Introduction
Cervical cancer is the fourth most frequently
diagnosed cancer and the fourth leading cause of cancer death in
women, with an estimated 570,000 cases and 311,000 deaths having
occurred worldwide in 2018 (1).
Persistent human papilloma virus (HPV) infection is a direct cause
for CC (2). With the development of
DNA examination and cytological screening for high-risk HPV and the
promotion of the cervical cancer vaccine, the morbidity rate of
cervical cancer has been declining (3). However, the vaccines only cover some of
the cancer-causing (‘high-risk’) types of HPV, such as HPV-16 and
HPV-18 (4). Women should do regular
Pap smear screening even after vaccination (5). Inspite of advances in the treatment of
CC, the 5-year survival rate remains <50% in China (6). Hence, it is particularly necessary to
find a new and effective treatment strategy targeting CC.
Long non-coding RNAs (lncRNAs) are a group of
non-coding RNAs with >200 nucleotides and are involved in
various biological activities including cell proliferation,
migration, invasion, apoptosis and inflammatory responses (7,8). LncRNA
is a kind of non-coding RNA which modulates gene expression through
epigenetic control, transcriptional regulation, and
post-transcriptional regulation (9).
Multiple reports have verified that abnormal expression of lncRNAs
could affect the expression or function of oncogenes, tumor
suppressors, metabolic enzyme genes as well as transcription
factors and signaling pathways (10–14). For
example, enforced expression of HOX transcript antisense RNA
(HOTAIR) increased invasiveness and metastasis in breast cancer
(11). Maternally expressed gene 3
was found as a tumor suppressor associated with the pathogenesis
and progression in human meningiomas (12). LncRNA SNHG15 promotes colon cancer
cell invasion and metastasis through blocking degradation of
transcription factor Slug (13).
LncRNA PVT1 facilitates tumorigenesis and progression via
regulation of miR-128-3p/gremlin 1 axis and bone morphogenetic
protein (BMP) signaling pathway in glioma (14). A growing body of evidence has
indicated the importance of lncRNAs for the progression of CC. At
present, the mechanisms of metastasis associated lung
adenocarcinoma transcript 1 (MALAT1), HOTAIR and CDKN2B antisense
RNA 1 (ANRIL) lncRNAs in cervical cancer have been characterized in
detail (15–17). Guo et al (15) demonstrated that HOTAIR was
upregulated in cervical cancer tissues compared with peritumoral
tissues. Suppression of HOTAIR reduced autophagy and reversal of
epithelial-mesenchymal transition through the inhibition of the Wnt
signaling pathway, which consequently enhanced radiotherapy
sensitivity in CC. MALAT1 has been reported to be generally
upregulated in CC and reduces the efficiency of radiation treatment
on CC cell lines by interacting with miR-143 in vitro
(16). LncRNA ANRIL was
significantly increased both in CC tissues and cell lines and
regulated CC cell proliferation, migration and invasion through the
PI3K/Akt pathway (17).
The aforementioned studies demonstrated the impacts
of lncRNAs on CC progression and their potential as treatment
targets. lncRNA-AK001903 has been verified as an upregulated gene
in active ulcerative colitis tissues by non-coding (NC) RNA
microarray (18). However, the
expression and function of lncRNA-AK001903 in CC and its
correlation with prognosis of patients with CC remains to be
elucidated. Hence, the present study aimed to investigate the roles
of lncRNA-AK001903 in the progression of cervical cancer, which may
provide a new target for the treatment of CC.
Materials and methods
Cell lines and tissue samples
Cervical cancer cell lines: Ca Ski, Hela, Siha, and
C33a were purchased from Procell Life Science & Technology Co.,
Ltd. and the normal cell line H8 was purchased from Shanghai Yu Bo
Biotech Co., Ltd. Ca Ski and H8 were cultured in RPMI-1640
supplemented with 10% heat-inactivated FBS (fetal bovine serum)
(Gibco; Thermo Fisher Scientific Inc.). Hela, Siha and C33a were
cultured in Minimum Essential Medium (Gibco; Thermo Fisher
Scientific Inc.) supplemented with 10% heat-inactivated FBS.
Penicillin (100 U/ml)/streptomycin (100 µg/ml) (GE Healthcare Life
Sciences) was used in the culture medium and cells were grown in a
humidified atmosphere containing 5% CO2 at 37°C. Cells
were passed to the next generation every three days. In the present
study, tissues were collected from Department of Gynecological
Oncology in Sun Yat-sen Memorial Hospital (Guangzhou, China)
between June 2016 and May 2017. The inclusion criteria of the
patients were as follows: i) Females diagnosed with CC using the
International Federation of Gynecology and Obstetrics (FIGO, 2018)
system (19); and ii) underwent
biopsy or trachelectomy surgery. The exclusion criteria of the
patients were as follows: i) Diagnosed with other tumors; ii)
underwent radiotherapy or chemotherapy or any other treatment prior
to surgery; and iii) CC recurrence. Peritumoral tissue samples were
taken at least 1 cm distal to tumor margins. Tissue histology was
independently evaluated by two pathologists. A total of 29 CC
tissues (mean age, 37.6 years; age range, 21–77 years) and
peritumoral tissues were collected and immediately placed and
stored in liquid nitrogen. All of the 29 CC tissues and peritumoral
tissues were used for reverse-transcription quantitative (RT-q) PCR
analysis, 3 CC tissues and corresponding peritumoral tissues were
used for microarray analysis and 26 CC tissues were used for ISHH.
All patients underwent biopsy or trachelectomy surgery at the Sun
Yat-sen Memorial Hospital (Guangzhou, China). All specimens were
obtained with the approval of the Sun Yat-sen Memorial Hospital
Review Board (approval no. 2015132) (Guangzhou, China) and a
written informed content was obtained from each patient.
RNA microarray
Human 12×135k Long Non-coding RNA Array was
manufactured by NimbleGen Systems Inc. Each array represented all
long transcripts, both protein coding mRNAs and lncRNAs in the
human genome. In total, >23,000 lncRNAs were collected from the
authoritative data sources including NCBI RefSeq (GRCh37 (hg19);
http://www.ncbi.nlm.nih.gov/refseq/),
UCSC (GRCh37 (hg19); http://genome.ucsc.edu/cgi-bin/hgTables), RNAdb 2.0
(http://research.imb.uq.edu.au/rnadb/), lncRNAs from
literatures (20,21) and UCRs (NCBI Build 35 (hg17);
http://users.soe.ucsc.edu/~jill/ultra.html).
