Introduction
Lung carcinoma is one of the most common types of
cancer and one of the leading causes of cancer-associated deaths
worldwide (1,2). In 2018, there were 2.1 million new
cases and 1.8 million people died of lung cancer (3). The development and progression of lung
cancer is a complicated process that may be associated with
pleurisy or severe lung infections with bacteria and viruses or
fungi, making lung cancer difficult to treat (4–6). Lung
cancer can be divided into two categories, including non-small cell
lung cancer (NSCLC) and small cell lung cancer (7). Although the treatment outcome for lung
carcinoma have greatly improved due to advances in the technologies
and treatment strategies, the prognosis of patients with lung
carcinoma remains poor, since most patients are diagnosed at a late
stage (8,9). Alternative natural therapies, such as
β-himachalene, apigenin, geranial and Dracocephalum
kotschyi, have demonstrated anticancer properties, but with
disadvantages such as cytotoxic effects (10–14).
Therefore, research has focused on the understanding of the
molecular mechanisms of lung carcinoma and identifying novel
diagnostic biomarkers and therapeutic targets.
Epigenetic modifications by long non-coding RNAs
(lncRNAs) and microRNAs (miRNAs) are crucial for the development
and metastasis of lung cancer (15).
lncRNAs are a group of RNAs >200 nucleotides in length involved
in multiple biological processes including gene imprinting, histone
modification, chromatin remodeling, transcriptional activation,
transcriptional interference, nuclear transport, and cell cycle
regulation. lncRNAs are also involved in development of various
types of tumor (16–18). For example, NEAT1 (non-coding nuclear
enriched abundant transcript 1) is upregulated in breast cancer,
colorectal cancer and non-small cell lung cancer (19,20).
Small nucleolar RNA host gene 7 is upregulated in lung cancer
tissues and cells and promotes the proliferation, migration and
invasion of lung cancer cells by regulating Fas apoptotic
inhibitory molecule 2 expression (21). miRNAs are a group of small non-coding
RNAs that post-transcriptionally regulate gene expression and are
also involved in tumor development and metastasis (22). Notably, lncRNAs have been reported to
exert their functions as competing endogenous RNAs (ceRNAs) by
sponging miRNAs (23–25). Previous studies have revealed various
lncRNA-miRNA-mRNA interaction networks in lung carcinoma (26–28).
Cong et al (26) have
demonstrated that lncRNA LINC00665 functions as a ceRNA to sponge
miR-98, which regulates ERK signaling, and promotes the development
of lung cancer. lncRNA PVT1 has been reported to competitively bind
miR-424-5p, which is involved in regulating the
coactivator-associated arginine methyltransferase 1 in NSCLC
(29). Furthermore, lncRNAs can
exert oncogenic roles, as well as function as tumor inhibitors. For
example, lncRNA GACAT3 expression is decreased in NSCLC, and high
expression levels of lncRNA GACAT3 inhibit the invasion and
metastasis of NSCLC (30). Another
study has demonstrated that lncRNA BX357664 suppresses NSCLC
development by inhibiting cell proliferation and invasion (31). However, the roles of
lncRNA-miRNA-mRNA networks in lung carcinoma remain unclear.
Previously, LINC00887 has been identified to accelerate the
malignant transformation ability of NSCLC cells (32). Using high-throughput nascent RNA
capture sequencing, another study has identified LINC00887 as a
highly expressed lncRNA in lung adenocarcinoma and squamous cell
carcinoma (33). However, the
detailed underlying mechanisms require further clarification.
The present study aimed to investigate the role of
the LINC00887/miR-206/NRP1 axis in the development of lung cancer.
LINC00887 functioned as the sponge of miR-206 to upregulate NRP1
expression. A decrease in LINC00887 may serve as a prognostic and
diagnostic marker and also be used as a novel therapeutic target
for patients with lung cancer.
Materials and methods
Clinical specimens
Lung carcinoma tissues and adjacent normal tissues
(>5 cm from tumor) were obtained from 40 patients (age range,
35–70 years; mean age, 63±7.83 years) with lung cancer who
underwent surgical resection at Shaanxi Provincial People's
Hospital (Xi'an, China) between March 2017 and December 2018. Two
pathologists evaluated all specimens according to the World Health
Organization guidelines and the pTNM Union for International Cancer
Control pathological staging criteria (34). Inclusion criteria were: i) Primary
lung cancer diagnosed by pathological examination; and ii) No local
or systemic treatments were administered before surgery. All
tissues were frozen in liquid nitrogen until RNA isolation. Written
consent was obtained from all patients. The present study was
approved by the Ethics Committee of Shaanxi Provincial People's
Hospital (approval number, XJYYLL-2019287).
Cell culture and transfection
Three lung carcinoma cell lines, namely the
adenocarcinoma A549 cell line and the large cell carcinoma
NCI-H1299 and NCI-H460 cell lines, were obtained from the American
Type Culture Collection. The normal human bronchial epithelial
BEAS-2B cell line was obtained from the Chinese Cell Bank of the
Chinese Academy of Sciences. The cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific Inc.) with 10% FBS
(Hyclone; GE Healthcare Life Sciences), 1% penicillin and
streptomycin at 37°C with 5% CO2. Mycoplasma detection
was negative in all cell lines.
BEAS-2B, A549 or NCI-H460 cells were seeded in 6- or
96-well plates. When the confluency reached 60–70%, the medium was
changed to serum-free RPMI-1640, and transfection with small
interfering (si)RNAs targeting LINC00887 or a control siRNA
(si-Con). MiR-206 mimics, miR-206 inhibitors and their negative
controls were obtained from Guangzhou RiboBio Co., Ltd. LINC00887
was overexpressed using the expression plasmid pcDNA3.1(+)
(Invitrogen; Thermo Fisher Scientific Inc.). Empty vectors without
LINC00887 cDNA were used as negative controls. LINC00887 siRNA
(si-LINC00887, and negative control (si-NC) were purchased from
Shanghai GenePharma Co. Ltd. Cells were transfected with LINC00887
siRNA, miR-206 mimics, miR-206 inhibitors and their negative
controls at 50 nM concentration. All transfections were performed
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
sequences of all mimics and inhibitors and their negative controls
are presented in the Table I. Cells
were harvested 72 h after transfection for further experiments.
 | Table I.Sequence of si-LINC00887, miR-206
mimics, miR-inhibitors and their negative controls. |
Table I.
Sequence of si-LINC00887, miR-206
mimics, miR-inhibitors and their negative controls.
|
Oligonucleotides | Sequence
(5′→3′) |
|---|
| si-NC |
GGCCTTTGCGTCACGCCTTAG |
| si-LINC00887
1# |
GGCCTTTGCAGTTATTAGGAA |
| si-LINC00887
2# |
CCTGTTCTCTCTGGTTCTC |
| si-LINC00887
3# |
GTCCCTGTTCTCTCTGGTT |
| mimics-NC |
UUGUACUACACAAAAGUACUG |
| miR-206 mimics |
UGGAAUGUAAGGAAGUGUGUGG |
| inhibitor NC |
CAGUACUUUUGUGUAGUACAA |
| miR-206
inhibitor |
CCACACACUUCCUUACAUUCCA |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from tissues and cell lines was isolated
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was synthesized using the PrimeScript RT
reagent kit (Takara Bio, Inc.). qPCR was performed from cDNA using
the SYBR® Green PrimeScript™ PLUS RT-PCR kit (Takara
Bio, Inc.). The reaction conditions were 95°C for 30 sec, 60°C for
40 sec (40 cycles), and a final extension step at 72°C for 10 min.
Small RNA-rich samples were isolated from cells using
TRIzol® reagent or the mirVana miRNA Isolation kit
(Thermo Fisher Scientific, Inc.). U6 or GAPDH was used as an
endogenous control. The relative expression levels were analyzed
using the 2−ΔΔCq method (35). The primers used are listed in
Table II.
 | Table II.Primers used in the present
study. |
Table II.
