Introduction
Hepatocellular carcinoma (HCC) is the most common
type of primary liver cancer in adults, with an incidence of
~850,000 new cases worldwide annually (1). Therefore, early detection of HCC is
crucial for improving survival rates. Human HCC development is a
multistep process characterized by progression from cirrhosis, to
low-grade dysplastic nodule (LGDN), to high-grade dysplastic nodule
(HGDN), to early HCC (eHCC) and, finally, to progressed HCC
(2). A key step in early detection
of HCC in the clinical setting is to differentiate between eHCC and
dysplastic nodule (DN), as the transition from DN to eHCC denotes
the earliest step of malignant transformation in the cirrhotic
liver. The radiological observation of liver pathology has been
developed for >30 years and has achieved significant advances in
diagnosis (3). However, there are
several reasons due to which imaging methods cannot accurately
identify the stage of hepatocarcinogenesis. For example, the
pathological characteristics of eHCC closely resemble those of
HGDN, and a definitive pathological differentiation between the two
is currently lacking (4). Moreover,
most well-differentiated early-stage HCCs do not stain on
angiography or retain lipiodol within the tumor, thereby making
their diagnosis difficult (2).
Whether Gd-EOB-DTPA-enhanced magnetic resonance or computed
tomography imaging are sufficiently sensitive to detect eHCC is
subject to dispute (5,6). Although some morphological criteria
were put forward to distinguish DNs from eHCC, they are unreliable
as diagnostic indicators to discriminate between the two,
particularly for HGDN and eHCC, since they are close to each other
in the stepwise morphological progression. Conventional
immunohistochemistry (IHC) biomarkers, such as p53, E-cadherin and
CD34, are widely used to confirm the presence of liver cancer, but
cannot identify the specific HCC stage (7,8).
Glypican 3 (GPC3) is a cell surface protein linked to the cell
membrane via a glycosylphosphatidylinositol anchor and is highly
expressed in HCC and certain other human cancers, including
melanoma and neuroblastoma (9). GPC3
staining was found to be negative or focally weakly expressed in
HCC precursor lesions, but diffuse GPC3 staining is observed in the
majority of HCCs (10). A growing
body of evidence supports GPC3 as a novel biomarker for HCC, and
its protein expression is associated with poor prognosis of
patients with HCC (11,12). However, the sensitivity and
specificity of GPC3 must be further optimized. Therefore, there is
an urgent need for objective and effective markers that are
sensitive to the differences between HGDNs and eHCCs.
Heterogeneous nuclear ribonucleoproteins (hnRNPs)
are defined as nuclear RNA-binding proteins that form complexes
with pre-mRNA. It is widely accepted that the members of this
protein family are known regulators of cell cycle progression, cell
differentiation, cell cycle arrest and DNA damage. HNRNPA3 is a
protein encoded in humans by the HNRNPA3 gene and is a
member of the hnRNP A/B family. HNRNPA3 is involved in RNA binding,
mRNA transport and mRNA splicing via spliceosome. Although the
detailed association between tumorigenesis and HNRNPA3 has not been
fully elucidated, the HNRNP protein family members are closely
associated with cancer regulation. The expression of APOBEC3B, a
cytosine deaminase, increases with increasing levels of HNRNPA3 in
cancer cell lines, and the possible underlying mechanism may be
through telomere elongation (13).
Altered expression of hnRNP A/B members, including A3, was found to
antagonize alternative splicing factor/splicing factor 2 (ASF/SF2),
a prototypical SR protein, in patients with non-small cell lung
cancer (14). In light of these
findings, hnRNP protein family members may be considered as
valuable diagnostic/prognostic markers in cancer. Therefore, the
aim of the present study was to investigate the role of HNRNPA3 in
the process of hepatocarcinogenesis and to further validate its
diagnostic/prognostic value for determining the differentiation
degree of HCC. The results of the present study demonstrated that
HNRNPA3 expression gradually increased in non-tumor hepatic tissue,
DNs, eHCC and progressed HCC. It was confirmed that HNRNPA3
combined with GPC3 is a helpful diagnostic biomarker in the
differential diagnosis during the multistep process of
hepatocarcinogenesis, particularly in the differential diagnosis
between HGDN and eHCC. In addition, HNRNPA3 expression was
associated with HCC differentiation. High expression of HNRNPA3 was
also demonstrated to be associated with poor survival rates in
patients with HCC.
Materials and methods
Tissue samples
The study population comprised a total of 56
samples, including human DNs, well-differentiated, moderately and
poorly differentiated HCC, with corresponding paired cirrhotic
liver tissues. The samples were collected from patients undergoing
surgery at Qilu Hospital, Shandong University (Jinan, China). The
study protocol was approved by the Ethics Committee of Shandong
University (approval no. 2012028). Patient cases were recorded at
the Department of Pathology between March 2013 and June 2019. The
Hepatitis B Virus (HBV) infection status of patients with HCC or DN
were collected (Tables SI and
SII). The histological slides were
reviewed to confirm the diagnosis in all cases, which were
classified as follows: i) Clinicopathologically typical HCC (n=48),
tumors arising in men and women aged 43–89 years and displaying the
typical histological characteristics of HCC. In a total of 48
samples, there were 38 progressed and 10 well-differentiated HCCs
(including 7 eHCCs). eHCC was defined as being ≤2 cm in diameter,
displaying a vaguely nodular pattern with indistinct margins and no
tumor capsule (15). ii) DNs (n=8),
nodules differing from the surrounding liver parenchyma with
regards to size, color, texture and degree of bulging of the cut
surface. LGDN was defined as a nodule exhibiting a mild increase in
cell density with a monotonous pattern and/or clonal changes,
whereas HGDN was defined as a nodule exhibiting marked cytological
and architectural atypia. Few unpaired non-triadal arteries may be
seen. Stromal and vascular invasion are absent (16).
