Introduction
Pancreatic cancer is the seventh leading cause of
global cancer deaths in industrialized countries (1) and the third most common in the USA
(2). Chinese people increasingly
live in urban areas; when combined with other factors, such as
aging and environmental pollution, this has shifted the disease
spectrum in China from infectious to non-infectious diseases and
the health burden of cancer is increasing (3). In 2010, 34,509 men and 23,226 women
died from pancreatic cancer in China, with the number of deaths
exceeding that in the United States (4,5).
Pancreatic ductal adenocarcinoma (PDAC) is a type of exocrine
pancreatic cancer and is the most common type of pancreatic cancer
with 95% of all pancreatic cancer cases being PDAC (6).
Existing therapies for PDAC leave much to be
desired. Surgery is the most commonly used treatment for pancreatic
cancer; even so, only ~20% of patients with pancreatic cancer are
candidates for surgery as most pancreatic cancers are first
diagnosed after the disease has metastasized (7). In recent years, targeted therapy has
been employed in pancreatic cancer; for instance, erlotinib is
approved by the Food and Drug Administration authority for patients
with advanced pancreatic cancer in combination with gemcitabine
(8), and larotrectinib
(Vitrakvi®) is approved as a treatment for pancreatic
cancer that is metastatic or cannot be removed with surgery
(9). However, targeted therapy
options for pancreatic cancer remain very limited. Hence, the
identification of novel target molecules for PDAC is vitally
important.
A previous study of aggressive glioblastoma
identified rhophilin 2 (RHPN2) as a novel driver gene of
mesenchymal transformation (10).
The aforementioned study demonstrated that RHPN2 gene amplification
was associated with a dramatic decrease in the overall survival of
patients with glioma. RHPN2 has been described as a Ras homolog
family member A (RhoA)-binding protein, but its biological function
remains unclear (11,12).
The function of RHPN2 in PDAC remains unknown. The
aim of the present study was to elucidate the function of RHPN2 in
PDAC and identify the possible therapeutic target gene for PDAC
therapy.
Materials and methods
Bioinformatics analysis
cBio Cancer Genomics Portal (https://cbioportal.org) was used to explore the role
of RHPN2 in cancer genomics (13,14). The
frequency of alterations of RHPN2 and patients' survival data in
pan-cancer and pancreatic cancer were exported from the cBio Cancer
Genomics Portal. Gene Expression Omnibus (GEO) dataset values were
downloaded from the GDS4336/8035980 datasets (45 pairs of PDAC
tumor and adjacent normal tissue, Homo sapiens) (15). The protein network of RHPN2 was
analyzed using Cytoscape software version 3.8.0 (16).
Tissue samples
PDAC tissues and matching adjacent normal tissues
(61 pairs) were acquired from the Sichuan Provincial Cancer
Hospital (Chengdu, China). Patients were recruited from April 2013
to November 2015. The inclusion criteria were as follows: i) No
history of any other active cancer; ii) no active cancer treatment;
and iii) no past history of pancreatic cancer. The tissue sample
were acquired by resection. The distance between the PDAC tissues
and matching adjacent normal tissues was ~1 cm. The mean age of the
patients was 65.8 years, age range, 44–81 years and there were 27
males and 34 females. Tissue were stored at −80°C before fixation.
For long-term preservation, tissues were embedded in paraffin.
Patients were followed-up for at least 5 years. The Ethics
Committee of Sichuan University (Chengdu, China) approved the use
of the human samples (approval no. 20191109). All specimens were
used properly in accordance with the protocol of the Ethics
Committee. Written informed consent was obtained from all the
patients enrolled in the present study. The classification systems
for stage and grade used in the present study were from AJCC Cancer
Staging Manual 8th edition (17).
Immunohistochemistry (IHC)
RHPN2 protein levels were assessed by IHC. Tumor
tissue were immersed in 10% neutral buffered formalin (NBF) for
fixation for at least 3 days at room temperature. Then the whole
tissues were dehydrated in a gradients of ethanol (incubated 50%
ethanol for 10 min, 70% ethanol for 10 min, 80% ethanol for 10 min,
95% ethanol for 10 min, 100% ethanol for 10 min thrice). The
tissues were exchanged ethanol with xylene in room temperature in
the following sequence: 2:1 ethanol:xylene for 10–15 min, 1:1
ethanol:xylene for 10–15 min, 1:2 ethanol:xylene for 10–15 min,
100% xylene for 10–15 min thrice. Then xylene was exchanged with
paraffin in 58°C in the following sequence: 2:1 ×ylene:paraffin for
30 min, 1:1 ×ylene: paraffin for 30 min, 1:2 ×ylene:paraffin for 30
min, 100% paraffin for 2 h and 100% paraffin overnight, and then
embedded in a fresh new paraffin. Next, 4-µm thick paraffin
sections were cut. After deparaffinization and hydration (the
reverse sequence of paraffinization and dehydration), the slides
were microwaved for antigen retrieval. The slides were blocked in
BSA blocking buffer (cat. no. 37520; Thermo Fisher Scientific Inc.)
at room temperature for 1 h. The slides were then incubated at 4°C
overnight with anti-RHPN2 (1:1,000; cat. no. PA5-62469; Thermo
Fisher Scientific Inc.). After washing three times with
phosphate-buffered saline (PBS), the slides were incubated with
goat anti-rabbit poly-horseradish peroxidase-conjugated secondary
antibody (1:500; cat. no. 32260; Thermo Fisher Scientific Inc.) at
room temperature for 2 h. The slides were then developed using
diaminobenzidine (DAB) staining (18). The hematoxylin staining was performed
using the H&E stain kit (cat no. ab245880; Abcam). The slide
were incubated in hematoxylin, Mayer's (Lillie's Modification) for
5 min at 4°C. Then slides were rinsed with two changes of distilled
water to remove excess stain. Subsequently, the slides were
incubated with bluing reagent from the H&E stain kit for 10
sec. These slides were observed by light microscope Leica DM3000
(Leica Microsystems Ltd.) (magnification, ×200). Images were
assessed using the Aperio ImageScope software version 12.4.0.5043
(Leica Microsystems).