RNA labeling and array
hybridization
In total 3 CC tissues and 3 corresponding
peritumoral tissues were used to synthesize double-stranded
complementary DNA (cDNA). Double-strand cDNA (ds-cDNA) was
synthesized from 5 µg of total RNA using a SuperScript ds-cDNA
synthesis kit (Invitrogen; Thermo Fisher Scientific Inc.) in the
presence of 100 pmol oligo dT primers according to the
manufacturer's instructions. Ds-cDNA was cleaned and labeled in
accordance with the NimbleGen Gene Expression Analysis protocol
(NimbleGen Systems, Inc.). Briefly, ds-cDNA was incubated with 4 µg
RNase A at 37°C for 10 min and cleaned using
phenol:chloroform:isoamyl alcohol (25:24:1), followed by ice-cold
absolute ethanol precipitation. The purified cDNA was quantified
using NanoDrop ND-1000 (Thermo Fisher Scientific Inc.). For Cy3
labeling of cDNA, the NimbleGen One-Color DNA labeling kit
(NimbleGen Systems, Inc.) was used according to the manufacturer's
instructions detailed in the Gene Expression Analysis protocol
(NimbleGen Systems, Inc.). In brief, 1 µg ds-cDNA was incubated for
10 min at 98°C with 1 optical density (OD) of Cy3-9mer primer.
Next, 100 pmol of deoxynucleoside triphosphates and 100 U of the
Klenow fragment (New England Biolabs Inc.) were added and the
mixture incubated at 37°C for 2 h. The reaction was stopped by 0.5
mol/l EDTA (0.1 times the volume of the previous mixture), and the
labeled ds-cDNA was purified by isopropanol/ethanol precipitation.
Microarrays were hybridized at 42°C for 16–20 h with 4 µg of Cy3
labelled ds-cDNA in hybridization buffer/hybridization component A
(NimbleGen Systems, Inc.) in a hybridization chamber (hybridization
system; NimbleGen Systems, Inc.).
Microarray wash, scanning and data
extraction
Washing was performed three times using the SeqCap
EZ Hybridization and Wash kit (cat. no. 05634261001; NimbleGen
Systems, Inc.). After being washed in an ozone-free environment,
the slides were scanned using the Axon GenePix 4000B microarray
scanner (Molecular Devices, LLC.). Raw data were extracted as pair
files using NimbleScan software version 2.5 (Roche NimbleGen, Inc.)
and normalized through quantile normalization and the Robust
Multichip Average algorithm included in the NimbleScan software.
All gene level files were imported into GeneSpring GX software
version. 11.5.1 (Agilent Technologies Inc.) or further analysis.
Differentially expressed lncRNAs with statistical significance
between two groups were identified through volcano plot filtering.
Differentially expressed lncRNAs between two samples were
identified through fold change filtering. P-value was calculated
using the paired t-test. The threshold set for up- and
downregulated genes was a fold change ≥2.0 and a P-value ≤0.05.
Hierarchical clustering was performed using the GeneSpring GX
software version 11.5.1 (Agilent Technologies Inc.).
Transfection
A total of 4 AK001903-specific small interfering
(si)RNAs (si-AK001903-713, siRNA-AK001903-1306, siRNA-AK001903-1051
and siRNA-AK001903-1605) were used to knock down lncRNA-AK001903,
and a non-targeting siRNA (si-NC) oligonucleotide was used as a
negative control (all Shanghai Gene Pharma Co. Ltd.). Sequences
were listed in Table I. Ca Ski were
plated at 60% confluence (5×105 cells/well) on a 6-well
plate. The following day, the culture medium was replaced with 500
µl of Opti-MEM (Gibco; Thermo Fisher Scientific Inc.). A total of
250 µl of Opti-MEM and 5 µl of Lipofectamine® RNAiMAX
(Invitrogen; Thermo Fisher Scientific Inc.) were mixed in one tube,
20 pM siRNA (Shanghai GenePharma Co. Ltd.) and 250 µl of Opti-MEM
were mixed in the other tube. Then they were combined and incubated
for 5 min at room temperature. After 6 h incubation in 37°C, the
transfection medium was replaced with 2 ml of standard growth
medium. Cells were haversted 48 h post transfection and subsequent
experiments performed.
 | Table I.siRNA sequences used in the present
study. |
Table I.
siRNA sequences used in the present
study.
|
| Sequence |
|---|
|
|
|
|---|
| siRNA set | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
|
siRNA-AK001903-713 |
GCCCACACCAAUCUUAGAATT |
UUCUAAGAUUGGUGUGGGCTT |
|
siRNA-AK001903-1306 |
CCACAUGUCUCAGCUAUAUTT |
AUAUAGCUGAGACAUGUGGTT- |
|
siRNA-AK001903-1051 |
CGGACCCAUAUUAUCAUAUTT- |
AUAUGAUAAUAUGGGUCCGTT |
|
siRNA-AK001903-1605 |
GCAGUUCUUUAACCAAUGUTT |
ACAUUGGUUAAAGAACUGCTT |
| siRNA-NC |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
RT-qPCR
Total RNA was extracted from tissues or cell lines
(Ca Ski, Hela, Siha, C33a and H8) using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific Inc.) according to the manufacturer's
instructions. Total RNA (500 ng) was reverse transcribed into cDNA
using a PrimeScript® RT reagent kit with gDNA eraser
(Takara Bio Inc.). The protocol for RT was as follows: 37°C for 15
min, 85°C for 5 sec and termination at 4°C. Quantitative PCR was
performed using an ABI 7500 Real-Time PCR system and a
SYBR® Premix Ex TaqTM II kit (Takara Bio Inc.). PCR
thermocycling conditions were as follows: predenaturation at 95°C
for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec
and annealing at 60°C for 30 sec. β-actin was used as an endogenous
reference. Fold-changes of the relative expression of target genes
were calculated using 2−ΔΔCq method (22). All experiments were performed in
duplicate and repeated twice. Primers for quantitative PCR were
presented in Table II.
 | Table II.Reverse-transcription quantitative
PCR primers. |
Table II.
Reverse-transcription quantitative
PCR primers.