IHC
The streptavidin-peroxidase-biotin method was
employed (17). Tissue sections were
fixed with 4% paraformaldehyde at room temperature for 14 h.
Paraffin-embedded tissue sections were cut into 4-µm-thick
sections, dewaxed in dimethylbenzene (twice, 20 min each time) and
hydrated in gradient ethanol (100, 95, 85 and 75%, each for 5 min)
at room temperature. The sections were treated according to the
following method: Antigen retrieval with EDTA at 95°C for 45 min
(cat. no. ZLI-9069; OriGene Technologies, Inc.), endogenous
peroxidase removed at room temperature for 10 min (cat. no. SP9000;
OriGene Technologies, Inc), blocking with 10% goat serum at room
temperature for 10 min (cat. no. SP9000; OriGene Technologies,
Inc.). Tissue sections were subsequently incubated with primary
antibodies against HNRNPA3 (1:75; cat. no. 25142-1-AP; ProteinTech
Group, Inc.) or GPC3 (1.5 ml, ready-to-use; cat. no. MAB-0617;
Fuzhou Maixin Biotech Co., Ltd.) at 4°C for 12 h. Following the
primary incubation, tissue sections were incubated with
ready-to-use horseradish peroxidase-labeled secondary antibody
(cat. no. SP9000; OriGene Technologies, Inc.) for 20–30 min at room
temperature, DAB color reaction for 2 min or 45 sec at room
temperature, respectively, counterstained with hematoxylin for 20
sec at room temperature, differentiation, dehydration and
transparency. For negative controls, the primary antibody was
replaced with PBS. Tissue sections were observed under a light
microscope (magnification, ×40).
Scoring
All the sections were evaluated by two observers
blinded to the experimental groups. HNRNPA3 staining intensity was
scored as follows: i) 0, negative; ii) 1, weak; iii) 2, moderate;
and iv) 3, strong. The staining area (percentage of stained cells)
was scored as follows: i) 0, 0%; ii) 1, 1–25%; iii) 2, 26–50%; iv)
3, 51–75%; and v) 4, 76–100%. These two scores were added together
to produce the final IHC score in different samples. The suitable
cut-off points were confirmed by receiver operating characteristics
(ROC) curve analysis. GPC3 was considered as positive when moderate
to strong nuclear, cytoplasmic and/or membranous staining was seen
in ≥10% of tumor cells (18).
Statistical analysis
Statistical analysis was performed using Graphpad
Prism 5 (Graphpad Software, Inc.). The Fisher's exact test was used
to determine significant differences in expression of HNRNPA3
between HCC, DN and surrounding normal tissues. The Kruskal-Wallis
test (followed by post hoc Dunn's multiple comparison test) was
used to compare IHC sum scores among non-tumorous tissue, DN and