Primers used in the present
study.
| Gene | Forward
(5′→3′) | Reverse
(5′→3′) |
|---|
| miR-206 |
GCGTCTGGAATGTAAGGAAGTG |
GTGCAGGGTCCGAGGT |
| U6 |
TTGGTCTGATCTGGCACATATAC |
AAAAATATGGAGCGCTTCACG |
| NRP1 |
ATGCGAATGGCTGATTCAGG |
TCCATCGAAGACTTCCACGTAG |
| LINC00887 |
TCCTGCTTGGCAGGTAACAG |
AACGATGCCTCAGTCGAAGG |
| β-actin |
TGTCACCAACTGGGACGATA |
GGGGTGTTGAAGGTCTCAAA |
Cell proliferation assays
A549 or NCI-H460 cells (3×103 cells/well)
were seeded in 96-well plates. After transfection with si-NC or
si-LINc00887, cell proliferation was evaluated using a Cell
Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.)
according to the manufacturer's instructions. Cells were cultured
for 0, 24, 48 or 96 h at 37°C and incubated with 10 µl CCK-8
reagent per well at 37°C for 1 h. The absorbance was measured at
450 nm using an Exl 800 microplate reader (BioTek China). The
5-ethynyl-2-deoxyuridine (EdU) staining assay was performed to
determine DNA synthesis in proliferating cells using an EdU assay
kit (Invitrogen; Thermo Fisher Scientific, Inc.). After
transfection with si-NC or si-LINC00887, A549 or NCI-H460 cells
were cultured at 37°C for 48 h, fixed with 4% paraformaldehyde at
room temperature for 10 min and permeabilized with 0.3% Triton
X-100 at room temperature for 10 min. Subsequently, cells were
incubated with 10 µM EdU for 2 h at 37°C, and cell nuclei were
stained with DAPI (5 µg/ml) at room temperature for 10 min. The
number of EdU-positive cells was counted under a light microscope
in five random fields (magnification, ×100; Olympus Corporation).
All assays were independently performed in triplicate.
Wound healing assay
After transfection with si-NC or si-LINC00887, A549
or NCI-H460 cells (4×105 cells/well) were seeded in
6-well plates and cultured at 37°C until confluent. The wound was
created by scratching the cell layer with a sterile pipette tip.
The floating cells were washed away using PBS. The streaked cells
were cultured in serum-free RPMI-640 medium at 37°C for 48 h. An
inverted optical microscope (Olympus Corporation) was used to
monitor the closure of the wound at 0 and 48 h at ×50
magnification. The gap distance was quantitatively evaluated using
ImageJ software (version 1.49; National Institutes of Health).
Cell migration and invasion
assays
For invasion assays, 24-well Transwell chambers (8.0
µm pore size, Costar; Corning, Inc.) with Matrigel-precoated
membranes were used. A total of 1×105 A549 or NCI-H460
cells in 100 µl FBS-free RPMI-1640 were added to the upper
chambers, while 500 µl RPMI-1640 medium with 10% FBS was added to
the bottom chambers. After 48 h, the non-invaded cells on the upper
side of the membrane in the chamber were removed by swabs, and the
invading cells in the lower side of the chamber were stained with
0.1% crystal violet for 20 min at room temperature. The cells were
observed and counted under a light microscope (magnification, ×200;
Olympus Corporation). For migration detection, the Matrigel was not
used and all other steps were the same as the cell invasion
assay.
Western blotting
A549 or NCI-H460 cells were lysed using RIPA buffer
(Sigma-Aldrich; Merck KGaA) with 1 mM phenylmethylsulphonyl
fluoride. The concentration of obtained total protein was
quantified using a BCA protein assay kit (Thermo Fisher Scientific,
Inc.). Equal amounts (25 µg) of proteins were loaded and separated
via 10% SDS-PAGE and electro-transferred to a PVDF membrane (EMD
Millipore). After blocking with 5% bovine serum albumin (Beyotime
Institute of Biotechnology) at room temperature for 1 h, the
membrane was incubated with anti-neuropilin 1 (NRP1) (1:2,000; cat.
no. ab81321; Abcam) and anti-β-actin (1:5,000; ab179467; Abcam)
primary antibodies at 4°C overnight. After washing three times with
TBST (5 min per wash), the members were subsequently incubated with
an HRP-labelled goat anti-rabbit secondary antibody (ab6721;
1:10,000; Abcam) at room temperature for 1 h. The immunolabelling
was visualized using an ECL system (EMD Millipore) according to the
manufacturer's protocol. All assays were performed independently in
triplicate and the densitometric analysis was performed using
ImageJ (version 1.49; National Institutes of Health).
Dual-luciferase reporter assay
The reporter plasmid containing 3′UTR of LINC00887
or NRP1 with wild (Wt LINC00887 or Wt NRP1) or mutant (Mut
LINC00887 or Mut NRP1) miR-206 binding sites were constructed by
Guangzhou RiboBio Co. Ltd. For the reporter assay, lung cancer
cells or 293T cells in 24-well plates and co-transfected with the
reporter plasmid and miR-206 mimic or NC using
Lipofectamine® 2000 (Invitrogen, Thermo Fisher
Scientific, Inc.). Cells were harvested 48 h later and assayed with
a luciferase reporter assay system (Promega Corporation). The
relative luciferase activity was normalized to Renilla
luciferase activity.
Tumor xenograft model
BALB/c nude mice (age, 8 weeks; weight, 21–25 g)
were obtained from Beijing Vital River Laboratory Animal Technology
Co., Ltd., and housed at a room temperature of 25°C with a 12 h
light/dark cycle. The mice were maintained in an individually
ventilated cage system under specific pathogen-free conditions
(temperature; 25°C; humidity, 55%) with free access to food and
water. Then the posterior flank of 6-week-old male BALB/c nude mice
(n=10) were subcutaneously injected with NCI-H460
(4×106) cells transfected with si-NRP1 or si-negative
control (NC). Tumor volumes were examined every 4 days. After 17
days, the mice were euthanized by CO2 inhalation
(CO2 flow rate, 20% of cage volume) and tumor tissues
were dissected, photographed and weighed. The expression levels of
miR-206 in tumor tissues were detected by RT-qPCR as
aforementioned. The protein levels of NRP1 in tumor tissues were
measured by western blotting as aforementioned. The murine
experiments were conducted in July 2017. The animal experiment was
performed in compliance with the authenticated animal protocols of
the Ethical Committee of Animal Welfare of Shaanxi Provincial
People's Hospital (approval. No. IACUC-20190116).
Target prediction
Potential target miRNAs of LINC00887 were predicted
using LncBase V2(http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index).
The target genes of miR-206 were predicted using the bioinformatics
algorithms: TargetScanV7.2 (http://www.targetscan.org/vert_72/).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7.0 (GraphPad Software, Inc.). Data are presented as the mean
± SD for three independent experiments. The differences between two
groups were analyzed by unpaired Student's t-test, and one-way
ANOVA followed by the Tukey's post hoc test was performed to
analyze the differences among more than 2 groups. Pearson's
correlation analysis was performed to analyze the correlation
between LINC00887 and miR-206 expression as well as the clinical
variables. P<0.05 was considered to indicate a statistically
significant difference.
