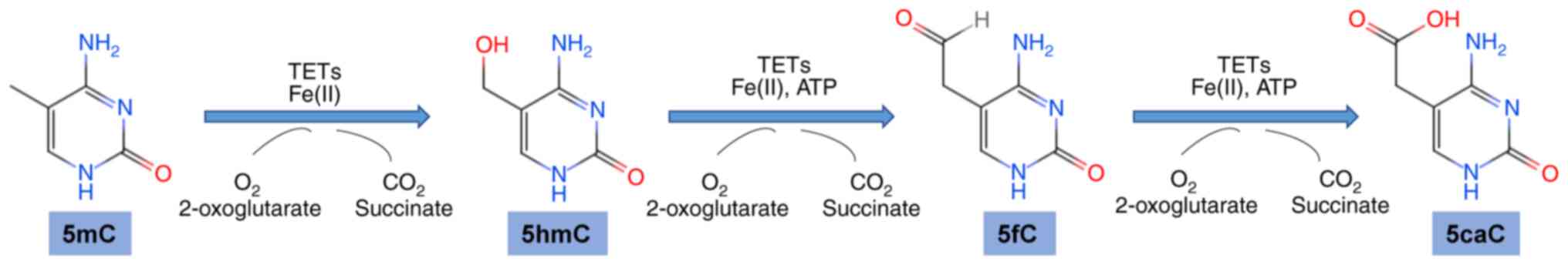
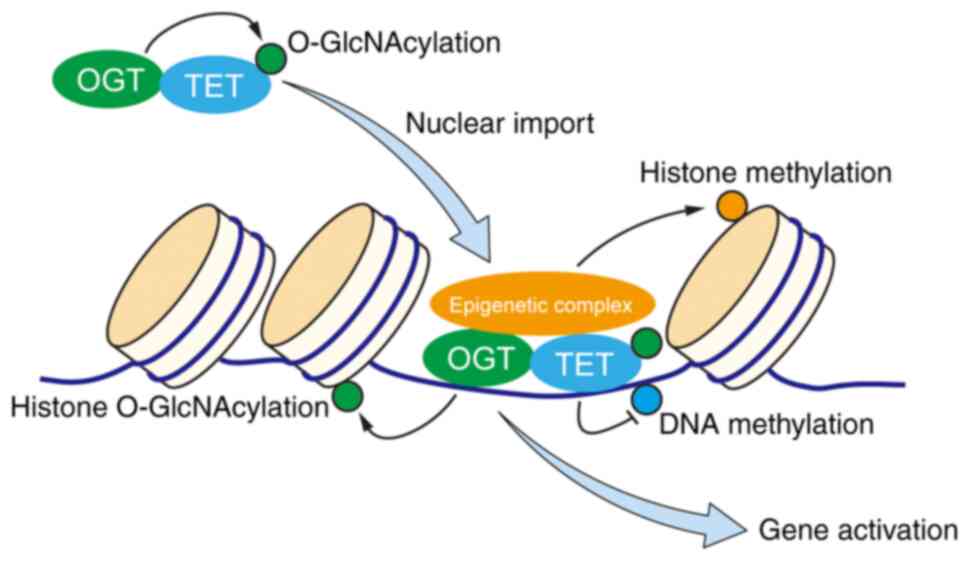
|
1
|
Schubeler D: Function and information
content of DNA methylation. Nature. 517:321–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scott-Browne JP, Lio CJ and Rao A: TET
proteins in natural and induced differentiation. Curr Opin Genet
Dev. 46:202–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang X and Qian K: Protein
O-GlcNAcylation: Emerging mechanisms and functions. Nat Rev Mol
Cell Biol. 18:452–465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wells L, Vosseller K and Hart GW:
Glycosylation of nucleocytoplasmic proteins: Signal transduction
and O-GlcNAc. Science. 291:2376–2378. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hart GW, Housley MP and Slawson C: Cycling
of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins.
Nature. 446:1017–1022. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vella P, Scelfo A, Jammula S, Chiacchiera
F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin
K and Pasini D: Tet proteins connect the O-linked
N-acetylglucosamine transferase Ogt to chromatin in embryonic stem
cells. Mol Cell. 49:645–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hrit J, Goodrich L, Li C, Wang BA, Nie J,
Cui X, Martin EA, Simental E, Fernandez J, Liu MY, et al: OGT binds
a conserved C-terminal domain of TET1 to regulate TET1 activity and
function in development. Elife. 7:e348702018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darılmaz Yüce G and Ortaç Ersoy E: Lung
cancer and epigenetic modifications. Tuberk Toraks. 64:163–170.
