamine
(S-GlcNAcylation), A new post-translational modification in
mammals. Mol Cell Proteomics. 15:3405–3411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Berthier A, Vinod M, Porez G, Steenackers
A, Alexandre J, Yamakawa N, Gheeraert C, Ploton M, Maréchal X,
Dubois-Chevalier J, et al: Combinatorial regulation of hepatic
cytoplasmic signaling and nuclear transcriptional events by the
OGT/REV-ERBα complex. Proc Natl Acad Sci USA. 115:E11033–E11042.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao J, Yang Y, Qiu R, Zhang K, Teng X, Liu
R and Wang Y: Proteomic analysis of the OGT interactome: Novel
links to epithelial-mesenchymal transition and metastasis of
cervical cancer. Carcinogenesis. 39:1222–1234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Biwi J, Clarisse C, Biot C, Kozak RP,
Madunic K, Mortuaire M, Wuhrer M, Spencer DIR, Schulz C, Guerardel
Y, et al: OGT Controls the expression and the glycosylation of
E-cadherin, and affects glycosphingolipid structures in human colon
cell lines. Proteomics. 19:e18004522019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi Y, Tomic J, Wen F, Shaha S, Bahlo A,
Harrison R, Dennis JW, Williams R, Gross BJ, Walker S, et al:
Aberrant O-GlcNAcylation characterizes chronic lymphocytic
leukemia. Leukemia. 24:1588–1598. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hayakawa K, Hirosawa M, Tabei Y, Arai D,
Tanaka S, Murakami N, Yagi S and Shiota K: Epigenetic switching by
the metabolism- sensing factors in the generation of orexin neurons
from mouse embryonic stem cells. J Biol Chem. 288:17099–17110.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Toleman C, Paterson AJ, Whisenhunt TR and
Kudlow JE: Characterization of the histone acetyltransferase (HAT)
domain of a bifunctional protein with activable O-GlcNAcase and HAT
activities. J Biol Chem. 279:53665–53673. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh JP, Qian K, Lee JS, Zhou J, Han X,
Zhang B, Ong Q, Ni W, Jiang M, Ruan HB, et al: O-GlcNAcase targets
pyruvate kinase M2 to regulate tumor growth. Oncogene. 39:560–573.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Macauley MS, Shan X, Yuzwa SA, Gloster TM
and Vocadlo DJ: Elevation of Global O-GlcNAc in rodents using a
selective O-GlcNAcase inhibitor does not cause insulin resistance
or perturb glucohomeostasis. Chem Biol. 17:949–958. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fuentes-García G, Castañeda-Patlan MC,
Vercoutter-Edouart AS, Lefebvre T and Robles-Flores M:
O-GlcNAcylation Is Involved in the regulation of stem cell markers
expression in colon cancer cells. Front Endocrinol (Lausanne).
10:2892019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jang H, Kim TW, Yoon S, Choi SY, Kang TW,
Kim SY, Kwon YW, Cho EJ and Youn HD: O-GlcNAc regulates
pluripotency and reprogramming by directly acting on core
components of the pluripotency network. Cell Stem Cell. 11:62–74.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Olivier-Van Stichelen S, Wang P, Comly M,
Love DC and Hanover JA: Nutrient-driven O-linked
N-acetylglucosamine (O-GlcNAc) cycling impacts neurodevelopmental
timing and metabolism. J Biol Chem. 292:6076–6085. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Abramowitz LK, Harly C, Das A, Bhandoola A
and Hanover JA: Blocked O-GlcNAc cycling disrupts mouse
hematopoeitic stem cell maintenance and early T cell development.
Sci Rep. 9:125692019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Delatte B and Fuks F: TET proteins: On the
frenetic hunt for new cytosine modifications. Brief Funct Genomics.
12:191–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ito R, Katsura S, Shimada H, Tsuchiya H,
Hada M, Okumura T, Sugawara A and Yokoyama A: TET3-OGT interaction
increases the stability and the presence of OGT in chromatin. Genes
Cells. 19:52–65. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shi FT, Kim H, Lu W, He Q, Liu D, Goodell
MA, Wan M and Songyang Z: Ten-eleven translocation 1 (Tet1) is
regulated by O-linked N-acetylglucosamine transferase (Ogt) for
target gene repression in mouse embryonic stem cells. J Biol Chem.
288:20776–20784. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang Q, Liu X, Gao W, Li P, Hou J, Li J
and Wong J: Differential regulation of the ten-eleven translocation
(TET) family of dioxygenases by O-linked β-N-acetylglucosamine
transferase (OGT). J Biol Chem. 289:5986–5996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bauer C, Gobel K, Nagaraj N, Colantuoni C,
Wang M, Müller U, Kremmer E, Rottach A and Leonhardt H:
Phosphorylation of TET proteins is regulated via O-GlcNAcylation by
the O-linked N-acetylglucosamine transferase (OGT). J Biol Chem.