| Primer Set | Sequence/Assay
ID |
|---|
| AK001903.1 | Forward:
5′-AATCTGCCCACACCAATCTT-3′ |
|
| Reverse:
5′-CAGTGTGCTGAAATTCACCTG-3′ |
| AI184890 | Forward:
5′-GTCTCACTCTGTTGCCTGGG-3′ |
|
| Reverse:
5′-TGGGGACATTTGCGGAAATTTAT-3′ |
| AK097842 | Forward:
5′-AGGGTCTACATCGGCTCCTT-3′ |
|
| Reverse:
5′-CGTTCATGGTGCCGTCAAAG-3′ |
| BG419628 | Forward:
5′-GCACTGTGACCTCCCTGATC-3′ |
|
| Reverse:
5′-TGGGTCCACTTCGCAAATGA-3′ |
| ASLNC01516 | Forward:
5′-CGGACTGTTCTCCTTCCCAC-3′ |
|
| Reverse:
5′-GGGATTGCAGGTGTGATCCA-3′ |
| ASLNC16271 | Forward:
5′-AGCCTCCCTGTACAAGCAAC-3′ |
|
| Reverse:
5′-GTAAGTTCCCCGCCCTGTAG-3′ |
| ASLNC14492 | Forward:
5′-GGTAACGAATGCCCCTCCAA-3′ |
|
| Reverse:
5′-AGTAGGGCTGACTCTCCCAG-3′ |
| ASLNC03532 | Forward:
5′-CCTCCTCTCACCCAGGATCA-3′ |
|
| Reverse:
5′-GTGTTCCTGAGAAGGCCCTC-3′ |
| BF675100 | Forward:
5′-CAATGCTCAAACCACAGGCC-3′ |
|
| Reverse:
5′-TGTCATTTTCTCCTCGCCCC-3′ |
| ASLNC17636 | Forward:
5′-TGAGCCGAGATTGTGCCATT-3′ |
|
| Reverse:
5′-AAATGAGGCAGGTGACAGGG-3′ |
| β-actin | Forward: 5′-
TGGCACCCAGCACAATGAA-3′ |
|
| Reverse:
5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ |
ISHH
Tissues were fixed with 4% paraformaldehyde (with
0.1% Diethyl pyrocarbonate) at room temperature for 24 h. Then they
were paraffin-embedded and sliced into 20-µm thick slices and
spread over PLL-coated glass slides. The paraffin sections were
deparaffinized and rehydrated and treated with 0.2 mol/l HCl at
37°C for 20 min, 3 µg/ml proteinase K (PCR grade; Roche
Diagnostics) at 37°C for 7 min followed by post-fixation with 4%
paraformaldehyde (with 0.1% diethyl pyrocarbonate) for 5 min at
room temperature. After acetylation in a solution consisting of
1.5% triethanolamine, 0.25% acetic anhydride, and 0.25% HCl at 40°C
for 4 h, the slides were washed by 5X SSC once for 15 min.
Pre-hybridization was performed using the pre-hybridization
solution (cat. no. AR0152; Boster Biological Technology Co. Ltd.)
at 37°C for 4 h. Subsequently, a digoxigenin-labeled
oligonucleotide probe (5′-CCACACCCACCTTCCCACATGCATGATA-3′; Shanghai
GeneBio Co., Ltd.) was diluted to 200 nM using hybridization buffer
(cat. no. AR0062; Boster Biological Technology Co. Ltd.) and
hybridized with tissues for 16 h at 55°C. Slides were washed twice
at 55°C for 30 min with a solution consisting of 50% formamide, 2X
SSC and 0.01% Tween 20, and then 10 µg/ml RNase A was added at 37°C
for 1 h in buffer (0.5 M NaCl, 10 mM Tris (pH 8.0), 1 mM EDTA, and
0.01% Tween 20). Subsequently, the samples were washed in 2X SSC
wash buffer with 0.01% Tween 20 at 55°C for 30 min, followed by
washing in 0.2X SSC wash buffer with 0.01% Tween 20 at 55°C for 30
min. After additional washing in Tris-buffered saline (TBS, pH 7.6)
and blocking in a buffer consisting of 10% Blocking Reagent (cat.
no. 11096176001; Roche Diagnostics), 0.1 M maleate, 0.15 M NaCl,
and 0.01% Tween 20 in TBS, DIG- labeled probes were detected by
biotinylated digoxin antibody (cat. no. AR0147; Boster Biological
Technology Co. Ltd.) at 37°C for 1 h, streptavidin-biotin
complex-peroxisome (SABC-POD; cat. no. AR0148; Boster Biological
Technology Co. Ltd.) at 37°C for 20 min and biotin-horseradish
peroxidase (HRP) (cat. no. AR0149; Boster Biological Technology Co.
Ltd.) at 37°C for 20 min. The samples were stained using DAB (cat.
no. ZLI-9019; Origene Technologies Inc.) and images were captured
using light microscopy (Nikon Corporation).
CCK-8 assay
Cell proliferation was assessed using the CCK-8
assay. Ca Ski were plated at 1×103 cells/well on 96-well
plates with three wells for each group. Cell viability was measured
over 5 days using a Cell Counting Kit-8 (CCK-8) (Dojindo Molecular
Technologies Inc.). A total of 10 µl of CCK-8 (5 mg/ml) was added
to each well. After incubating for 4 h at 37°C, the absorbance was
determined at 450 nm.
Transwell assays
Cell migation and invasion were examined using
Polycarbonate Membrane transwell inserts (Costar; Corning Inc.).
The upper compartment was pre-coated with RPMI-1640 medium without
serum for migation assay and Matrigel for 2 h in 37°C (Corning
Inc.) for invasion assay. After 48 h of transfection,
5×104 Ca Ski cells were placed into the upper
compartment. RPMI-1640 medium with 20% FBS was used in the lower
compartment. Cells were incubated at 37°C for 24 h for the migation
assay and 48 h for the invasion assay. Then the compartment were
fixed at room temperature with 4% paraformaldehyde for 20 min and
stained with crystal violet (1 mg/ml) for 20 min at room
temperature, and cells not crossing the membrane were cleaned.
Images were captured using light microscopy (Nikon
Corporationm).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 8.0 software (GraphPad Software, Inc.) and SPSS 20.0 software
(IBM Corp.). All experiments were performed in triplicate.
Comparisons between two groups were performed by paired sample
t-tests. Three or more experimental groups were compared by one-way
ANOVA followed by the post hoc Bonferroni test. The median value
[tissues' CT (cycle threshold) value-corresponding β-actin CT
value=7.2] of lncRNA-AK001903 expression level was used to divide
patients into high and low lncRNA-AK001903 expression level groups.
χ2 test was used to compare the association between
different clinical features and gene expression level. All data are
presented as mean ± standard deviation (SD). P<0.05 was
considered to indicate a statistically significant difference.
