Introduction
According to the new case data for cancer of
Globocan 2018 (http://gco.iarc.fr.), combined with
the statistics of patients in America with tumors among male and
female in 2020, colorectal cancer (CRC) has become the third most
common cancer in the world, and the incidence of CRC is expected to
increase (1–3). Data from Globocan reports that compared
with 2018 by 2040, the incidence of colon cancer will increase 1.75
times, while rectal cancer will increase 1.67 times (https://gco.iarc.fr/tomorrow/home). A number of
potential molecular targets or markers for CRC have been identified
by The Cancer Genome Atlas (TCGA) project, including tumor necrosis
factor α-induced protein 8 (TNFAIP8) (2,4). This
protein belongs to the TNFAIP8-like (TIPE) family, which contains
three other members, TIPE1, TIPE2 and TIPE3 (5,6).
Although the members of the TIPE family have similar amino acid
sequences, they are involved in different biological activities
(6,7).
TNFAIP8 was the first protein of this family to be
discovered, and its expression is limited in various normal
tissues, but upregulated in malignant cells of various cancers,
such as chronic myelogenous leukemia, lymphoblastic leukemia, and
lung carcinoma (8–10). Previous studies have suggested an
association between high levels of TNFAIP8 expression and
tumorigenesis (5,11,12).
TNFAIP8 has several variants and different protein isoforms that
coexist in the same cells (https://www.ncbi.nlm.nih.gov/gene/25816, accessed on
23 December 2019). Recent studies have demonstrated that in certain
TCGA data sets, TNFAIP8 variant 2 (v2) mRNA (encoding isoform b) is
upregulated in eight types of human cancer, but no changes have
been identified in colon adenocarcinoma; TNFAIP8 v1 mRNA is
downregulated in seven types of human cancer, including colon
adenocarcinoma; additionally, low levels of TNFAIP8 v3-v6 mRNA are
expressed in both cancer or normal tissue (4,12). A
previous study has suggested that the TNFAIP8 protein that is
upregulated in cancer is encoded by TNFAIP8 v2 mRNA, and that
TNFAIP8 v2 promotes carcinogenesis by suppressing p53 activity in
human cancer cells (4). The
functions of TNFAIP8 in promoting cell proliferation and inhibiting
apoptosis are considered to facilitate its cancer-promoting role
(13,14).
In contrast to TNFAIP8 expression, TIPE1 expression
is low in certain types of cancer, including hepatocellular
carcinoma (HCC), gastric and lung cancer, and CRC (15–18).
However, high expression levels of TIPE1 are detected in most human
carcinoma cell lines, including cervical and ovarian cancer cell
lines (5,19). Different posttranslational
modifications of TIPE1 have also been predicted to exist (12,20).
TIPE1 has two transcript variants, both of which encode the same
protein (https://www.ncbi.nlm.nih.gov/gene/126282, accessed on
23 December 2019). TIPE1 inhibits the p65 and c-Jun N-terminal
kinase pathways by suppressing Rac1 activity to increase
caspase-mediated apoptosis in HCC cells (15). In gastric cancer, TIPE1 suppresses
cell invasion and migration by inhibiting the Wnt/β-catenin
signaling pathway (18). TIPE1 is
regarded as a proapoptotic factor with the ability to suppress cell
migration (15,18).
Unlike other TIPE family members, TIPE2 has no
transcript variants (https://www.ncbi.nlm.nih.gov/gene/79626). TIPE2 has
been reported to be expressed at lower levels in certain types of
cancer, such as breast and gastric cancer, HCC and lung cancer,
compared with those in adjacent normal tissues (21–24).
TIPE2 negatively regulates Toll-like and T cell receptors to avoid
excessive immune response and maintain immune homeostasis (7). In addition, TIPE2 is an inhibitor of
the NF-κB and MAPK signaling pathways and helps reduce the
activation of activator protein-1 and NF-κB (25,26).
TIPE2 is a negative regulator of immunity and inflammation that can
promote apoptosis and inhibit cell migration (7,24).
TIPE3 specifically binds the second messenger of
phospholipids and promotes cell proliferation, and the expression
of TIPE3 is upregulated in lung, esophageal and cervical cancer, as
well as colon adenocarcinoma (27).
TIPE3 has two transcript variants (v1 and v2) that encode two
isoforms, and isoform 2 has a shorter N-terminus compared with that
of isoform 1 (https://www.ncbi.nlm.nih.gov/gene/388121, accessed on
21 December 2019). TIPE3 mRNA v1 and protein are highly expressed
in glioblastoma (GBM), where its protein expression is negatively
associated with p38 phosphorylation and apoptosis, and only TIPE3
isoform 1 interacts with p38 (28).