Cell culture
Pancreatic cancer cell lines PANC1 (cat. no.
CRL-1469™) and SW1990 (cat. no. CRL-2172™) were purchased from
ATCC. PANC1 cells were cultured in DMEM medium (cat. no. 11965092;
Thermo Fisher Scientific Inc.) with 10% fetal bovine serum (cat.
no. 12483020; Thermo Fisher Scientific Inc.). SW1990 cells were
cultured in Leibovitz's L-15 Medium (cat. no. 11415056; Thermo
Fisher Scientific Inc.) with 10% fetal bovine serum (cat. no.
12483020; Thermo Fisher Scientific Inc.). Cells and medium were
cultured at 37°C with 5% CO2. The exponentially growing
cells were used for subsequent experiments.
Detection of RHPN2 and CEP78 by
reverse-transcription quantitative (RT-q)PCR
Total RNA from tumor tissues and PANC1 and SW1990
cells was extracted with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific Inc.) following the manufacturer's instructions.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an
internal control. cDNA synthesis were performed by QuantiTect
Reverse Transcription Kit (cat. no. 205311; Qiagen GmbH). Reactions
were incubated at 42°C for 50 min followed by heat inactivation for
5 min at 80°C for reverse transcription. The gene expression levels
were assessed via qPCR using the 2−ΔΔCq method (19). The PCR amplification was performed
using SYBR™-Green PCR Master Mix (cat. no. 4334973; Thermo Fisher
Scientific, Inc.). The primers used were as follows: RHPN2,
forward, 5′-AAGGGCTGTAATCCCCTTGC-3′ and reverse,
5′-CCGCACCTTTGAGTTTGTGG-3′; centrosomal protein 78 (CEP78),
forward, 5′-TGGCAGGGAGCAGATCACA-3′ and reverse,
5′-AAGCCAGCCATACAGTCAAGA-3′; and GAPDH, forward,
5′-CTGACTTCAACAGCGACACC-3′ and reverse,
5′-TAGCCAAATTCGTTGTCATACC-3′. Thermocycling conditions consisted of
50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C
for 15 sec and 60°C for 60 sec. RHPN2 mRNA expression levels in the
control were arbitrarily defined as 100%. The relative RHPN2 levels
[log2 (Tumor/Normal)] were calculated.
Transfection
Small interfering (si) RNA-RHPN2 and pcDNA3.1-RHPN2
transfection were performed. siRNA-RHPN2 and pcDNA3.1-RHPN2 were
designed and constructed by Shanghai Shengong Biology Engineering
Technology Service., Ltd. Scrambled siRNA (si-NC) and pcDNA3.1 were
used as controls. The sequences used were as follows: RHPN2,
5′-AAGCTGCGGAGCATTGAGGTG-3′ and scrambled siRNA,
5′-GGTGCCGAATTGAGGTGACGA-3′. Cells were seeded into 24-well plates
at a density of 5×104 cells/well overnight. Transfection
was performed using Lipofectamine® 2000 (cat. no.
11668027; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. SiRHPN2 (0.6 µg) or pcDNA3.1-RHPN2 (1
µg) were used separately. Si-NC (0.6 μg) or empty plasmid (1 µg)
were used as control. The cells were transfected at 37°C in a
CO2 incubator for 24 h. RHPN2 mRNA expression levels of
the si-NC transfection group and those of the pcDNA3.1 transfection
group were arbitrarily defined as 100%. Cell proliferation analysis
was performed at 0, 24, 48 and 72 h following transfection, and
cell apoptosis analysis was performed 24 h following transfection.
The untransfected cells were also used as blank controls.
Cell proliferation assay
Cellular growth was analyzed by the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT)-based colorimetric assay (9–13).
Briefly, cells were placed into 96-well plates at a density of
5×105/well. MTT reagent was added to the medium at a
final concentration of 0.1 mg/ml. After the formation of insoluble
formazan, 100 µl of dimethyl sulfoxide was added to each well to
solubilize the formazan. The optical density was measured on a
microplate reader equipped with a 570 nm filter.
Cell apoptosis analysis
Transfected PANC1 cells and SW1990 cells were
suspended at 5×105/ml in Annexin V-fluorescein
isothiocyanate (FITC) (Abcam) in Annexin binding buffer for flow
cytometry (cat. no. V13246; Thermo Fisher Scientific Inc.). The
suspension was incubated for 15 min at room temperature followed by
the addition of 0.5 µl propidium iodide (PI; Abcam) to each sample.
Samples were analyzed using a FACSCalibur instrument (BD
Biosciences) using a 488 nm excitation line (argon ion laser or
solid-state laser) and emission was detected at 530 nm (for FITC)
and 575–610 nm (for PI). The data were analyzed using the BD
FACSuite™ version 1.01 (BD Biosciences). Early stage apoptosis was
assessed.