2016.(In Turkish). View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sasanakietkul T, Murtha TD, Javid M, Korah
R and Carling T: Epigenetic modifications in poorly differentiated
and anaplastic thyroid cancer. Mol Cell Endocrinol. 469:23–37.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alam R, Abdolmaleky HM and Zhou JR:
Microbiome, inflammation, epigenetic alterations, and mental
diseases. Am J Med Genet B Neuropsychiatr Genet. 174:651–660. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ciesielski P, Jozwiak P and Krzeslak A:
TET proteins and epigenetic modifications in cancers. Postepy Hig
Med Dosw (Online). 69:1371–1383. 2015.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li D and Zeng Z: Epigenetic regulation of
histone H3 in the process of hepatocellular tumorigenesis. Biosci
Rep. 39:BSR201918152019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Losi L, Lauriola A, Tazzioli E, Gozzi G,
Scurani L, D'Arca D and Benhattar J: Involvement of epigenetic
modification of TERT promoter in response to all-trans retinoic
acid in ovarian cancer cell lines. J Ovarian Res. 12:622019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L and
Rao A: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song J, Moscinski L, Zhang H, Zhang X and
Hussaini M: Does SF3B1/TET2 double mutation portend better or worse
prognosis Than Isolated SF3B1 or TET2 Mutation? Cancer Genomics
Proteomics. 16:91–98. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen L, Wu H, Diep D, Yamaguchi S,
D'Alessio AC, Fung HL, Zhang K and Zhang Y: Genome-wide analysis
reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics.
Cell. 153:692–706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ko M, An J, Bandukwala HS, Chavez L, Aijö
T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al:
Modulation of TET2 expression and 5-methylcytosine oxidation by the
CXXC domain protein IDAX. Nature. 497:122–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Good CR, Madzo J, Patel B, Maegawa S,
Engel N, Jelinek J and Issa JJ: A novel isoform of TET1 that lacks
a CXXC domain is overexpressed in cancer. Nucleic Acids Res.
45:8269–8281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh KP, Yabuuchi A, Rao S, Huang Y,
Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky
G, et al: Tet1 and Tet2 regulate 5-hydroxymethylcytosine production
and cell lineage specification in mouse embryonic stem cells. Cell
Stem Cell. 8:200–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dawlaty MM, Breiling A, Le T, Barrasa MI,
Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F and
Jaenisch R: Loss of Tet enzymes compromises proper differentiation
of embryonic stem cells. Dev Cell. 29:102–111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W,
Xie ZG, Shi L, He X, Jin SG, et al: The role of Tet3 DNA
dioxygenase in epigenetic reprogramming by oocytes. Nature.
477:606–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ito S, Shen L, Dai Q, Wu SC, Collins LB,
Swenberg JA, He C and Zhang Y: Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Science. 333:1300–1303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng J, Guo S, Chen S, Mastriano SJ, Liu
C, D'Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al: An
extensive network of TET2-targeting microRNAs regulates malignant
hematopoiesis. Cell Rep. 5:471–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu
J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al: Tumor development is
associated with decrease of TET gene expression and
5-methylcytosine hydroxylation. Oncogene. 32:663–669. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haferlach T, Nagata Y, Grossmann V, Okuno
Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T,
et al: Landscape of genetic lesions in 944 patients with
myelodysplastic syndromes. Leukemia. 28:241–247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fernandez-Mercado M, Yip BH, Pellagatti A,
Davies C, Larrayoz MJ, Kondo T, Pérez C, Killick S, McDonald EJ,
Odero MD, et al: Mutation patterns of 16 genes in primary and
secondary acute myeloid leukemia (AML) with normal cytogenetics.