Results
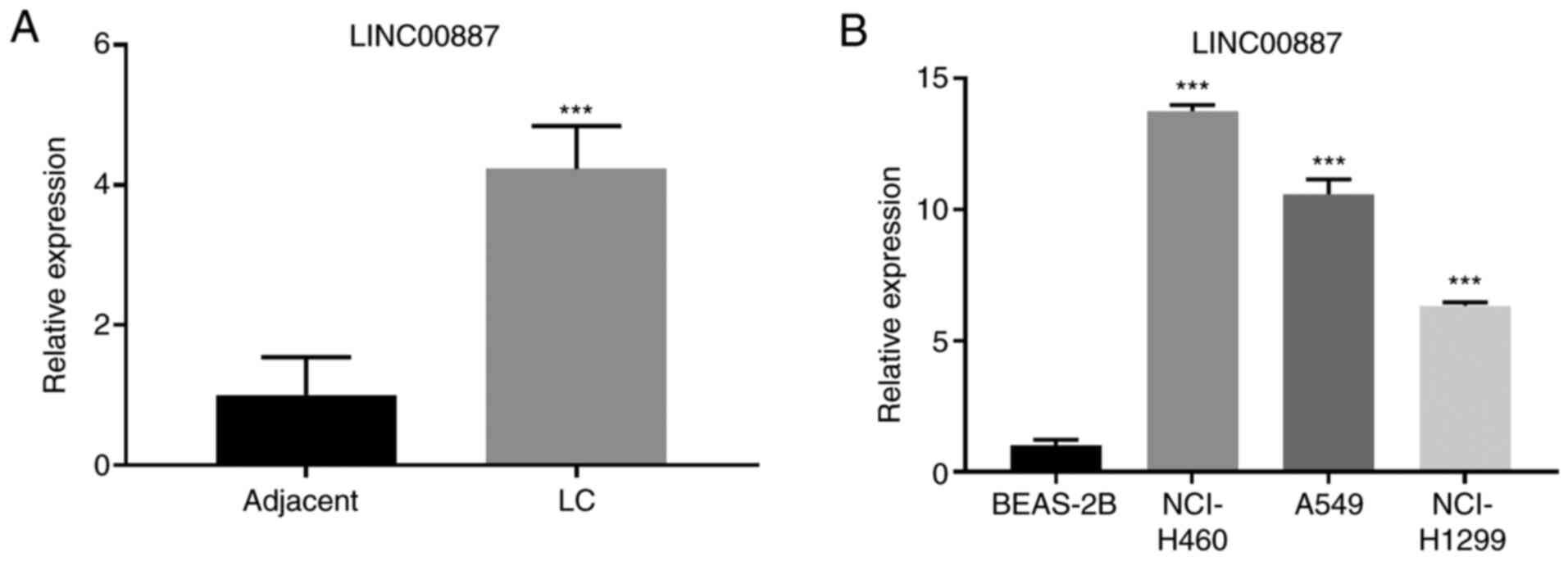
LINC00887 is upregulated in lung
carcinoma tissues and cell lines
RT-qPCR was performed to analyze the expression
levels of LINC00887 in the collected 40 pairs of lung carcinoma and
adjacent normal tissues. As presented in Fig. 1A, LINC00887 expression levels were
significantly upregulated in lung carcinoma tissues compared with
those in adjacent normal tissues (P<0.001). To assess the
association of LINC00887 expression with clinicopathologic
characteristics, the expression levels of LINC00887 were
categorized as low (n=20) or high (n=20) in relation to the median
value (cut-off, 2.12). High LINC00887 expression was significantly
associated with advanced TNM stage and lymph node metastasis
(Table III). Additionally,
LINC00887 expression levels were significantly higher in three lung
cancer cell lines (NCI-H460, A549 and NCI-H1299) compared with
those in the normal human bronchial epithelium BEAS-2B cell line
(Fig. 1B). NCI-H460 and A549, which
exhibited high expression levels of LINC00887, were used for
subsequent in vitro assays.
 | Table III.Association between LINC00887 and
clinical features of patients. |
Table III.
Association between LINC00887 and
clinical features of patients.
|
| LINC00887
expression |
|
|---|
|
|
|
|
|---|
|
Characteristics | Low (n=20) | High (n=20) | P-value |
|---|
| Age, years |
|
≤65 | 9 | 10 | 0.752 |
|
>65 | 11 | 10 |
|
| Sex |
|
Male | 12 | 13 | 0.744 |
|
Female | 8 | 7 |
|
| TNM stage |
|
I+II | 15 | 7 | 0.011 |
|
III+IV | 5 | 13 |
|
| Lymph node
metastasis |
|
Negative | 10 | 3 | 0.018 |
|
Positive | 10 | 17 |
|
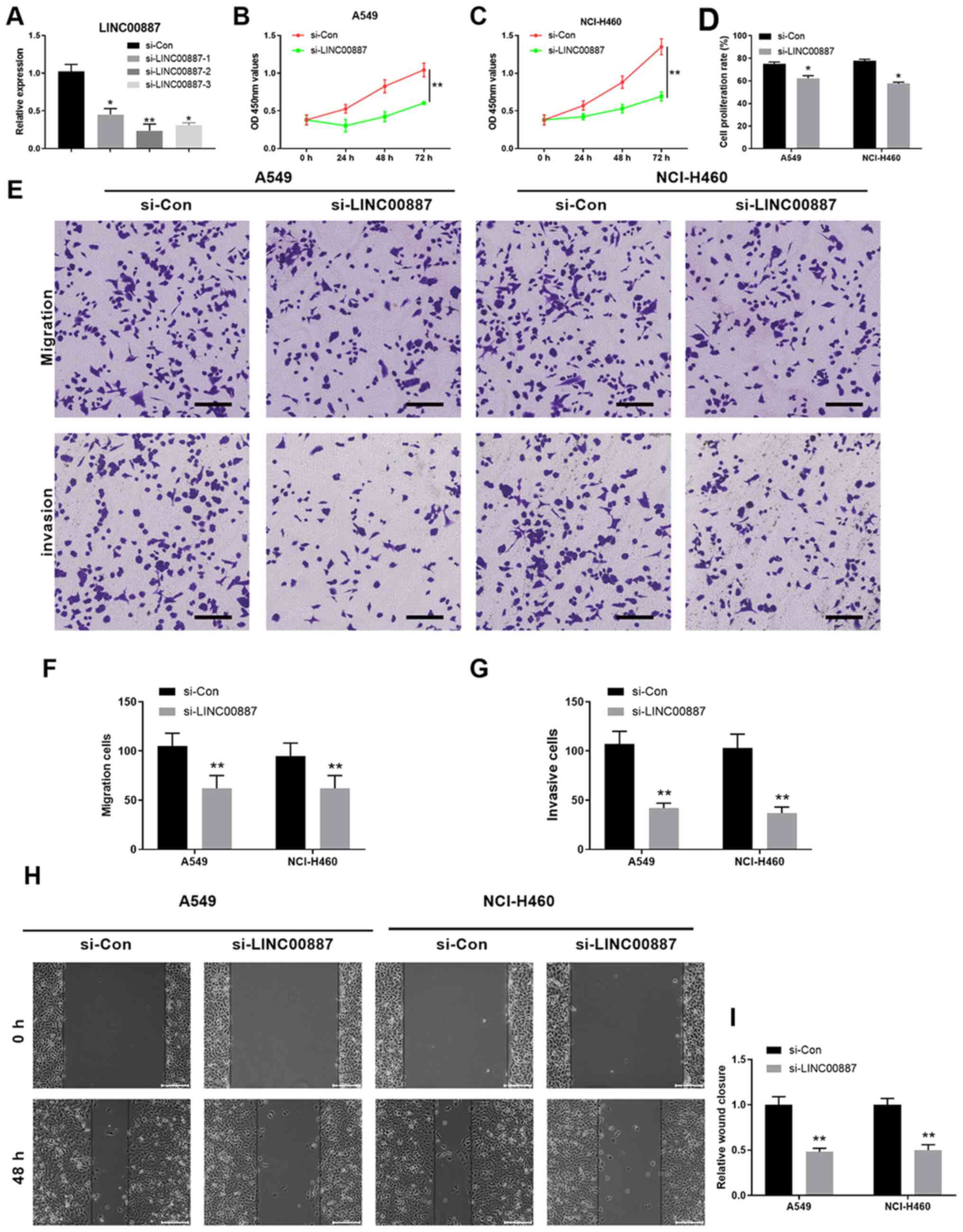
LINC00887 knockdown suppresses
proliferation, migration and invasion of lung carcinoma cells in
vitro
To investigate the function of LINC00887, the
expression of LINC00887 was knocked down using siRNAs. All three
siRNAs mentioned in Table I
significantly silenced LINC00887 expression compared with that in
the si-Con group, with si-LINC00887-2 exhibiting the strongest
knockdown efficiency (Fig. 2A).
Therefore, subsequent experiments were performed using
si-LINC00887-2. The CCK-8 assay demonstrated that cell
proliferation was significantly decreased in A549 and NCI-H460 lung
carcinoma cells after si-LINC00887 transfection compared with that
in cells transfected with si-Con (P<0.01; Fig. 2B and C). Consistently, EdU and DAPI
double staining confirmed that LINC00887 knockdown significantly
inhibited the proliferation of A549 and NCI-H460 cells (P<0.05;
Figs. 2D and S1). Furthermore, Transwell assays revealed
that silencing LINC00887 significantly inhibited the migratory and
invasive capabilities of A549 and NCI-H460 lung carcinoma cells
(P<0.01; Fig. 2E-G). In addition,
wound healing assays demonstrated that LINC00887 knockdown
significantly decreased the relative wound closure in A549 and
NCI-H460 cells (P<0.01; Fig. 2H and
I). To further validate the function of LINC00887, LINC00887
was overexpressed in normal human lung epithelial BEAS-2B cells
(Fig. S2A). LINC00887
overexpression did not alter cell proliferation, migration and
invasion in normal BEAS-2B cells (Fig.