Statistical analyses
All the experiments were repeated three times. Data
are presented as mean ± standard deviation (SD). Survival data were
analyzed using the log-rank test. Kaplan-Meier analysis was used
for generation of the survival curves. Paired two-tailed Student's
t-tests were used to analyze the mean values of paired groups
(tumor and normal tissue from the same patient). Unpaired
two-tailed Student's t-tests were used to analyze the mean values
of the two PDAC cell lines. One-way ANOVA was used followed by the
post hoc Tukey's test was used for multiple comparisons. All
calculations were performed using SPSS software (version 16.0; SPSS
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
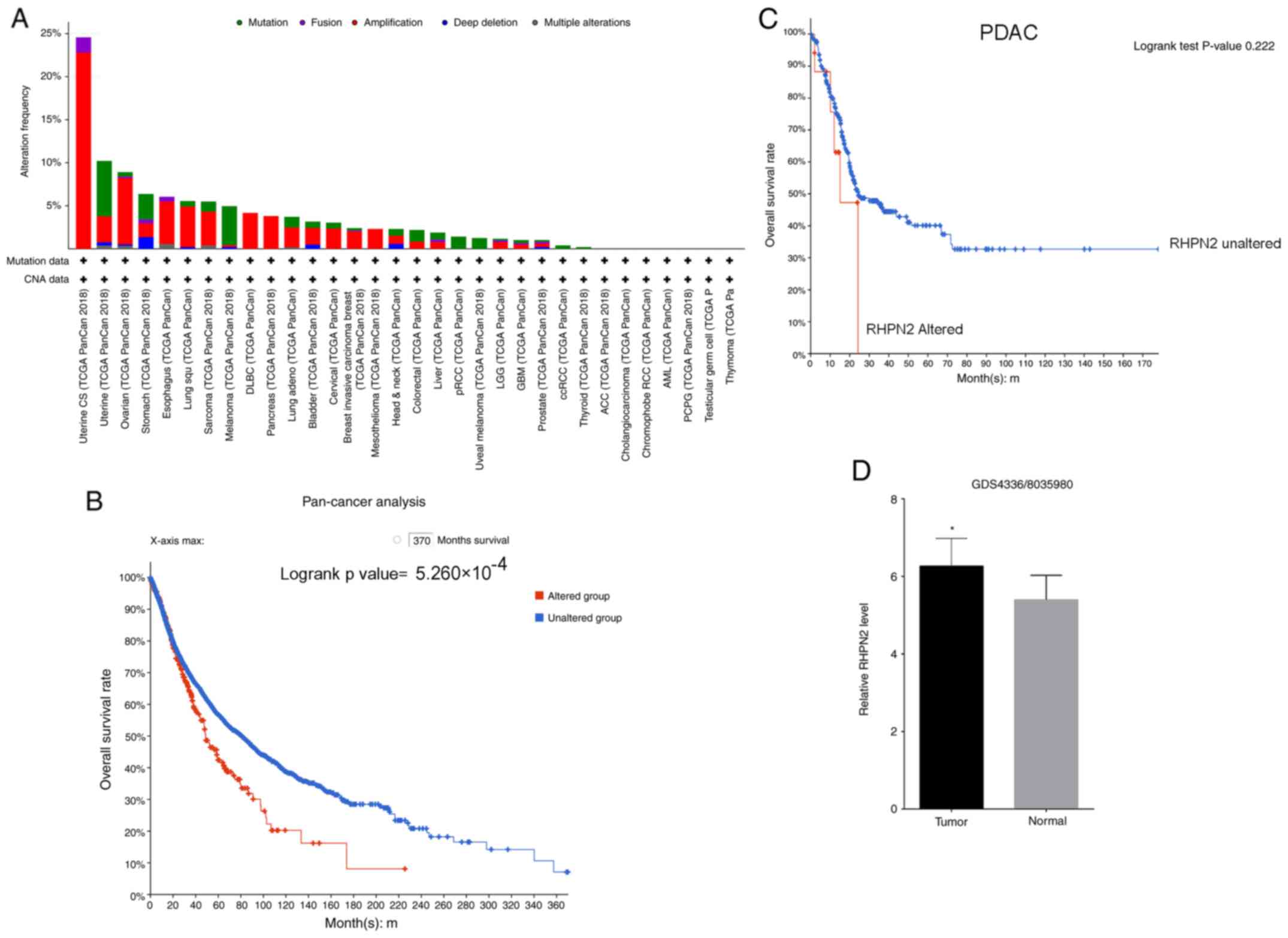
Bioinformatics analysis of RHPN2 in
pan-cancer and patients with pancreatic cancer
Investigation of the frequency of alterations of
RHPN2 in pan-cancer analysis revealed that the predominant
alteration was amplification (Fig.
1A). Pan-cancer patients were divided into two groups,
RHPN2-altered and RHPN2-unaltered, and the patients' survival
according to their RHPN2 alteration status was tested. The results
revealed that patients with pan-cancer in the RHPN2-altered group
had a lower overall survival rate compared with the RHPN2-unaltered
group (Fig. 1B). Next, the survival
rate of patients with pancreatic cancer was tested according to
whether RHPN2 was altered or not (Fig.
1C), and it was demonstrated that the difference between the
RHPN2-altered and RHPN2-unaltered groups was not significant; which
was may be due to the limited number of patients. Finally, the GEO
dataset for RHPN2 expression in 45 pairs of tumor tissues and
adjacent normal tissues (GDS4336/8035980) was assessed and it was
demonstrated that tumor tissues showed higher RHPN2 levels compared
with adjacent normal tissues (Fig.
1D).