TIPE3 is upregulated in lung cancer compared with adjacent
non-tumor tissues, and different subcellular localization of TIPE3
induces different effects; TIPE3 was highly expressed in plasma
membrane of the lung cancer cells with long pseudopodia,
particularly in the position of protrusion, indicating that it was
involved in cell motility and contributed to the growth and
migration of lung cancer cells (29). However, in the cytoplasm, both
isoform 1 and 2 of exogenous TIPE3 can attenuate the proliferation
and migration of lung cancer cells by inhibiting AKT and ERK
activation (29). Another study has
demonstrated that the mRNA expression of TIPE3 is downregulated in
certain types of human cancer, such as nasopharyngeal carcinoma
(NPC), bladder, head and neck, lung and prostate cancer, as well as
CRC, and is associated with DNA CpG island hypermethylation; in
addition, TIPE3 inhibits NPC cell proliferation, migration and
invasion (30). Therefore, the
biological role of TIPE3 remains unknown, and only a limited number
of studies have demonstrated that TIPE3 is a carcinogen (28,29).
In previous studies, TNFAIP8, TIPE2 and TIPE3 levels
have been reported to be higher in CRC compared with those in
adjacent non-tumor tissues (5,6). In
addition, TIPE1 is expressed at lower levels in CRC compared with
those in adjacent non-tumor tissues. However, there are limited
reports on the transcription levels of the TIPE family members in
CRC, and it is unclear whether the expression levels of TIPE family
genes within each sample are interrelated. The present study aimed
to further explore the co-expression patterns of the TIPE family
members within individual CRC samples and to assess the possible
associations between the mRNA expression levels of the TIPE family
members and the clinicopathological characteristics of patients
with CRC.
Materials and methods
Tissue samples
Tumor and paired adjacent tissues from 49 patients
with colorectal adenocarcinoma who had not received radiotherapy
and chemotherapy prior to the surgery were collected from Zhongshan
Hospital affiliated with Xiamen University (Xiamen, China) from
January 2017 to December 2018. The age range was 34 to 87 years
old, with an average age of 60 years old. Among them, there were 35
males and 14 females. The clinical specimens were histologically
confirmed in the pathology department. Besides, there were 4
clinical cases without differentiation related information. The
adjacent tissues were collected 3–5 cm away from the tumor edge;
they may have included adenoma tissue adjacent to the carcinoma
rather than heterogeneous samples composed of mucosa, muscularis
and serosa. The samples were frozen and stored at −80°C until
further analysis. All samples were obtained with patient written
consent and approval of the Committee on Medical Ethics of
Zhongshan Hospital affiliated with Xiamen University. For western
blot analysis, regardless of clinicopathological characteristics
such as age, sex, tumor size and metastasis, five samples were
randomly selected from the patients.
RNA isolation and reverse
transcription
Total RNA was extracted from paired tumor and normal
tissues from 49 patients with CRC using TransZol Up reagent
(TransGen Biotech Co., Ltd.) according to the manufacturer's
instructions. The RNA concentration was determined using a Pultton
P100 Micro Volume Spectrophotometer (Pultton Technology, Ltd.).
Every 1 µg of the total RNA was reverse-transcribed into cDNA in a
20 µl system of TransScript® All-in-One First-Strand
cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) (TransGen
Biotech Co., Ltd.) according to the manufacturer's
instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
The expression levels of TNFAIP8, TIPE1, TIPE2 and
TIPE3 mRNA were evaluated by RT-qPCR using β-actin as the internal
control. The generated cDNAs were used for qPCR using TransStart
Top Green qPCR SuperMix (TransGen Biotech Co., Ltd.) according to
the manufacturer's protocol. The qPCR process was performed using a
two-step method in a 20-µl total reaction volume using a CFX
Connect™ Real-Time system (Bio-Rad Laboratories, Inc.). The
following thermocycling conditions were used: 94°C for 30 sec,
followed by 40 cycles of 94°C for 5 sec, 60°C for 15 sec, and 72°C
for 10 sec followed by final extension at 72°C for 40 sec. The
primer sequences were as follows: TNFAIP8 forward,
5′-ATGCACTCCGAAGCAGAAGAATCC-3′ and reverse,
5′-CGTCTATTAAGGTGGTGGCGATGG-3′; TIPE1 forward,
5′-GGACACCTTCAGCACCAAGAGC-3′ and reverse,
5′-GTGGCGCGGTACAGCTCATC-3′; TIPE2 forward,
5′-ATGTGCTGCTAGAGTTGGTGGAAC-3′ and reverse,
5′-TTGCCAAGGTGCTGAGTGAAGTC-3′; TIPE3 forward,
5′-GAACCAGACCGCCATGACCATTG-3′ and reverse,
5′-CCAGTTCATGCACCAGGTCCTTG-3′; and β-actin forward,
5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse, 5′-GGGCACGAAGGCTCATCATT-3′.
