Introduction
Breast cancer is one of the most common malignant
tumors among females. According to statistics in 2018, ~9.57
million individuals died of cancer worldwide, of which ~627,000
died from breast cancer (1).
However, the current prevention and treatment methods need to be
improved. Clinical, epidemiological and biological studies have
indicated that exogenous estrogen is associated with the occurrence
and development of breast cancer (2). Bisphenol A (BPA) is an exogenous
estrogen, which is one of the most widely used industrial compounds
in human daily life. BPA is similar to estrogen in structure and
has stable chemical properties. It is widely used in plastic
products, food containers, beverage bottles and dental fillings
materials (3). When heated in
mi-crowave ovens, plastic residues, such as bisphenol A may
penetrate into the food (4). BPA is
difficult to metabolise and excrete, thus interfering with the
endocrine system of the human body (5). In previous years, a large number of
experiments have confirmed that BPA can interfere with the normal
functions of the human reproductive, nervous and immune systems and
embryonic development. In addition, BPA promotes the occurrence and
development of various tumors, such as breast and prostate cancer
and children's reproductive system tumors (6,7). Liu
et al (8) reported that
bisphenol can regulate the expression of EMT-related protein
markers by promoting the expression of Snail, thereby enhancing the
migration ability of breast cancer MCF-7 cells.
Ginger (Zingiber officinale) is the fresh
rhizome of ginger, a perennial herb of the Zingiberaceae family.
Ginger is a traditional Chinese medicine that is used for both food
and medicine (9). Previous studies
have reported that ginger serves an antitumor role in various types
of malignant tumors (10–17). For example, 6-gingerol, the main
active component of ginger, can induce the apoptosis of gastric
cancer cells via different mechanisms (11). (6)-gingerol can effectively inhibit colon
tumor growth in nude mice (12) and
(6)-paradol has antitumor-promoting
properties (13). Ginger extract can
inhibit growth and induce apoptosis in prostate cancer models in
vivo and in vitro (14).
Volatile ginger oil has strong cytotoxicity in cervical cancer
cells (15) and treatment of
gastroin-testinal tumors (16).
Karki has demonstrated that ginger oil inhibits hallmarks [cyclin
D1, cyclin dependent kinase (Cdk)-2, Cdk-4 and Bcl-2] of breast
cancer cells (17). The purpose of
the present study was to investigate the function of ginger
essential oil (GEO) on breast cancer cells induced by bisphenol A,
and provide an experimental and theoretical basis of potential
therapeutic targets and the development of novel drugs.
MCF-7, a widely studied epithelial cancer cell line
derived from breast adenocarcinoma, has characteristics of
differentiated mammary epithelium (18). In the present study, relative and
absolute quantitative isobaric labeling (iTRAQ) technology was for
comprehensive proteomic analysis of MCF-7 cells, which were treated
with single or a therapeutic trace estrogen combination (BSA). In
addition, bioinformatics and function-al analysis, including Gene
Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG),
cluster analysis and protein-protein interaction (PPI) network
analysis, were used. The present study may provide an experimental
basis to improve our under-standing of the underlying molecular
mechanisms of GEO-induced apoptosis in breast cancer cells.
Materials and methods
Cell culture
The human breast cancer cell line MCF-7 was obtained
from Fuheng Bio-logical Company (https://www.fudancell.com). Cell culture was completed
at the Laboratory Animal Science and Technology Center of Jiangxi
University of Traditional Chinese Medicine. MCF-7 cells were
cultured in MEM medium (Solibao, China, http://www.solarbio.com) with 10% fetal bovine serum
(FBS; Serana Europe) and 1% penicillin-streptomycin (Solibao,
China) and incubated in a humid CO2 incubator at 37°C.
MCF-7 is an estrogen receptor breast cancer cell line (18).
Preparation of GEO
In total, 500 g fresh ginger was crushed, put into a
5,000-ml round-bottom flask, mixed with 3,000 ml distilled water,
heated and refluxed in a heating hood for 6 h. The volatile oil was
extracted using n-hexane and dried over anhydrous sodium sulfate.
Weight the ginger oil and stored at 4°C. Additionally, 200 mg/l was
determined as the optimal concentration of ginger essential oil to
treat MCF-7 cells through cytotoxicity tests (19).
Cell treatment
BPA was purchased from Macklin Inc., and dissolved
with DMSO (Solibao, China, http://www.solarbio.com) solution. MCF-7 cells were
plated at 1×106 cells per well in a 6-well plate. Then MCF-7 cells
were incubated for 24 h at 37°C to allow adhesion, and then the
minimum Eagle's medium (MEM; Hyclone, Cytiva) was removed and new
media with GEO (25, 50, 100, 150, 200 and 250 mg/l) or BPA (10-5,
10-6, 10-7, 10-8 and 10-9 mol/l) or GEO-BPA (GEO 200 mg/l, BPA
10-5, 10-6, 10-7, 10-8 and 10-9 mol/l) at the different
concentrations were added, MCF-7 cells were again incubated at 37°C
in a 5% CO2 incubator. After 48 h, the cells were
collected for further analysis. All experiments were performed in
triplicate.
Cell viability analysis
Cell viability was determined using an MTT assay.
MCF-7 cells were inoculated in a 96-well plate with 1×104 cells per
well. The cells were incubated at 37°C for 24 h to allow them to
adhere to the bottom of the plate, then MEM was removed and new
media of GEO (25, 50, 100, 150, 200 and 250 mg/l) or BPA (10-5,
10-6, 10-7, 10-8 and 10-9 mol/l) or GEO-BPA (GEO 200 mg/l, BPA
10-5, 10-6, 10-7, 10-8, and 10-9 mol/l) at the different
concentrations were added. After reaching the treatment time (24,
48 or 72 h), 20 µl of 5 mg/ml MTT was added to each well. The cells
were incubated in a 5% CO2 incubator at 37°C for 4 h.
The medium was then removed and MTT was dissolved with 150 µl DMSO
solvent per well. The cells were stirred in the dark for 15 min on
a shaking table at 75 rpm, and then the absorbance was measured at
490 nm using spectrophotometry.
Protein extraction
An appropriate amount (10–15 µl) of cell sample was
treated with liquid nitrogen, then protein lysate (8 M urea + 1%
SDS, including protease inhibitor cocktail (Thermo Fisher
Scientific, Inc.) at a ratio of 1:5 was added. Cells were
centri-fuged at 16,000 × g at 4°C for 30 min to collect the
supernatant and 5× volume of pre-cooled acetone was added and
precipitated overnight at −20°C. The next day the supernatant was
centrifuged at 12,000 × g at 4°C for 30 min and the precipitate was
collected. The precipitate was washed with 90% pre-cooled acetone
and air dried. The precipitate was then dissolved it with 200 µl of
protein lysate (8M urea + 1% SDS, with cocktail) and finally
centrifuged at 12,000 × g at 4°C for 30 min to collect the protein
supernatant.
SDS-PAGE
The protein sample (20 µg) was mixed with a 5X
loading buffer and boiled for 5 min. Then the SDS-PAGE was
performed on a 12.5% (v/w) polyacrylamide gel for quantification.
Biologically repeated in triplicate.
Reductive alkylation and enzymatic
hydrolysis
Triethylammonium bicarbonate buffer (TEAB) (1 mol/l)
was added to a 100-µg protein sample of different treatment groups
to make a TEAB final concentration 100 mM in tubes. Then tris
(2-carboxyethyl) phosphine (TCEP) was added to each tube to make a
final TCEP concentration of 100 mM, and reacted at 37°C for 60 min.
After, iodoacetamide was added to each tube to make a final
concentration of 40 mM. The reaction solution was incubated at room
temperature in the dark for 40 min, then pre-cooled acetone
(acetone: sample v/v =6:1) was added to each tube. The solution was
precipitated at −20°C for 4 h, then centrifuged at 10,000 × g for
20 min at 4°C. The precipitate was collected and fully dissolved
with 100 µl of 100 mM TEAB. Trypsin was then added according to the
enzyme: protein (m/m) = 1:50, and incubated at 37°C overnight.
iTRAQ labeling
The samples were digested with trypsin, the peptides
were dried using a vacuum pump, and reconstituted with 0.5 M TEAB.
According to the manufacturer's instructions, peptides (~100 µg) in
each group were labeled using the iTRAQ Labeling kit (cat. no.
4390812; Shanghai AB SCIEX Analytical Instrument Trading Co.).
High pH ultra high-performance liquid
chromatography (UPLC) first dimension separation
The peptide samples (100 mM/l) of MCF-7 cells in
different treatment groups were reconstituted with UPLC loading
buffer [2% acetonitrile (adjusted to pH 10.0 with ammonia)], and a
high pH liquid phase separation was performed with a reverse phase
ACQUITY UPLC BEH C18 column (1.7 µm, 2.1 mm X 150; Waters
Coporation) using the Vanquish Flex Binary UHPLC system (Thermo
Fisher Scientific, Inc.). Mobile phase A: 2% Acetonitrile (adjusted
to pH 10.0 with ammonia), Mobile phase B: 80% Acetonitrile
(adjusted to pH 10.0 with ammonia). The ultraviolet detection
wavelength was 214 nm. The flow rate was 200 µl/min and the
gradient was 47 min.