S2B-F). In summary, the present results suggested that
LINC00887 knockdown suppressed the proliferation, migration and
invasion of lung carcinoma cells in vitro.
LINC00887 directly interacts with
miR-206
Multiple studies have demonstrated that lncRNAs can
exert their functions as competing endogenous RNAs (ceRNAs) by
competitively binding miRNAs involved in regulating target gene
expression (36,37). However, whether LINC00887 has a
similar function in lung carcinoma remains unknown. Using the
bioinformatics DIANA tool lncbase V2 (38), miR-206 was identified to have
putative binding sites with LINC00887 (Fig. 3A). To verify whether LINC00887
directly interacted with miR-206, Dual-Luciferase reporter assays
were performed, and the results revealed that the miR-206 mimic
significantly inhibited the luciferase activity of the LINC00887-wt
reporter, but not that of the LINC00887-mut reporter (Fig. 3B). In addition, the regulation
between miR-206 and LINC00887 was further examined; as presented in
Fig. 3C, LINC00887 knockdown
significantly upregulated miR-206 expression, whereas miR-206
overexpression using the miR-206 mimic significantly downregulated
LINC00887 expression in A549 and NCI-H460 lung carcinoma cells
(Fig. 3D). Notably, significantly
lower levels of miR-206 were detected in lung carcinoma tissues
compared with those in adjacent normal tissues (Fig. 3E). Pearson's correlation analysis
revealed that miR-206 expression was negatively correlated with
LINC00887 expression (P<0.001; Fig.
3F).
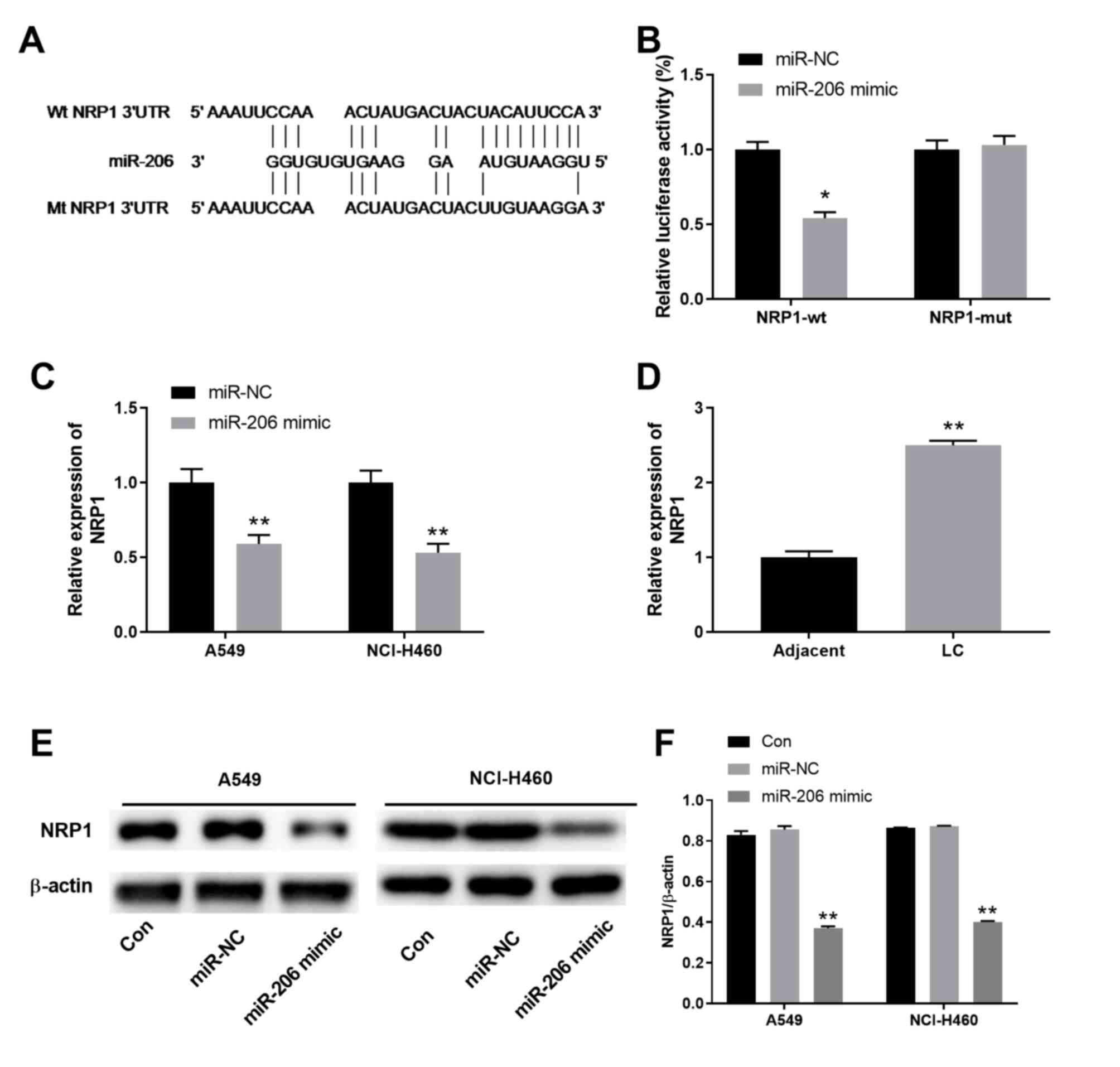
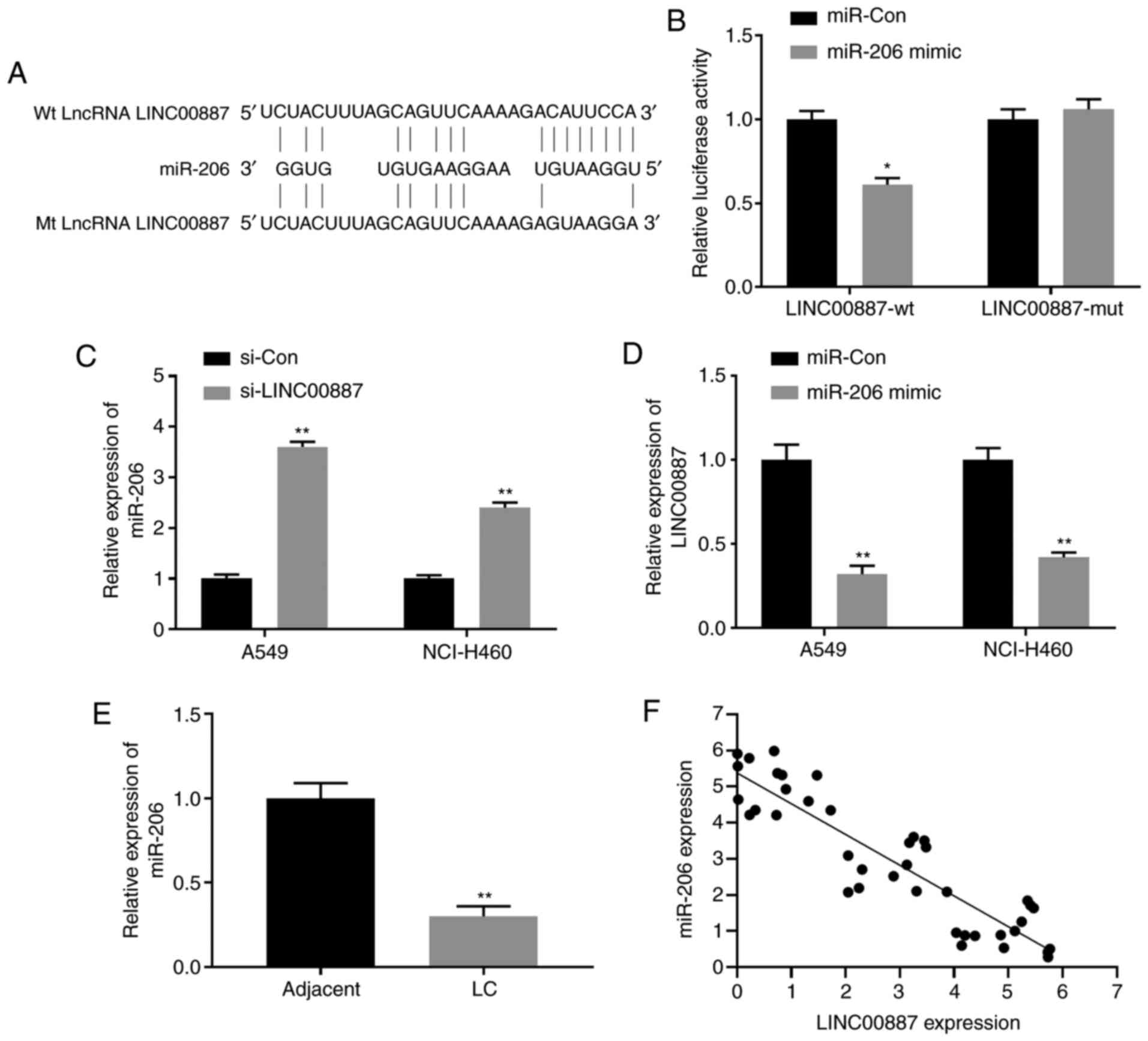 | Figure 3.LINC00887 directly interacts with
miR-206. (A) Putative binding site between lncRNA LINC00887 and
miR-206 revealed by the DIANA-LncBase. (B) 293 cells were
transfected with luciferase reporter vectors containing lncRNA
LINC00887 WT or mutant sequences (pmirGLO-LINC00887-wt or
pmirGLO-LINC00887-mut, respectively), in the presence of miR-206
mimic or miR-Con. Relative luciferase activity was analyzed 72 h
later. (C) Expression levels of miR-206 in LC A549 and NCI-H460
cells transfected with si-Con or si-LINC00887 were analyzed via
RT-qPCR 72 h post transfection. (D) Expression levels of LINC00887
in LC A549 and NCI-H460 cells transfected with miR-Con or miR-206
mimic were analyzed by RT-qPCR 72 h post transfection. (E)
Expression levels of miR-206 in LC tissues and adjacent normal
tissues were analyzed via RT-qPCR. (F) Pearson correlation analysis
between miR-206 expression and LINC00887 expression in LC tissues.