HCC. The Fisher's exact test was also used to analyze the
association between HNRNPA3 expression and clinicopathological
variables in DN and HCC (the χ2 test was applied to the
differentiation part due to limitations of statistical approach).
The χ2 test was used to analyze the association between
HNRNPA3 expression and indicators that were subdivided into three
groups, such as serum enzymes and tumor biomarkers in HCC. The
Fisher's exact test was applied to find an association between
HNRNPA3 expression and indicators that were subdivided into two
groups. ROC curves were constructed and the area under the curve
(AUC) was calculated to assess the ability of HNRNPA3 expression to
discriminate these three different stages during progression of
liver cancer, based on the highest Youden's index (sensitivity and
1-specificity). The prognosis rates of patients were calculated
using the Kaplan-Meier method and differences between survival
curves were examined using a log-rank test (19). P<0.05 was considered to indicate a
statistically significant difference.
Database
The survival analysis of HNRNPA3 in patients with
HCC according to The Human Protein Atlas (https://www.proteinatlas.org/). Different patient
samples in The Human Protein Atlas were originally derived from The
Cancer Genome Atlas (TCGA) database established by National Cancer
Institute. Specific analysis methods are outlined in previous
literature (19).
Results
HNRNPA3 expression increases in a
stepwise trend during multistep hepatocarcinogenesis
To the best of our knowledge, the expression of
HNRNPA3 in liver cancer has not been investigated before and the
present study was the first to investigate the protein expression
of HNRNPA3 using IHC in a total of 56 paraffin-embedded liver
samples, including 8 cases of DNs, 48 cases of HCC and their paired
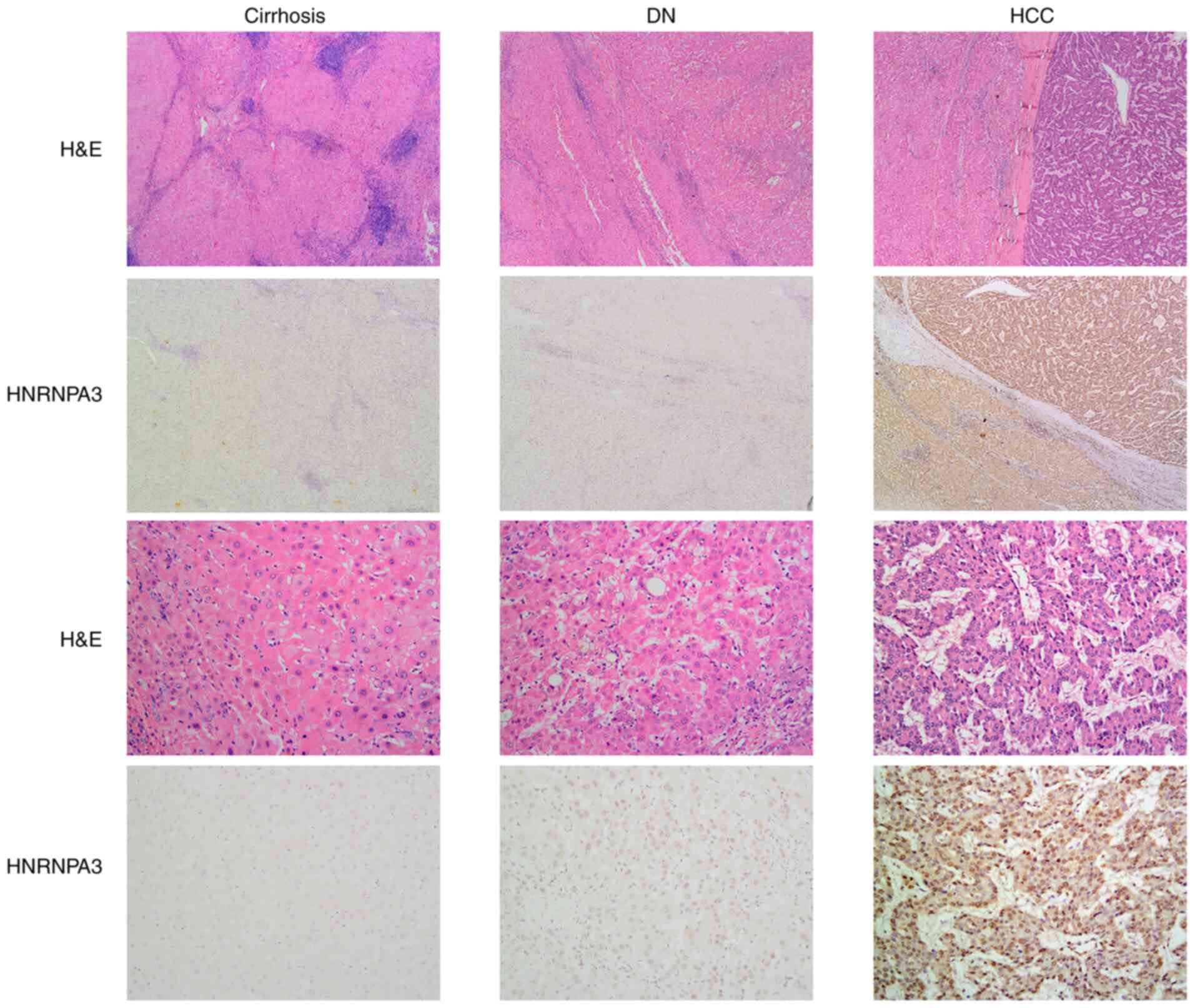
non-tumor hepatic tissues. The immunostaining profiles of HNRNPA3
in liver samples (cirrhotic, DN and HCC), with HNRNPA3 staining the
nucleus, are shown in Fig. 1.
HNRNPA3 expression was found to be negative or low in non-tumor
hepatic tissue, whereas its expression was high in DNs and highest
in HCC (Fig. 1). Of the 48 cases of
non-tumor hepatic tissue, only 1 case (2.08%) displayed positive
HNRNPA3 expression. By contrast, 5 out of 8 cases of DNs were
negative and the remaining cases (37.50%) were classified as
positive (Table I). Intriguingly, 40
out of 48 HCC cases (83.33%) exhibited positive expression, whereas
the remaining samples (16.67%) were negative.