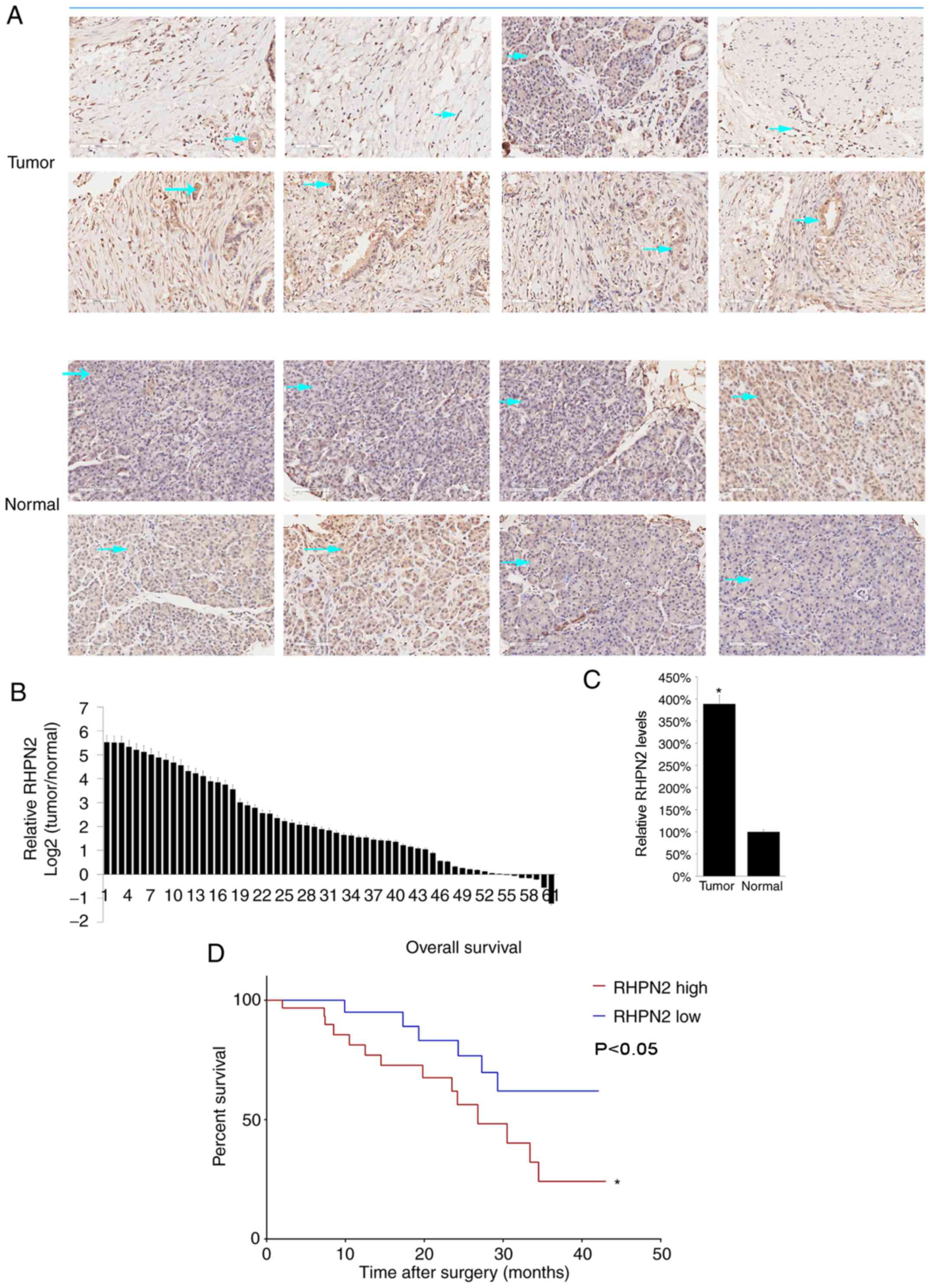
High RHPN2 levels in PDAC tissues are
correlated with low survival rate
To further study the role of RHPN2, PDAC tissues
with matched adjacent normal tissues from 61 patients from the
Sichuan Provincial Cancer Hospital (Chengdu, China) were collected.
The clinical information of the 61 patients with PDAC are listed in
Table SI. Majority of the patients
had stage IIB PDAC (Table SI).
RHPN2 protein levels were assessed by immunohistochemistry (IHC).
Eight representative cases were presented and it was demonstrated
that RHPN2 is mostly expressed in the cytoplasm as denoted by the
arrows (Fig. 2A). Next, RHPN2 mRNA
levels were determined by RT-qPCR. The results demonstrated that
RHPN2 mRNA levels were higher in tumor tissue compared with normal
tissues (Fig. 2B), and the mean
value of mRNA RHPN2 levels in tumor tissues was higher compared
with that in normal tissues (Fig.
2C). In addition, the relationship between RHPN2 and the
survival of patients with PDAC was assessed by dividing patients
into RHPN2-high vs. RHPN2-low groups according to the median value
of RHPN2 (3.59). Patient survival was followed across 50 months and