High-performance liquid
chromatography-mass spectrometry (HPLC-MS)
Samples were analyzed using HPLC-MS in positive and
negative ion mode, a reverse phase C18 column (75 µm × 25 cm) and
an Easy-1200 chromatography instrument (both Thermo Fisher
Scientific, Inc.). The MS instrument was Q_Exactive HF-X (Thermo
Fisher Scientific, Inc.) and chromatographic separation time was
120 min. Mobile phase A: 2% Acetonitrile 0.1% formic acid and B:
80% acetonitrile 0.1% formic acid. The flow rate was 300 nl/min,
and the MS scanning range (m/z) was 350-1,300. Nitrogen gas
tem-perature was 35°C and nebulizer pressure was 5500 psi.
Proteomics data analysis
The original MS data were extracted from the
original files. These files were submitted to the Proteome
discoverer server (https://www.thermofisher.com/order/catalog/product/OPTON-30810#/OPTON-30810)
and the NCBInr database (https://www.ncbi.nlm.nih.gov) was searched.
Bioinformatics analysis
For proteins obtained by MS, proteins that were
upregulated by >1.2-fold, downregulated by >0.8-fold and
P<0.05 were considered to be differentially expressed (20,21).
Cluster version 3.0 was used for hierarchical cluster analysis, and
the cellular components and molecules of the different proteins
were analyzed by BLAST2GO version 2.5.0 (https://www.blast2go.com), KOBAS version 2.1.1
(http://kobas.cbi.pku.edu.cn) and Goa
tools version 0.6.5 (https://www.ebi.ac.uk/GOA). The functional and
biological processes were classified by clustering, and the
differences in protein functions were discussed based on annotation
information. The gene symbols obtained from the target protein
sequence database were used to study the target protein and
experimental evidence using Cytoscape version 3.7.2 and the STRING
database (https://string-db.org/) for network
analysis of protein-protein interactions (PPIs) (22).
Statistical analysis
For one-variable experiments, P-values were
calculated using the paired Student's t-test, while P-values from
two-variable experiments were calculated using the two-way ANOVA
and Dunnett's multiple comparisons test. Data are expressed as mean
± standard deviation and mean ± SEs of three independent biological
replicates. P<0.05 was considered to indicate a statistically
significant difference.
Results
Cell viability
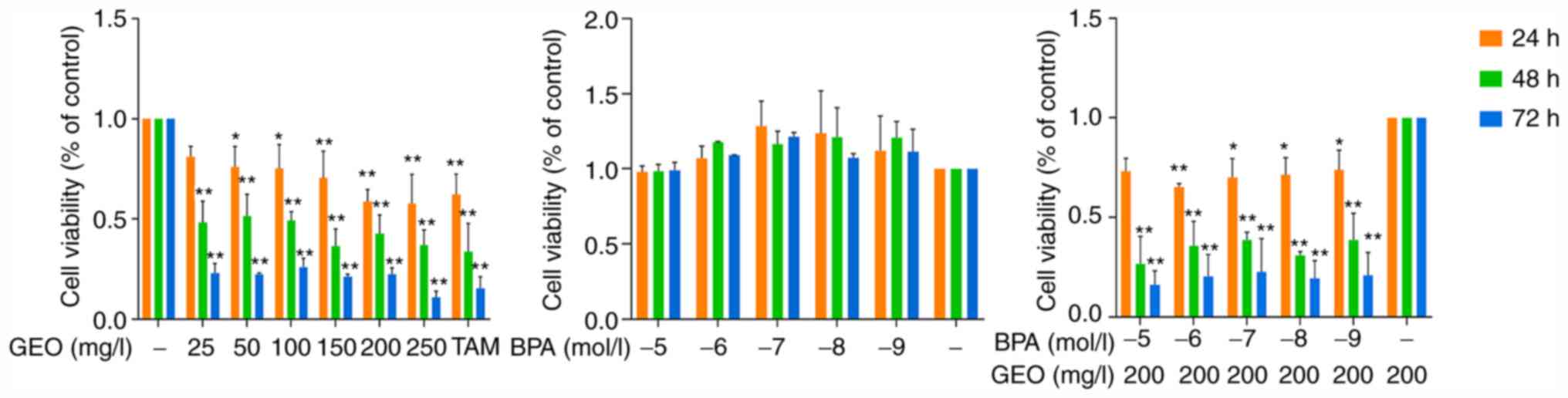
As shown in Fig. 1,
when the treatment time was constant, the viability MCF-7 cells
decreased with increasing GEO concentration. This was most notable
when the treatment concentration was 250 mg/l at all treatment
durations. When the GEO concentration was constant, the viability
of MCF-7 cells decreased with the prolongation of the treatment
time, and reached the lowest at 72 h. Compared with the control
group, BPA improved cell viability and the cell viability reached
its maximum when the BPA concentration was 10-7 mol/l. Meanwhile,
following GEO-BPA treatment, the cell viability decreased compared
with the BPA group. In short, GEO inhibited the proliferation of
breast cancer MCF-7 cells, while BPA promoted this; however, GEO
overcame BPA-induced promotion of proliferation.
LC-MS/MS
The present study used iTRAQ technology to analyze
the proteomic characteristics of MCF-7 cells, which were treated
with different concentrations of GEO or BPA alone or GEO mixed with
BPA. The experimental procedure is shown in Fig. 2. Overall, a total of 5,084 proteins
were detected (Table SI). The
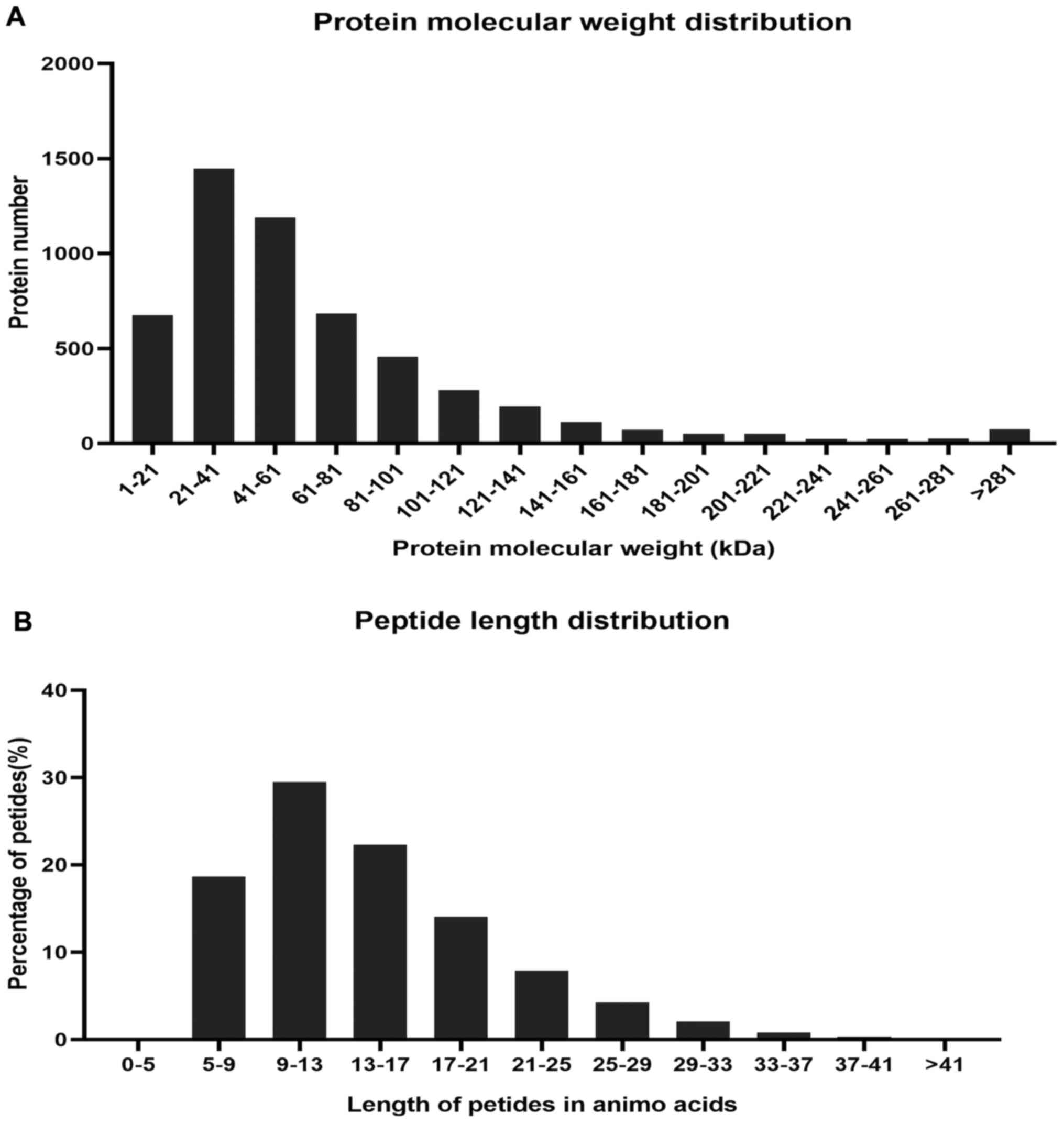
quality control of protein data showed that the molecular mass of
the protein was in the range of 0–100 kDa (Fig. 3A), and the length of most peptides
were between 7 and 17 amino acids (Fig.
3B), which is similar to the properties of known trypsin
peptides (23).
