Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most
common pathological type of Non-Hodgkin lymphoma (NHL), which
accounts for ~25 to 35% of NHL in western developed countries and
up to 60% in developing countries (1). The average age of DLBCL onset is 67–70
years. DLBCL is an aggressive B-cell lymphoma that can be cured in
~50% of patients, however ~30% of DLBCL patients are refractory or
eventually relapse (2). DLBCL is the
subtype of lymphoma with the worst prognosis (3). Due to the high recurrence and the
limitations of current treatment of DLBCL, it is urgent to identify
new DLBCL drivers, therapeutic targets and efficacy predictors.
Long non-coding RNAs (lncRNAs) are a type of RNA in
eukaryotic cells with a transcript length greater than 200
nucleotides, which are located in the nucleus or cytoplasm, and
cannot encode any proteins (4).
LncRNAs can bind to proteins, DNA and RNA, and play a wide range of
regulatory roles at epigenetic, transcriptional and
post-transcriptional levels (5).
With the in-depth study of lncRNAs, a growing amount of evidence
has revealed that lncRNAs are abnormally expressed in various human
tumors and involved in the tumorigenesis and development of human
tumors (6,7). Therefore, lncRNAs have been attracting
more and more attention as new tumor biomarkers and regulators
(8). The role of lncRNAs in the
tumorigenesis and development of DLBCL has become a focus of
researchers in recent years. LncRNA RP11-513G11.1 was discovered to
be highly expressed in DCBCL, and closely associated with the
prognosis of DLBCL patients (9).
Zhao et al (10) revealed
that SNHG14 was upregulated in DLBCL and drove the development and
immune evasion by interacting with microRNA (miR)-5590-3p,
regulating ZEB1 and PD-L1 checkpoints. Furthermore, SMAD5-AS1 was
downregulated in DLBCL, and upregulation of SMAD5-AS1 suppressed
cell proliferation by inhibiting miR-135-5p and upregulating APC
expression (11).
Urothelial carcinoma-associated 1 (UCA1) was first
identified in bladder transitional cell carcinoma, and the entire
sequence consisted of three exons with a length of 1.4 kb (12). Scientists have revealed that UCA1 is
dysregulated in several human cancers and plays an important role
in cancer progression (13). In
2006, upregulation of UCA1 was revealed in bladder cancer, and
enhanced cell growth and invasion viability (14). In addition, UCA1 was revealed to be
closely associated to tumorigenesis and progression in breast
cancer (15), oral squamous cell
carcinoma (16,17) and ovarian cancer (18). However, to date, the expression and
biological function of UCA1 in DLBCL have not been reported.
The aim of the present study was to determine the
expression level of UCA1 in DLBCL tissues and cell lines, as well
as the specific functions of UCA1 and miR-331-3p in the progression
of DLBCL.
Materials and methods
Clinical information and cell
lines
In total, 38 DLBCL samples (18 males and 20 females;
age range: 24–74 years; mean age: 53 years) and 38 normal samples
of adjacent lymph nodes were collected during biopsy from July 2017
to January 2019 at Chengwu People's Hospital (Heze, China). The
DLBCL diagnosis was confirmed by histopathological examination, and
patients were not treated with radiotherapy or chemotherapy before
surgery. The samples were placed in liquid nitrogen for a short
time, and then stored at −80°C until use. The inclusion criteria
were as follows: i) Patients that were diagnosed with DLBCL
according to the WHO classification criteria; ii) patients that had
not received radiotherapy or chemotherapy prior to surgery; and
iii) patients with complete clinical data and long-term follow-up
data. The exclusion criteria were as follows: i) Patients with
serious function damage of the heart, liver and kidney; ii)
pregnant and lactating women; and iii) previous history of any
malignancy. The present study was approved by the Institutional
Human Experimentation and Ethics Committee of Chengwu People's
Hospital. Moreover, the guidelines of the declaration of Helsinki
were strictly adhered to during the study. Prior to the start of
the study, all patients signed written informed consent.
In the present study, GM12878 and JeKo-1 cell lines
were purchased from the Type Culture of the Chinese Academy of
Sciences (Shanghai, China). Other cell lines TMD8, U2932, OCI-Ly-10
and OCI-Ly-7 were obtained from BeNa Culture Collection (Beijing,
China). All cells were cultured in a modified Roswell Park Memorial
Institute-1640 medium (RPMI-1640), which contained 10% fetal bovine
serum (FBS), 100 g/l penicillin and 100 g/l streptomycin at 37°C
with 5% CO2. Then, 0.25% trypsin was used to passage the
cells every other day.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR analysis
Total RNA was isolated from DLBCL tissues and cells
according to the manufacturer's instructions using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Then, the total
extracted RNA was used as the template for reverse transcriptional
cDNA via Transcript cDNA Synthesis kit (Beijing TransGen Co., Ltd.)
according to the manufacturer's protocol. The PCR reaction
conditions were initial denaturation at 95°C for 10 min, followed
by 40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C
for 30 sec, and extension at 72°C for 30 sec. The primer sequences
used were as follows: UCA1 forward, 5′-GCACCCTAGACCCGAAACTT-3′ and
reverse, 5′-CCGGACTGCTTCAAGTGTGA-3′; miR-331-3p forward,
5′-GAGCTGAAAGCACTCCCAA-3′ and reverse, 5′-CACACTCTTGATGTTCCAGGA-3′;
GAPDH forward, 5′-CCTGACCTGCGTGTGGACT-3′ and reverse,
5′-GCTGTGGATGGGGAGGTGTC-3′; U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′
and reverse, 5′-TTCACGAATTTGCGTGTCAT-3′. GAPDH was used as the
endogenous control of UCA1, while U6 was the endogenous control of
miR-331-3p. The expression of UCA1 and miR-331-3p were detected by
RT-qPCR with SYBP Premix Ex Taq II (Takara Bio, Inc.) and
LightCycler® 96 thermocycler (Roche Diagnostics). The
2−ΔΔCq (19) method was
used to calculate the experimental results.
Cell transfection
U2932 cells (4×105) in the logarithmic
growth phase were seeded into a six-well plate. When cell
confluence reached 70–80%, U2932 cells were transfected with
pcDNA3.1-UCA1(UCA1 vector, 40 nM), small interfering (si)-UCA1 (40
nM), miR-331-3p mimics (40 nM), miR-331-3p inhibitor (80 nM), or
their corresponding negative control (40 nM) by Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After transfection at
37°C for 1 h, the cells were incubated with complete medium at
37°C, 5% CO2 and saturated humidity for another 48 h.