Results
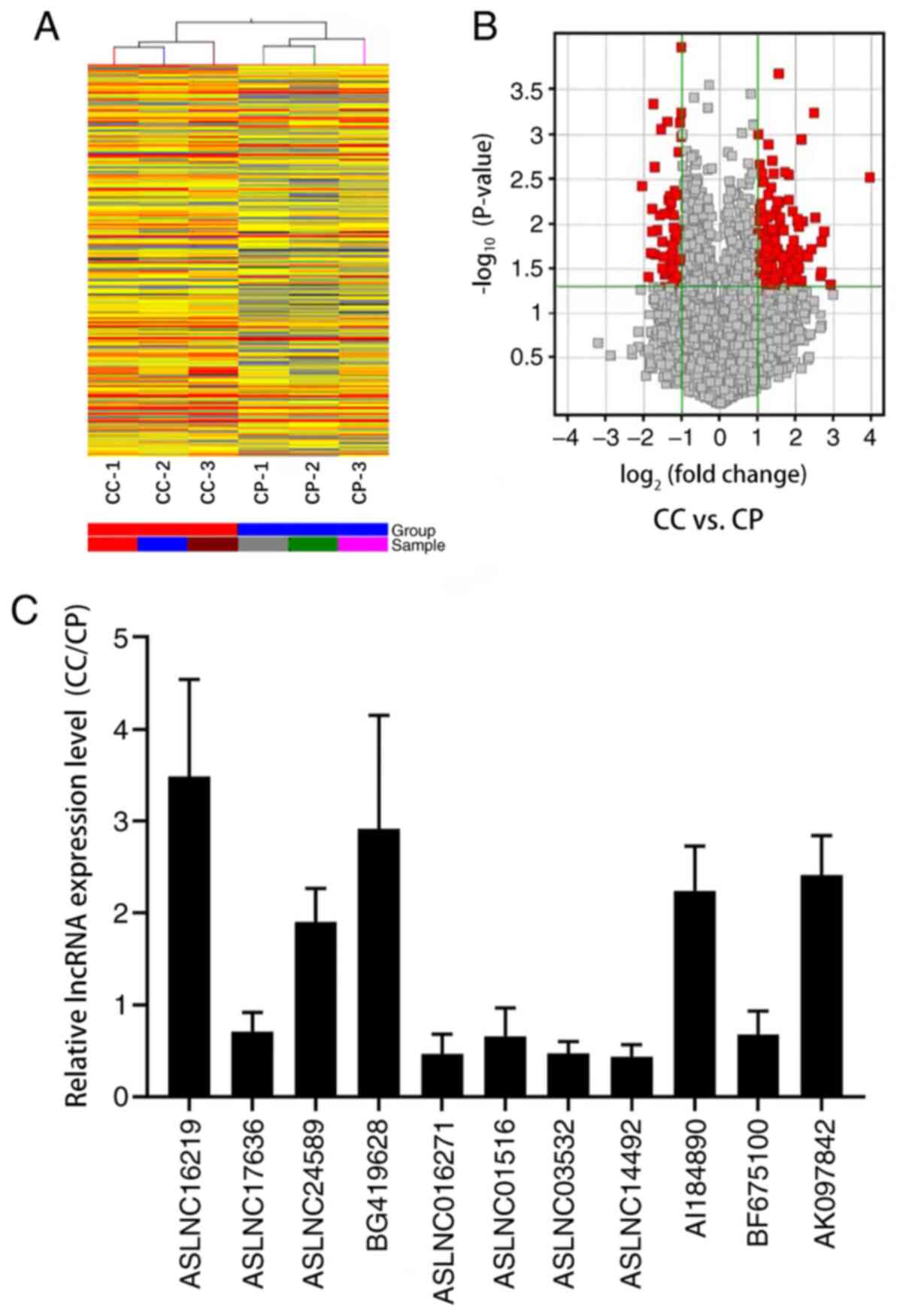
LncRNAs expression profile in CC
Hierarchical clustering demonstrated systematic
variations in the expression of lncRNAs between CC tissues and
corresponding peritumoral tissues (Fig.
1A). Volcano Plots were used to visualize the relationship
between fold-change (magnitude of change) and statistical
significance (which took both magnitude of change and variability
into consideration) (Fig. 1B).
According to the microarray expression profiling data, 453
distinctively dysregulated lncRNAs were detected in CC tissues
compared to the controls (fold change, ≥2.0 and P<0.05)
(Fig. 1A and B). The top 10 of 324
significantly upregulated and 129 significantly downregulated
lncRNA transcripts are summarized in Table III. To validate the microarray
analysis findings, 5 upregulated and 6 downregulated lncRNAs
expression were detected and compared between 9 pairs of CC tissues
and their corresponding peritumoral tissues using RT-qPCR. These
data confirmed that ASLNC16219 (seqname, AK001903), ASLNC24589
(seqname, U88892), BG419628 (seqname, chr20:1594825-1615675),
AI184890 (seqname, HMlincRNA1030), AK097842 (seqname, HMlincRNA810)
were upregulated in CC tissues compared to peritumoral tissues,
whereas the expression of ASLNC17636 (seqname, AK055280),
ASLNC016271 (seqname, AK021467), ASLNC01516 (seqname, NR_024345),
ASLNC03532 (seqname, uc001esn), ASLNC14492 (seqname, AY927461) and
BF675100 (seqname, chr12:92933675-92975575) was decreased (all
P<0.05; Fig. 1C). Thus, the
results strongly revealed that expression changes of lncRNAs were
involved in the development of cervical cancer.
 | Table III.Top 10 significantly up- and
downregulated lncRNAs in CCa vs. CPb. |
Table III.
Top 10 significantly up- and
downregulated lncRNAs in CCa vs. CPb.
| Seq name | P-value | Fold changec | MORd | CC | CP | Associated | Source | Relationship |
|---|
| AK001903 | 0.003 | 15.547 | Up | 1,570.119 | 105.897 |
| misc_lncRNA | Intergenic |
| HMlincRNA1030 | 0.046 | 7.559 | Up | 1,336.257 | 151.717 |
| lincRNA | Intergenic |
| U88892 | 0.012 | 6.743 | Up | 657.814 | 97.620 |
| misc_lncRNA | Intron
sense-overlapping |
| HMlincRNA810 | 0.015 | 6.467 | Up | 796.874 | 136.080 |
| lincRNA | Exon
sense-overlapping |
|
chr20:1594825-1615675 | 0.033 | 6.414 | Up | 1,263.708 | 220.417 |
| lincRNA | Intron
sense-overlapping |
| HMlincRNA1030 | 0.037 | 6.212 | Up | 1,382.065 | 221.246 |
| lincRNA | Intergenic |
| uc010ldj | 0.019 | 5.369 | Up | 2,010.306 | 385.026 | HSP27e | UCSC_knowngene | Exon
sense-overlapping |
| uc010dld | 0.023 | 5.025 | Up | 397.059 | 81.809 | TUBB6f | UCSC_knowngene | Exon
sense-overlapping |
| NR_024204 | 0.009 | 4.565 | Up | 3,247.554 | 680.789 | NCRNA00152g | RefSeq_NR | Intergenic |
| DQ786233 | 0.043 | 4.469 | Up | 603.588 | 113.588 |
| misc_lncRNA | Intergenic |
| NR_024345 | 0.004 | 4.153 | Down | 1,615.112 | 7,101.768 | C12orf27h | RefSeq_NR | Intergenic |