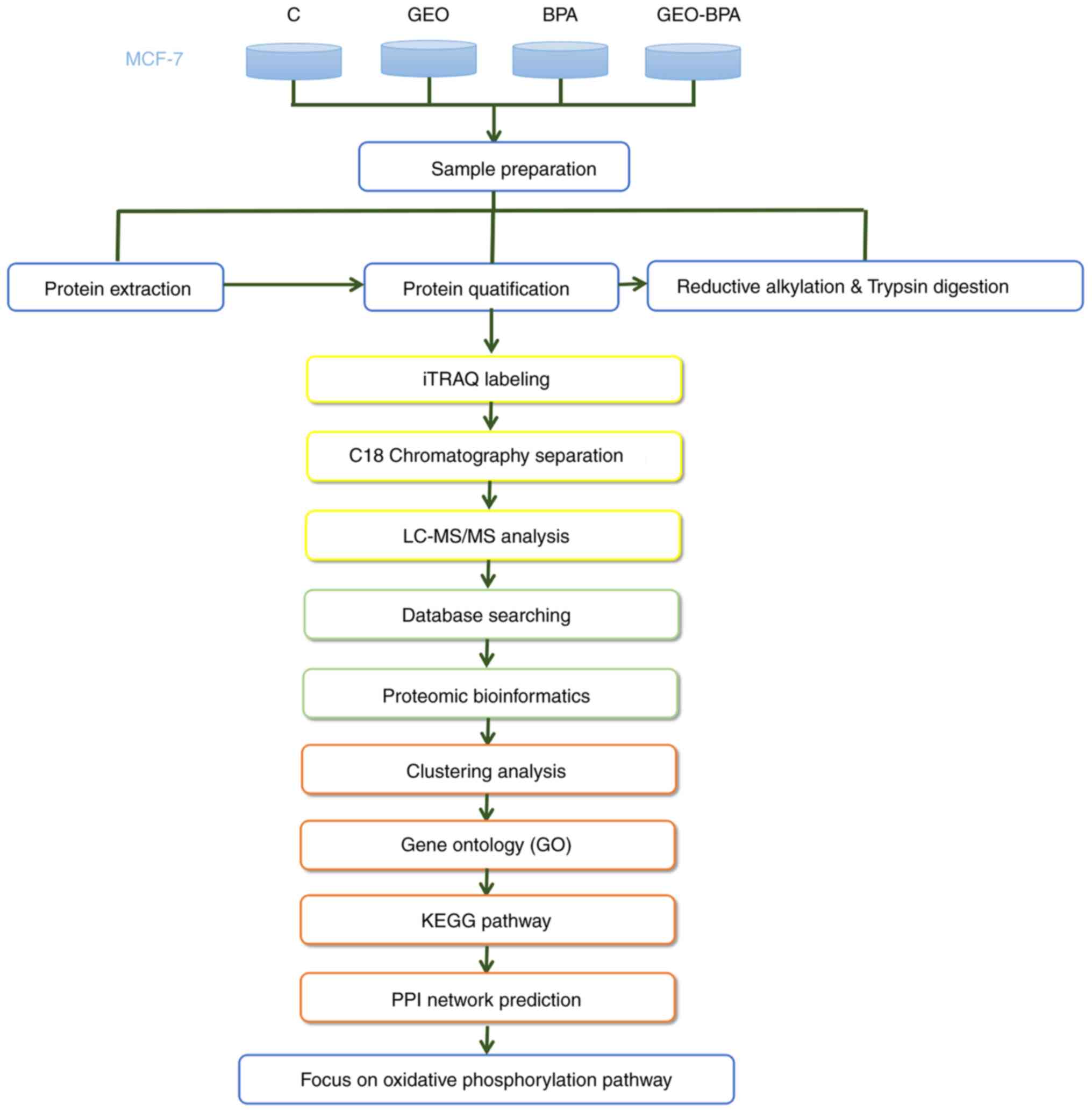 | Figure 2.Experimental process. For
quantitative proteomic analysis of design of exper-iment, the
experiment was divided into four groups (control, GEO, BPA and
GEO-BPA), and each experiment was performed in triplicate. The
extracted proteins were prepared by reductive alkylation, digested
with trypsin and labeled with Iraqi reagents. Analysis was
performed using reversed-phase LC-MS/MS. Bioinformatics tools were
further used to analyze the resulting data. GEO, ginger essential
oil; BPA, bisphenol A; LC, liquid chromatography; MS, mass
spectrometry; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI,
protein-protein interaction. |
Compared with the control group, MCF-7 cells treated
with GEO, BPA and GEO-BPA showed 45 (14 up- and 31 downregulated)
and 481 (141 up- and 340 downregulated) differentially expressed
proteins. Compared with the BPA group, MCF-7 cells treated with
GEO-BPA showed 210 (117 up- and 93 downregulated) differentially
expressed proteins, respectively (Tables
I and SII–IV). According to the optimal selection
criterion for differentially expressed proteins, compared with
cells treated with BPA alone, some proteins in cells treated with
GEO and BPA were significantly upregulated or downregulated
(P<0.05). According to the aforementioned criteria, a total of
34 differentially expressed proteins were further processed (Table
II), of which 13 were significantly upregulated and 21 were
significantly downregulated (P<0.05).
 | Table I.Protein quantification in MCF-7 cells
treated with GEO or BPA alone and GEO-BPA combined. |
Table I.
Protein quantification in MCF-7 cells
treated with GEO or BPA alone and GEO-BPA combined.
| Comparison between
groups | Upregulated
proteins, n | Downregulated
pro-teins, n | Protein count,
n |
|---|
| GEO vs. C | 14 | 31 | 45 |
| BPA vs. C | 141 | 340 | 481 |
| GEO-BPA vs. C | 13 | 21 | 34 |
| GEO-BPA vs.
BPA | 117 | 93 | 210 |
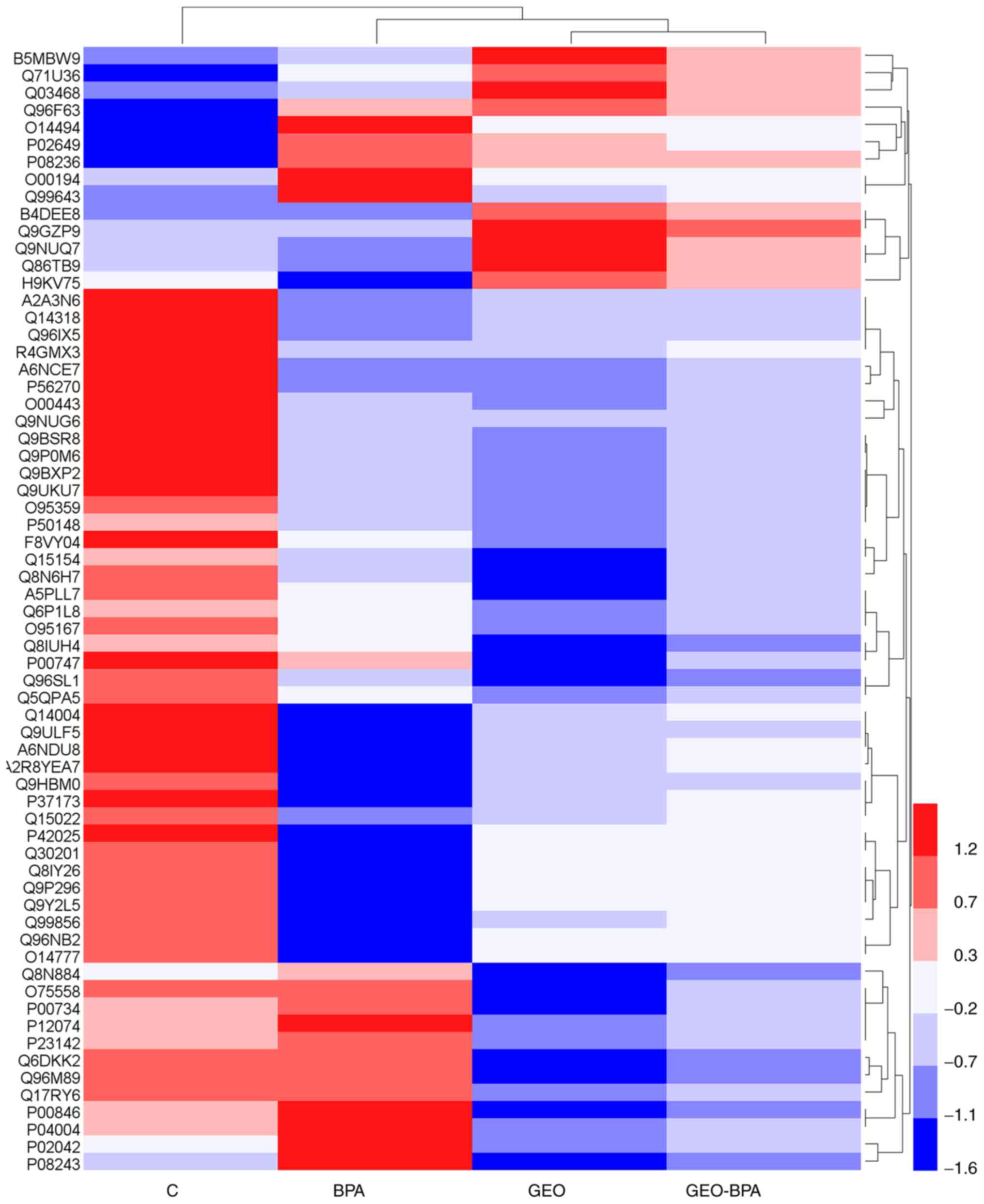
Clustering analysis
The results of hierarchical clustering are displayed
in the form of a heat map, in which red indicates upregulation and
blue indicates downregulation. The observed difference in protein
expression between the groups are shown in Fig. 4. It was observed the overall
expression pattern of genes in the GEO, BPA and GEO-BPA groups was
different compared with those in the control group. In the control
group, most genes showed a highly upregulated expression patterns
(red bands), while in the GEO and GEO-BPA groups, most genes were
downregulated.