Vector plasmid, pcDNA3.1-UCA1, miR-331-3p mimics, miR-331-3p
inhibitor and the corresponding NC were obtained from Shanghai
GenePharma Co., Ltd.. The sequences were as follows: si-UCA1,
5′-GAGCCGAUCAGACAAACAAUU-3′; si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′;
miR-331-3p mimics, 5′-GCCCCUGGGCCUAUCCUAGAA-3′; miR-331-3p
inhibitor, 5′-UUCUAGGAUAGGCCCAGGGGC-3′; mimic NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; inhibitor NC:
5′-CAGUACUUUUGUGUAGUACAA-3′.
Dual luciferase reporter assay
The interactions between UCA1 and miR-331-3p were
predicted by starBase (http://www.starbase.sysu.edu.cn). The pMirReporter
plasmid (Sigma-Aldrich; Merck KGaA) was used to produce the
UCA1-mutated (mut) and UCA1-wild (wt). UCA1-mut or UCA1-wt, miR-NC
or miR-331-3p mimics were co-transfected into U2932 cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
After transfection at 37°C for 48 h, the relative luciferase
activities were detected by Dual-Luciferase reporter assay kit
(Promega Corporation), according to the manufacturer's protocol.
Luciferase activity was measured and normalized to Renilla
luciferase activity.
MTT assay
DLBCL cells were cultured in a 96-well plate
(1×104 cells/well) and cultured for 24, 48, 72 and 96 h.
Then MTT reagent (10 µl; 5 mg/ml) was added to each well and
incubated for another 4 h at 37°C. After the supernatant was
removed, dimethyl sulfoxide (DMSO) was added (100 µl/well). The
absorbance value at a wavelength of 490 nm was measured with a
microplate reader.
Transwell assays
DLBCL cell migration and invasion were assessed
using Transwell chambers (8 µm; BD Biosciences). Cell invasion was
detected in the upper chamber with Matrigel (BD Falcon; BD
Biosciences). Cell migration was performed without Matrigel. First,
1×105 cells were inoculated in the upper Transwell
compartment. Then, 10% FBS was added into the lower compartment.
After culture at 37°C for 24 h, the cells were fixed with 4%
formaldehyde for 30 min and stained with 0.5% crystal violet for 20
min at room temperature. Finally, the number of migrated and
invasive cells were counted under an optical microscope
(magnification, ×100; Olympus Corporation).
Statistical analysis
SPSS 22.0 (IBM Corp.) and GraphPad Prism 7.0
(GraphPad Software, Inc.) were used for statistical analysis. All
data are presented as the mean ± standard deviation (SD). Each
experiment was performed at least three times. The paired Student's
t-test was used to evaluate the differences between two groups.
One-way ANOVA and Tukey's post hoc test were performed to detect
the differences in multiple groups. Kaplan-Meier method and
log-rank test were used to plot the survival curve of DLBCL
patients. P<0.05 was considered to indicate a statistically
significant difference.
Results
UCA1 and miR-331-3p are aberrantly
expressed in DLBCL
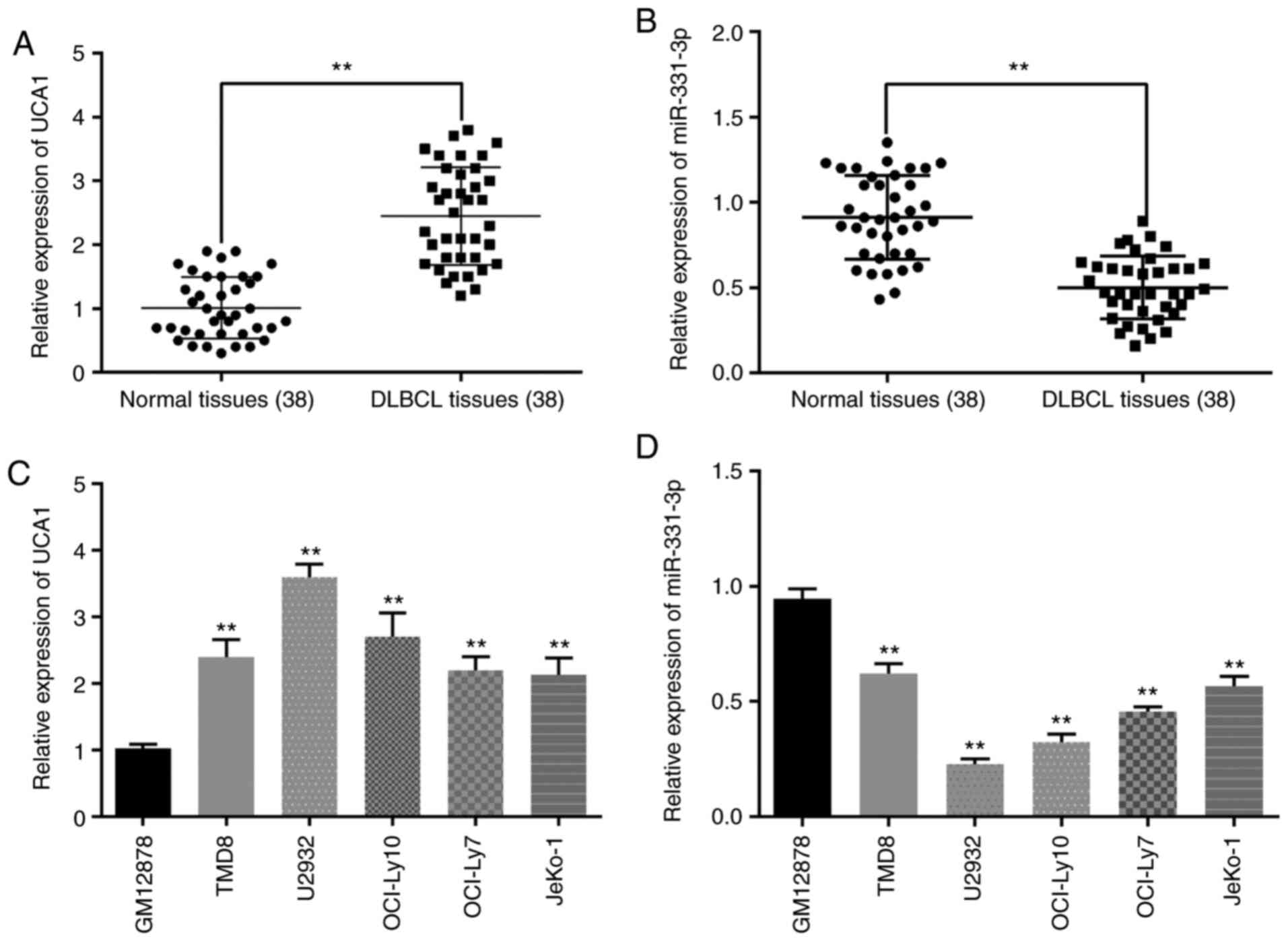
First, the expression pattern of UCA1 and miR-331-3p
in DLBCL tissues was detected by RT-qPCR analysis. The results
revealed that the expression of UCA1 was higher in DLBCL tissues
than in normal tissues (Fig. 1A).