| AK021467 | 0.039 | 3.649 | Down | 267.197 | 1,035.963 |
| misc_lncRNA | Intron
sense-overlapping |
| AY927461 | 0.021 | 3.490 | Down | 496.854 | 1,600.906 |
| misc_lncRNA | Intron
sense-overlapping |
| uc001esn | 0.012 | 3.480 | Down | 93.238 | 318.969 | AX746564 | UCSC_knowngene | Intergenic |
|
chr12:92933675-92975575 | 0.007 | 3.476 | Down | 109.867 | 383.899 |
| lincRNA | Exon
sense-overlapping |
| AK055280 | 0.000 | 3.327 | Down | 273.747 | 910.918 |
| misc_lncRNA | Intron
sense-overlapping |
| uc003wog | 0.021 | 3.321 | Down | 236.854 | 840.568 | LOC154822 | UCSC_knowngene | Natural
antisense |
|
chr3:64987550-65012925 | 0.002 | 3.274 | Down | 81.832 | 274.405 |
| lincRNA | Intergenic |
| LIT1571 | 0.012 | 3.110 | Down | 257.196 | 806.208 |
| RNAdb | Intergenic |
| HMlincRNA1338 | 0.008 | 2.965 | Down | 115.321 | 347.049 |
| lincRNA | Intergenic |
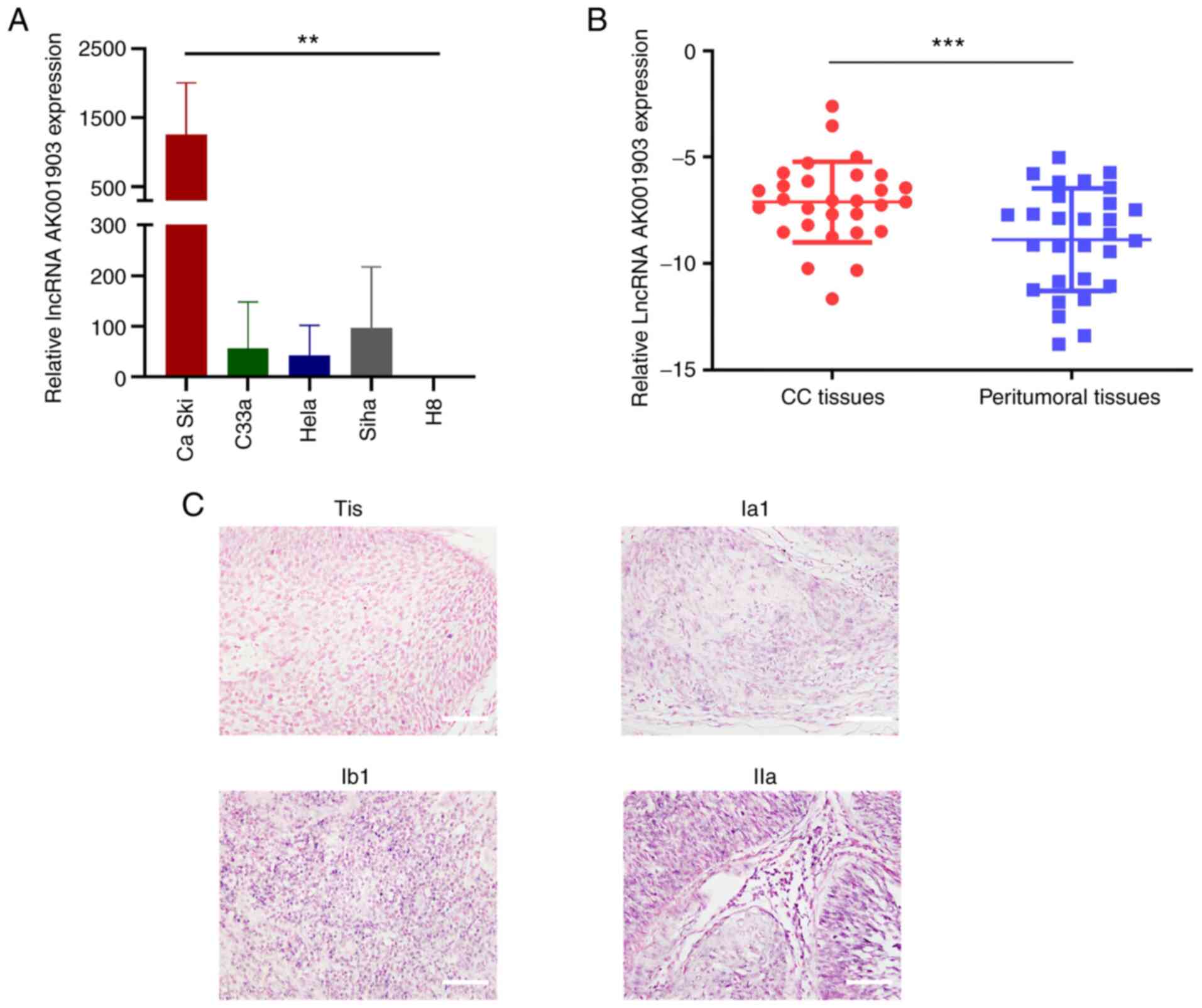
LncRNA-AK001903 is upregulated in CC
tissues and cells and related to FIGO stage in patients with
CC
Among the 20 most significantly differentially
expressed lncRNAs (DE lncRNAs) in the CC tissues vs. CP
(peritumoral tissues), the most notably upregulated one was
lncRNA-AK001903 (fold change, 15.547; Table III). To verify the roles of
lncRNA-AK001903 in CC cells, expression of lncRNA-AK001903 in CC
cell lines and the normal cell line H8 were determined by RT-qPCR.
The results demonstrated that Ca Ski exhibited significantly higher
expression level of lncRNA-AK001903 compared with C33a, Hela and
Siha cells. Besides, all these cervical cancer cell lines expressed
higher level of lncRNA-AK001903 compared with H8 cells (Fig. 2A). In addition, lncRNA-AK001903
expression was assessed in 29 CC tissues and their corresponding
peritumoral tissues to confirm the expression change of
lncRNA-AK001903 in CC tissues. The result revealed significantly
higher levels of lncRNA-AK001903 gene expression compared with
corresponding peritumoral tissues (Fig.
2B). Clinicopathologic features of the 29 patients with CC were
shown in Table IV. However,
according to the results of RT-qPCR, the high and low expression of
AK001903 in tumor tissues appeared to be independent of the
patient's age, tumor size, FIGO stage, lymph node metastasis,
differentiation, and whether the tumor was confined to the cervix
(Table IV). These results suggested
the probable oncogenic role of lncRNA-AK001903 in cervical cancer
tumorigenesis. In addition, an ISHH probe of lncRNA-AK001903 was
designed and synthesized to examine the expression differences
among CC tissues. The results demonstrated that lncRNA-AK001903 was
mostly located in the nucleus of cells and that high
lncRNA-AK001903 expression was associated with advanced FIGO stage
(Fig. 2C).
 | Table IV.Clinicopathologic features of CC
tissues and corresponding peritumoral tissues (n=29). |
Table IV.