PLoS One. 7:e423342012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shih AH, Abdel-Wahab O, Patel JP and
Levine RL: The role of mutations in epigenetic regulators in
myeloid malignancies. Nat Rev Cancer. 12:599–612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li R, Zhou Y, Cao Z, Liu L, Wang J, Chen
Z, Xing W, Chen S, Bai J, Yuan W, et al: TET2 loss dysregulates the
behavior of bone marrow mesenchymal stromal cells and accelerates
Tet2−/−Driven myeloid malignancy progression. Stem Cell
Reports. 10:166–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Cai X, Cai CL, Wang J, Zhang W,
Petersen BE, Yang FC and Xu M: Deletion of Tet2 in mice leads to
dysregulated hematopoietic stem cells and subsequent development of
myeloid malignancies. Blood. 118:4509–4518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Ozark PA, Smith ER, Zhao Z,
Marshall SA, Rendleman EJ, Piunti A, Ryan C, Whelan AL, Helmin KA,
et al: TET2 coactivates gene expression through demethylation of
enhancers. Sci Adv. 4:eaau69862018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan W, Zhu S, Qu K, Meeth K, Cheng J, He
K, Ma H, Liao Y, Wen X, Roden C, et al: The DNA Methylcytosine
Dioxygenase Tet2 sustains immunosuppressive function of
Tumor-infiltrating myeloid cells to promote melanoma progression.
Immunity. 47:284–297.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Itoh H, Kadomatsu T, Tanoue H, Yugami M,
Miyata K, Endo M, Morinaga J, Kobayashi E, Miyamoto T, Kurahashi R,
et al: TET2-dependent IL-6 induction mediated by the tumor
microenvironment promotes tumor metastasis in osteosarcoma.
Oncogene. 37:2903–2920. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Levine ZG and Walker S: The Biochemistry
of O-GlcNAc Transferase: Which functions make it essential in
mammalian cells? Annu Rev Biochem. 85:631–657. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma J, Banerjee P, Whelan SA, Liu T, Wei
AC, Ramirez-Correa G, McComb ME, Costello CE, O'Rourke B, Murphy A
and Hart GW: Comparative proteomics reveals dysregulated
mitochondrial O-GlcNAcylation in diabetic hearts. J Proteome Res.
15:2254–2264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hart GW, Slawson C, Ramirez-Correa G and
Lagerlof O: Cross talk between O-GlcNAcylation and phosphorylation:
Roles in signaling, transcription, and chronic disease. Annu Rev
Biochem. 80:825–858. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Love DC and Hanover JA: The hexosamine
signaling pathway: Deciphering the ‘O-GlcNAc code’. Sci STKE.
2005:re132005.PubMed/NCBI
|
|
38
|
Gambetta MC and Muller J: A critical
perspective of the diverse roles of O-GlcNAc transferase in
chromatin. Chromosoma. 124:429–442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bond MR and Hanover JA: O-GlcNAc cycling:
A link between metabolism and chronic disease. Annu Rev Nutr.
33:205–229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hanover JA, Krause MW and Love DC:
Bittersweet memories: Linking metabolism to epigenetics through
O-GlcNAcylation. Nat Rev Mol Cell Biol. 13:312–321. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mulloy B, Dell A, Stanley P and James HP:
Structural analysis of glycans. In: Essentials of Glycobiology 3rd.
Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, et al: Cold
Spring Harbor; NY: pp. 639–652. 2015, PubMed/NCBI
|
|
42
|
Maynard JC, Burlingame AL and
Medzihradszky KF: Cysteine S-linked N-acetylglucosamine
(S-GlcNAcylation), A new post-translational modification in
mammals. Mol Cell Proteomics. 15:3405–3411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Berthier A, Vinod M, Porez G, Steenackers
A, Alexandre J, Yamakawa N, Gheeraert C, Ploton M, Maréchal X,
Dubois-Chevalier J, et al: Combinatorial regulation of hepatic
cytoplasmic signaling and nuclear transcriptional events by the
OGT/REV-ERBα complex. Proc Natl Acad Sci USA. 115:E11033–E11042.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao J, Yang Y, Qiu R, Zhang K, Teng X, Liu
R and Wang Y: Proteomic analysis of the OGT interactome: Novel
links to epithelial-mesenchymal transition and metastasis of
cervical cancer. Carcinogenesis. 39:1222–1234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Biwi J, Clarisse C, Biot C, Kozak RP,
Madunic K, Mortuaire M, Wuhrer M, Spencer DIR, Schulz C, Guerardel
Y, et al: OGT Controls the expression and the glycosylation of