GO function annotation and
analysis
GO is a type of functional classification system.
The GO database provides a standardized description of gene
products from the per-spectives of function, participating in
biological pathways and localization in cells. The GO database
showed 53, 59 and 57 functional annotations for differentially
expressed proteins in cells treated with GEO (n=45), BPA (n=481)
and GEO-BPA (n=210) (Tables
SV–SVII). In addition, the GO
functional annotations of differentially expressed proteins in each
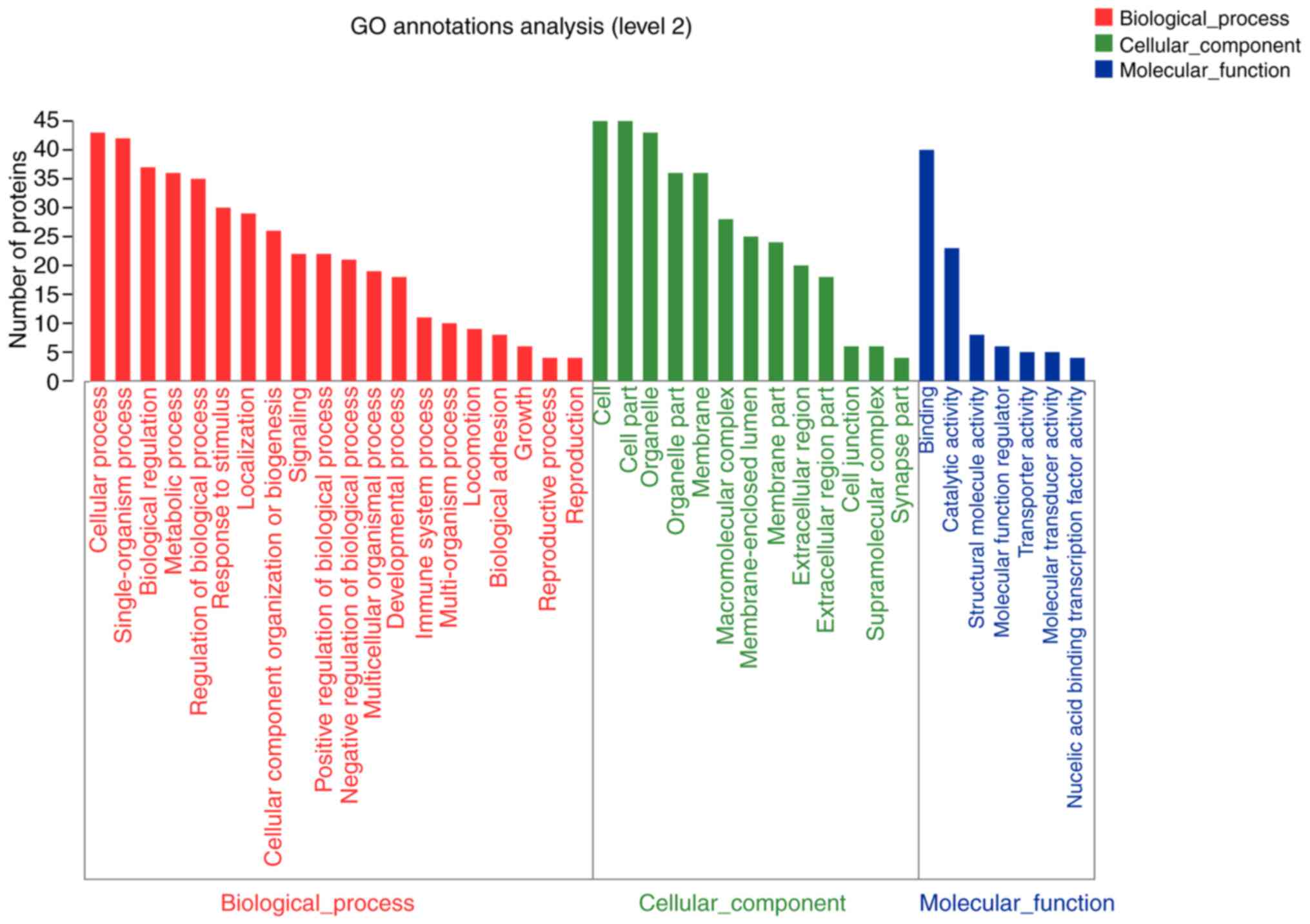
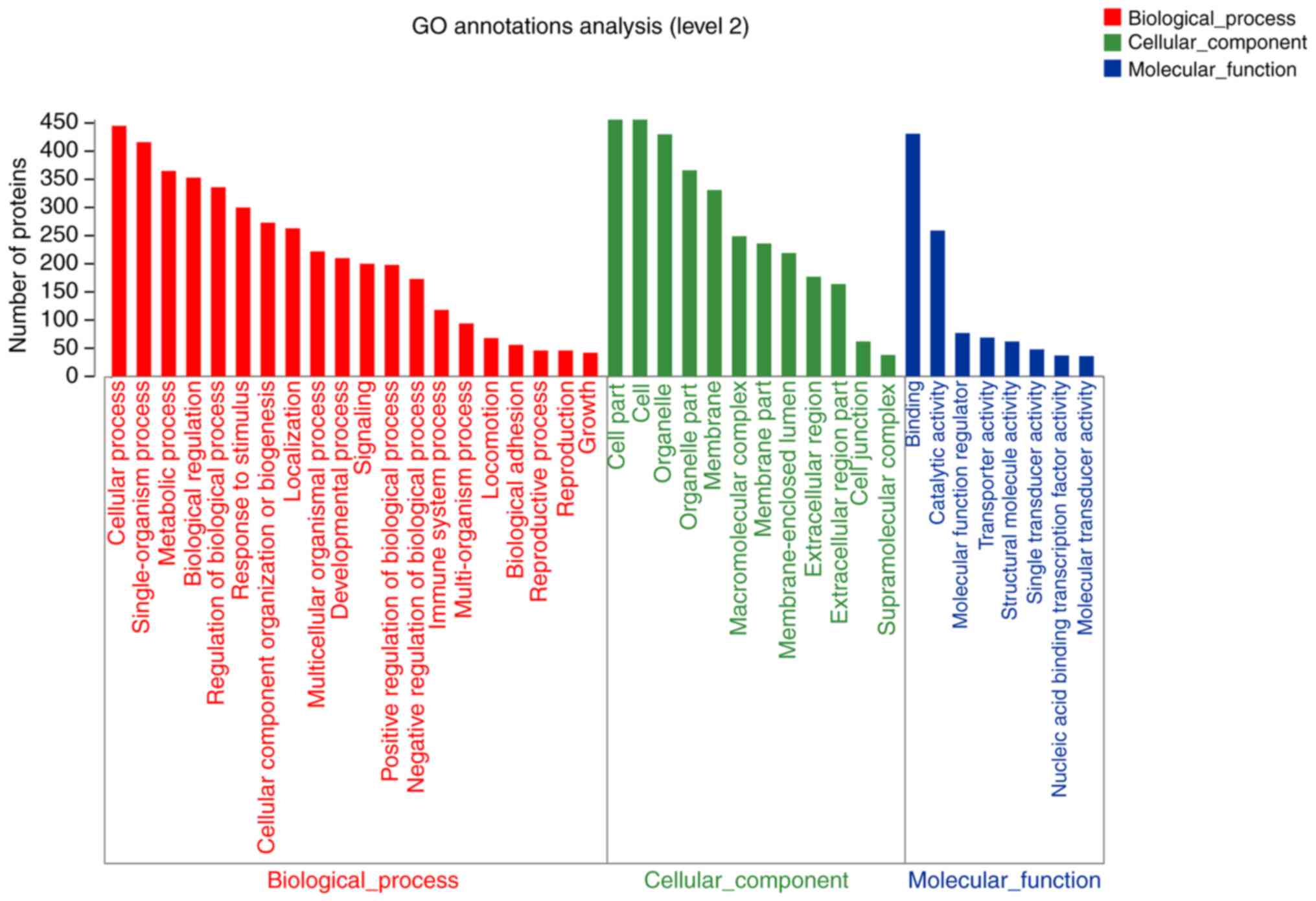
group were analyzed. The results showed that compared with the
con-trol group, 45 differentially expressed proteins were more
likely to be located at the cell parts and macromolecular complex
in GEO alone group, and were closely associated with catalytic
activity and protein binding activity. These proteins are involved
in cellu-lar and metabolic processes and responses to stimuli
(Fig. 5). Only 111 diffeentially
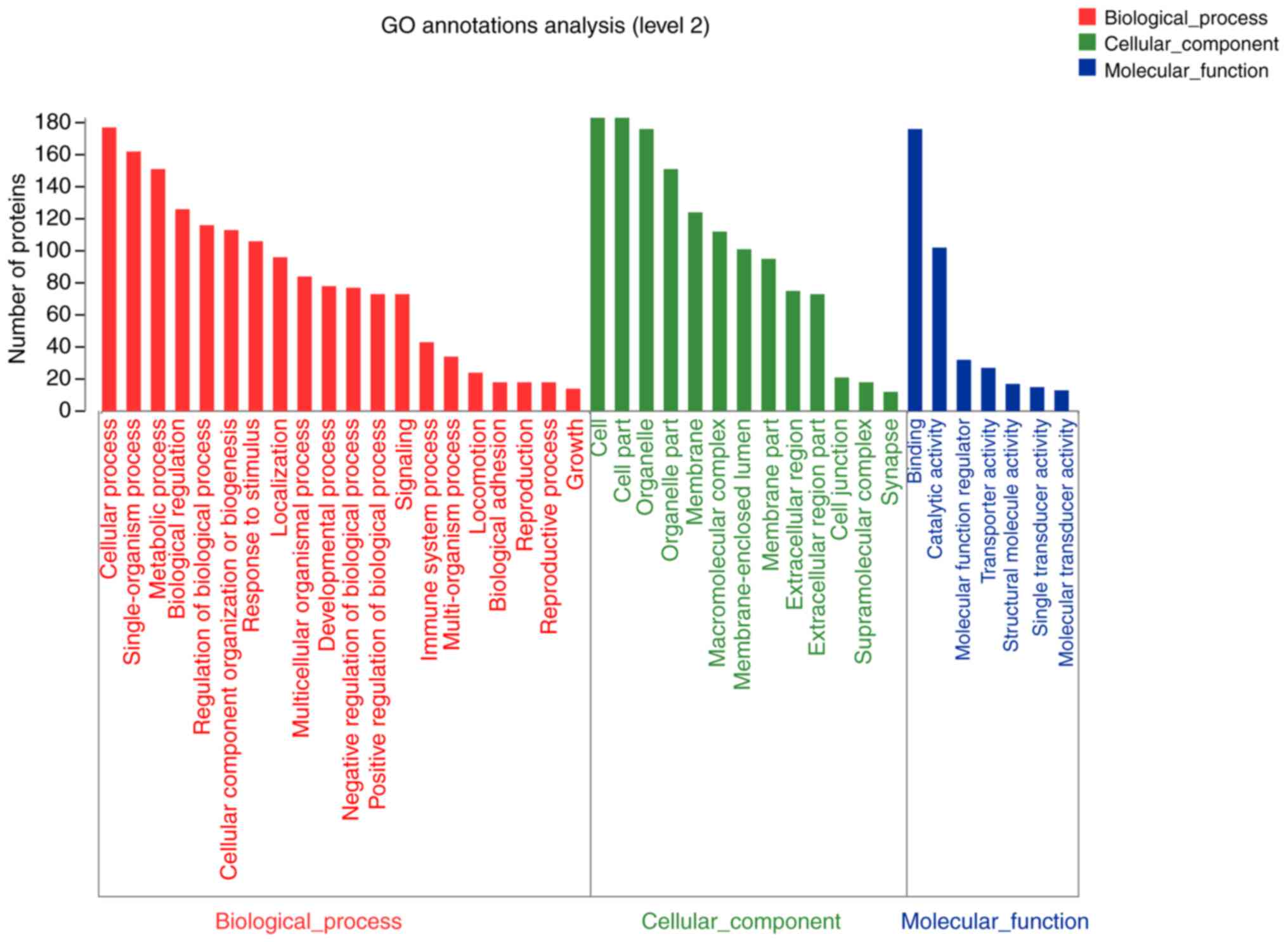
expressed proteins in the BPA alone group were reported in the
organelles and membrane, which were associated with structural and
molecular activity. These proteins were involved in various
biological processes, such as metabolic processes, organization of
cellular component or biogenesis and localization (Fig. 6). The 192 differentially expressed
proteins in the GEO-BPA combination group were primarily located in
the cell part, single-organism process, organelle part and were
involved in catalytic activity and structural molecule activity,
which could affect signaling, cellular component organ-ization or