the RHPN2-high group had a lower survival rate compared with the
RHPN2-low group (Fig. 2D).
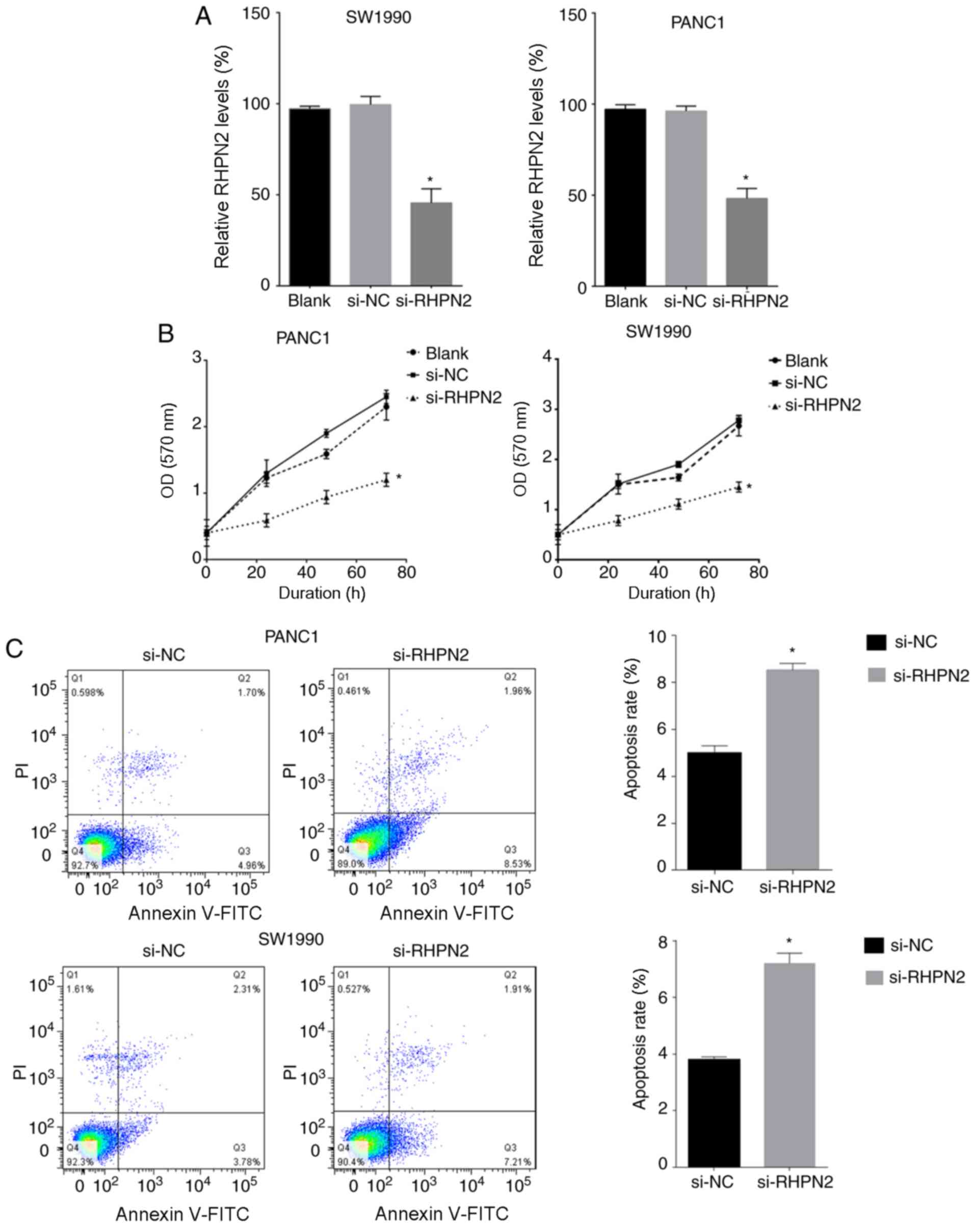
RHPN2 inhibition reduces PDAC cell
proliferation and increases apoptosis rate
PANC1 and SW1990 cell lines were used for testing
the effects of RHPN2 on cultured PDAC cells. PANC1 and SW1990 cells
were transfected with si-RHPN2. The downregulation of RHPN2 mRNA
levels was confirmed by RT-qPCR in both cell lines (Fig. 3A). The proliferation of PANC1 and
SW1990 cells was tested by the MTT assay, which demonstrated that
inhibition of RHPN2 reduced PDAC cell proliferation (Fig. 3B). Cell apoptosis was assayed using
Annexin V/propidium iodide (PI) double-staining. Downregulation of
RHPN2 increased the apoptosis rate of PANC1 and SW1990 cells
compared with the negative control (Fig.
3C).
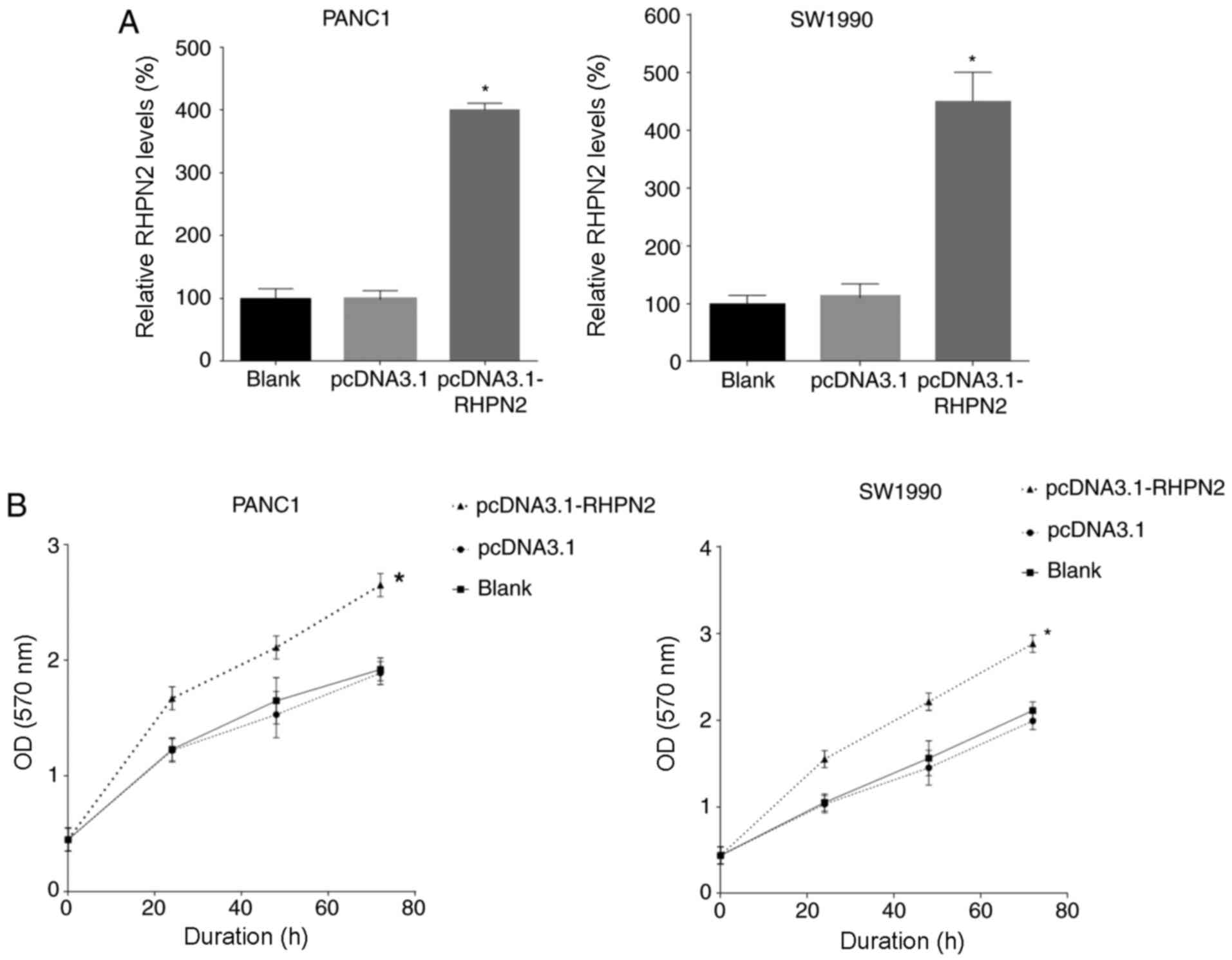
Overexpression of RHPN2 promotes PDAC
cell proliferation
Next, RHPN2 was overexpressed in PANC1 and SW1990
cells by transfecting with RHPN2 overexpression plasmid
(pcDNA3.1-RHPN2). RHPN2 levels were tested 12 h later by RT-qPCR
and the results demonstrated that RHPN2 expression was upregulated
by plasmid transfection (Fig. 4A).