 | Table I.Expression of HNRNPA3 in non-tumorous
hepatic tissue, DN and HCC. |
Table I.
Expression of HNRNPA3 in non-tumorous
hepatic tissue, DN and HCC.
|
|
| HNRNPA3
expression |
|
|---|
|
|
|
|
|
|---|
| Tissue samples | n | Negative, n (%) | Positive, n (%) | P-value |
|---|
| Non-tumorous hepatic
tissued | 48 | 47 (97.92) | 1 (2.08) | =0.0075a |
| DNd | 8 | 5 (62.50) | 3 (37.50) | =0.0123b |
| HCCd | 48 | 8 (16.67) | 40 (83.33) |
<0.0001c |
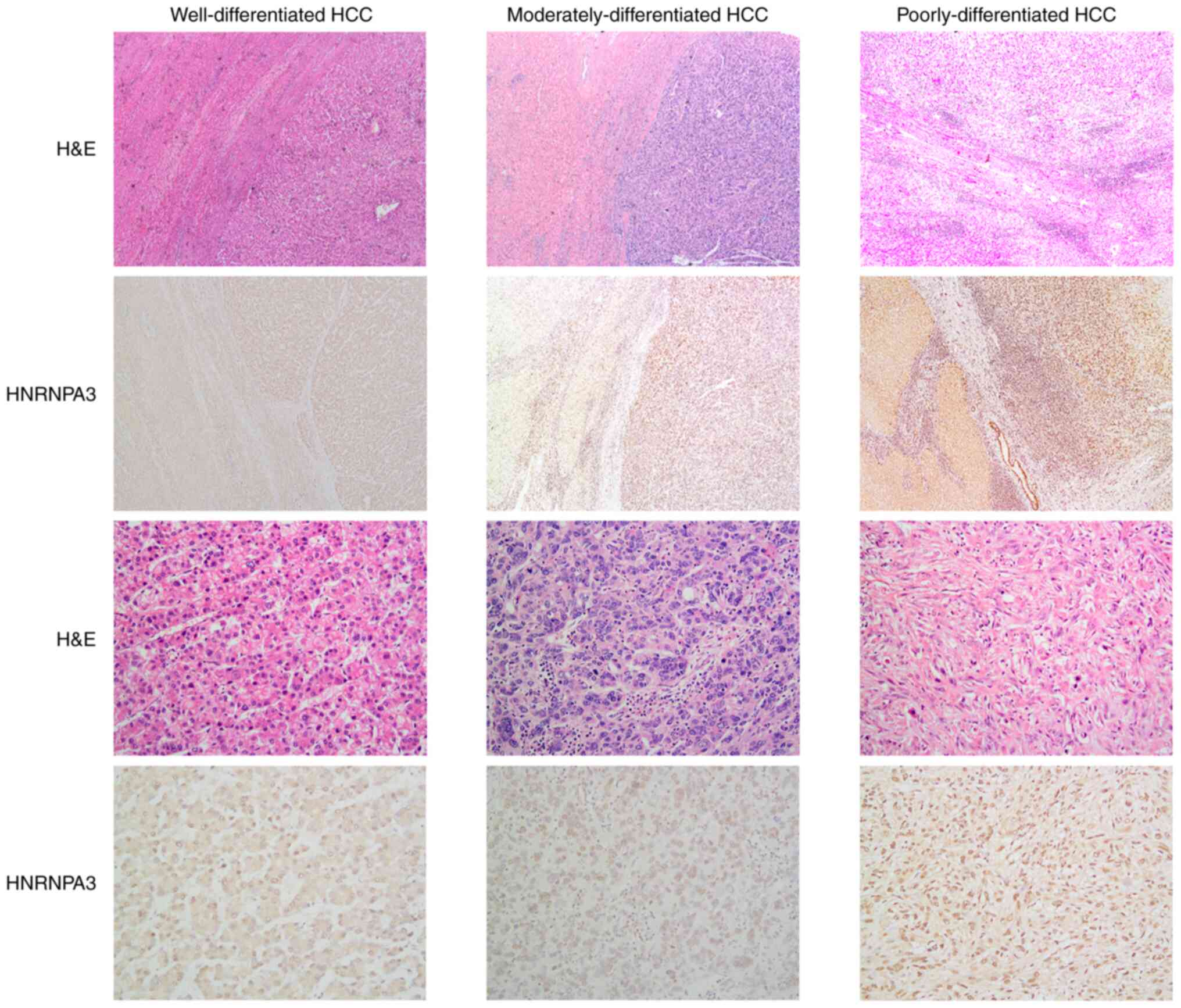
Immunostaining for HNRNPA3 was performed on
well-differentiated, moderately and poorly differentiated HCCs
(Fig. 2). Well-differentiated HCC
cases included 8 (80.00%) positive for HNRNPA3 expression and only
2 negative cases. In progressed HCC cases, HNRNPA3 immunoreactivity
was predominately positive (84.21%) among the 38 samples. In
addition, the present data revealed that HNRNPA3 expression
increased in a stepwise trend from non-tumor tissue via DNs to
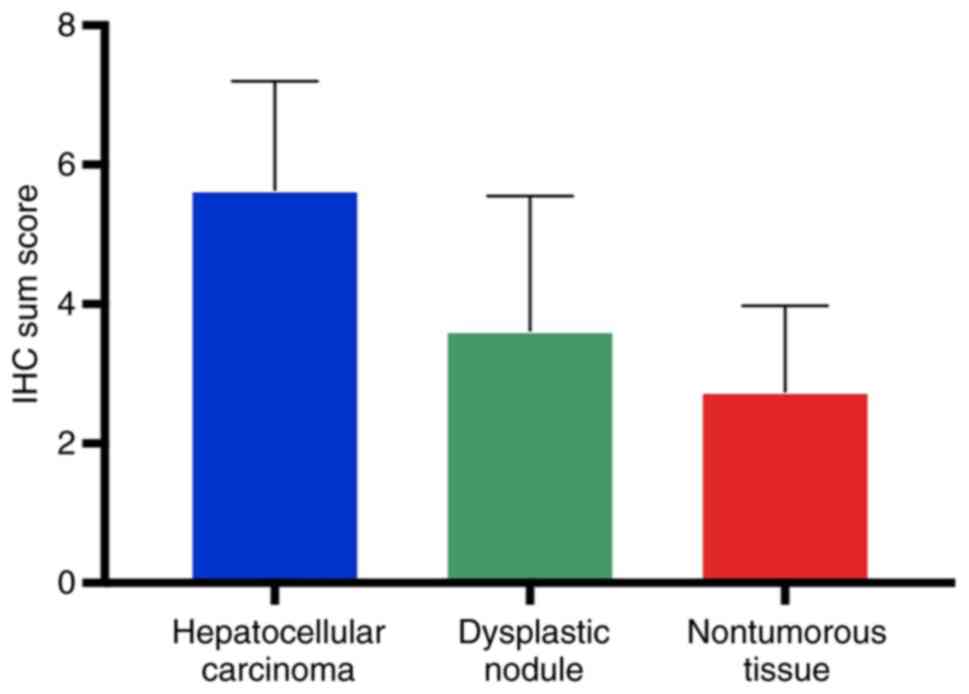
well-differentiated HCC and advanced HCC (Table I, Fisher's exact test; Fig. 3, Kruskal-Wallis test; P<0.0001).
It was also observed that the expression level of HNRNPA3 increased
along with an increasing degree of tumor malignancy and was highly
expressed in undifferentiated HCC samples (Fig. 2).
Clinical value of HNRNPA3 expression
in the differential diagnosis between HGDN and eHCC
ROC curves were used to evaluate the ability of
HNRNPA3 expression to differentiate between DN and HCC. The ROC
curves revealed that the AUC value was up to 0.8216 (95% CI,
0.6896–0.9537; P=0.0038; Fig. S1A).
ROC curves were also constructed to test the ability of HNRNPA3
expression to differentiate between non-tumor tissues and HCC. The
ROC curves revealed that the AUC value was up to 0.9225 (95% CI,
0.8625–0.9826; P<0.0001; Fig.
S1B). The same method was further applied to determine the
ability of HNRNPA3 expression to discriminate between DN and
well-differentiated HCC, with an AUC value of up to 0.8188 (95% CI,
0.6244–1.013; P=0.0235; Fig. S1C).
A noteworthy finding was that well-differentiated HCC could be
distinguished from HGDN by HNRNPA3 expression with an AUC value of
0.8083 (95% CI, 0.5934–1.023; P=0.0448; Fig. S1D). These data supported the value
of the expression level of HNRNPA3 in distinguishing specific
stages of hepatocarcinogenesis, from cirrhotic tissue to DN and
HCC. The differentially upregulated HNRNPA3 expression between HGDN
and well-differentiated HCC may therefore represent a specific
biomarker for this developmental stage of hepatocarcinogenesis.