Data are presented as the mean ± SD. *P<0.05; **P<0.01 vs.
miR-Con, si-Con or adjacent tissues. RT-qPCR, reverse
transcription-quantitative PCR; LC, lung carcinoma; miR, microRNA;
Con, control; si, small interfering RNA; wt, wild-type; mt,
mutant. |
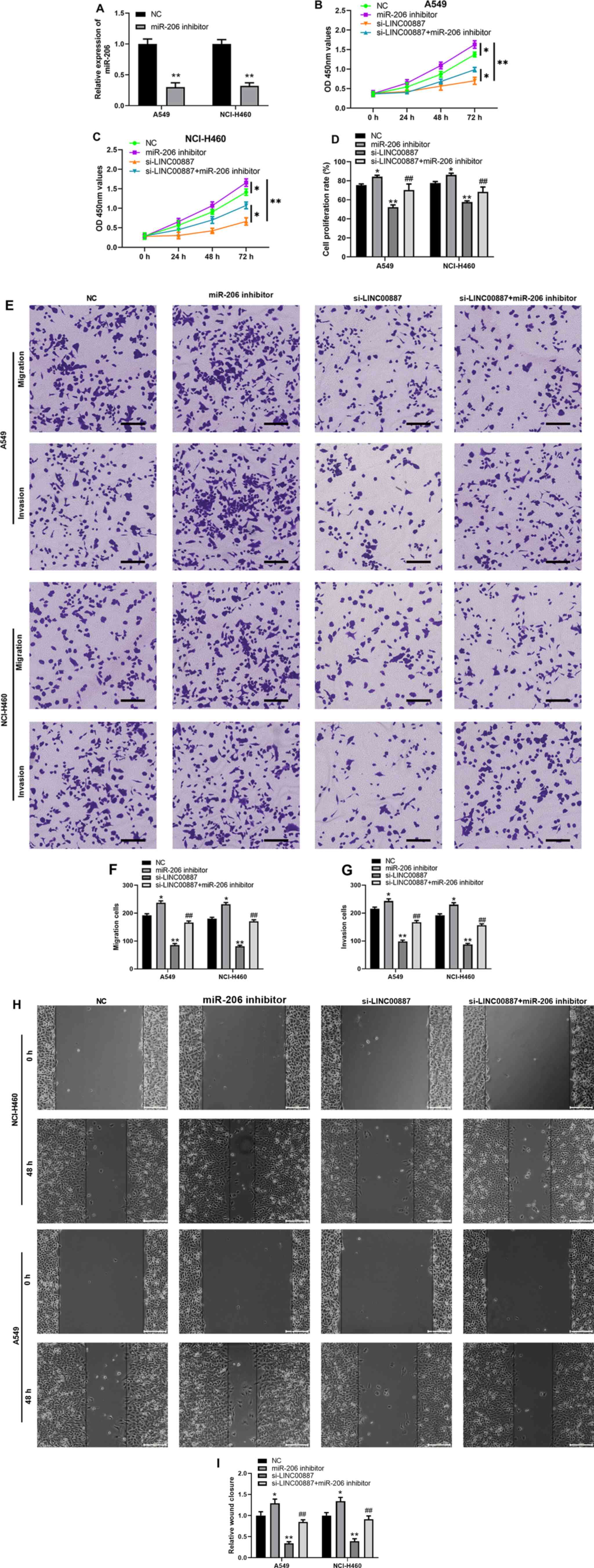
LINC00887 regulates lung carcinoma
cell proliferation, migration and invasion via miR-206
To further investigate the functional association
between LINC00887 and miR-206, and test whether LINC00887 exerts
its function via miR-206, a miR-206 inhibitor was used to
downregulate miR-206 expression in lung carcinoma A549 and NCI-H460
cells (Fig. 4A). As LINC00887
knockdown enhanced miR-206 expression (Fig. 3C), A549 and NCI-H460 cells
transfected with si-LINC00887 were further transfected with the
miR-206 inhibitor or inhibitor-NC. As presented in Figs. 4B-D and S3, the results of the CCK-8 and EdU/DAPI
staining assays demonstrated that LINC00887 knockdown significantly
inhibited cell proliferation, whereas miR-206 inhibitor ameliorated
the inhibition of cell proliferation induced by LINC00887 knockdown
in lung carcinoma A549 and NCI-H460 cells. Consistently, as
demonstrated by Transwell and wound healing assays, silencing
LINC00887 significantly inhibited cell migration or invasion,
whereas miR-206 inhibitor ameliorated the inhibition of cell
migration or invasion induced by LINC00887 knockdown in lung
carcinoma A549 and NCI-H460 cells (Fig.
4E-I).
miR-206 regulates NRP1 expression by
targeting NRP1 3′-UTR
To determine the target of miR-206 in lung
carcinoma, bioinformatics prediction was performed using
TargetScan, which revealed that miR-206 targeted the 3′-UTR of NRP1
with 17 complementary binding sites (Fig. 5A). To validate the prediction,
Dual-Luciferase reporter assay was performed, and the results
demonstrated that the relative luciferase activities in 293T cells
transfected with the NRP1-wt reporter vector, but not with
NRP1-mut, were inhibited by co-transfection with the miR-206 mimic
(Fig. 5B). Furthermore, miR-206
overexpression significantly downregulated both the mRNA and
protein levels of NRP1 in lung carcinoma A549 and NCI-H460 cells
(Figs. 5C, E and F, and S5). In
addition, significantly higher expression levels of NRP1 were
detected in lung carcinoma tissues compared with those in the
adjacent normal control tissues (Fig.
5D).