Conversely, the expression level of miR-331-3p in DLBCL tissues was
decreased compared with normal tissues (Fig. 1B). Moreover, compared with normal
cell line GM12878, UCA1 was revealed to be upregulated in DLBCL
cell lines (TMD8, U2932, OCI-Ly10, OCI-Ly7 and JeKo-1), especially
in U2932 cells (Fig. 1C). Hence, in
the following study, U2932 cells were selected as the research
representative of DLBCL cells. On the contrary, miR-331-3p
expression was lower in the DLBCL cell lines than in the GM12878
cells (Fig. 1D). All these data
indicated that UCA1 and miR-331-3p may participate in the
progression of DLBCL.
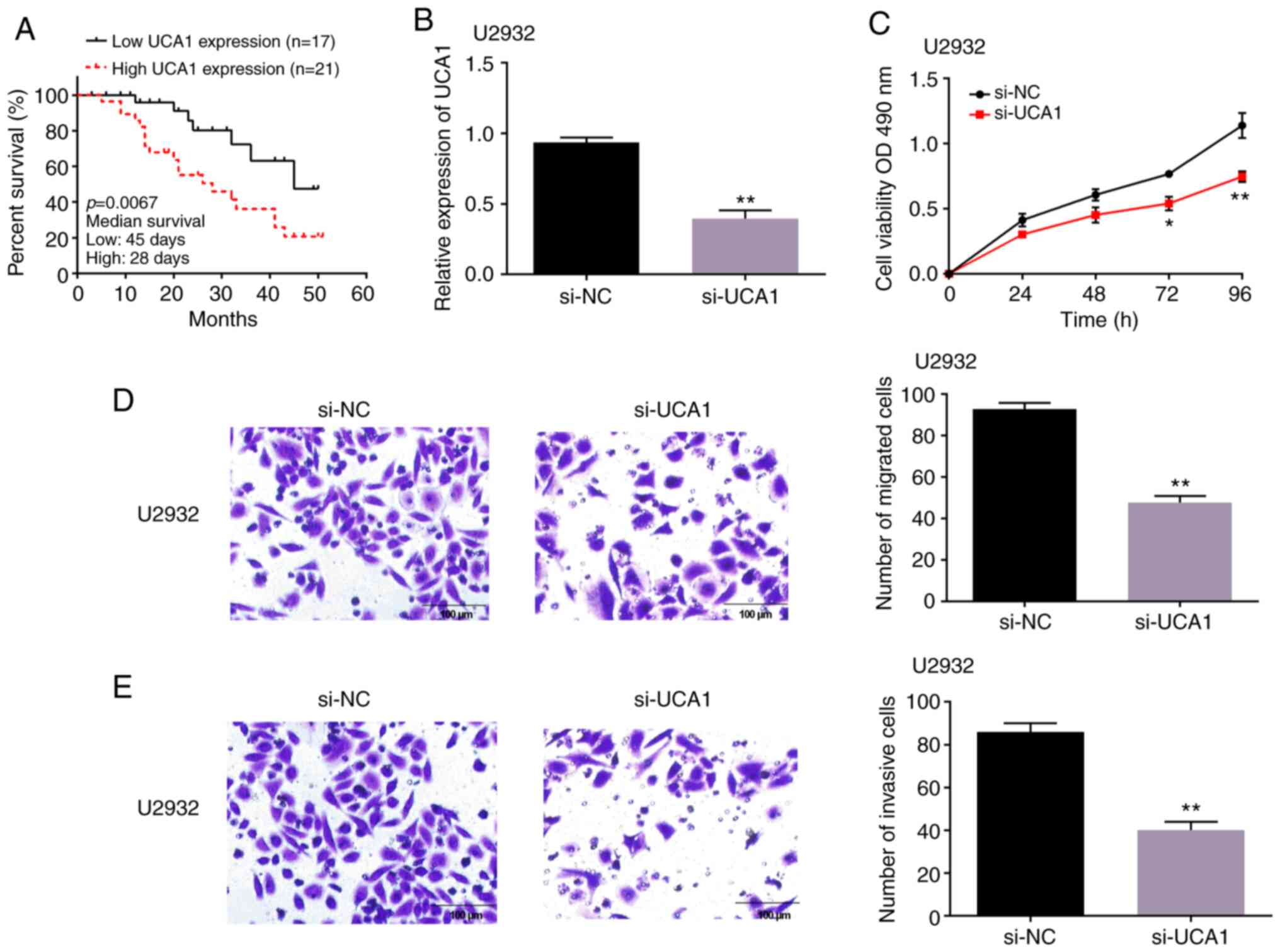
Knockdown of UCA1 inhibits cell
proliferation, migration and invasion in DLBCL
Next, the specific role of UCA1 in the progression
of DLBCL was investigated. According to the average expression of
UCA1, the DLBCL patients were divided into a high-UCA1 expression
group (n=21) and a low-UCA1 expression group (n=17). The survival
time in both groups was investigated by Kaplan-Meier. The results
revealed that DLBCL patients with high UCA1 expression had shorter
survival time than the patients in the low-expression group
(Fig. 2A). Moreover, it was
determined that UCA1 was closely associated with IPI score
(Table I). Then, si-UCA1 was
transfected into U2932 cells, and it was revealed that the
expression level of UCA1 was significantly decreased by si-UCA1
transfection (Fig. 2B).