biogenesis and negative regulation of biological processes
(Fig. 7). Changes in biological
processes indicated that GEO affects BPA-treated breast cancer
cells
KEGG pathway analysis
The KEGG database can be used to associate the gene
catalogue in the whole genomes with higher levels of system
function at the cell, species and ecosystem levels (24). Through KEGG pathway analysis, the key
signaling path-ways and related regulatory processes of each group
were obtained (Tables
SVIII–SX). The KEGG secondary
category of each group is presented in Figs. 8–10.
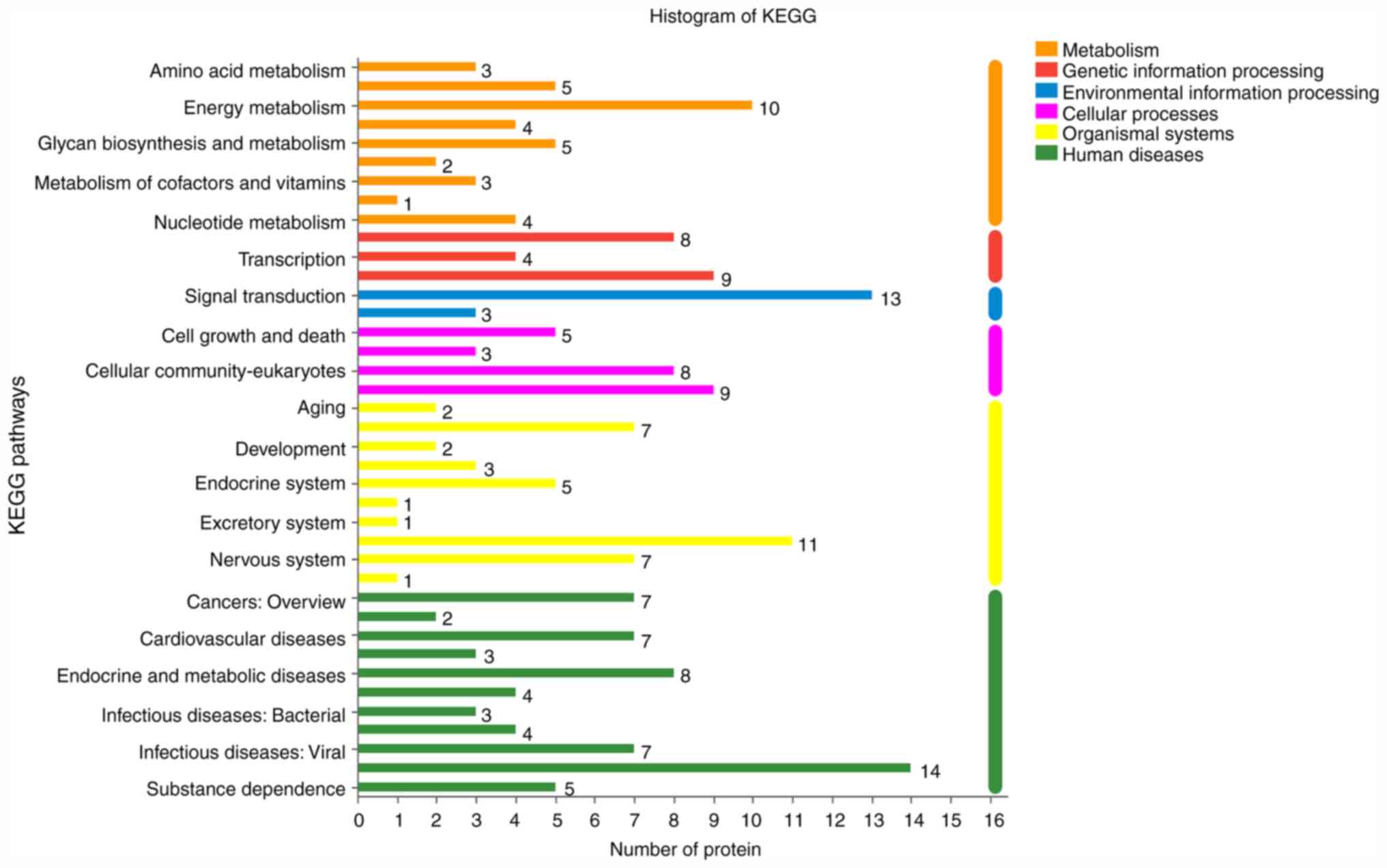
KEGG analy-sis showed that the signaling pathways identified in the
GEO group were primarily associated with energy metabolism,
transportation and catabolism, immune system and cell motility
(Fig. 8). KEGG signaling pathways in
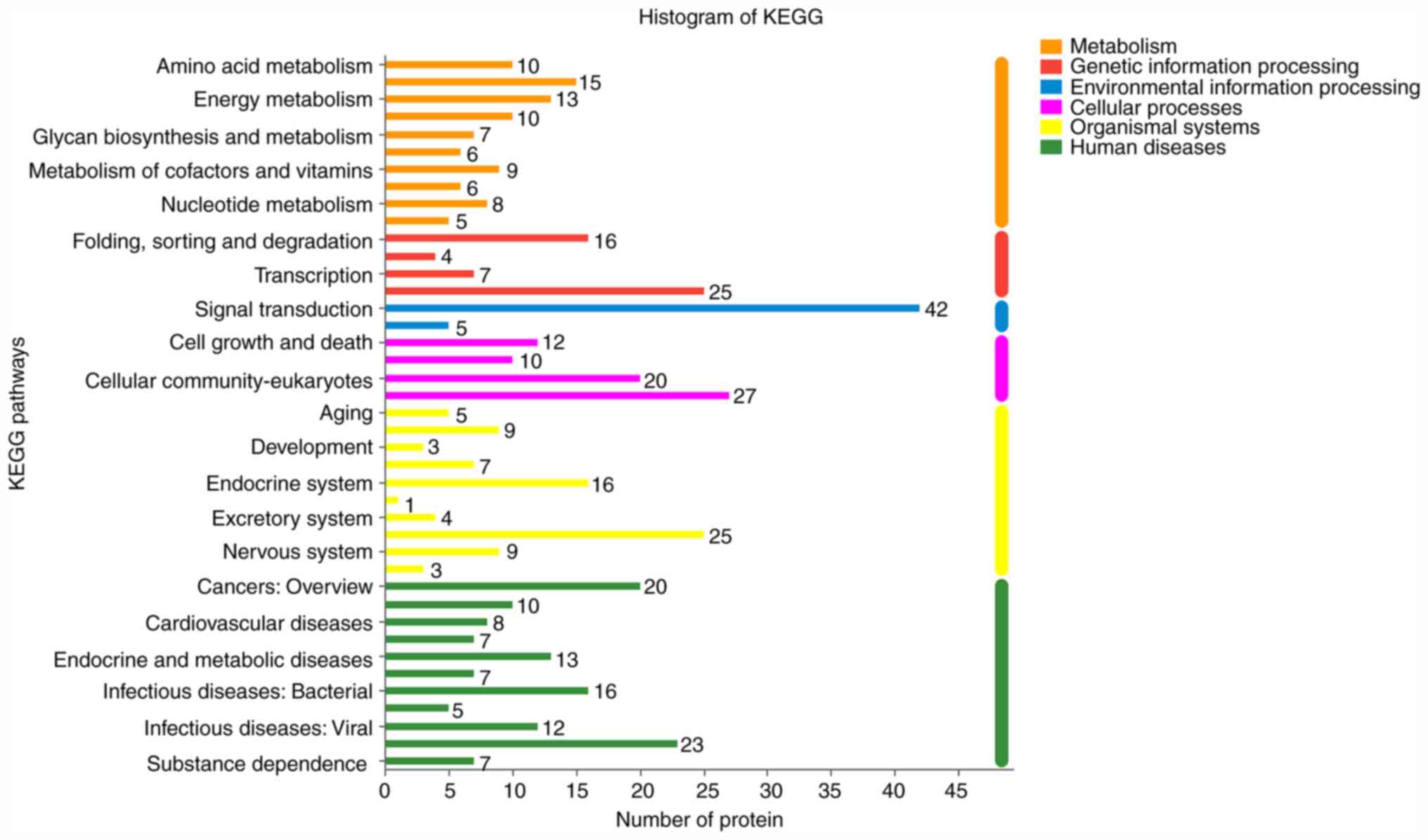
the BPA group were mainly associated with the oncogenes signaling
pathway, cellular community-eukaryotes and the endocrine system
(Fig. 9). The KEGG signaling pathway
in the GEO-BPA combination group was associated with signal
transduction, immune system, energy metabolism, cancer and
endocrine and metabolic diseases (Fig.
10), and from the description of the pathway, it can be seen
that the main pathways of these three groups are associated with
oxidative phosphorylation, endocytosis, focal adhesions and
ribosomes (Tables SVIII–SX; Fig.
10).
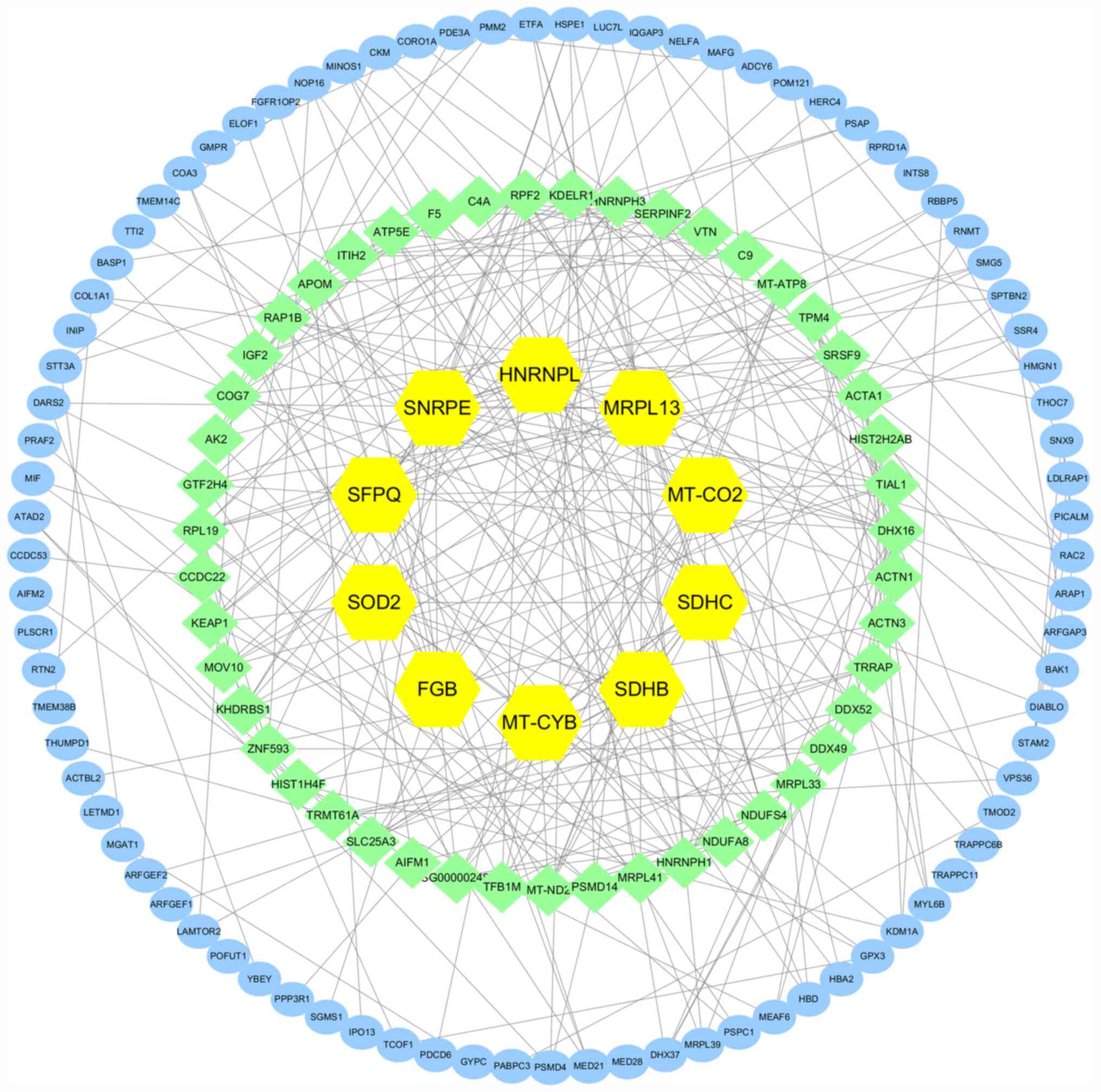
PPI network analysis
Differentially expressed proteins between direct
interaction mod-els may help in gaining important information about
the target protein (25). Analysis
of PPI network is shown in Fig. 11,
the yellow represents the protein with highest degree value, while