GPC3 immunostaining was negative in 87.5% (7/8) of
DN samples, and only one sample exhibited positive expression. By
contrast, 81.25% of HCCs (39/48) displayed positive expression of
GPC3. The expression of GPC3 was markedly different between DN and
HCC. The sensitivity and specificity of GPC3 expression for
diagnosing HCC stage were 81.25 and 87.50%, respectively (Table SIII). Furthermore, HNRNPA3 and GPC3
were combined as a two-marker set in the differential diagnosis.
The positive expression of both markers was not observed in DNs.
Therefore, the two-marker set was used to improve the detection of
HCC, with a sensitivity of 70.83% and a specificity of 100% in
serial tests. The ROC yielded a larger AUC of 0.854 (95% CI,
0.734–0.934; P<0.0001) for HNRNPA3+/GPC3+
compared with that of GPC3 staining alone (AUC=0.844; 95% CI,
0.722–0.927; P<0.0001) in the diagnosis of HCC (Fig. S2A). When distinguishing between DN
and well-differentiated HCC, a larger AUC of 0.850 (95% CI,
0.605–0.972; P<0.0001; Fig. S2B
and Table SIII) and optimal
specificity (100%) were obtained from the combination of the two
markers compared with GPC3 staining alone (AUC=0.838; 95% CI;
P<0.0001). In addition, HNRNPA3 or GPC3 staining alone did not
have the highest diagnostic accuracy when distinguishing eHCC from
DN. The HNRNPA3+/GPC3+ serial test exhibited
an AUC of 0.857 (95% CI, 0.584–0.980; P<0.001; Fig. S2C and Table SIII), a sensitivity of 71.43% and a
specificity of 100.0% in distinguishing eHCC from DN. Furthermore,
the two-marker set may prove to be an effective biomarker for
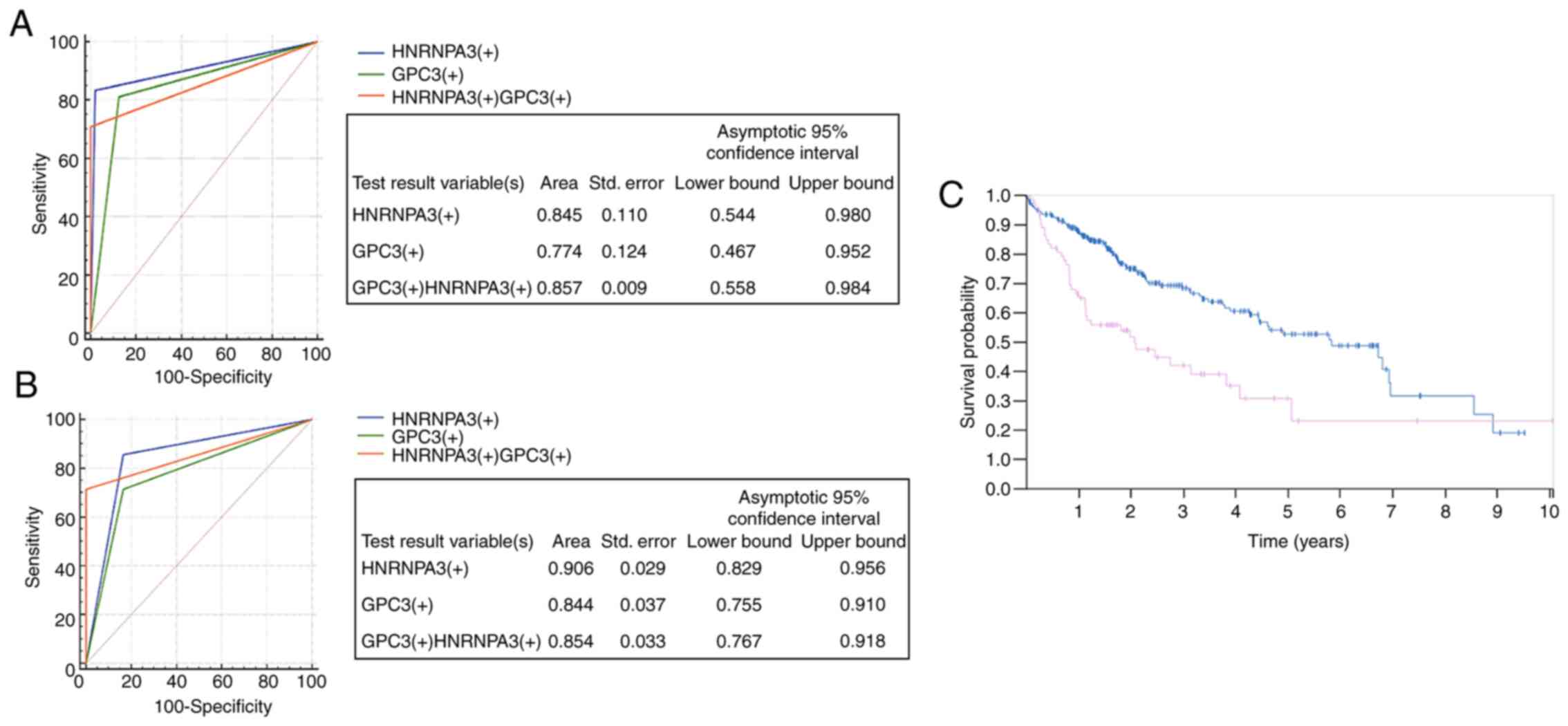
differentiating between eHCC and HGDN. The specificity of HNRNPA3
staining alone was 83.33%, whereas this increased to 100% with
HNRNPA3+/GPC3+, with the AUC of the combined
test at 0.857 (95% CI, 0.558–0.984; P=0.0001; Fig. 4A and Table II). Another notable discovery was
that HNRNPA3 staining alone (AUC=0.906) was superior compared with
GPC3 staining (AUC=0.844) or the combined assay (AUC=0.854) for
distinguishing HCC from non-tumor tissue, and the sensitivity and
specificity of HNRNPA3 staining were 83.33 and 97.92%, respectively
(95% CI, 0.829–0.956; P<0.0001; Fig.