LINC00887-miR-206-NRP1 interaction
network regulates lung carcinoma cell proliferation, migration and
invasion
To further validate the function of the
LINC00887-miR-206-NRP1 interaction network, lung carcinoma A549 and
NCI-H460 cells were transfected with si-LINC00887, miR-206 mimic,
si-NRP1 or the si-NC. CCK-8 assays revealed that compared with the
NC, transfection with si-LINC00887, miR-206 mimic or si-NRP1
significantly inhibited A549 and NCI-H460 cell proliferation
(Figs. 6A and B, and S5). The effect of si-LINC00887, miR-206
mimic on cell proliferation was further confirmed using EdU/DAPI
staining (Figs. 6C and S4). Transwell assays demonstrated that
LINC00887 knockdown, miR-206 overexpression or NRP1 silencing
significantly inhibited migration and invasion of A549 and NCI-H460
cells (Fig. 6D-F). In addition,
wound healing assays revealed that si-LINC00887, miR-206 mimic
significantly inhibited the relative migration distance of A549 and
NCI-H460 (Fig. 6G and H). NRP1
protein levels in A549 and NCI-H460 cells transfected with
si-LINC00887, the miR-206 mimic, si-NRP1 or the NC were further
analyzed. Compared with the NC group, LINC00887 knockdown, miR-206
overexpression or NRP1 silencing significantly inhibited NRP1
protein expression, with si-NRP1 transfection exhibiting the lowest
expression levels of NRP1 protein among all groups (Fig. 6I and J).
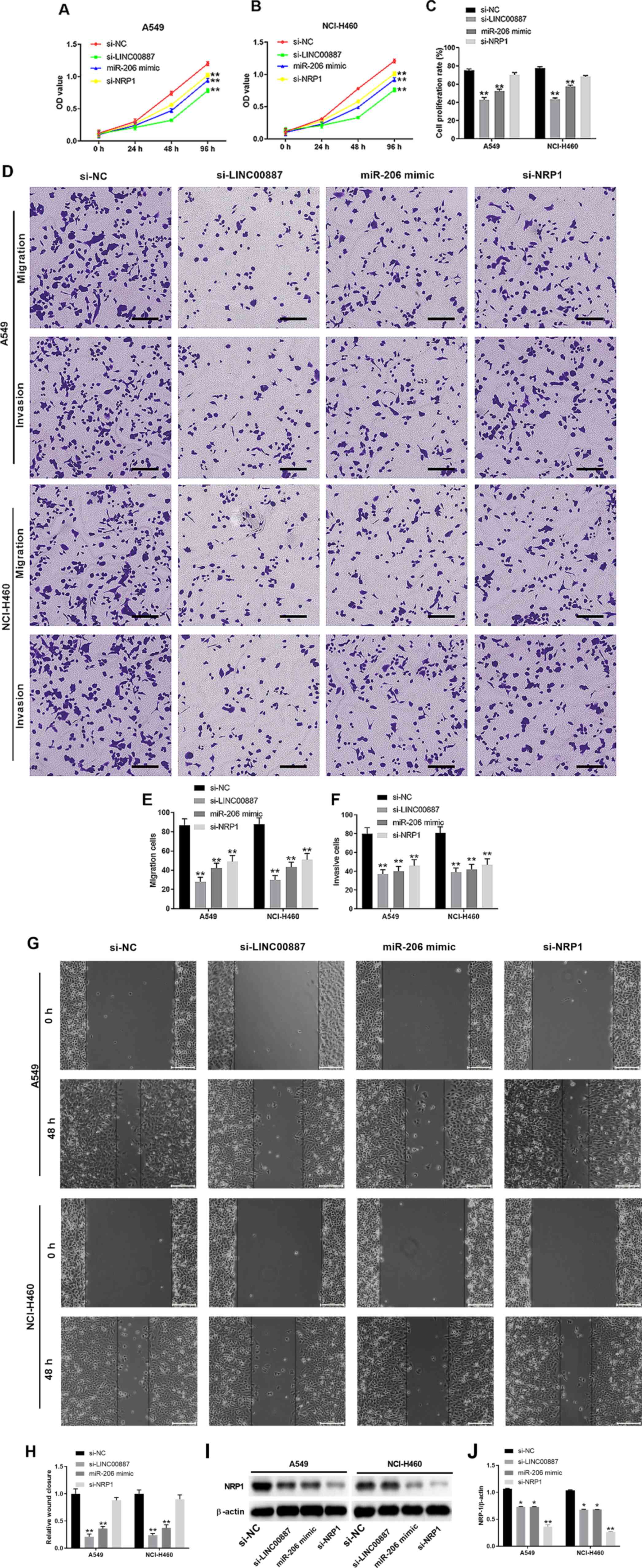 | Figure 6.LINC00887-miR-206-NRP1 interaction
network regulates lung carcinoma cell proliferation, migration and
invasion. Lung carcinoma A549 or NCI-H460 cells were transfected
with si-LINC00887, miR-206 mimic, si-NRP1 or si-NC. (A-C) Cell
proliferation of (A) A549 and (B) NCI-H460 cells was assessed using
the Cell Counting Kit-8 assay at the indicated time points, or (C)
assessed by EdU and DAPI staining. (D) Transwell assays were used
to analyze (E) migration and (F) invasion of A549 or NCI-H460
cells. Scale bar, 200 µm. (G and H) Cell migration capability was
analyzed via wound-healing assay. Scale bar, 200 µm. (I and J) NRP1
protein expression was analyzed 72 h post transfection. The western
blot experiments were repeated ≥3 times independently. NRP1
expression was normalized to the expression levels of β-actin. Data
are presented as the mean ± SD. *P<0.05; **P<0.01 vs. NC. NC,
negative control; miR, microRNA; NRP1, neuropilin 1; si, small
interfering RNA; OD, optical density; EdU,
5′-ethynyl-2′-deoxyuridine. |
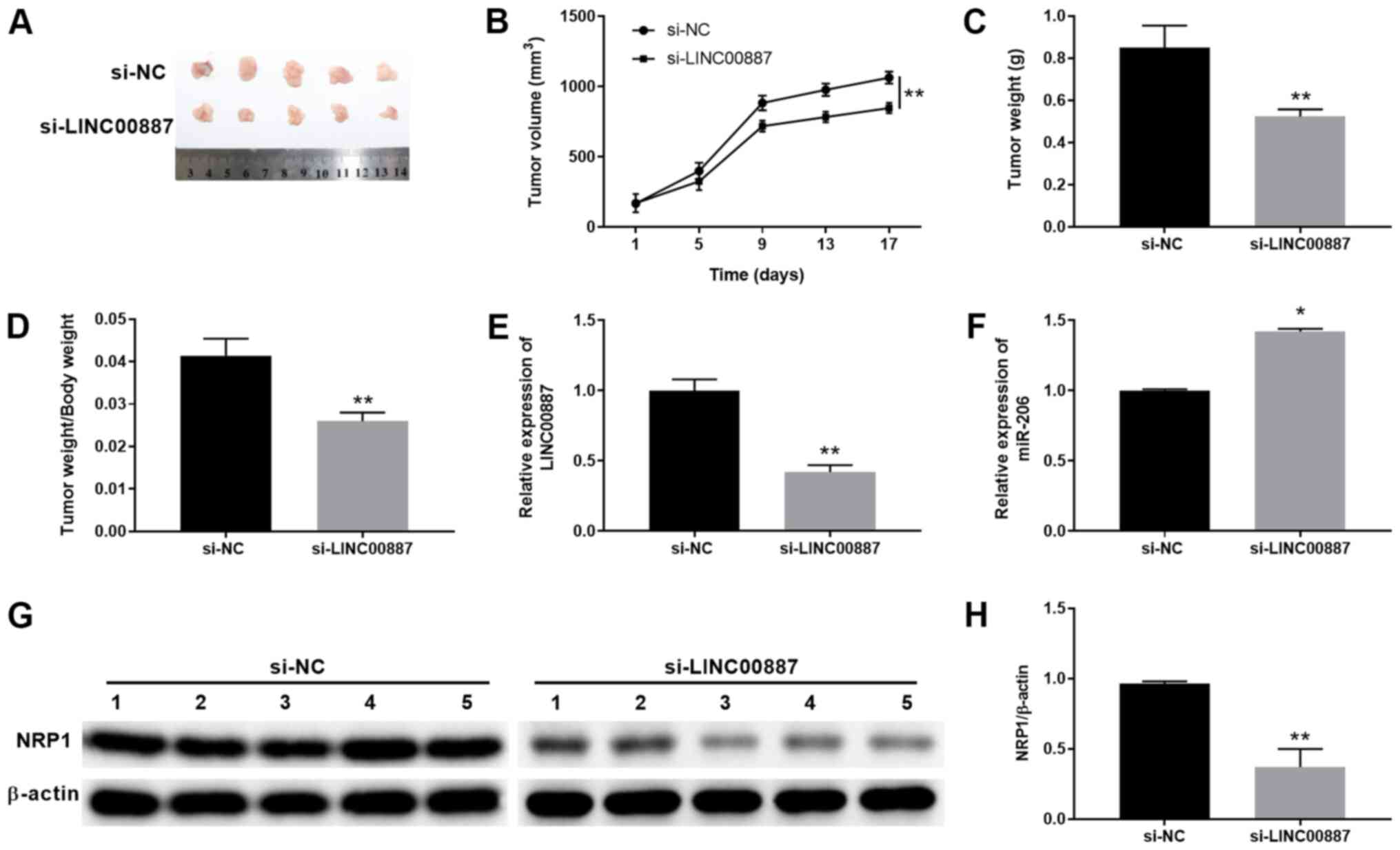
LINC00887 knockdown inhibits lung
carcinoma tumor growth in vivo
To investigate the function of LINC00887 on tumor
growth in vivo, a xenograft tumor model was established
using NCI-H460 cells transfected with si-NC or si-LINC00887
(Fig. 7A). The tumor volume was
measured every 4 days after implantation and mice were euthanized
on day 17 (Fig. 7B). Compared with
the si-NC group, the tumor growth was significantly inhibited in
the si-LINC00887 group (Fig. 7B),
with significantly lower xenograft tumor sizes and weights
(Fig. 7A-D). Additionally, LINC00887
and miR-206 expression in tumor tissues were examined. As presented
in Fig. 7E and F, tumor tissues from
the mice in the si-LINC00887 group exhibited significantly lower
expression levels of LINC00887 and significantly higher expression
levels of miR-206 compared with those in tissues from mice in the
si-NC group. NRP1 protein levels were significantly downregulated
in tumor tissues from the si-LINC00887 group compared with in
tissues from the si-NC group (Fig. 7G
and H). Overall, these results suggested that LINC00887
knockdown suppressed lung carcinoma growth in a xenograft
model.