other colors represent other proteins that interact with these
differentially expressed target proteins. The PPI analysis revealed
the connection degree of the pro-tein interactions. A higher degree
of connectivity may show more protein complexes. SNRPE, SDHB, SDHC,
MRPL13, SOD2, MT-CYB, MT-CO2, HNRNPL, SFPQ and FGB showed a high
degree of connectivity by comparing the differentially expressed
proteins between GEO-BPA and BPA treatments and these targets
marked as yellow. The middle-degree targets such as NDUFA 8, NDUFA
4, ND 2 and IGF 2 are marked in light green, and blue marked
targets with low degree values.
Discussion
iTRAQ is one of the most advanced technologies in
modern quantitative proteomics (26). It combines stable isotope labeling
and tandem mass spectrometry, and can compare the relative content
of protein in normal and diseased samples in one experiment
(27). The present study
systematically identified and analyzed the differences of proteome
expression in breast cancer cells treated with GEO-BPA and BPA
alone. The re-sults showed that GEO effectively inhibited the
viability of breast cancer cells, which is in accordance with the
findings of Karkihave, GEO inhibits CD44/ALDH1, the hall-marks of
breast cancer cells (17), and BPA
promoted the proliferation of MCF-7 cells, which is consistent with
previous research (28). The
GEO-BPA, BPA and GEO treat-ment groups showed 34, 481 and 210
differentially expressed proteins, respectively, compared with the
control group. These differentially expressed proteins could be
used as biomarkers to evaluate the effect of GEO on breast cancer
induced by BPA, and to guide future treatment strategies for breast
cancer.