Clinicopathologic features of CC
tissues and corresponding peritumoral tissues (n=29).
| Features | Low expression | High
expression | P-value |
|---|
| Age, years |
|
| 0.139 |
|
>50 | 4 | 9 |
|
|
≤50 | 10 | 6 |
|
| Tumor size, cm |
|
| 0.893 |
| ≥3 | 9 | 10 |
|
|
<3 | 5 | 5 |
|
| FIGO stage, R |
|
| 0.264 |
|
≥II | 4 | 8 |
|
|
<II | 10 | 7 |
|
| Lymphatic
metastasis |
|
| 0.924 |
|
Yes | 3 | 3 |
|
| No | 11 | 12 |
|
|
Differentiation |
|
| 0.700 |
|
Low | 4 | 6 |
|
|
Moderate or high | 10 | 9 |
|
| Confined to the
cervix |
|
| 0.837 |
| No | 8 | 8 |
|
|
Yes | 6 | 7 |
|
LncRNA-AK001903 regulates the cell
proliferation, migration, and invasion in Ca Ski cells
As lncRNA-AK001903 was highly expressed in CC cells
and tissues compared with H8 cells and peritumoral tissues and
related to the FIGO stage of CC, further experimentation was
performed to investigate whether lncRNA AK001903 may be a potential
oncogene during the progression of CC. Four siRNAs targeting
lncRNA-AK001903 at different sites (Table I) were designed and transfected into
Ca Ski cells due to the highest expression of lncRNA-AK001903 in Ca
Ski. The results demonstrated that of the 4 siRNAs used
siRNA-lncRNA-AK001903-1605 produced the most effective interference
of lncRNA-AK001903 expression and was then used for further
experimentation (Fig. 3A). Next, the
effects of knockdown of lncRNA-AK001903 on the proliferation,
invasion and migration of Ca Ski cells was investigated. The CCK-8
assay demonstrated that lncRNA-AK001903 knockdown significantly
inhibited the proliferation of lncRNA-AK001903 compared to the
si-NC group (Fig. 3B). Transwell
assay results demonstrated that the ability of migration and
invasion were suppressed by knockdown of lncRNA-AK001903 (Fig. 3C).
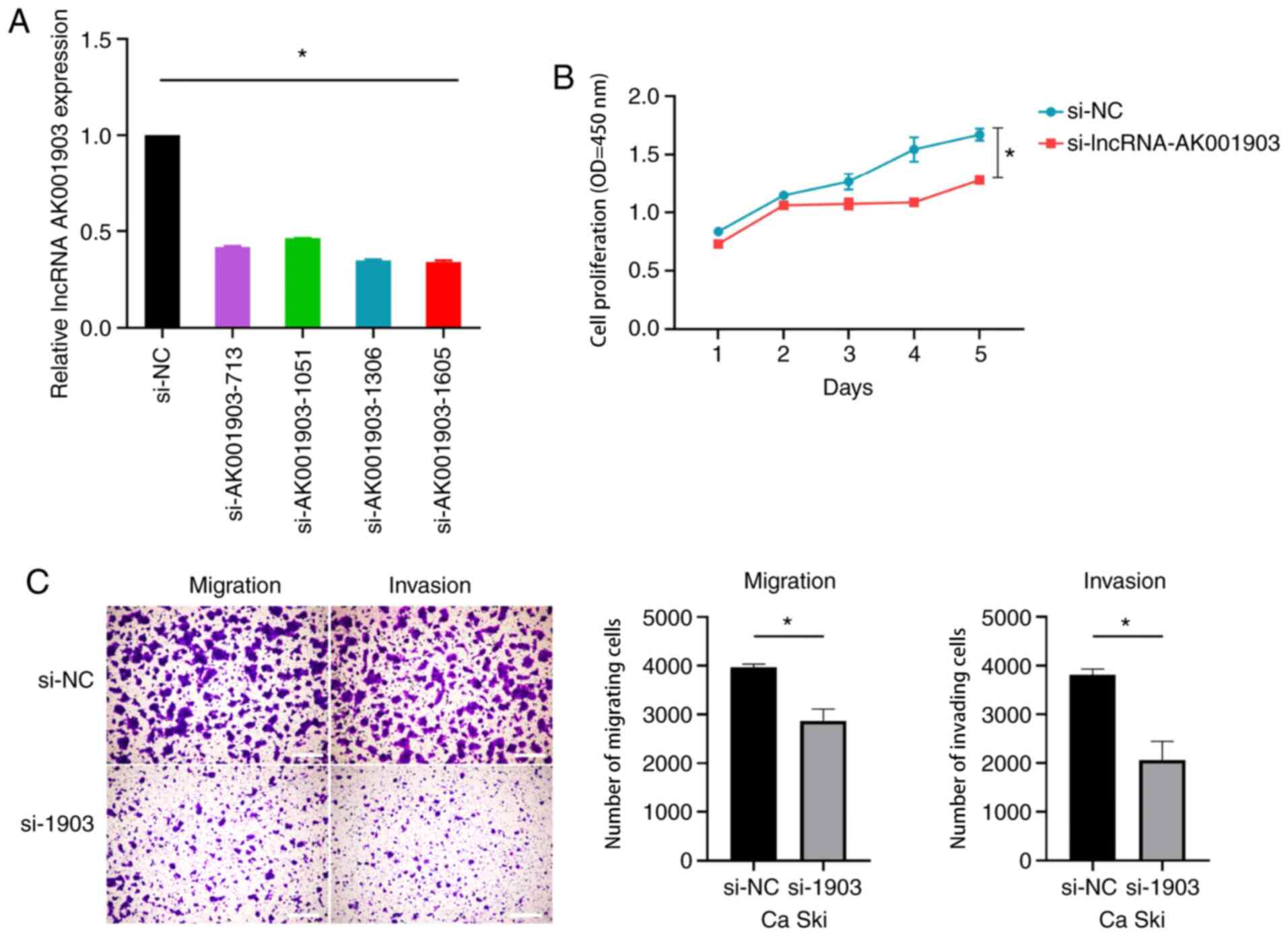 | Figure 3.Effects of lncRNA-AK001903 silencing
on CC progression. (A) Ca Ski cells were transfected with
siRNA-lncRNA-AK001903-713, siRNA-lncRNA-AK001903-1306,
siRNA-lncRNA-AK001903-1051, siRNA-lncRNA-AK001903-1605 or siRNA-NC.
LncRNA-AK001903 expression was analyzed by reverse-transcription
quantitative PCR at 48 h post-infection. β-actin was an internal
control. (B and C) Ca Ski cells were transfected with
siRNA-lncRNA-AK001903-1605 or siRNA-NC and then analyzed by the
CCK-8 assay and transwell assay, respectively. Scale bar, 1,000 µm.
Data are shown as mean ± SD. *P<0.05. CC, cervical cancer;
lncRNA, long non-coding RNA; si, small interfering; NC, negative
control; OD, optical density. |
Discussion
Cancer is a complex disease, involving various
changes in gene expression (23,24).
These gene expression changes cause cancer development, including
metastasis (25), cell proliferation
(26), invasion (27), and angiogenesis (28). Numerous large-scale discovery studies
have demonstrated the prospect of using lncRNAs as diagnostic and
prognostic biomarkers, even in successful development of RNAi-based
and oligo-based drugs (29,30). For example, lncRNA highly
up-regulated in liver cancer level in blood and tissues may be used
to detect liver cancer (31,32). HOTAIR in tissues can used as a
prognostic marker for overall survival in breast cancer (11). Treatment with MTL-CEBPA (a small
activating RNA drug) in mice lowered hepatocellular carcinoma tumor
burden and improved clinically relevant parameters of liver
function (29). As for the lncRNAs
in cervical cancer, increasing numbers of reports have verified
that lncRNAs are identified as potential biomarkers for cancer
prognosis, invasion, metastasis, chemo-resistance and
radio-resistance (33). Functional
analysis of lncRNAs associated with CC progression may provide in
depth understanding on the progression of CC (34).