The proliferation of PANC1 and SW1990 cells was tested by the MTT
assay and the results demonstrated that overexpression of RHPN2
promoted PANC1 and SW1990 cell proliferation (Fig. 4B).
CEP78 expression is negatively
associated with RHPN2 expression
A previous study demonstrated that RHPN2 activated
RhoA (10). In the present study,
the network of RHPN2 was constructed and it was revealed that CEP78
was one of the proteins that interacted with RHPN2. The alteration
frequency of CEP78 in pan-cancer analysis was assessed and the
findings revealed that the most frequent alteration of CEP78 was
mutation (Fig. 5B). Subsequently,
the CEP78 mRNA levels were analyzed by RT-qPCR, and relative CEP78
levels [log2 Tumor/Normal)] were calculated. The present
study demonstrated that CEP78 mRNA levels were lower in tumor
tissues (Fig. 5C), and the mean
value of CEP78 levels in tumor tissues was lower compared with
CEP78 levels in normal tissues (Fig.
5D). In addition, the significance of CEP78 in patients with
PDAC survival was assessed after dividing the 61 patients into
CEP78-high vs. CEP78-low groups according to the median value of
CEP78 (2.78). Patient survival was followed across 50 months, and
we found that the CEP78-low group had a lower survival rate
compared with the CEP-78 high group (Fig. 5E), which is consistent with a
previous study that demonstrated that low expression of CEP78 is
associated with poor prognosis of patients with colorectal cancer
(20). Next, to evaluate whether the
expression of RHPN2 is related with that of CEP78, RHPN2 levels
were overexpressed or silenced in cultured PDAC cells by
transfection of pcDNA3.1-RHPN2 and si-RHPN2, respectively and it
was demonstrated that overexpression of RHPN2 decreased CEP78
levels (Fig. 5F) and silencing of
RHPN2 increased CEP78 levels (Fig.
5G) in both PANC1 and SW1990 cells. The aforementioned results
indicate that CEP78 is involved in RNPN2 function.
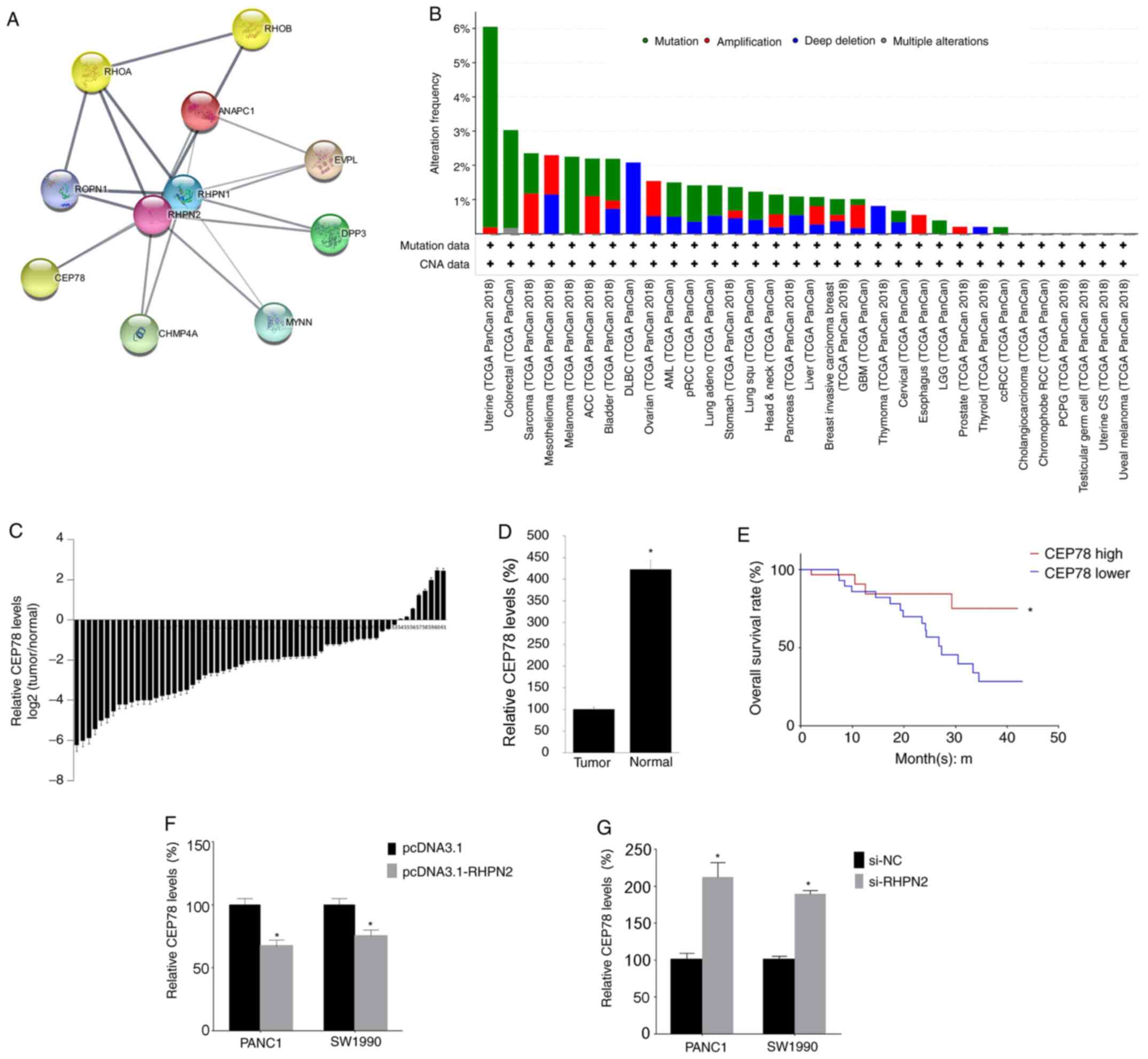 | Figure 5.RHPN2 network and correlation
analysis. (A) The RHPN2 network as revealed by Cytoscape software.