Through GO annotation and KEGG pathway analysis, the
specific regulation function and signal transduction pathways
during GEO-BPA treatment were determined, which provided new
insights into breast cancer development and proposed potential
treatment strategies. GO functional annotations showed that,
compared with the control group, 210 proteins were differentially
expressed in the GEO-BPA combined treatment group. These
differentially expressed proteins were mainly located in organelles
and membranes, which can affect signal transduction, tissue or
biogenesis of cellular com-ponents and negative regulation of
biological processes. KEGG signal pathway analysis showed that
oxidative phosphorylation, cAMP signaling pathway, focal adhesions
and ribosomes in the GEO-BPA combination treatment group was richer
compared with other pathways. The oxidative phosphorylation pathway
contains the largest number of differential proteins, which is
related to energy metabolism and is the key to regulating the
content of ATP and oxygen in cells (29).
Tumor cells obtain energy through the glycolysis and
oxidative phosphorylation pathways. Under normoxic conditions,
normally differentiated cells obtain energy primarily through
oxidative phosphorylation, which is generally ≥40% of the total
ATP. However, cancer cells tend to rely on aerobic glycolysis (a
phenomenon of converting glucose into lactic acid under aerobic
conditions) to generate energy, and use the inter-mediate products
of glycolysis as building blocks of cell proliferation (30). The dysregulation of mitochondrial
metabolism is a characteristic of the metabolic reprogramming of
cancer cells (31). However, aerobic
glycolysis is not enough to completely replace the energy produced
by oxidative phosphorylation (32).
Even in an anoxic environment, continuous mitochondrial oxidative
phosphorylation is still indispensable for the biological
activities of tumor cells. In recent years, research on the
antitumor effect of energy metabolism pathways has become
increasingly popular (33,34). Sharma and Singh reported that
dichloroacetate can stimulate oxidative phosphorylation by changing
mitochondrial morphology for glycolysis, inducing apoptosis and
inhibiting proliferation of breast cancer cells (33). Fan et al (34) reported that Jar-TA, a natural
diterpenoid derivative, induces apoptosis of esophageal cancer
cells by double inhibition of glycolysis/oxidative
phosphorylation.
KEGG analysis showed that there were 10
differentially expressed proteins in the GEO-BPA combined treatment
group, which regulated the oxidative phosphorylation pathway and
possibly regulated intracellular energy metabolism and oxygen
transport in breast cells. Meanwhile, the top five degrees of PPI
network interaction results were SNRPE, SDHB, SDHC, MRPL13 and
SOD2, among which SDHB, SDHC and SOD2 were associated with energy
metabolism (35). The present study
showed that down-regulation of the protein of SDHB, SDHC, SOD2 and
COX-2 may be an important factor related to oxidative damage that
promotes GEO to treat BPA-induced breast cancer cells. SDH is a key
enzyme in the tricarboxylic acid cycle and oxidative
phosphorylation of the respiratory chain. The lack of SDH inhibits
the tricarboxylic acid cycle and the transmission of electrons in
the respiratory chain, resulting in the production of ROS and
oxidative damage to cells. Therefore, whether the functions of the
SDHB and SDHC genes are normal or not will affect the function of
SDH, which will eventually lead to excessive production of ROS in
mitochondria and induce apoptosis (35). Functional studies of yeast models
have shown that deletion of the SDHA or SDHB genes and point
mutations of SDHB, SDHC and SDHD are associated with SDH
dysfunction and increased production of ROS (36). In human cell lines, SDHB-silencing,
drug inhibition or RNA interference also eliminates the activity of
complex II, and increases production of ROS and nuclear stability
of HIF1α under normoxic conditions (37). Compared with parent cells, SDHC
mutations also increase the level of ROS production, genomic
instability and tumorigenesis of mutant cells (38). The present study also demonstrated
that the expression of SOD2 protein was downregulated. SOD2 is a
type of superoxide dismutase, which catalyzes the conversion of
superoxide to oxygen and hydrogen peroxide through a
disproportionate reaction, thus removing superoxide and protecting
cells from oxidative damage (39).
SOD2-downregulation in the present study indicated that there may
have been an oxidative stress reaction, releasing peroxide and
inducing cell death.
The current results demonstrated that the expression
of COX-2 protein was de-creased in the GEO-BPA treatment group,
which is consistent with the results of previous studies. For
example, Han et al showed that downregulation of COX-2 can
make survivin and Bcl-2 mRNA and protein expression levels
decrease, Bax mRNA and pro-tein expression levels increase, thus
affect proliferation and apoptosis of human breast cancer MCF-7
cells (40). Meanwhile, malonate and
3-nitropropionic acid are com-pounds that specifically inhibit SDH
activity and then induce ROS production and apoptosis (41). However, as anticancer drugs, both of
these compounds have secondary toxicity (neurological diseases), so
their value for in vivo treatment is limited (42). Therefore, the active ingredients of
traditional Chinese medicine that are used for medi-cine and food
are worthy of in-depth study. Besides, decreased expression of
enzymes involved in oxidative phosphorylation, including NADH
dehydrogenase (ubiquinone) 1 β subcomplex subunit 8 (NDUFA8), NADH
dehydrogenase (ubiquinone) iron-sulfur protein 4 (NDUFS4) and NADH
dehydrogenase 2 (ND2), may cause mitochondrial defects and affect
the energy metabolism of breast cancer cells (43).
Overall, the iTRAQ-based proteomics data presented
in the current study may be useful to clarify the proteomics
profile of MCF-7 cells treated with GEO and BPA, and these data may
help improve our understanding of the molecular mechanisms
underlying the effects of these treatments. It was reported that,
in MCF-7 cells treated with GEO-BPA, seven differentially expressed
proteins were associated with oxidative phosphorylation (COX-2,
NDUFA8, NDUFS4, ND2, SDHB, SDHC and SOD2) were significantly
reduced (P<0.05). KEGG analysis highlighted that oxidative
phos-phorylation may be one of the mechanisms by which GEO
inhibited BPA-induced MCF-7 breast cancer cell proliferation. PPI
analysis showed that SNRPE, SDHB, SDHC, MRPL13 and SOD2 had high
connectivity and were in the center of the network. These
differentially expressed proteins may be the key to the inhibitory
effect of GEO on BPA-induced MCF-7 cells and showed that the
molecular function about energy metabolism underlying this effect.
The limitation of the present study lies in screening
differentially expressed proteins through in vitro
experiments. In future research, in vivo and in vitro
experiments must be used to further analyze the role and mechanisms
of ginger against breast cancer.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Double-First
Class Discipline Construction Project (grant nos. JXSYLXK-ZHYAO115
and ZHYI025) and The Program of Health Commission Jiangxi Province
(grant nos. 20195635 and 2019A262).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
DL, TH and ZL carried out the experiments and
drafted the manuscript. DL and LL participated in the statistical
analyses. LC, XL and QW conceived and designed the study and helped
to draft the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
iTRAQ
|
isobaric tags for relative and
absolute quantitation
|
|
GEO
|
ginger essential oil
|
|
BPA
|
bisphenol A
|
|
GEO-BPA
|
ginger essential oil and bisphenol
A
|
|
COX2
|
cytochrome c oxidase subunit 2
|
|
TEAB
|
triethylammonium bicarbonate
buffer
|
|
TCEP
|
tris (2-carboxyethyl) phosphine
|
|
LC
|
liquid chromatography
|
|
MS
|
mass spectrometry
|
|
DMSO
|
dimethylsulfoxide
|
|
TAM
|
tamoxifen
|
|
NDUFA8
|
NADH dehydrogenase (ubiquinone) 1
alpha subcomplex subunit 8
|
|
NDUFS4
|
NADH dehydrogenase (ubiquinone)
iron-sulfur protein 4, mitochondrial
|
|
ND2
|
NADH-ubiquinone oxidoreductase chain
2
|
|
SDHB
|
succinate dehydrogenase (ubiquinone)
iron-sulfur subunit, mitochondrial
|
|
SDHC
|
succinate dehydrogenase cytochrome
b560 subunit, mitochondrial
|
|
SOD2
|
superoxide dismutase (Mn),
mitochondrial
|
|
SNRPE
|
small nuclear ribonucleoprotein E
|
|
MRPL13
|
39S ribosomal protein L13,
mitochondrial
|
|
ROS
|
39S ribosomal protein L13,
mitochondrial
|
|
OXPHOS
|
oxidative phosphorylation
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Derouiche S, Warnier M, Mariot P, Gosset
P, Mauroy B, Bonnal JL, Slomianny C, Delcourt P, Prevarskaya N and
Roudbaraki M: Bisphenol A stimulates human prostate cancer cell
migration via remodelling of calcium signalling. Springerplus.
2:542013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kloukos D, Pandis N and Eliades T: In vivo
bisphenol - a release from dental pit and fissure sealants: A
systematic review. J Dent. 41:659–667. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krishnan AV, Stathis P, Permuth SF, Tokes
L and Feldman D: Bisphenol-A: An estrogenic substance is released
from polycarbonate flasks during autoclaving. Endocrinology.