Subsequently, MTT and Transwell assays were used to assess DLBCL
cell activity. The results revealed that cell proliferation of
U2932 cells was inhibited by depletion of UCA1 (Fig. 2C). Similarly, it was observed that
depletion of UCA1 decreased the number of migrated and invaded
U2932 cells (Fig. 2D and E). The
present findings demonstrated that UCA1 expression affected the
survival time of DLBCL patients and silencing of UCA1 suppressed
cell proliferation, migration and invasion of DLBCL cells.
 | Table I.Associations between the expression
level of UCA1 and clinical characteristics of DLBCL patients
(n=38). |
Table I.
Associations between the expression
level of UCA1 and clinical characteristics of DLBCL patients
(n=38).
|
|
| UCA1 expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristics | Number of cases
n=38 | Low (n=17) | High (n=21) | P-value |
|---|
| Age (years) |
|
|
| 0.389 |
|
>60 | 15 | 8 | 7 |
|
|
≤60 | 23 | 9 | 14 |
|
| Sex |
|
|
| 0.973 |
|
Male | 18 | 8 | 10 |
|
|
Female | 20 | 9 | 11 |
|
| Ann Arbor
stage |
|
|
| 0.618 |
|
I–II | 24 | 10 | 14 |
|
|
III–IV | 14 | 7 | 7 |
|
| Clinical stage |
|
|
| 0.899 |
|
I–II | 13 | 6 | 7 |
|
|
III–IV | 25 | 11 | 14 |
|
| LDH ratio |
|
|
| 0.054 |
| ≤1 | 20 | 6 | 14 |
|
|
>1 | 18 | 11 | 7 |
|
| IPI score |
|
|
| 0.021a |
|
0-2 | 26 | 8 | 18 |
|
|
3-5 | 12 | 9 | 3 |
|
UCA1 can act as a sponge of miR-331-3p
in DLBCL
Starbase database predicted that there were
complementary sequences between UCA1 and miR-331-3p (Fig. 3A). To verify this prediction,
miR-331-3p mimics or miR-331-3p inhibitor was transfected into
U2932 cells. RT-qPCR results revealed that the expression of
miR-331-3p was significantly increased by miR-331-3p mimics, but
reduced by miR-331-3p inhibitor (Fig.
3B). Then, a Dual Luciferase Reporter assay was performed to
assess the relative luciferase activity of UCA1. It was observed
that the luciferase activity of UCA1-wt was decreased by
miR-331-3p, but there was no change in UCA1-mut (Fig. 3C). Moreover, the relationship between
UCA1 and miR-331-3p was explored. The results revealed that