(B) The amplification of CEP78 across various types of cancer. (C)
CEP78 mRNAs levels in the 61 tumor samples and matching normal
adjacent tissues were quantified by RT-qPCR. (D) The mean value of
CEP78 mRNA expression in tumor tissue and normal adjacent tissues.
(E) CEP78-high vs. CEP78-low groups and patient survival across 50
months, with survival analysis performed by log-rank test. (F)
PANC1 and SW1990 cells were transfected with pcDNA3.1-RHPN2
separately; 24 h later, CEP78 levels were tested by RT-qPCR. (G)
PANC1 and SW1990 cells were transfected with si-RHPN2 separately;
24 h later, the CEP78 levels were tested by RT-qPCR. Data are
presented as mean ± SD, and each experiment was repeated at least
three times. *P<0.05. RHPN2, rhophilin 2; PDAC, pancreatic
ductal adenocarcinoma; pcDNA3.1, empty vector; CEP78, centrosomal
protein 78; NC, negative control; si, small interfering; RT-q,
reverse-transcription quantitative; TCGA, The Cancer Genome
Atlas. |
RHPN2 is the target gene of
miR-144-3p, miR-141-3p, miR-96-5p, miR-200a and miR-183-5p
Bioinformatics analysis in the present study
indicated RHPN2 is the target of some important microRNAs (miR)
including miR-144-3p, miR-141-3p, miR-96-5p, miR-200a and
miR-183-5p (Fig. S1).
Discussion
In the present study, the function of RHPN2 in PDAC
was assessed and the findings revealed that the level of RHPN2 was
higher in PDAC tumor tissues compared with adjacent normal tissue.
Notably, high RHPN2 levels in PDAC tissues were associated with a
low survival rate in patients with PDAC. As the overall 5-year
survival rate of PDAC is ~7.1% (21), identification of a novel gene related
to the survival of PDAC patients is critically important. In
addition, in the present study RHPN2 overexpression promoted the
growth of PDAC cells and RHPN2 inhibition promoted cell apoptosis.
Hence, PDAC growth promoted by RHPN2 may increase the death rate
among patients with PDAC.
To the best of our knowledge, this is the first
report of the function of RHPN2 in PDAC. Until now, the oncogenic
function of RHPN2 has been studied only in malignant glioma; the
aforementioned study demonstrated that RHPN2 drives mesenchymal
transformation by triggering RhoA activation (10).
Notably, the data from network analysis in the
present study demonstrated that both RHPN1 and CEP78 have a close
relationship with RHPN2. The role of RHPN1 in cancer is still
unknown (22); and it is
hypothesized that RHPN1 may play a role similar to RHPN2. The
present study demonstrated that the levels of CEP78 are low in PDAC
tumor tissues compared with normal tissue, and that higher levels
of CEP78 in tumor tissue were associated with increased survival
rate in patients with PDAC. The molecular interaction between RHPN2
and CEP78 remains unknown; it is possible that RHPN2 may inhibit
CEP78, which could be reintroduced in PDAC cells resulting in G2/M
phase arrest. Future studies can investigate the aforementioned
phenomenon.
Bioinformatics analysis in the present study
indicated RHPN2 is the target of some important microRNAs (miR)
including miR-144-3p, miR-141-3p, miR-96-5p, miR-200a and
miR-183-5p. These microRNAs serve important roles in the molecular
mechanisms of some types of cancer. For example, miR-144-3p acts as
a suppressive factor in laryngeal squamous cell carcinoma (23), gastric cancer (24), hepatocellular carcinoma (25), and pancreatic cancer (26). miR-141-3p inhibits colorectal cancer,
its overexpression significantly delayed colorectal cancer
progression (27). miR-96-5p and
miR-200a also have a suppressive role in certain types of cancer,
such as breast cancer, ovarian cancer, oral carcinoma and gastric
adenocarcinoma (28–34). On the other hand, miR-183-5p promotes
cell proliferation, migration, and cell cycling in non-small cell
lung cancer (35).
A limitation of the present study was that the
relationship between microRNAs and RHPN2 and the downstream
molecular of RHPN2 was not investigated. Future studies can
investigate this.
In conclusion, the findings of the present study
demonstrated that RHPN2 may promote PDAC and is therefore a
promising candidate for targeted therapy, which is vitally needed
for patients with PDAC whose prospects for survival are dismal.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Bo Yuan
(Department of General Practice, West China Hospital, Sichuan
University) for discussions of the present study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WB collected the patient data and performed the
bioinformatics analysis. XF performed PCR and transfection. XT and
WB performed the apoptosis analysis. XT contributed to the study
design and manuscript writing. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Sichuan University (Chengdu, China; approval no.
20191109) and written informed consent was provided by all patients
enrolled.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Ervik M, Lam F, Colombet M, Mery
L, Piñeros M, Znaor A, Soerjomataram I and Bray F: Global Cancer
Observatory: Cancer Today. International Agency for Research on
Cancer; Lyon: 2018
|
|
3
|
Yang J, Siri JG, Remais JV, Cheng Q, Zhang
H, Chan KK, Sun Z, Zhao Y, Cong N, Li X, et al: The Tsinghua-Lancet
Commission on Healthy Cities in China: Unlocking the power of
cities for a healthy China. Lancet. 391:2140–2184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yadav D and Lowenfels AB: The Epidemiology
of Pancreatitis and Pancreatic Cancer. Gastroenterology.
144:1252–1261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:612014.PubMed/NCBI
|
|
6
|
Hidalgo M, Cascinu S, Kleeff J, Labianca
R, Löhr JM, Neoptolemos J, Real FX, Van Laethem JL and Heinemann V:
Addressing the challenges of pancreatic cancer: Future directions
for improving outcomes. Pancreatology. 15:8–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lambert A, Schwarz L, Borbath I, Henry A,
Van Laethem JL, Malka D, Ducreux M and Conroy T: An update on
treatment options for pancreatic adenocarcinoma. Ther Adv Med
Oncol. Sep 25–2019.(Epub ahead of print). doi:
10.1177/1758835919875568. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hammel P, Huguet F, van Laethem JL,
Goldstein D, Glimelius B, Artru P, Borbath I, Bouché O, Shannon J,
André T, et al: LAP07 Trial Group: effect of chemoradiotherapy vs.
chemotherapy on survival in patients with locally advanced
pancreatic cancer controlled after 4 months of gemcitabine with or
without erlotinib: The LAP07 Randomized Clinical Trial. JAMA.