The relative mRNA expression of the target gene in each sample was
calculated using the quantification cycle (Cq) value based on the
2−∆∆Cq method, where ∆Cq=Cq (target)-Cq (control)
(31).
Western blot analysis
The tissue samples were lysed using RIPA buffer
(Sigma-Aldrich; Merck KGaA) with 1% protease inhibitor cocktail and
1% phenylmethanesulfonyl fluoride (Gold Biotechnology, Inc.) at 4°C
and cleared by centrifugation for 10 min at 10,000 × g at 4°C. The
protein concentration was determined by the Bradford assay (Bio-Rad
Laboratories, Inc.). The protein samples were separated by 12% or
15% SDS-PAGE and transferred to polyvinylidene difluoride membranes
(EMD Millipore). The membranes were blocked by 5% skimmed milk at
room temperature for 1 h and incubated with the following specific
primary antibodies at 4°C overnight: Rabbit polyclonal antibodies
against TNFAIP8 (1:500; cat. no. ab195810; Abcam), TIPE1 (1:1,000;
cat. no. SAB2102488; Sigma-Aldrich; Merck KGaA), TIPE2 (1:500; cat.
no. 15940-1-AP; ProteinTech Group, Inc.) and TIPE3 (1:1,000; cat.
no. AP11822c; Abgent, Inc.), and a rabbit monoclonal antibody
against β-actin (1:5,000; cat. no. AB0035; Abways Technology,
Inc.). Subsequently, the membranes were washed by TBST which
contained 0.05% Tween 20 (cat. no. T1085; Solarbio, Inc.), and
incubated with a horseradish peroxidase-conjugated goat anti-rabbit
IgG antibody (1:2,000; ZF-0311; OriGene Technologies, Inc.) for 1 h
at room temperature. The protein bands were visualized using
enhanced chemiluminescence reagents (cat. no. P10300; NCM Biotech).
The grayscale values of the protein band images were analyzed using
ImageJ software (v.1.51; National Institutes of Health), and the
relative protein expression levels were calculated as the ratio of
the grayscale value of the protein band to that of the β-actin
band.
TCGA gene expression analysis
TIPE family mRNA data from normal colon and CRC
samples in TCGA were analyzed by using the UALCAN web server
(http://ualcan.path.uab.edu/) (32). Since the UALCAN Web server was
produced using data from the TCGA, its analysis results are
equivalent to the data provided by TCGA (32), so the expression and correlation
analyses were performed on 243 adenocarcinoma, 37 mucinous
adenocarcinoma and 41 normal samples from TCGA (32).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 8.0 (GraphPad Software, USA). Each experiment was
repeated at least 3 times and data were expressed as the means ±
standard error of mean (SEM). A paired Student's t-test was
performed to examine the differences in relative mRNA expression
levels of the target genes between CRC and paired adjacent tissues.
For multiple comparisons, one-way ANOVA followed by the post hoc
Bonferroni correction was used to adjust the significance levels
and the corrected P-value was 0.0166. The associations between the
expression levels of TIPE family members and the
clinicopathological features of patients with CRC, including age,
sex, tumor location, Tumor-Node-Metastasis (TNM) stage defined
using the 8th Edition for TNM Classification of Malignant Tumors
(33), degree of differentiation and
lymph node metastasis, were analyzed by the χ2 test (all
data satisfied the conditions of the χ2 test, and there
was no need to use Fisher's exact test). The patients were divided
into two groups (high or low expression) using the median mRNA
expression value as the cut-off point. Linear regression analysis
was performed to determine whether significant linear relationships
existed among the TIPE family members. P<0.05 was considered to
indicate a statistically significant difference.