4B, Table III). These results
indicated that HNRNPA3 may be superior to conventional markers,
such as GPC3, for differentiating HCC from non-tumor tissue.
 | Table II.Sensitivity, specificity, PPV and NPV
for detection of early HCC (n=7) from HGDN (n=6) using
HNRNPA3+, GPC3+ and combined
HNRNPA3+/GPC3+. |
Table II.
Sensitivity, specificity, PPV and NPV
for detection of early HCC (n=7) from HGDN (n=6) using
HNRNPA3+, GPC3+ and combined
HNRNPA3+/GPC3+.
| Phenotype | Early HCC, n | HGDN, n | Sensitivity | Specificity | PPV | NPV |
|---|
|
HNRNPA3+ | 6 | 1 | 85.71 | 83.33 | 85.7 | 83.3 |
|
GPC3+ | 5 | 1 | 71.43 | 83.33 | 83.3 | 71.4 |
|
HNRNPA3+/GPC3+ | 5 | 0 | 71.43 | 100.00 | 100.0 | 75.0 |
 | Table III.Sensitivity, specificity, PPV and NPV
for detection of HCC (n=48) from non-tumorous tissue (n=48) using
HNRNPA3+, GPC3+ and combined
HNRNPA3+/GPC3+. |
Table III.
Sensitivity, specificity, PPV and NPV
for detection of HCC (n=48) from non-tumorous tissue (n=48) using
HNRNPA3+, GPC3+ and combined
HNRNPA3+/GPC3+.
| Phenotype | HCC, n | Non-tumorous
tissue, n | Sensitivity | Specificity | PPV | NPV |
|---|
|
HNRNPA3+ | 40 | 1 | 83.33 | 97.92 | 97.6 | 85.5 |
|
GPC3+ | 39 | 6 | 81.25 | 87.50 | 86.7 | 82.4 |
|
HNRNPA3+/GPC3+ | 34 | 0 | 70.83 | 100.00 | 100.0 | 77.4 |
HNRNPA3 expression is associated with
tumor differentiation in HCC
To further investigate the clinical significance of
HNRNPA3 in liver cancer, the association between HNRNPA3 expression
and clinicopathological factors in HCC was investigated (Table IV). HNRNPA3 expression was found to
be significantly associated with tumor differentiation (P=0.0472).
However, no significant association was found between HNRNPA3
expression and other clinicopathological factors, including age,
sex, cirrhosis, tumor number, tumor size, tumor biomarker levels
and specific serum enzymes (Tables
IV and SIV). There was also no
significant association between HNRNPA3 expression and
clinicopathological factors in DN (Table SV).
 | Table IV.Association between HNRNPA3
expression and clinicopathological factors in HCC. |
Table IV.
Association between HNRNPA3
expression and clinicopathological factors in HCC.
|
|
| HNRNPA3
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | n | Negative | Positive | P-value |
|---|
| Agea |
|
|
|
|
|
≤55 | 14 | 3 | 11 |
|
|
>55 | 34 | 5 | 29 | 0.6757 |
| Gendera |
|
|
|
|
|
Male | 41 | 6 | 35 |
|
|
Female | 7 | 2 | 5 | 0.3297 |
| Tumor
sizea |
|
|
|
|
| ≤5 | 29 | 5 | 24 |
|
|
>5 | 19 | 3 | 16 | >0.9999 |
| Tumor number
(>1)a |
|
|
|
|
|
Yes | 6 | 1 | 5 |
|
| No | 42 | 7 | 35 | >0.9999 |
|
Cirrhosisa |
|
|
|
|
|
Yes | 32 | 5 | 27 |
|
| No | 16 | 3 | 13 | >0.9999 |
|
Differentiationb |
|
|
|
|
|
Well | 10 | 2 | 8 |
|
|
Moderate | 32 | 3 | 29 |
|
|
Poor | 6 | 3 | 3 | 0.0472 |
HNRNPA3 expression is associated with
survival rate
According to the Human Protein Atlas, high
expression of HNRNPA3 indicates a lower survival rate compared with
that of patients with liver cancer exhibiting low expression of
HNRNPA3 (Fig. 4C).
Discussion
HCC is the third most frequent cause of
cancer-related mortality, and patients who are diagnosed at an
earlier stage and receive effective treatment have improved overall
survival (20). However, in the
majority of cases, HCC is diagnosed at a late stage. HBV infection
and Hepatitis C Virus infection are the most important risk factors
for HCC. Due to the high infection rate of chronic HBV in China,
the present study focused on HBV-related HCC. HBV-induced
hepatocarcinogenesis is a multistep process progressing from
cirrhosis, to LGDN, to HGDN, and finally to eHCC and progressed HCC
(2). LGDN hepatocytes display
minimal abnormalities, with well-defined boundaries, normal or
slightly increased nuclear-cytoplasmic ratio and less nuclear
atypia. Mitotic activity is absent and the portal tract is present.
The thickness of the liver plate is 1–2 cell layers, it does not
include pseudoglandular arrangements, and there is no obvious
thickening. HGDNs display increased nuclear-cytoplasmic ratio, and
more pronounced nuclear atypia and basophilic cytoplasm, the
density of the cells is >twice the normal density, occasional
mitotic figures may be seen, and the thickness of the liver plate
is ≤3 cell layers. On microscopic observation there is a relative
HGDN boundary, but this boundary is not clear on high
magnification. Irregular trabecular hepatocytes are frequently
arranged in HGDNs. Occasionally, pseudoadenoid arrangement may be
seen (16). It is difficult to
confirm whether the diagnosis is HGDN or well-differentiated HCC in
the clinical setting, as they share similar pathological
characteristics. The applicability of ultrasonography for
diagnosing HCC in clinical practice is limited by the morphological
similarity with tumors, operator dependency and deficient
diagnostic accuracy (21).