315:1844–1853. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
O'Reilly EM and Hechtman JF: Tumour
response to TRK inhibition in a patient with pancreatic
adenocarcinoma harbouring an NTRK gene fusion. Ann Oncol.
30:viii36–viii40. 2019. View Article : Google Scholar
|
|
10
|
Danussi C, Akavia UD, Niola F, Jovic A,
Lasorella A, Pe'er D and Iavarone A: RHPN2 drives mesenchymal
transformation in malignant glioma by triggering RhoA activation.
Cancer Res. 73:5140–5150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Sheng R, Källberg M, Silkov A, Tun
MP, Bhardwaj N, Kurilova S, Hall RA, Honig B, Lu H, et al:
Genome-wide functional annotation of dual-specificity protein- and
lipid-binding modules that regulate protein interactions. Mol Cell.
46:226–237. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Steuve S, Devosse T, Lauwers E,
Vanderwinden JM, André B, Courtoy PJ and Pirson I: Rhophilin-2 is
targeted to late-endosomal structures of the vesicular machinery in
the presence of activated RhoB. Exp Cell Res. 312:3981–3989. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang G, He P, Tan H, Budhu A, Gaedcke J,
Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A, et al:
Integration of metabolomics and transcriptomics revealed a fatty
acid network exerting growth inhibitory effects in human pancreatic
cancer. Clin Cancer Res. 19:4983–4993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: software for visualization and analysis of biological
networks. Data Mining in Proteomics. Springer; pp. 291–303.
2011
|
|
17
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
et al: AJCC Cancer Staging Manual. 8th edition. Springer; 2017
|
|
18
|
Fahimi HD: Cytochemical detection of
peroxisomes in light and electron microscopy with
3,3-diaminobenzidine. Methods Mol Biol. 1595:93–100. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang M, Duan T, Wang L, Tang J, Luo R,
Zhang R and Kang T: Low expression of centrosomal protein 78
(CEP78) is associated with poor prognosis of colorectal cancer
patients. Chin J Cancer. 35:622016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stark AP, Sacks GD, Rochefort MM, Donahue
TR, Reber HA, Tomlinson JS, Dawson DW, Eibl G and Hines OJ:
Long-term survival in patients with pancreatic ductal
adenocarcinoma. Surgery. 159:1520–1527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lal MA, Andersson AC, Katayama K, Xiao Z,
Nukui M, Hultenby K, Wernerson A and Tryggvason K: Rhophilin-1 is a
key regulator of the podocyte cytoskeleton and is essential for
glomerular filtration. J Am Soc Nephrol. 26:647–662. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang SY, Lu ZM, Lin YF, Chen LS, Luo XN,
Song XH, Chen SH and Wu YL: miR-144-3p, a tumor suppressive
microRNA targeting ETS-1 in laryngeal squamous cell carcinoma.
Oncotarget. 7:11637–11650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li B, Zhang S, Shen H and Li C:
MicroRNA-144-3p suppresses gastric cancer progression by inhibiting
epithelial-to-mesenchymal transition through targeting PBX3.
Biochem Biophys Res Commun. 484:241–247. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang HW, Ye ZH, Yin SY, Mo WJ, Wang HL,
Zhao JC, Liang GM, Feng ZB, Chen G and Luo DZ: A comprehensive
insight into the clinicopathologic significance of miR-144-3p in
hepatocellular carcinoma. OncoTargets Ther. 10:3405–3419. 2017.
View Article : Google Scholar
|
|
26
|
Li J, Sun P, Yue Z, Zhang D, You K and
Wang J: miR-144-3p induces cell cycle arrest and apoptosis in
pancreatic cancer cells by targeting proline-rich protein 11
expression via the mitogen-activated protein kinase signaling
pathway. DNA Cell Biol. 36:619–626. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: MiR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi Y, Zhao Y, Shao N, Ye R, Lin Y, Zhang
N, Li W, Zhang Y and Wang S: Overexpression of microRNA-96-5p
inhibits autophagy and apoptosis and enhances the proliferation,
migration and invasiveness of human breast cancer cells. Oncol
Lett. 13:4402–4412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu B, Zhang J and Yang D: miR-96-5p
promotes the proliferation and migration of ovarian cancer cells by
suppressing Caveolae1. J Ovarian Res. 12:572019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang H, Ma N, Li W and Wang Z:
MicroRNA-96-5p promotes proliferation, invasion and EMT of oral
carcinoma cells by directly targeting FOXF2. Biol Open. 9:92020.
View Article : Google Scholar
|
|
31
|
Zhou HY, Wu CQ and Bi EX: MiR-96-5p
inhibition induces cell apoptosis in gastric adenocarcinoma. World
J Gastroenterol. 25:6823–6834. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi C, Yang Y, Zhang L, Yu J, Qin S, Xu H
and Gao Y: MiR-200a-3p promoted the malignant behaviors of ovarian
cancer cells through regulating PCDH9. OncoTargets Ther.
12:8329–8338. 2019. View Article : Google Scholar
|
|
33
|
Tang W, Yu X, Zeng R and Chen L:
LncRNA-ATB promotes cisplatin resistance in lung adenocarcinoma
cells by targeting the miR-200a/β-catenin pathway. Cancer Manag
Res. 12:2001–2014. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu J, Ke F, Chen T, Zhou Q, Weng L, Tan
J, Shen W, Li L, Zhou J, Xu C, et al: MicroRNAs that regulate PTEN
as potential biomarkers in colorectal cancer: A systematic review.
J Cancer Res Clin Oncol. 146:809–820. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang H, Ma Z, Liu X, Zhang C, Hu Y, Ding
L, Qi P, Wang J, Lu S and Li Y: MiR-183-5p is required for
non-small cell lung cancer progression by repressing PTEN. Biomed
Pharmacother. 111:1103–1111. 2019. View Article : Google Scholar : PubMed/NCBI
|