Results
TIPE family member mRNA expression in
CRC
RT-qPCR analysis was performed to determine the
relative mRNA levels of TIPE family members in the paired tumor and
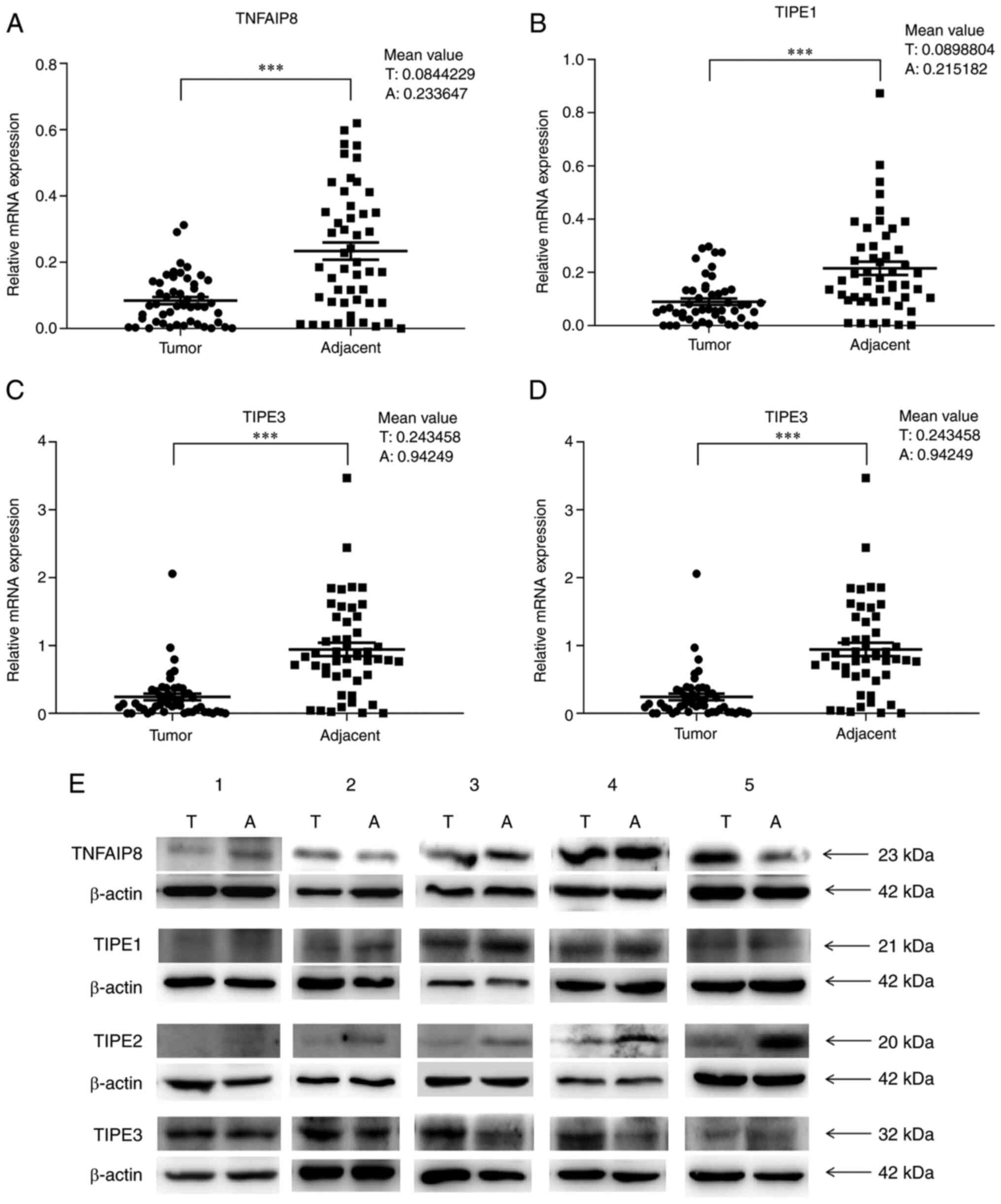
adjacent tissues from patients with CRC. The expression levels of
TNFAIP8, TIPE1, TIPE2 and TIPE3 were significantly lower in the
tumor tissues compared with those in the adjacent tissues (all
P<0.0001; Fig. 1A-D). The RT-qPCR
results demonstrated that the mRNA expression levels of TNFAIP8,
TIPE1, TIPE2 and TIPE3 in the adjacent tissues were ~2.8-, 2.4-,
2.5- and 3.9-fold higher, respectively, compared with those in the
tumor tissues (Fig. 1A-D). The most
notable fold-change was observed in the expression levels of
TIPE3.