Furthermore, some widely accepted serum tumor markers, including
α-fetoprotein, have not demonstrated satisfactory specificity and
sensitivity in the diagnosis of HCC (22).
HnRNPs are located at the border regions of
chromatin to interact with newly synthesized nuclear RNAs (23). It is widely accepted that members of
this protein family are known to act as regulators in cell cycle
progression, cell differentiation, cell cycle arrest and DNA
damage. HNRNP K has been demonstrated to coordinate with P53 in a
mutually dependent manner under conditions of DNA damage, with
knockdown of HNRNP K leading to defects in cell-cycle checkpoint
arrest (24).
HNRNPA3 is a protein encoded in humans by the
HNRNPA3 gene and belongs to the hnRNP A/B family, along with
other candidate transcription factors that interact with the
regulatory region of the HOXC8 gene (25). HNRNPA3 is located on
chromosome 2, which is well known for its ability to regulate
telomere length (26). It plays key
roles in RNA binding, mRNA transport and mRNA splicing via
spliceosome. Although the detailed association between
tumorigenesis and HNRNPA3 has not been fully elucidated, the HNRNP
protein family members are closely associated with cancer
regulation (14,27). Moreover, HNRNPA3 has potential roles
in carcinogenesis that have yet to be extensively investigated.
Overexpression of another family member, HNRNPA1/A2, was found to
control alternative splicing of the pyruvate kinase M gene and,
thus, favor aerobic glycolysis to enhance tumorigenesis (27–29).
HNRNPA3 expression has not been investigated in the
context of hepatocarcinogenesis before, and evaluation methods for
HNRNPA3 using IHC are also unknown. Therefore, in the present study
a semi-quantitative scoring criterion was employed in the IHC
evaluation for HNRNPA3, considering both the staining intensity and
percentage of positively stained cells. The subsequent
verifications including ROC curves confirmed that this evaluation
system is appropriate for HNRNPA3 analysis and diagnosing specific
HCC stage. Based on the data and IHC results of the present study,
HNRNPA3 expression was shown to increase in a stepwise manner from
non-tumorous hepatic tissue via DN to HCC. It was verified by AUC
that HCC was distinguishable from non-tumor tissue based on the
changes in the expression of HNRNPA3, which appeared to be more
effective compared with the currently available biomarkers.
Moreover, HGDN and eHCC can be differentiated by the expression of
HNRNPA3. GPC3 is highly expressed in HCC and is commonly used as a
marker for differential diagnosis between HGDN and eHCC; however,
it lacks sufficient sensitivity and specificity (9,10,12). Of
note, the combined two-marker set
(HNRNPA3+/GPC3+) was shown to increase the
diagnostic accuracy between HGDN and eHCC, with a larger AUC and
100% specificity. Of note, more aggressive tumors, such as
undifferentiated HCC, may be accompanied by high levels of HNRNPA3
expression. The Human Protein Atlas also supported that high
expression of HNRNPA3 was associated with poor survival rate of
patients with HCC. Therefore, HNRNPA3 may be considered as a
potential diagnostic marker to identify specific stages during HCC
development, and its overexpression may indicate a poor
prognosis.
In regards to the association between HNRNPA3
expression and clinicopathological factors in HCC, the
χ2 test indicated that HNRNPA3 was associated with tumor
differentiation. No such association was observed with serum enzyme
levels, tumor biomarkers and HCC. However, the small sample size
may limit the ability to evaluate the potential association between
HNRNPA3 and clinicopathological factors in DN.
Collectively, the data of the present study revealed
that the expression of HNRNPA3 increased in a stepwise manner from
non-tumor cirrhotic tissue to DN and was the highest in HCC.
HNRNPA3 combined with GPC3 may prove to be of value as a diagnostic
tool to distinguish between HGDN and eHCC, which could provide a
novel therapeutic strategy for pathologists to diagnose HCC with
high accuracy. A high level of HNRNPA3 expression was also found to
be associated with lower tumor differentiation and poor survival of
patients with HCC. However, there is a limitation in the present
study. Only the role of HNRNPA3 in the progression of HBV-related
HCC was investigated. Further studies are warranted to reveal the
potential significance and prognostic value of HNRNPA3 in
HCV-related HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81672842, 81872362
and 81802914) and The Taishan Scholars Program of Shandong Province
(grant no. ts201511096).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PG and HZ conceived and designed the present study.
XR, MD and YD performed the experiments and drafted the initial
manuscript. All authors have read and approved the final
manuscript, and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shandong University (Jinan, China; approval no.
2012028). Written informed consent was provided by all participants
prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kudo M: Multistep human
hepatocarcinogenesis: Correlation of imaging with pathology. J
Gastroenterol. 44 (Suppl 19):S112–S118. 2009. View Article : Google Scholar
|
|
3
|
Saito K, Kotake F, Ito N, Ozuki T, Mikami
R, Abe K and Shimazaki Y: Gd-EOB-DTPA enhanced MRI for
hepatocellular carcinoma: Quantitative evaluation of tumor
enhancement in hepatobiliary phase. Magn Reson Med Sci. 4:1–9.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Inchingolo R, Faletti R, Grazioli L,
Tricarico E, Gatti M, Pecorelli A and Ippolito D: MR with
Gd-EOB-DTPA in assessment of liver nodules in cirrhotic patients.