Discussion
Emerging studies have demonstrated that lncRNAs are
important regulators that act as oncogenic or tumor suppressor
molecules in various tumors (17,39). For
example, the metastasis-associated lung adenocarcinoma transcript 1
(MALAT1), also called NEAT2, is an abundant and highly conserved
lncRNA across vertebrates. MALAT1 induces an EMT switch via the
PI3K/AKT pathway in epithelial ovarian cancer (40). Moreover, MALAT1 expression has been
reported to be a potential predictor of tumor metastasis and
prognosis in CRC (41). Maternally
expressed 3 was considered as a tumor suppressor and a potential
therapeutic candidate in cervical tumors (42). lncRNAs exert their functions by
sponging miRNAs via lncRNA-miRNA-mRNA regulatory axes (23–25). The
present study extended the understanding of the lncRNA-miRNA-mRNA
networks by revealing that LINC00887 directly interacted with
miR-206, whereas miR-206 targeted NRP1 to promote the progression
of lung carcinoma.
LINC00887 is a newly identified lncRNA involved in
multiple types of cancer. Low LINC00887 expression levels have been
observed in an invasive follicular thyroid carcinoma (43). A microarray analysis conducted by Zhu
et al (44) has revealed that
LINC00887 is one of the top 10 downregulated lncRNAs between stage
II and stage III colorectal cancer. In human papillary thyroid
cancer, a previous study has demonstrated that the expression
levels of LINC00887 are upregulated compared with those in
non-tumor thyroid tissues (45).
Therefore, LINC00887 may function as an oncogenic lncRNA or a tumor
suppressor in different types of tumors. In the present study, the
expression levels of LINC00887 were upregulated in lung carcinoma
tissues and cells compared with those in healthy tissues and cells;
therefore, LINC00887 may act as an oncogene in lung cancer.
Consistently, LINC00887 knockdown suppressed lung carcinoma cell
proliferation, migration and invasion in vitro, as well as
lung xenograft tumor growth in vivo.
Multiple miRNAs have been predicted as the potential
targets of LINC00887, such as miR-138, miR-181 and miR-204
(45). In the present study,
LINC00887 was identified to directly interact with miR-206. miR-206
has been reported as a tumor suppressor inhibiting cell
proliferation, migration and invasion in gastric, colorectal,
breast and laryngeal cancer (46–49).
Furthermore, Samaeekia et al (50) have reported that miR-206 inhibits
stemness and metastasis of breast cancer by regulating the
myocardin-related transcription factor A/interleukin-11 signaling
pathway. In the present study, the expression levels of miR-206
were downregulated in lung carcinoma tissues compared with those in
healthy tissues, suggesting that it may be a tumor suppressor.
Additionally, miR-206 overexpression inhibited lung carcinoma A549
or NCI-H460 cell proliferation, migration and invasion.
In the present study, NRP1 was identified as a
direct target gene controlled by miR-206. NRP1 is a transmembrane
glycoprotein that acts as a co-receptor for vascular endothelial
growth factor and transforming growth factor-β, and is a promising
novel target for chronic lymphocytic leukaemia therapy (51). NRP1 has been reported to be regulated
by multiple miRNAs, such as miR-130, miR-141 and miR-338 in ovarian
cancer, gastric cancer and pancreatic cancer (52–54).
Additionally, a previous study has demonstrated that miR-206
regulates NRP1 expression in breast cancer (55). Consistently, the present study
revealed that miR-206 regulated NRP1 expression by targeting the
NRP1 3′-UTR in lung carcinoma cells. However, future studies are
required to determine whether miR-206 has other targets and whether
NRP1 is also regulated by multiple miRNAs in lung carcinoma.
Bioinformatics analysis should be performed to analyze the
association between the expression levels of LINC00887 and
miR-206/NRP1 in patients with lung cancer in The Cancer Genome
Atlas database.
In conclusion, the results of the present study
revealed that LINC00887 was upregulated in lung carcinoma tissues
and cell lines. Furthermore, LINC00887 promoted lung carcinoma
progression and metastasis by sponging miR-206 to regulate NRP1
expression. The xenograft tumor model experiment demonstrated that
LINC00887 knockdown inhibited lung tumor growth in vivo.
Overall, the LINC00887-miR-206-NRP1 axis may reveal a novel insight
for lung tumorigenesis and may provide potential therapeutic
strategies for patients with lung carcinoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study available from the corresponding author on reasonable
request.
Authors' contributions
LBX and XPR conceived and designed the experiments.
BXB, JX and YJR performed the experiments. DH and SHW analyzed and
interpreted the data. LBX wrote the manuscript. XPR revised the
manuscript. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients. The present study was approved by the Ethics Committee of
Shaanxi Provincial People's Hospital. The animal experiment was
approved by the Institutional Animal Care and Use Committee of
Shaanxi Provincial People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tanase A, Colita A, Ianosi G, Neagoe D,
Branisteanu DE, Calina D, Docea AO, Tsatsakis A and Ianosi SL: Rare
case of disseminated fusariosis in a young patient with graft vs.
host disease following an allogeneic transplant. Exp Ther Med.
12:2078–2082. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ungureanu A, Zlatian O, Mitroi G, Drocaş
A, Ţîrcă T, Călina D, Dehelean C, Docea AO, Izotov BN, Rakitskii
VN, et al: Staphylococcus aureus colonisation in patients from a
primary regional hospital. Mol Med Rep. 16:8771–8780. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calina D, Rosu L, Roșu AF, Ianoşi G,
Ianoşi S, Zlatian O, Mitruț R, Docea AO, Rogoveanu O, Mitruț P, et
al: Etiological diagnosis and pharmacotherapeutic management of
parapneumonic pleurisy. Farmacia. 64:946–952. 2016.
|
|
7
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
8
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Knight SB, Crosbie PA, Balata H, Chudziak
J, Hussell T and Dive C: Progress and prospects of early detection
in lung cancer. Open Biol. 7:1700702017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Daphedar A and Taranath TC:
Characterization and cytotoxic effect of biogenic silver
nanoparticles on mitotic chromosomes of drimia polyantha (Blatt.