miR-331-3p was downregulated by UCA1 vector, while it was
upregulated by si-UCA1 transfection (Fig. 3D). Furthermore, miR-331-3p mimics
decreased the expression level of UCA1, while miR-331-3p inhibitor
increased the expression level of UCA1 (Fig. 3E). Furthermore, Pearson's correlation
analysis revealed that there was a negative correlation between
UCA1 and miR-331-3p (Fig. 3F). These
experimental results indicated that UCA1 can be a sponge to
regulate miR-331-3p expression in DLBCL.
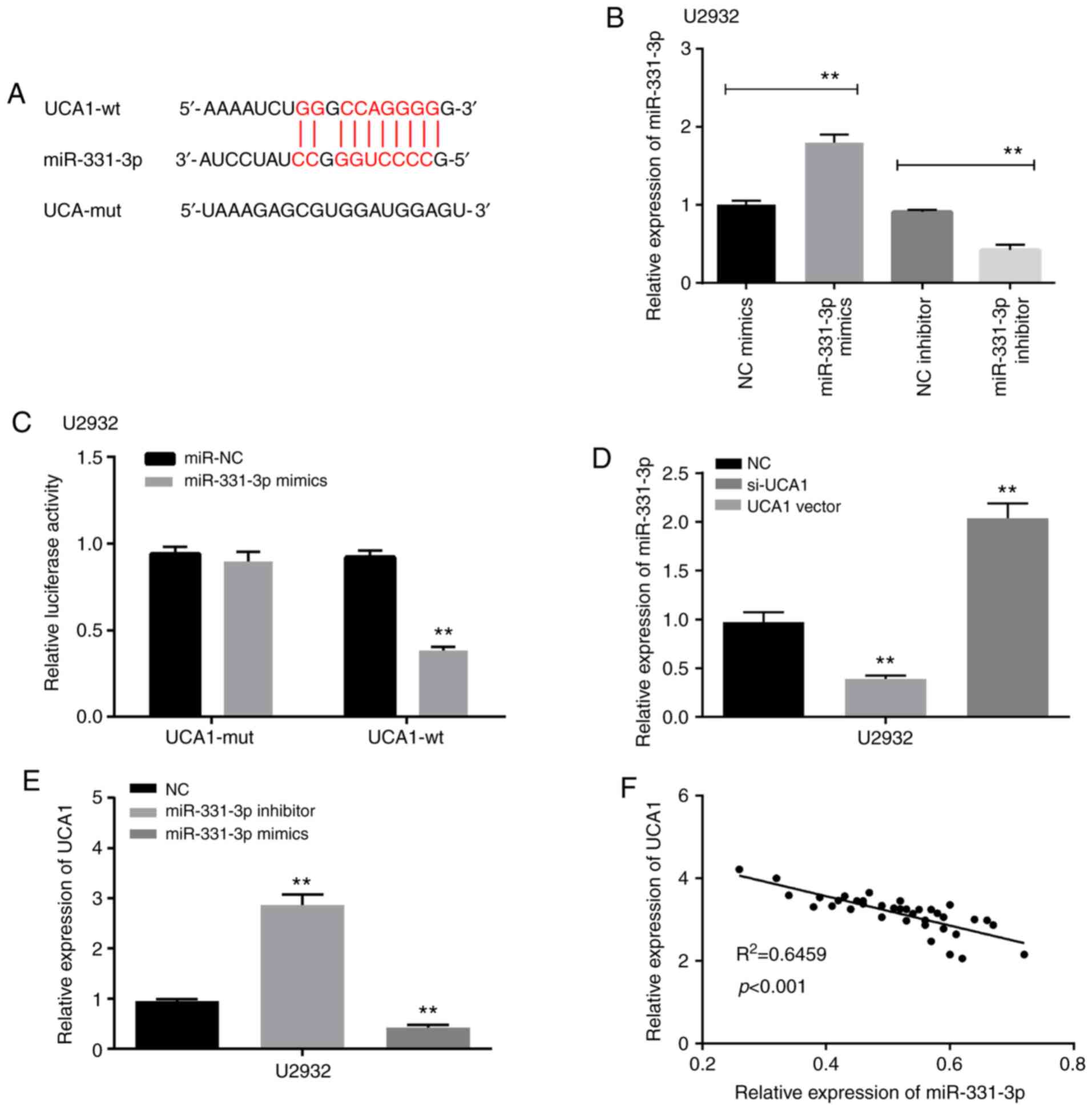 | Figure 3.UCA1 can act as a sponge of miR-331-3p
in DLBCL. (A) Binding sites between UCA1 and miR-331-3p. (B) The
expression of miR-331-3p was increased by miR-331-3p mimics, while
it was reduced by miR-331-3p inhibitor. (C) The luciferase activity
of UCA1-wt was significantly reduced by miR-331-3p mimics, while
there was no change on UCA1-mut. (D) The expression of miR-331-3p
was increased by si-UCA1, but reduced by UCA1 vector. (E) The
expression of UCA1 was increased by miR-331-3p inhibitor, while it
was decreased by miR-331-3p mimics. (F) There was a negative
correlation between UCA1 and miR-331-3p. **P<0.01. UCA1,
urothelial cancer associated 1; miR-331-3p, microRNA-331-3p; DLBCL,
diffuse large B-cell lymphoma; wt, wild-type; mut, mutated; NC,
negative control; si-, small interfering. |
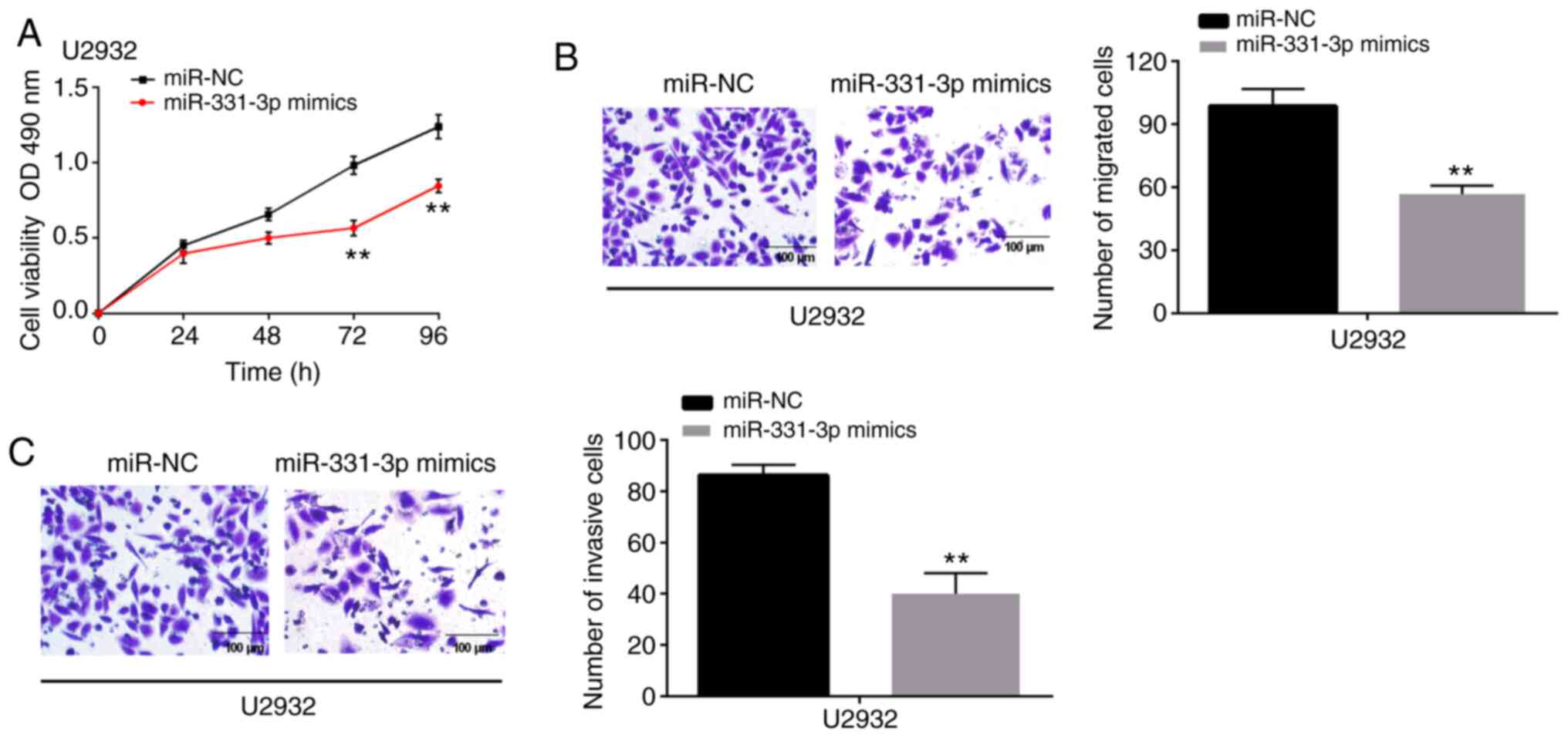
miR-331-3p overexpression inhibits
cell proliferation, migration and invasion in DLBCL cells
To investigate the role of miR-331-3p on the
progression of DLBCL cells, miR-331-3p mimics were transfected into
U2932 cells. Functionally, it was revealed that miR-331-3p mimics
inhibited the cell proliferation capability of U2932 cells
(Fig. 4A). Moreover, when U2932
cells were transfected with miR-331-3p mimics, a significant
decrease of migrated and invasive cells were observed in the
Transwell migration and invasion assays, compared with miR-NC
(Fig. 4B and C). Additionally,
downregulation of miR-331-3p was revealed to be associated with LDH
ratio in DLBCL patients (Table II).