World J Hepatol. 10:462–473. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saito J, Kim SR, Kudo M, Imoto S, Ando K,
Nakajima T, Fukuda K, Otono Y, Kim SK, Komaki T, et al:
Well-differentiated hepatocellular carcinoma detected as
hypovascularity by only CT during hepatic arteriography. Intern
Med. 51:885–890. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sugimoto K, Kim SR, Imoto S, Tohyama M,
Kim SK, Matsuoka T, Yano Y, Kudo M and Hayashi Y: Characteristics
of hypovascular versus hypervascular Well-differentiated
hepatocellular carcinoma smaller than 2 cm-Focus on tumor size,
markers and imaging detectability. Dig Dis. 33:721–727. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borentain P, Carmona S, Mathieu S, Jouve
E, El-Battari A and Gérolami R: Inhibition of E-selectin expression
on the surface of endothelial cells inhibits hepatocellular
carcinoma growth by preventing tumor angiogenesis. Cancer Chemother
Pharmacol. 77:847–856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng WG, Fu Y, Li YL and Sugiyama T:
Potential role of p53 mutation in chemical hepatocarcinogenesis of
rats. World J Gastroenterol. 10:46–52. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ho M and Kim H: Glypican-3: A new target
for cancer immunotherapy. Eur J Cancer. 47:333–338. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamauchi N, Watanabe A, Hishinuma M,
Ohashi K, Midorikawa Y, Morishita Y, Niki T, Shibahara J, Mori M,
Makuuchi M, et al: The glypican 3 oncofetal protein is a promising
diagnostic marker for hepatocellular carcinoma. Mod Pathol.
18:1591–1598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bakheet AMH, Zhao C, Chen JN, Zhang JY,
Huang JT, Du Y, Gong LP, Bi YH and Shao CK: Improving pathological
early diagnosis and differential biomarker value for hepatocellular
carcinoma via RNAscope technology. Hepatol Int. 14:96–104. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Tommaso L, Franchi G, Park YN, Fiamengo
B, Destro A, Morenghi E, Montorsi M, Torzilli G, Tommasini M,
Terracciano L, et al: Diagnostic value of HSP70, glypican 3, and
glutamine synthetase in hepatocellular nodules in cirrhosis.
Hepatology. 45:725–734. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mishra N, Reddy KS, Timilsina U, Gaur D
and Gaur R: Human APOBEC3B interacts with the heterogenous nuclear
ribonucleoprotein A3 in cancer cells. J Cell Biochem.
119:6695–6703. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boukakis G, Patrinou-Georgoula M,
Lekarakou M, Valavanis C and Guialis A: Deregulated expression of
hnRNP A/B proteins in human non-small cell lung cancer: Parallel
assessment of protein and mRNA levels in paired tumour/non-tumour
tissues. BMC Cancer. 10:4342010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagtegaal I and Washington M:
WHO_Classification_of_Tumours_Digestive.pdf. Nagtegaal I, Klimstra
D and Washington M: World Health Organization; 2019
|
|
16
|
Nascimento C, Bottino A, Nogueira C and
Pannain V: Analysis of morphological variables and arterialization
in the differential diagnosis of hepatic nodules in explanted
cirrhotic livers. Diagn Pathol. 2:512007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shao S, Li Z, Gao W, Yu G, Liu D and Pan
F: ADAM-12 as a diagnostic marker for the proliferation, migration
and invasion in patients with small cell lung cancer. PLoS One.
9:e859362014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nguyen TB, Roncalli M, Di Tommaso L and
Kakar S: Combined use of heat-shock protein 70 and glutamine
synthetase is useful in the distinction of typical hepatocellular
adenoma from atypical hepatocellular neoplasms and
well-differentiated hepatocellular carcinoma. Mod Pathol.
29:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:P1301–P1314. 2018. View Article : Google Scholar
|
|
21
|
Singal AG, Nehra M, Adams-Huet B, Yopp AC,
Tiro JA, Marrero JA, Lok AS and Lee WM: Detection of hepatocellular
carcinoma at advanced stages among patients in the HALT-C trial:
Where did surveillance fail. Am J Gastroenterol. 108:425–432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marrero JA, Feng Z, Wang Y, Nguyen MH,
Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D,
et al: Alpha-fetoprotein, Des-gamma carboxyprothrombin, and
Lectin-bound Alpha-fetoprotein in early hepatocellular carcinoma.
Gastroenterology. 137:110–118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fakan S, Leser G and Martin TE:
Ultrastructural distribution of nuclear ribonucleoproteins as
visualized by immunocytochemistry on thin sections. J Cell Biol.
98:358–363. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moumen A, Masterson P, O'Connor MJ and
Jackson SP: hnRNP K: An HDM2 target and transcriptional coactivator
of p53 in response to DNA damage. Cell. 123:1065–1078. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Makeyev AV, Kim CB, Ruddle FH, Enkhmandakh
B, Erdenechimeg L and Bayarsaihan D: HnRNP A3 genes and pseudogenes
in the vertebrate genomes. J Exp Zool Part A Comp Exp Biol.
303A:259–271. 2005. View Article : Google Scholar
|
|
26
|
Tanaka E, Fukuda H, Nakashima K, Tsuchiya
N, Seimiya H and Nakagama H: HnRNP A3 binds to and protects
mammalian telomeric repeats in vitro. Biochem Biophys Res Commun.
358:608–614. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
David CJ, Chen M, Assanah M, Canoll P and
Manley JL: HnRNP proteins controlled by c-Myc deregulate pyruvate
kinase mRNA splicing in cancer. Nature. 463:364–368. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen M and Manley JL: Mechanisms of
alternative splicing regulation: Insights from molecular and
genomics approaches. Nat Rev Mol Cell Biol. 10:741–754. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harada Y, Nakamura M and Asano A:
Temporally distinctive changes of alternative splicing patterns
during myogenic differentiation of C2C12 cells. J Biochem.
118:780–790. 1995. View Article : Google Scholar : PubMed/NCBI
|