& McCann) stearn. Toxicol Rep. 5:910–918. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sani TA, Mohammadpour E, Mohammadi A,
Memariani T, Yazd MV, Ramin R, Daniela C, Anca OD, Marina G, Etemad
L and Shahsavand S: Cytotoxic and apoptogenic properties of
Dracocephalum kotschyi aerial part different fractions on calu-6
and mehr-80 lung cancer cell lines. Farmacia. 65:189–199. 2017.
|
|
12
|
Saab AM, Guerrini A, Sacchetti G, Maietti
S, Zeino M, Arend J, Gambari R, Bernardi F and Efferth T:
Phytochemical analysis and cytotoxicity towards multidrug-resistant
leukemia cells of essential oils derived from lebanese medicinal
plants. Planta Med. 78:1927–1931. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shao H, Jing K, Mahmoud E, Huang H, Fang X
and Yu C: Apigenin sensitizes colon cancer cells to antitumor
activity of ABT-263. Mol Cancer Ther. 12:2640–2650. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng S, Kapur A, Patankar MS and Xiong MP:
Formulation, characterization, and antitumor properties of trans-
and cis-citral in the 4T1 breast cancer xenograft mouse model.
Pharm Res. 32:2548–2558. 2015.PubMed/NCBI
|
|
15
|
Forde PM, Brahmer JR and Kelly RJ: New
strategies in lung cancer: Epigenetic therapy for non-small cell
lung cancer. Clin Cancer Res. 20:2244–2248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calle AS, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun H, Huang Z, Sheng W and Xu MD:
Emerging roles of long non-coding RNAs in tumor metabolism. J
Hematol Oncol. 11:1062018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun M, Gadad SS, Kim DS and Kraus WL:
Discovery, annotation, and functional analysis of long noncoding
rnas controlling cell-cycle gene expression and proliferation in
breast cancer cells. Mol Cell. 59:698–711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
She K, Huang J, Zhou H, Huang T, Chen G
and He J: lncRNA-SNHG7 promotes the proliferation, migration and
invasion and inhibits apoptosis of lung cancer cells by enhancing
the FAIM2 expression. Oncol Rep. 36:2673–2680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin C and Yang L: Long noncoding RNA in
Cancer: Wiring signaling circuitry. Trends Cell Biol. 28:287–301.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang C, Wang C, Jia Z, Tong W, Liu D, He
C, Huang X and Xu W: Differentially expressed mRNAs, lncRNAs, and
miRNAs with associated co-expression and ceRNA networks in
ankylosing spondylitis. Oncotarget. 8:113543–113557. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan CN, Ma L and Liu N: Systematic
analysis of lncRNA-miRNA-mRNA competing endogenous RNA network
identifies four-lncRNA signature as a prognostic biomarker for
breast cancer. J Transl Med. 16:2642018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao
C, Ji S, Shen Y, De W and Qiang Y: Long non-coding RNA linc00665
promotes lung adenocarcinoma progression and functions as ceRNA to
regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis.
10:842019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo T, Li J, Zhang L, Hou W, Wang R, Zhang
J and Gao P: Multidimensional communication of microRNAs and long
non-coding RNAs in lung cancer. J Cancer Res Clin Oncol. 145:31–48.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu S, Yan G, Zhang J and Yu L: Knockdown
of long noncoding RNA (lncRNA) metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) inhibits proliferation,
migration, and invasion and promotes apoptosis by targeting miR-124
in retinoblastoma. Oncol Res. 26:581–591. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang D and Hu Y: Long non-coding RNA PVT1
competitively binds microRNA-424-5p to regulate CARM1 in
radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic
Acids. 16:130–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang X, Zhang W, Cheng SQ and Yang RL:
High expression of lncRNA GACAT3 inhibits invasion and metastasis
of non-small cell lung cancer to enhance the effect of
radiotherapy. Eur Rev Med Pharmacol Sci. 22:1315–1322.
2018.PubMed/NCBI
|
|
31
|
Yang J, Du YM and Li B: LncRNA BX357664
inhibits the proliferation and invasion of non-small cell lung
cancer cells. Eur Rev Med Pharmacol Sci. 23:660–669.
2019.PubMed/NCBI
|
|
32
|
Tian Y, Yu M, Sun L, Liu L, Huo S, Shang
W, Sheng S, Wang J, Sun J, Hu Q, et al: Long noncoding RNA00887
reduces the invasion and metastasis of nonsmall cell lung cancer by
causing the degradation of miRNAs. Oncol Rep. 42:1173–1182.
2019.PubMed/NCBI
|
|
33
|
Ali MM, Akhade VS, Kosalai ST, Subhash S,
Statello L, Meryet-Figuiere M, Abrahamsson J, Mondal T and Kanduri
C: PAN-Cancer analysis of S-phase enriched lncRNAs identifies
oncogenic drivers and biomarkers. Nat Commun. 9:8832018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kartha RV and Subramanian S: Competing
endogenous RNAs (ceRNAs): New entrants to the intricacies of gene
regulation. Front Genet. 5:82014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Balas MM and Johnson AM: Exploring the
mechanisms behind long noncoding RNAs and cancer. Noncoding RNA
Res. 3:108–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jin Y, Feng SJ, Qiu S, Shao N and Zheng
JH: LncRNA MALAT1 promotes proliferation and metastasis in
epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med
Pharmacol Sci. 21:3176–3184. 2017.PubMed/NCBI
|
|
41
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Biol Ther. 17:104–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Celestino R, Nome T, Pestana A, Hoff AM,
Gonçalves AP, Pereira L, Cavadas B, Eloy C, Bjøro T,
Sobrinho-Simões M, et al: CRABP1, C1QL1 and LCN2 are biomarkers of
differentiated thyroid carcinoma, and predict extrathyroidal
extension. BMC Cancer. 18:682018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu H, Yu J, Zhu H, Guo Y and Feng S:
Identification of key lncRNAs in colorectal cancer progression
based on associated protein-protein interaction analysis. World J
Surg Oncol. 15:1532017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
You X, Zhao Y, Sui J, Shi X, Sun Y, Xu J,
Liang G, Xu Q and Yao Y: Integrated analysis of long noncoding RNA
interactions reveals the potential role in progression of human
papillary thyroid cancer. Cancer Med. 7:5394–5410. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Deng M, Qin Y, Chen X, Wang Q and Wang J:
MiR-206 inhibits proliferation, migration, and invasion of gastric
cancer cells by targeting the MUC1 gene. Onco Targets Ther.
12:849–859. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sun P, Sun D, Wang X, Liu T, Ma Z and Duan
L: MiR-206 is an independent prognostic factor and inhibits tumor
invasion and migration in colorectal cancer. Cancer Biomark.
15:391–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu Y, Shao ZM, He QZ, Jiang BQ, Wu Y and
Zhuang ZG: Hsa-MiR-206 represses the proliferation and invasion of
breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci.
19:2091–2104. 2015.PubMed/NCBI
|
|
49
|
Zhang T, Liu M, Wang C, Lin C, Sun Y and
Jin D: Down-Regulation of miR-206 promotes proliferation and
invasion of laryngeal cancer by regulating VEGF expression.
Anticancer Res. 31:3859–3863. 2011.PubMed/NCBI
|
|
50
|
Samaeekia R, Adorno-Cruz V, Bockhorn J,
Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, et al:
MiR-206 inhibits stemness and metastasis of breast cancer by
targeting MKL1/IL11 pathway. Clin Cancer Res. 23:1091–1103. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chaudhary B, Khaled YS, Ammori BJ and
Elkord E: Neuropilin 1: Function and therapeutic potential in
cancer. Cancer Immunol Immunother. 63:81–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen C, Hu Y and Li L: NRP1 is targeted by
miR-130a and miR-130b, and is associated with multidrug resistance
in epithelial ovarian cancer based on integrated gene network
analysis. Mol Med Rep. 13:188–196. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peng Y, Liu YM, Li LC, Wang LL and Wu XL:
MicroRNA-338 inhibits growth, invasion and metastasis of gastric
cancer by targeting NRP1 expression. PLoS One. 9:e944222014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma L, Zhai B, Zhu H, Li W, Jiang W, Lei L,
Zhang S, Qiao H, Jiang X and Sun X: The miR-141/neuropilin-1 axis
is associated with the clinicopathology and contributes to the
growth and metastasis of pancreatic cancer. Cancer Cell Int.
19:2482019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Seifi-Alan M, Shams R, Bandehpour M,
Mirfakhraie R and Ghafouri-Fard S: Neuropilin-1 expression is
associated with lymph node metastasis in breast cancer tissues.
Cancer Manag Res. 10:1969–1974. 2018. View Article : Google Scholar : PubMed/NCBI
|