132:2279–2286. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang YQ, Wong CK, Zheng JS, Bouwman H,
Barra R, Wahlström B, Neretin L and Wong MH: Bisphenol A (BPA) in
China: A review of sources, environmental levels, and potential
human health impacts. Environ Int. 42:91–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rogers JA, Metz L and Yong VW: Review:
Endocrine disrupting chemicals and immune responses: a focus on
bisphenol-A and its potential mechanisms. Mol Immunol. 53:421–430.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vom Saal FS, Nagel SC, Coe BL, Angle BM
and Taylor JA: The estrogenic endocrine disrupting chemical
bisphenol A (BPA) and obesity. Mol Cell Endocrinol. 354:74–84.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lui YZ, Zeng GQ, Ge LC, Liu H, Du J and
Wang HS: Bisphenol A induces epithelial mesenchymalization of human
breast cancer MCF-7 cells. Acta Scientiae Circumstantiae.
35:608–612. 2015.(In Chinese).
|
|
9
|
White B: Ginger: An overview. Am Fam
Physician. 75:1689–1691. 2007.PubMed/NCBI
|
|
10
|
Ali BH, Blunden G, Tanira MO and Nemmar A:
Some phytochemical, pharmacological and toxicological properties of
ginger (Zingiber officinale Roscoe): A review of recent
research. Food Chem Toxicol. 46:409–420. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ishiguro K, Ando T, Maeda O, Ohmiya N,
Niwa Y, Kadomatsu K and Goto H: Ginger ingredients reduce viability
of gastric cancer cells via distinct mechanisms. Biochem Biophys
Res Commun. 362:218–223. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeong C-H, Bode AM, Pugliese A, Cho YY,
Kim HG, Shim JH, Jeon YJ, Li H, Jiang H and Dong Z: [6]-Gingerol
suppresses colon cancer growth by targeting leukotriene A4
hydrolase. Cancer Res. 69:5584–5591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Surh YJ, Park KK, Chun KS, Lee LJ, Lee E
and Lee SS: Anti-tumor-promoting activities of selected pungent
phenolic substances present in ginger. J Environ Pathol Toxicol
Oncol. 18:131–139. 1999.PubMed/NCBI
|
|
14
|
Brahmbhatt M, Gundala SR, Asif G, Shamsi
SA and Aneja R: Ginger phytochemicals exhibit synergy to inhibit
prostate cancer cell proliferation. Nutr Cancer. 65:263–272. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Santos PA SR, Avanço GB, Nerilo SB,
Marcelino RIA, V. Janeiro V, Valadares MC and Machinski M:
Assessment of Cytotoxic Activity of Rosemary (Rosmarinus
officinalis L.), Turmeric (Curcuma longa L.), and Ginger
(Zingiber officinale R.) Essential Oils in Cervical Cancer
Cells (HeLa). ScientificWorldJournal. 2016:92730782016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prasad S and Tyagi AK: Ginger and its
constituents: role in prevention and treatment of gastrointestinal
cancer. Gastroenterol Res Pract. 2015:1429792015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karki N: Inhibitory activity of ginger oil
against breast cancer cells (2011). LSU Master's Theses. 1714.
simplehttps://digitalcommons.lsu.edu/gradschool_theses/1714
|
|
18
|
Brooks SC, Locke ER and Soule HD: Estrogen
receptor in a human cell line (MCF-7) from breast carcinoma. J Biol
Chem. 248:6251–6253. 1973.PubMed/NCBI
|
|
19
|
Ninth Chinese Pharmacopoeia Commission:
Pharmacopoeia of the People's Republic of China (1). China Medical
Science and Technology Press; China: pp. 932010
|
|
20
|
Kan Y, Lyu Q, Jiang N, Han S, Li J,
Burdman S and Luo L: iTRAQ-based proteomic analyses of the
plant-pathogenic bacterium Acidovorax citrulli during entrance into
and resuscitation from the viable but nonculturable state. J
Proteomics. 211:1035472020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao Z, Wang J, Bai Y, Bao J and Dal E:
Identification and Verification of the Main Differentially
Expressed Proteins in Gastric Cancer via iTRAQ Combined with Liquid
Chromatography-Mass Spectrometry. Anal Cell Pathol.
2019:53106842019. View Article : Google Scholar
|
|
22
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47D:D607–D613. 2019.
View Article : Google Scholar
|
|
23
|
Swaney DL, Wenger CD and Coon JJ: Value of
using multiple proteases for large-scale mass spectrometry-based
proteomics. J Proteome Res. 9:1323–1329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu LR, Zeng R, Shao XX, Wang N, Xu YH and
Xia QC: Identification of differentially expressed proteins between
human hepatoma and normal liver cell lines by two-dimensional
electrophoresis and liquid chromatography-ion trap mass
spectrometry. Electrophoresis. 21:3058–3068. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Unwin RD, Griffiths JR and Whetton AD:
Simultaneous analysis of relative protein expression levels across
multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS.
Nat Protoc. 5:1574–1582. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jamaluddin MFB, Nagendra PB, Nahar P,
Oldmeadow C and Tanwar PS: Proteomic Analysis Identifies Tenascin-C
Expression Is Upregulated in Uterine Fibroids. Reprod Sci.
26:476–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feifei S and Yinyin W: Research status and
progress of relationship between environmental bisphenol A and
reproductive system malignant tumors. Health Res. 38:532–537.
2018.
|
|
29
|
Lui Q, Hu LG, Zhou QF and Jiang GB:
Tetrabromobisphenol A bis(2-hydroxyethylether) induced dysfunction
of respiratory chain oxidative phosphorylation and energy
metabolism rat pheochromocytoma cells by high performance liquid
chromatography-electrospray ionization-tandem mass spectrometry.
Chinese J Chromatography. 35:1–7. 2017. View Article : Google Scholar
|
|
30
|
Li QX, Zhang P and Liu H: Characteristics
and progress of energy metabolism of tumor cells. Chinese
Pharmacological Bulletin. 33:1499–1502. 2017.
|
|
31
|
Liberti MV and Locasale JW: The Warburg
Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci.
41:2872016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jose C, Bellance N and Rossignol R:
Choosing between glycolysis and oxidative phosphorylation: A
tumor's dilemma? Biochim Biophys Acta. 1807:552–561. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sharma P and Singh S: Combinatorial Effect
of DCA and Let-7a on Triple-Negative MDA-MB-231 Cells: A Metabolic
Approach of Treatment. Integr Cancer Ther. 19:15347354209114372020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan XX, Su N, Huang XJ, Li YF, Jia A, Fan
JP, Wang AF, Zhao NM and Ma YC: Jar-TTA, a natural diterpenoid
derivatives, induces apoptosis in human esophageal cancer cells
through dual inhibition of glycolysis and oxidative
phosphorylation. Chinese Pharmacological Bulletin. 35:950–957.
2019.
|
|
35
|
Guzy RD, Sharma B, Bell E, Chandel NS and
Schumacker PT: Loss of the SdhB, but Not the SdhA, subunit of
complex II triggers reactive oxygen species-dependent
hypoxia-inducible factor activation and tumorigenesis. Mol Cell
Biol. 28:718–731. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goffrini P, Ercolino T, Panizza E, Giachè
V, Cavone L, Chiarugi A, Dima V, Ferrero I and Mannelli M:
Functional study in a yeast model of a novel succinate
dehydrogenase subunit B gene germline missense mutation (C191Y)
diagnosed in a patient af-fected by a glomus tumor. Hum Mol Genet.
18:1860–1868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guzy RD, Sharma B, Bell E, Chandel NS and
Schumacker PT: Loss of the SdhB, but Not the SdhA, subunit of
complex II triggers reactive oxygen species-dependent
hypoxia-inducible factor activation and tumorigenesis. Mol Cell
Biol. 28:718–731. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Slane BG, Aykin-Burns N, Smith BJ, Kalen
AL, Goswami PC, Domann FE and Spitz DR: Mutation of succinate
dehydrogenase subunit C results in increased O2.-,
oxidative stress, and genomic instability. Cancer Res.
66:7615–7620. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zelko IN, Mariani TJ and Folz RJ:
Superoxide dismutase multigene family: A comparison of the CuZn-SOD
(SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures,
evolution, and expression. Free Radic Biol Med. 33:337–349. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han H, Yang S, Lin SG, Xu CS and Han ZH:
Effects and mechanism of downregulation of COX 2 expression by RNA
interference on proliferation and apoptosis of hu-man breast cancer
MCF 7 cells. Mol Med Rep. 10:3092–3098. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gomez-Lazaro M, Galindo MF,
Melero-Fernandez de Mera RM, Fernandez-Gómez FJ, Concannon CG,
Segura MF, Comella JX, Prehn JH and Jordan J: Reactive oxygen
species and p38 mitogen-activated protein kinase activate Bax to
induce mito-chondrial cytochrome c release and apoptosis in
response to malonate. Mol Pharmacol. 71:736–743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liot G, Bossy B, Lubitz S, Kushnareva Y,
Sejbuk N and Bossy-Wetzel E: Com plex II inhibition by 3-NP causes
mitochondrial fragmentation and neuronal cell death via an NMDA-
and ROS-dependent pathway. Cell Death Differ. 16:899–909. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Balsa E, Marco R, Perales-Clemente E,
Szklarczyk R, Calvo E, Landázuri MO and Enríquez JA: NDUFA4 is a
subunit of complex IV of the mammalian electron transport chain.
Cell Metab. 16:378–386. 2012. View Article : Google Scholar : PubMed/NCBI
|