E-cadherin, and affects glycosphingolipid structures in human colon
cell lines. Proteomics. 19:e18004522019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi Y, Tomic J, Wen F, Shaha S, Bahlo A,
Harrison R, Dennis JW, Williams R, Gross BJ, Walker S, et al:
Aberrant O-GlcNAcylation characterizes chronic lymphocytic
leukemia. Leukemia. 24:1588–1598. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hayakawa K, Hirosawa M, Tabei Y, Arai D,
Tanaka S, Murakami N, Yagi S and Shiota K: Epigenetic switching by
the metabolism- sensing factors in the generation of orexin neurons
from mouse embryonic stem cells. J Biol Chem. 288:17099–17110.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Toleman C, Paterson AJ, Whisenhunt TR and
Kudlow JE: Characterization of the histone acetyltransferase (HAT)
domain of a bifunctional protein with activable O-GlcNAcase and HAT
activities. J Biol Chem. 279:53665–53673. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh JP, Qian K, Lee JS, Zhou J, Han X,
Zhang B, Ong Q, Ni W, Jiang M, Ruan HB, et al: O-GlcNAcase targets
pyruvate kinase M2 to regulate tumor growth. Oncogene. 39:560–573.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Macauley MS, Shan X, Yuzwa SA, Gloster TM
and Vocadlo DJ: Elevation of Global O-GlcNAc in rodents using a
selective O-GlcNAcase inhibitor does not cause insulin resistance
or perturb glucohomeostasis. Chem Biol. 17:949–958. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fuentes-García G, Castañeda-Patlan MC,
Vercoutter-Edouart AS, Lefebvre T and Robles-Flores M:
O-GlcNAcylation Is Involved in the regulation of stem cell markers
expression in colon cancer cells. Front Endocrinol (Lausanne).
10:2892019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jang H, Kim TW, Yoon S, Choi SY, Kang TW,
Kim SY, Kwon YW, Cho EJ and Youn HD: O-GlcNAc regulates
pluripotency and reprogramming by directly acting on core
components of the pluripotency network. Cell Stem Cell. 11:62–74.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Olivier-Van Stichelen S, Wang P, Comly M,
Love DC and Hanover JA: Nutrient-driven O-linked
N-acetylglucosamine (O-GlcNAc) cycling impacts neurodevelopmental
timing and metabolism. J Biol Chem. 292:6076–6085. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Abramowitz LK, Harly C, Das A, Bhandoola A
and Hanover JA: Blocked O-GlcNAc cycling disrupts mouse
hematopoeitic stem cell maintenance and early T cell development.
Sci Rep. 9:125692019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Delatte B and Fuks F: TET proteins: On the
frenetic hunt for new cytosine modifications. Brief Funct Genomics.
12:191–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ito R, Katsura S, Shimada H, Tsuchiya H,
Hada M, Okumura T, Sugawara A and Yokoyama A: TET3-OGT interaction
increases the stability and the presence of OGT in chromatin. Genes
Cells. 19:52–65. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shi FT, Kim H, Lu W, He Q, Liu D, Goodell
MA, Wan M and Songyang Z: Ten-eleven translocation 1 (Tet1) is
regulated by O-linked N-acetylglucosamine transferase (Ogt) for
target gene repression in mouse embryonic stem cells. J Biol Chem.
288:20776–20784. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang Q, Liu X, Gao W, Li P, Hou J, Li J
and Wong J: Differential regulation of the ten-eleven translocation
(TET) family of dioxygenases by O-linked β-N-acetylglucosamine
transferase (OGT). J Biol Chem. 289:5986–5996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bauer C, Gobel K, Nagaraj N, Colantuoni C,
Wang M, Müller U, Kremmer E, Rottach A and Leonhardt H:
Phosphorylation of TET proteins is regulated via O-GlcNAcylation by
the O-linked N-acetylglucosamine transferase (OGT). J Biol Chem.
290:4801–4812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Singh JP, Zhang K, Wu J and Yang X:
O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer
Lett. 356:244–250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fujiki R, Hashiba W, Sekine H, Yokoyama A,
Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, et al:
GlcNAcylation of histone H2B facilitates its monoubiquitination.
Nature. 480:557–560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen Q, Chen Y, Bian C, Fujiki R and Yu X:
TET2 promotes histone O-GlcNAcylation during gene transcription.