Genome-wide microarray analysis has been used to
identify the differentially expressed genes with higher diagnostic
ability in tissues and cells, which facilitates the exploration of
molecular mechanisms of tumor development (35). In the present study, microarray
analysis was used to screen the aberrant lncRNAs in 3 CC patients
to distinguish them in cancer tissues from corresponding
peritumoral tissues. The results demonstrated that 453 lncRNAs (324
upregulated and 129 downregulated lncRNAs) were differentially
expressed between CC tissues and peritumoral tissues from the same
patients. A total of 6 downregulated and 5 upregulated lncRNAs were
further verified by RT-qPCR in the present study and the results in
9 pairs of CC and peritumoral tissues were consistent with the
microarray analysis.
It was recently reported that abnormal expression of
lncRNAs, such as HAND2-AS1 and DLX6-AS1 serve an important role in
the occurrence and development of CC (36–38). The
change of cell phenotype depends on the influence of gene
expression regulation (10). In the
present study the most significantly upregulated lncRNA
lncRNA-AK001903 was used to perform further experiments. The
findings of the present study revealed that lncRNA-AK001903 was
significantly upregulated in CC cell lines compared to normal cell
line H8 and CC tissues compared to peritumoral tissues by RT-qPCR,
and the level of lncRNA-AK001903 in CC tissues by ISHH was
associated with FIGO (2018) stage. The aforementioned findings of
the present study indicated that lncRNA-AK001903 may be a novel and
effective biomarker in CC.
In the present study, the biological function of
lncRNA-AK001903 in CC was explored. The CCK-8 assay demonstrated
that knockdown of lncRNA AK001903 inhibited proliferation of Ca Ski
cells, which indicated that lncRNA-AK001903 serves a crucial role
in CC development. In the present study, transwell assays
demonstrated that lncRNA-AK001903 promoted cell invasion and
migration in CC. Considering Ca Ski is a cervical cancer cell line
which was established from cells from a metastasis in the small
bowel mesentery (39),
lncRNA-AK001903 may act as an oncogene in the progression of CC and
is a promising therapeutic target for the treatment of patients
with CC.
In addition, the origin of lncRNA-AK001903 indicated
that it was a long intergenic noncoding RNA (lincRNA) (20,40).
LincRNAs, transcribed from intergenic regions of the genome, are
the most abundant class of lncRNAs found in over 10,000 species so
far (41). The potential mechanisms
of lincRNA function include co-transcriptional regulation,
regulation of gene expression in cis or in trans
through recruitment of proteins or molecular complexes, titration
of RNA-binding factors, and activation of posttranscriptional
regulation by pairing with other RNAs (42). Atianand et al (43) discovered that erythroid prosurvival;
also known as Ttc39aos1 was associated with chromatin at regulatory
regions of immune response genes to control nucleosome positioning
and repress transcription. TNFα and hnRNPL related immunoregulatory
LincRNA exerted important roles in the innate immune response and
inflammatory diseases in humans through combining with heterogenous
nuclear ribonucleoprotein L to form a ribonucleoprotein complex to
regulate tumor necrosis factor α induction (44). However, the mechanism of how
lncRNA-AK001903 promotes cell proliferation, invasion and migration
and possibly signal pathways remains unclear. Recently, increasing
studies have reported lncRNAs were able to act as competing
endogenous RNAs (ceRNAs) to competitively binding with microRNAs
(miRNAs) to relieve the translation repression of targeted mRNAs
induced by the common miRNAs and their downstream pathways
(45–47). Nevertheless, whether lncRNA-AK001903
also regulates the progression of cervical cancer by acting as a
ceRNA remains to be further explored.
The clinical application of lncRNA-AK001903 needs to
be further explored due to the limitation of the present study of
not tracking the prognostic differences among patients with
different expression levels.
In summary, the present study demonstrated that
lncRNA AK001903 is a potential oncogene in CC. LncRNA AK001903 was
able to promote CC tumor proliferation, migration and invasion and
may act as therapeutic target and auxiliary criteria for evaluating
FIGO stage.
Acknowledgments
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81572575), Guangdong
province Natural Scientific Grant (grant no. 2016A020215059),
Special Support for Guangdong College Students' innovation and
entrepreneurship training program (grant no. 1055813194), National
College Students' innovation and entrepreneurship training program
(grant no. 201310558097) and Guangdong clinical teaching base
teaching program (grant no. 2018JD004).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TY, XF and GZ contributed to the conception of the
study and designed the experiments. YW, QX, XF and GZ performed the
experiments. GZ, XF and RL contributed significantly to analysis
and manuscript preparation. ZL and YW made substantial
contributions to the acquisition of patient tissues and patient
data. ZL analyzed and interpreted the patient data. GZ, TY and ZL
revised the manuscript critically for important intellectual
content. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All specimens were obtained with the approval of the
Sun Yat-sen Memorial Hospital Review Board (approval number,
2015132) (Guangzhou, China). Written informed consent was obtained
from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
2
|
Walboomers JM, Jacobs MV, Manos MM, Bosch
FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ and Muñoz N:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar
|
|
3
|
Moon JY, Song IC, Ko YB and Lee HJ: The
combination of cisplatin and topotecan as a second-line treatment
for patients with advanced/recurrent uterine cervix cancer.
Medicine (Baltimore). 97:e03402018. View Article : Google Scholar
|
|
4
|
Jabbar B, Rafique S, Salo-Ahen OMH, Ali A,
Munir M, Idrees M, Mirza MU, Vanmeert M, Shah SZ, Jabbar I and Rana
MA: Antigenic peptide prediction from E6 and E7 oncoproteins of HPV
types 16 and 18 for therapeutic vaccine design using
immunoinformatics and MD simulation analysis. Front Immunol.
9:30002018. View Article : Google Scholar
|
|
5
|
US Preventive Services Task Force, ; Curry
SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, Davidson KW, Doubeni
CA, Epling JW Jr, Kemper AR, et al: Screening for Cervical Cancer:
US Preventive Services Task Force Recommendation Statement. JAMA.
320:674–686. 2018. View Article : Google Scholar
|
|
6
|
Zheng R, Zeng H, Zhang S, Chen T and Chen
W: National estimates of cancer prevalence in China, 2011. Cancer
Lett. 370:33–38. 2016. View Article : Google Scholar
|
|
7
|
Di Gesualdo F, Capaccioli S and Lulli M: A
pathophysiological view of the long non-coding RNA world.
Oncotarget. 5:10976–10996. 2014. View Article : Google Scholar
|
|
8
|
Sun L, Luo H, Liao Q, Bu D, Zhao G, Liu C,
Liu Y and Zhao Y: Systematic study of human long intergenic
non-coding RNAs and their impact on cancer. Sci China Life Sci.