290:4801–4812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Singh JP, Zhang K, Wu J and Yang X:
O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer
Lett. 356:244–250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fujiki R, Hashiba W, Sekine H, Yokoyama A,
Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, et al:
GlcNAcylation of histone H2B facilitates its monoubiquitination.
Nature. 480:557–560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen Q, Chen Y, Bian C, Fujiki R and Yu X:
TET2 promotes histone O-GlcNAcylation during gene transcription.
Nature. 493:561–564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Deplus R, Delatte B, Schwinn MK, Defrance
M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E,
et al: TET2 and TET3 regulate GlcNAcylation and H3K4 methylation
through OGT and SET1/COMPASS. EMBO J. 32:645–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hsu CH, Peng KL, Kang ML, Chen YR, Yang
YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al: TET1
suppresses cancer invasion by activating the tissue inhibitors of
metalloproteinases. Cell Rep. 2:568–579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guan W, Guyot R, Samarut J, Flamant F,
Wong J and Gauthier KC: Methylcytosine dioxygenase TET3 interacts
with thyroid hormone nuclear receptors and stabilizes their
association to chromatin. Proc Natl Acad Sci USA. 114:8229–8234.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Phoomak C, Silsirivanit A, Park D,
Sawanyawisuth K, Vaeteewoottacharn K, Wongkham C, Lam EW, Pairojkul
C, Lebrilla CB and Wongkham S: O-GlcNAcylation mediates metastasis
of cholangiocarcinoma through FOXO3 and MAN1A1. Oncogene.
37:5648–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ma Z and Vosseller K: Cancer metabolism
and elevated O-GlcNAc in oncogenic signaling. J Biol Chem.
289:34457–34465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yang WH, Kim JE, Nam HW, Ju JW, Kim HS,
Kim YS and Cho JW: Modification of p53 with O-linked
N-acetylglucosamine regulates p53 activity and stability. Nat Cell
Biol. 8:1074–1083. 2026. View Article : Google Scholar
|
|
70
|
Itkonen HM, Minner S, Guldvik IJ, Sandmann
MJ, Tsourlakis MC, Berge V, Svindland A, Schlomm T and Mills IG:
O-GlcNAc transferase integrates metabolic pathways to regulate the
stability of c-MYC in human prostate cancer cells. Cancer Res.
73:5277–5287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Olivier-Van Stichelen S, Guinez C, Mir AM,
Perez-Cervera Y, Liu C, Michalski JC and Lefebvre T: The hexosamine
biosynthetic pathway and O-GlcNAcylation drive the expression of
β-catenin and cell proliferation. Am J Physiol Endocrinol Metab.
302:E417–E424. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Thomson JP, Ottaviano R, Unterberger EB,
Lempiäinen H, Muller A, Terranova R, Illingworth RS, Webb S, Kerr
AR, Lyall MJ, et al: Loss of Tet1-Associated
5-hydroxymethylcytosine is concomitant with aberrant promoter
hypermethylation in liver cancer. Cancer Res. 76:3097–3108. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Delhommeau F, Dupont S, Della Valle V,
James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F,
Alberdi A, et al: Mutation in TET2 in myeloid cancers. N Engl J
Med. 360:2289–2301. 2029. View Article : Google Scholar
|
|
74
|
Itzykson R, Kosmider O, Renneville A,
Gelsi-Boyer V, Meggendorfer M, Morabito M, Berthon C, Adès L,
Fenaux P, Beyne-Rauzy O, et al: Prognostic score including gene
mutations in chronic myelomonocytic leukemia. J Clin Oncol.
31:2428–2436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Nibourel O, Kosmider O, Cheok M, Boissel
N, Renneville A, Philippe N, Dombret H, Dreyfus F, Quesnel B,
Geffroy S, et al: Incidence and prognostic value of TET2
alterations in de novo acute myeloid leukemia achieving complete
remission. Blood. 116:1132–1135. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dominguez PM, Ghamlouch H, Rosikiewicz W,
Kumar P, Béguelin W, Fontán L, Rivas MA, Pawlikowska P, Armand M,
Mouly E, et al: TET2 deficiency causes germinal center hyperplasia,
impairs plasma cell differentiation, and promotes B-cell
lymphomagenesis. Cancer Discov. 8:1632–1653. 2018.PubMed/NCBI
|
|
77
|
Cao T, Pan W, Sun X and Shen H: Increased
expression of TET3 predicts unfavorable prognosis in patients with
ovarian cancer-a bioinformatics integrative analysis. J Ovarian
Res. 12:1012019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Carella A, Tejedor JR, García MG,
Urdinguio RG, Bayón GF, Sierra M, López V, García-Toraño E,
Santamarina-Ojeda P, Pérez RF, et al: Epigenetic downregulation of
TET3 reduces genome-wide 5hmC levels and promotes glioblastoma
tumorigenesis. Int J Cancer. 146:373–387. 2020. View Article : Google Scholar : PubMed/NCBI
|