Collectively the data demonstrated that miR-331-3p upregulation
prevented DLBCL cell progression.
 | Table II.Associations between the expression
level of miR-331-3p and clinical characteristics of DLBCL patients
(n=38). |
Table II.
Associations between the expression
level of miR-331-3p and clinical characteristics of DLBCL patients
(n=38).
|
|
| miR-331-3p
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristics | Number of cases
n=38 | Low (n=18) | High (n=20) | P-value |
|---|
| Age (years) |
|
|
| 0.944 |
|
>60 | 15 | 7 | 8 |
|
|
≤60 | 23 | 11 | 12 |
|
| Sex |
|
|
| 0.732 |
|
Male | 18 | 8 | 10 |
|
|
Female | 20 | 10 | 10 |
|
| Ann Arbor
stage |
|
|
| 0.671 |
|
I–II | 24 | 12 | 12 |
|
|
III–IV | 14 | 6 | 8 |
|
| Clinical stage |
|
|
| 0.139 |
|
I–II | 13 | 4 | 9 |
|
|
III–IV | 25 | 14 | 11 |
|
| LDH ratio |
|
|
| 0.022a |
| ≤1 | 20 | 13 | 7 |
|
|
>1 | 18 | 5 | 13 |
|
| IPI score |
|
|
| 0.061 |
|
0-2 | 26 | 15 | 11 |
|
|
3-5 | 12 | 3 | 9 |
|
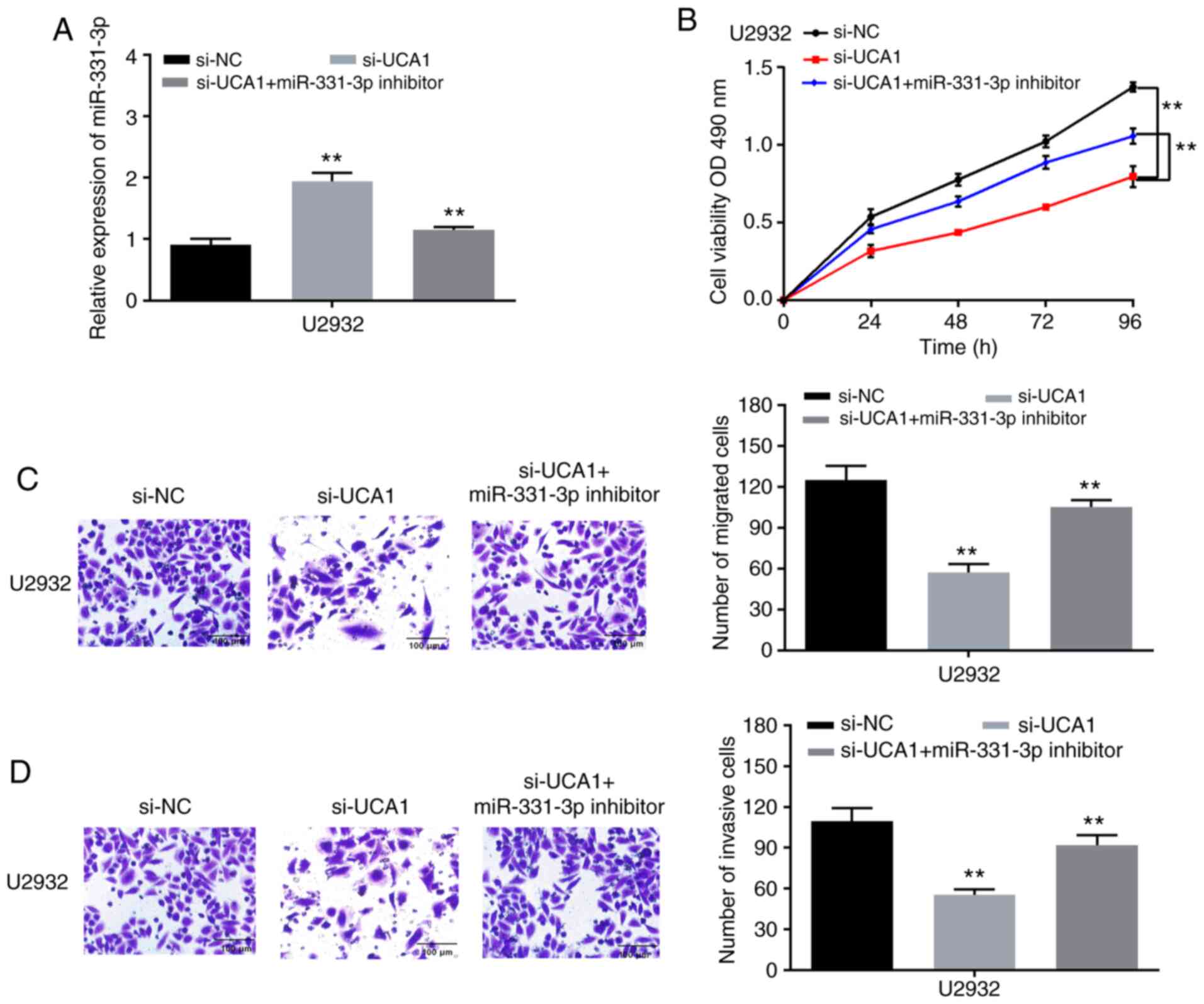
UCA1 regulates DLBCL cell progression
by competitively binding with miR-331-3p
To explore the role of DLBCL/miR-331-3p on the
progression of DLBCL, si-UCA1 were transfected into U2932 cells
with miR-331-3p inhibitor. As revealed in Fig. 5A, the expression of miR-331-3p was
increased by si-UCA1, while it was reduced by miR-331-3p inhibitor
with UCA1 knockdown. Next, functional experiments on the
UCA1/miR-331-3p axis were conducted. It was revealed that the
inhibitory effect of si-UCA1 on cell proliferation was restored by
miR-331-3p inhibitor (Fig. 5B).
Moreover, the suppression of cell migration ability was reversed by
miR-331-3p inhibitor (Fig. 5C).
Similarly, miR-331-3p inhibitor had the same effect on cell
invasion ability (Fig. 5D).
Consequently, the present data indicated that knockdown of UCA1 had
a negative effect on DLBCL cell progression by inhibiting
miR-331-3p expression.