56:324–334. 2013. View Article : Google Scholar
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar
|
|
10
|
Liu H, Luo J, Luan S, He C and Li Z: Long
non-coding RNAs involved in cancer metabolic reprogramming. Cell
Mol Life Sci. 76:495–504. 2019. View Article : Google Scholar
|
|
11
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, et al: Long non-coding RNA
HOTAIR reprograms chromatin state to promote cancer metastasis.
Nature. 464:1071–1076. 2010. View Article : Google Scholar
|
|
12
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010. View Article : Google Scholar
|
|
13
|
Jiang H, Li T, Qu Y, Wang X, Li B, Song J,
Sun X, Tang Y, Wan J, Yu Y, et al: Long non-coding RNA SNHG15
interacts with and stabilizes transcription factor Slug and
promotes colon cancer progression. Cancer Lett. 425:78–87. 2018.
View Article : Google Scholar
|
|
14
|
Fu C, Li D, Zhang X, Liu N, Chi G and Jin
X: LncRNA PVT1 facilitates tumorigenesis and progression of glioma
via Regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway.
Neurotherapeutics. 15:1139–1157. 2018. View Article : Google Scholar
|
|
15
|
Guo X, Xiao H, Guo S, Li J, Wang Y, Chen J
and Lou G: Long noncoding RNA HOTAIR knockdown inhibits autophagy
and epithelial-mesenchymal transition through the Wnt signaling
pathway in radioresistant human cervical cancer HeLa cells. J Cell
Physiol. 234:3478–3489. 2019. View Article : Google Scholar
|
|
16
|
Zhu P, Wang FQ and Li QR: Correlation
study between long non-coding RNA MALAT1 and radiotherapy
efficiency on cervical carcinoma and generation of radiotherapy
resistant model of cancer. Eur Rev Med Pharmacol Sci. 22:5140–5148.
2018.
|
|
17
|
Zhang D, Sun G, Zhang H, Tian J and Li Y:
Long non-coding RNA ANRIL indicates a poor prognosis of cervical
cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed
Pharmacother. 85:511–516. 2017. View Article : Google Scholar
|
|
18
|
Wu F, Huang Y, Dong F and Kwon JH:
Ulcerative colitis-associated long noncoding RNA, BC012900,
regulates intestinal epithelial cell apoptosis. Inflamm Bowel Dis.
22:782–795. 2016. View Article : Google Scholar
|
|
19
|
Bhatla N, Aoki D, Sharma DN and
Sankaranarayanan R: Cancer of the cervix uteri. Int J Gynaecol
Obstet. 143 (Suppl 2):S22–S36. 2018. View Article : Google Scholar
|
|
20
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar
|
|
21
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, et al: Many human large intergenic
noncoding RNAs associate with chromatin-modifying complexes and
affect gene expression. Proc Natl Acad Sci USA. 106:11667–11672.
2009. View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar
|
|
24
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar
|
|
25
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar
|
|
26
|
Zhuang C, Ma Q, Zhuang C, Ye J, Zhang F
and Gui Y: LncRNA GClnc1 promotes proliferation and invasion of
bladder cancer through activation of MYC. FASEB J. 33:11045–11059.
2019. View Article : Google Scholar
|
|
27
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar
|
|
28
|
Xu Y, Leng K, Yao Y, Kang P, Liao G, Han
Y, Shi G, Ji D, Huang P, Zheng W, et al: A novel circular RNA,
circ-CCAC1, contributes to CCA progression, induces angiogenesis,
and disrupts vascular endothelial barriers. Hepatology. Aug
4–2020.(Epub ahead of print). View Article : Google Scholar
|
|
29
|
Setten RL, Rossi JJ and Han SP: The
current state and future directions of RNAi-based therapeutics. Nat
Rev Drug Discov. 18:421–46. 2019. View Article : Google Scholar
|
|
30
|
Lavorgna G, Vago R, Sarmini M, Montorsi F,
Salonia A and Bellone M: Long non-coding RNAs as novel therapeutic
targets in cancer. Pharmacol Res. 110:131–138. 2016. View Article : Google Scholar
|
|
31
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar
|
|
32
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. Biomed Res Int. 2013:1361062013. View Article : Google Scholar
|
|
33
|
Dong J, Su M, Chang W, Zhang K, Wu S and
Xu T: Long non-coding RNAs on the stage of cervical cancer
(Review). Oncol Rep. 38:1923–1931. 2017. View Article : Google Scholar
|
|
34
|
Galvão M and Coimbra EC: Long noncoding
RNAs (lncRNAs) in cervical carcinogenesis: New molecular targets,
current prospects. Crit Rev Oncol Hematol. 156:1031112020.
View Article : Google Scholar
|
|
35
|
Schmidt U and Begley CG: Cancer diagnosis
and microarrays. Int J Biochem Cell Biol. 35:119–124. 2003.
View Article : Google Scholar
|
|
36
|
Slack FJ and Chinnaiyan AM: The Role of
Non-coding RNAs in Oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar
|
|
37
|
Tian Y, Wang YR and Jia SH: Knockdown of
long noncoding RNA DLX6-AS1 inhibits cell proliferation and
invasion of cervical cancer cells by downregulating FUS. Eur Rev
Med Pharmacol Sci. 23:7307–7313. 2019.
|
|
38
|
Jin L, Ji J, Shi L, Jin S and Pei L:
lncRNA HAND2-AS1 inhibits cancer cell proliferation, migration and
invasion by downregulating ROCK1 in HPV-positive and negative
cervical squamous cell carcinoma. Exp Ther Med. 18:2512–2518.
2019.
|
|
39
|
Pattillo RA, Hussa RO, Story MT, Ruckert
AC, Shalaby MR and Mattingly RF: Tumor antigen and human chorionic
gonadotropin in CaSki cells: A new epidermoid cervical cancer cell
line. Science. 196:1456–1458. 1977. View Article : Google Scholar
|
|
40
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar
|
|
41
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar
|
|
42
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar
|
|
43
|
Atianand MK, Hu W, Satpathy AT, Shen Y,
Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD,
Blin J, et al: A Long Noncoding RNA lincRNA-EPS acts as a
transcriptional brake to restrain inflammation. Cell.
165:1672–1685. 2016. View Article : Google Scholar
|
|
44
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014. View Article : Google Scholar
|
|
45
|
Franco-Zorrilla JM, Valli A, Todesco M,
Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA and
Paz-Ares J: Target mimicry provides a new mechanism for regulation
of microRNA activity. Nat Genet. 39:1033–1037. 2007. View Article : Google Scholar
|
|
46
|
Song J, Ye A, Jiang E, Yin X, Chen Z, Bai
G, Zhou Y and Liu J: Reconstruction and analysis of the aberrant
lncRNA-miRNA-mRNA network based on competitive endogenous RNA in
CESC. J Cell Biochem. 119:6665–6673. 2018. View Article : Google Scholar
|
|
47
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar
|