Nature. 493:561–564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Deplus R, Delatte B, Schwinn MK, Defrance
M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E,
et al: TET2 and TET3 regulate GlcNAcylation and H3K4 methylation
through OGT and SET1/COMPASS. EMBO J. 32:645–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hsu CH, Peng KL, Kang ML, Chen YR, Yang
YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al: TET1
suppresses cancer invasion by activating the tissue inhibitors of
metalloproteinases. Cell Rep. 2:568–579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guan W, Guyot R, Samarut J, Flamant F,
Wong J and Gauthier KC: Methylcytosine dioxygenase TET3 interacts
with thyroid hormone nuclear receptors and stabilizes their
association to chromatin. Proc Natl Acad Sci USA. 114:8229–8234.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Phoomak C, Silsirivanit A, Park D,
Sawanyawisuth K, Vaeteewoottacharn K, Wongkham C, Lam EW, Pairojkul
C, Lebrilla CB and Wongkham S: O-GlcNAcylation mediates metastasis
of cholangiocarcinoma through FOXO3 and MAN1A1. Oncogene.
37:5648–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ma Z and Vosseller K: Cancer metabolism
and elevated O-GlcNAc in oncogenic signaling. J Biol Chem.
289:34457–34465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yang WH, Kim JE, Nam HW, Ju JW, Kim HS,
Kim YS and Cho JW: Modification of p53 with O-linked
N-acetylglucosamine regulates p53 activity and stability. Nat Cell
Biol. 8:1074–1083. 2026. View Article : Google Scholar
|
|
70
|
Itkonen HM, Minner S, Guldvik IJ, Sandmann
MJ, Tsourlakis MC, Berge V, Svindland A, Schlomm T and Mills IG:
O-GlcNAc transferase integrates metabolic pathways to regulate the
stability of c-MYC in human prostate cancer cells. Cancer Res.
73:5277–5287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Olivier-Van Stichelen S, Guinez C, Mir AM,
Perez-Cervera Y, Liu C, Michalski JC and Lefebvre T: The hexosamine
biosynthetic pathway and O-GlcNAcylation drive the expression of
β-catenin and cell proliferation. Am J Physiol Endocrinol Metab.
302:E417–E424. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Thomson JP, Ottaviano R, Unterberger EB,
Lempiäinen H, Muller A, Terranova R, Illingworth RS, Webb S, Kerr
AR, Lyall MJ, et al: Loss of Tet1-Associated
5-hydroxymethylcytosine is concomitant with aberrant promoter
hypermethylation in liver cancer. Cancer Res. 76:3097–3108. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Delhommeau F, Dupont S, Della Valle V,
James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F,
Alberdi A, et al: Mutation in TET2 in myeloid cancers. N Engl J
Med. 360:2289–2301. 2029. View Article : Google Scholar
|
|
74
|
Itzykson R, Kosmider O, Renneville A,
Gelsi-Boyer V, Meggendorfer M, Morabito M, Berthon C, Adès L,
Fenaux P, Beyne-Rauzy O, et al: Prognostic score including gene
mutations in chronic myelomonocytic leukemia. J Clin Oncol.
31:2428–2436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Nibourel O, Kosmider O, Cheok M, Boissel
N, Renneville A, Philippe N, Dombret H, Dreyfus F, Quesnel B,
Geffroy S, et al: Incidence and prognostic value of TET2
alterations in de novo acute myeloid leukemia achieving complete
remission. Blood. 116:1132–1135. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dominguez PM, Ghamlouch H, Rosikiewicz W,
Kumar P, Béguelin W, Fontán L, Rivas MA, Pawlikowska P, Armand M,
Mouly E, et al: TET2 deficiency causes germinal center hyperplasia,
impairs plasma cell differentiation, and promotes B-cell
lymphomagenesis. Cancer Discov. 8:1632–1653. 2018.PubMed/NCBI
|
|
77
|
Cao T, Pan W, Sun X and Shen H: Increased
expression of TET3 predicts unfavorable prognosis in patients with
ovarian cancer-a bioinformatics integrative analysis. J Ovarian
Res. 12:1012019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Carella A, Tejedor JR, García MG,
Urdinguio RG, Bayón GF, Sierra M, López V, García-Toraño E,
Santamarina-Ojeda P, Pérez RF, et al: Epigenetic downregulation of
TET3 reduces genome-wide 5hmC levels and promotes glioblastoma
tumorigenesis. Int J Cancer. 146:373–387. 2020. View Article : Google Scholar : PubMed/NCBI
|