Discussion
Studies have revealed that various lncRNAs are
involved in the tumorigenesis and development of DLBCL, and act as
tumor suppressors or tumor carcinogens (20). For example, DBH-AS1 was confirmed to
drive DLBCL cell progression, and could be a therapeutic target for
the treatment of DLBCL (21).
Conversely, NONHSAG026900 suppressed DLBCL cell proliferation and
cell activity, and could be a favorable biomarker in DLBCL. LncUCA1
has been revealed to be aberrantly expressed in several human
tumors, and to participate in tumor tumorigenesis and development.
However, the role of UCA1 remains unclear. In the present research,
it was revealed that the expression of UCA1 was significantly
upregulated in DLBCL tissues and cell lines. Consistent with our
results, UCA1 was also upregulated in gastric (22) and adrenocortical cancer (23). In glioma, UCA1 was detected to drive
cell proliferation, migration and EMT (24). In agreement with previous studies,
functional experiments revealed that silencing of UCA1 suppressed
U2932 cell proliferation, and increased the number of migrated and
invaded cells. Similarly, UCA1 was demonstrated to be actively
involved in tumor initiation and progression, and act as an
oncogene in multiple myeloma (25)
and prostate cancer (26). The
present results indicated that UCA1 knockdown inhibited cell
proliferation, migration and invasion in DLBCL.
miRNAs are a type of endogenous non-coding
regulatory RNAs with a length of 20–25 nucleotides. Researches have
revealed that miRNAs regulate gene expression through
sequence-specific translation or mRNA cleavage, and are involved in
a series of important biological processes such as cell
proliferation, differentiation, and apoptosis (27,28). In
recent years, research on the relationship between DLBCL and miRNAs
has made great advances. For example, Ting et al (29) systematically analyzed a large number
of studies and found that miR-155, miR-334, miR-21, miR-146b-5p and
miR-17/92 clusters have certain value in predicting the
chemotherapy response in DLBCL. In previous research, UCA1 was
revealed to competitively bind with several miRNAs (13,30). Gao
et al (31) confirmed that
UCA1 facilitated thyroid cancer cell ability and metastasis by
decreasing the expression of miR-497-3p. In addition, UCA1 was
revealed to directly target miR-495 (32,33) and
miR-185-5p (34). In the present
study, UCA1 was revealed to act as a molecular sponge of
miR-331-3p, which was consistent with previous findings in multiple
myeloma (35). In addition, there
was a negative relationship between UCA1 and miR-331-3p in DLBCL.
Moreover, the expression level and role of miR-331-3p in DLBCL were
detected. The results revealed that miR-331-3p expression was
decreased in DLBCL tissues and cell lines. Furthermore, functional
experiments revealed that overexpression of miR-331-3p inhibited
U2932 cell proliferation, migration and invasion. In addition,
miR-331-3p inhibitor was revealed to reverse the inhibitory effect
on cell proliferation and metastasis which was stimulated by
si-UCA1. According to the present findings, UCA1 had a negative
effect on DLBCL cell progression by inhibiting miR-331-3p
expression.
In conclusion, upregulation of UCA1 and
downregulation of miR-331-3p were revealed in DLBCL tissues and
cell lines. Moreover, the present findings firstly indicated that
knockdown of UCA1 suppressed cell proliferation, migration and
invasion by competitively binding with miR-331-3p in DLBCL.
Combined with all findings, it is suggested that UCA1 could be a
target for treatment and prognosis of DLBCL.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Development
Plan of Medical and Health Science and Technology of Shandong
province (grant no. 2016WS0472) and the Key R&D Program
Projects of Shandong Province (grant no. 2018GSF118203).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
MZ, YD, JS and HC designed and conceived the
experiments. MZ, DZ and XD performed most of the experiments. MZ
collected and analyzed the clinical and pathological data. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Human Experimentation and Ethics Committee of Chengwu People's
Hospital (Heze, China). Moreover, the guidelines of the declaration
of Helsinki were strictly adhered to during the study. Prior to the
start of the study, all patients signed written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
García-Morales E, Lázaro-Martínez JL,
Martínez-Hernández D, Aragón-Sánchez J, Beneit-Montesinos JV and
González-Jurado MA: Impact of diabetic foot related complications
on the Health Related Quality of Life (HRQol) of patients-a
regional study in Spain. Int J Low Extrem Wounds. 10:6–11. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patterson E: The spending power after
NFIB: New direction, or medicaid exception? SMU Law Rev.
68:385–426. 2015.PubMed/NCBI
|
|
3
|
Miao Y, Medeiros LJ, Xu-Monette ZY, Li J
and Young KH: Dysregulation of cell survival in diffuse large B
cell lymphoma: Mechanisms and therapeutic targets. Front Oncol.
9:1072019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gil N and Ulitsky I: Regulation of gene
expression by cis-acting long non-coding RNAs. Nat Rev Genet.
21:102–117. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Xu R, Mai SJ, Ma YS, Zhang MY, Cao
PS, Weng NQ, Wang RQ, Cao D, Wei W, et al: LncRNA CSMD1-1 promotes
the progression of hepatocellular carcinoma by activating MYC
signaling. Theranostics. 10:7527–7544. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang D, Chen J, Yang L, Ouyang Q, Li J,
Lao L, Zhao J, Liu J, Lu Y, Xing Y, et al: NKILA lncRNA promotes
tumor immune evasion by sensitizing T cells to activation-induced
cell death. Nat Immunol. 19:1112–1125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mehrpour Layeghi S, Arabpour M, Esmaeili
R, Naghizadeh MM, Tavakkoly Bazzaz J and Shakoori A: Evaluation of
the potential role of long non-coding RNA LINC00961 in luminal
breast cancer: A case-control and systems biology study. Cancer
Cell Int. 20:4782020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goie TT and Naidoo M: Awareness of
diabetic foot disease amongst patients with type 2 diabetes
mellitus attending the chronic outpatients department at a regional
hospital in Durban, South Africa. Afr J Prim Health Care Fam Med.
8:e1–e8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao L, Liu Y, Zhang J, Liu Y and Qi Q:
LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted
diffuse large B cell lymphoma progression and immune evasion
through regulating PD-1/PD-L1 checkpoint. Cell Death Dis,.
10:7312019. View Article : Google Scholar
|
|
11
|
Pichu S, Patel BM, Apparsundaram S and
Goyal RK: Role of biomarkers in predicting diabetes complications
with special reference to diabetic foot ulcers. Biomark Med.
11:377–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chalatsa I, Arvanitis DA, Koulakiotis NS,
Giagini A, Skaltsounis AL, Papadopoulou-Daifoti Z, Tsarbopoulos A
and Sanoudou D: The Crocus sativus compounds trans-crocin 4
and trans-crocetin modulate the amyloidogenic pathway and tau
misprocessing in Alzheimer disease neuronal cell culture models.
Front Neurosci. 13:2492019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu M and Yang J: Down-regulation of lncRNA
UCA1 enhances radiosensitivity in prostate cancer by suppressing
EIF4G1 expression via sponging miR-331-3p. Cancer Cell Int.
20:4492020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kocaman G, Altinoz E, Erdemli ME, Gul M,
Erdemli Z, Gul S and Bag HG: Protective effects of crocin on
biochemistry and histopathology of experimental periodontitis in
rats. Biotech Histochem. 94:366–373. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu A, Lao C, Wang Z, Chen Y and Bai C:
Characterization of crocetin-monoglucuronide as a neuron-protective
metabolite of crocin-1. Mol Nutr Food Res. Apr 29–2019.(Epub ahead
of print). View Article : Google Scholar
|
|
16
|
Khalatbari-Mohseni A, Banafshe HR,
Mirhosseini N, Asemi Z, Ghaderi A and Omidi A: The effects of
crocin on psychological parameters in patients under methadone
maintenance treatment: A randomized clinical trial. Subst Abuse
Treat Prev Policy. 14:92019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deng L, Li J, Lu S and Su Y: Crocin
inhibits proliferation and induces apoptosis through suppressing
MYCN expression in retinoblastoma. J Biochem Mol Toxicol.
33:e222922019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Colapietro A, Mancini A, D'Alessandro AM
and Festuccia C: Crocetin and crocin from saffron in cancer
chemotherapy and chemoprevention. Anticancer Agents Med Chem.
19:38–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang X, Qian W and Ye X: Long noncoding
RNAs in diffuse large B-cell lymphoma: Current advances and
perspectives. Onco Targets Ther. 13:4295–4303. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Demurtas OC, Frusciante S, Ferrante P,
Diretto G, Azad NH, Pietrella M, Aprea G, Taddei AR, Romano E, Mi
J, et al: Candidate enzymes for saffron crocin biosynthesis are
localized in multiple cellular compartments. Plant Physiol.
177:990–1006. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taherkhani T, Asghari Zakaria R, Omidi M
and Zare N: Effect of ultrasonic waves on crocin and safranal
content and expression of their controlling genes in suspension
culture of saffron (Crocus sativus L). Nat Prod Res.
33:486–493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Altinoz E, Erdemli ME, Gul M, Aksungur Z,
Gul S, Bag HG, Kaya GB and Turkoz Y: Neuroprotection against CCl4
induced brain damage with crocin in Wistar rats. Biotech Histochem.
93:623–631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yorgun MA: Effects of crocin on diabetic
maculopathy: A placebo-controlled randomized clinical trial. Am J
Ophthalmol. 204:141–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roshankhah S, Jalili C and Salahshoor MR:
Effects of crocin on sperm parameters and seminiferous tubules in
diabetic rats. Adv Biomed Res. 8:42019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mozaffari S, Ramezany Yasuj S,
Motaghinejad M, Motevalian M and Kheiri R: Crocin acting as a
neuroprotective agent against methamphetamine-induced
neurodegeneration via CREB-BDNF signaling pathway. Iran J Pharm
Res. 18:745–758. 2019.PubMed/NCBI
|
|
27
|
Komoll RM, Hu Q, Olarewaju O, von Döhlen
L, Yuan Q, Xie Y, Tsay HC, Daon J, Qin R, Manns MP, et al:
MicroRNA-342-3p is a potent tumour suppressor in hepatocellular
carcinoma. J Hepatol. Jul 30–2020.(Online ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fuertes T, Ramiro AR and de Yebenes VG:
miRNA-based therapies in B cell non-hodgkin lymphoma. Trends
Immunol. 41:932–947. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ting CY, Liew SM, Price A, Gan GG, Bee-Lan
Ong D, Tan SY and Bee PC: Clinical significance of aberrant
microRNAs expression in predicting disease relapse/refractoriness
to treatment in diffuse large B-cell lymphoma: A meta-analysis.
Critical Reviews in Oncology/Hematology,. 144:1028182019.
View Article : Google Scholar
|
|
30
|
Guo Z, Wang X, Yang Y, Chen W, Zhang K,
Teng B, Huang C, Zhao Q and Qiu Z: Hypoxic tumor-derived exosomal
long noncoding RNA UCA1 promotes angiogenesis via miR-96-5p/AMOTL2
in pancreatic cancer. Mol Ther Nucleic Acids. Aug 25–2020.(Epub
ahead of print). View Article : Google Scholar
|
|
31
|
Gao H, Yang JY, Tong LX, Jin H and Liu CZ:
Long noncoding RNA UCA1 promotes proliferation and metastasis of
thyroid cancer cells by sponging miR-497-3p. Eur Rev Med Pharmacol
Sci,. 24:75552020.
|
|
32
|
Mohammadzadeh L, Hosseinzadeh H, Abnous K
and Razavi BM: Neuroprotective potential of crocin against
malathion-induced motor deficit and neurochemical alterations in
rats. Environ Sci Pollut Res Int. 25:4904–4914. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin L, Liu G and Yang L: Crocin improves
cognitive behavior in rats with Alzheimer's disease by regulating
endoplasmic reticulum stress and apoptosis. Biomed Res Int.
2019:94549132019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sebastin Santhosh M, Hemshekhar M,
Thushara RM, Devaraja S, Kemparaju K and Girish KS: Vipera russelli
venom-induced oxidative stress and hematological alterations:
Amelioration by crocin a dietary colorant. Cell Biochem Funct.
31:41–50. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Asanad K: Radiographic evolution of a
simple renal cyst to clear cell renal cell carcinoma in three
years. Urol Case Rep. 32:1012122020. View Article : Google Scholar : PubMed/NCBI
|