TIPE family protein expression in
CRC
It has been reported that the protein expression
levels of TNFAIP8, TIPE2 and TIPE3 are usually higher, and the
protein expression of TIPE1 is lower in CRC tissues compared with
those in adjacent or normal tissues (16,27,34,35). To
verify the protein expression levels of the TIPE family members,
western blot analyses was performed using protein extracts from
five randomly selected pairs of cancer and adjacent tissue samples.
The results demonstrated that the protein and mRNA expression
patterns of TIPE1 and TIPE2 were consistent in the five pairs of
tissues, and the expression levels of these two proteins were
appeared to be lower in the tumor tissues compared with those in
the adjacent tissues (Fig. 1E;
Table SI). In addition, the protein
expression patterns of TNFAIP8 and TIPE3 were partially consistent
with the mRNA expression patterns in these samples; the TNFAIP8
protein expression levels were not consistent with the mRNA
expression levels in four pairs of tissue samples, and the protein
expression levels of TNFAIP8 in three cancer tissue samples were
higher compared with those in the corresponding adjacent tissues
(Fig. 1E; Table SI). In addition, the expression
levels of TIPE3 in four cancer tissue samples were higher compared
with those in the corresponding adjacent tissues, whereas the mRNA
expression pattern exhibited the opposite trend (Fig. 1E; Table
SI).
Association between TIPE family
expression and patient clinicopathological characteristics
The associations between TIPE family member
expression levels and the clinicopathological features of patients
with CRC were analyzed. As presented in Tables I and II, the levels of TNFAIP8 mRNA in tumor
tissues exhibited a significant association with the tumor
differentiation grade (P=0.0204, Table
I), and the levels of TIPE2 mRNA in the tumor tissues were
weakly associated with sex (P=0.0468; Table II). No associations were identified
between TNFAIP8 or TIPE2 mRNA levels and other clinicopathological
characteristics in patients with CRC (all P>0.05; Tables I and II). The levels of TIPE1 and TIPE3 mRNA in
tumor tissues exhibited no significant associations with sex, age,
tumor site, TNM stage, differentiation degree or lymph node
metastasis in patients with CRC (all P>0.05; Tables III and IV).
 | Table I.Analysis of the association between
TNFAIP8 expression and patient clinicopathological
characteristics. |
Table I.
Analysis of the association between
TNFAIP8 expression and patient clinicopathological
characteristics.
|
|
| TNFAIP8
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | n | High, n (%) | Low, n (%) | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
|
Male | 35 | 20 (80.0) | 15 (62.5) | 1.838 | 0.1752 |
|
Female | 14 | 5 (20.0) | 9 (37.5) |
|
|
| Age |
|
|
|
|
|
|
≥60 | 35 | 20 (80.0) | 15 (62.5) | 1.838 | 0.1752 |
|
<60 | 14 | 5 (20.0) | 9 (37.5) |
|
|
| TNM stage |
|
|
|
|
|
|
I–II | 30 | 15 (60.0) | 15 (62.5) | 0.032 | 0.8575 |
|
III–IV | 19 | 10 (40.0) | 9 (37.5) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 19 | 10 (40.0) | 9 (37.5) | 0.03224 | 0.8575 |
|
Negative | 30 | 15 (60.0) | 15 (62.5) |
|
|
| Location |
|
|
|
|
|
|
Colon | 32 | 17 (68.0) | 15 (62.5) | 0.1635 | 0.6860 |
|
Rectum | 17 | 8 (32.0) | 9 (37.5) |
|
|
| Differentiation
grade |
|
|
|
|
|
|
Low | 5 | 5 (21.7) | 0 (0.0) | 5.38 | 0.0204a |
| Medium
and high | 40 | 18 (78.3) | 22 (100.0) |
|
|
 | Table II.Analysis of the association between
TIPE2 expression and patient clinicopathological
characteristics. |
Table II.
Analysis of the association between
TIPE2 expression and patient clinicopathological
characteristics.
|
|
| TIPE2
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | n | High, n (%) | Low, n (%) | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
|
Male | 35 | 21 (84.0) | 14 (58.3) | 3.953 | 0.0468a |
|
Female | 14 | 4 (16.0) | 10 (41.7) |
|
|
| Age |
|
|
|
|
|
|
≥60 | 35 | 19 (76.0) | 16 (66.7) | 0.523 | 0.4697 |
|
<60 | 14 | 6 (24.0) | 8 (33.3) |
|
|
| TNM stage |
|
|
|
|
|
|
I–II | 30 | 12 (48.0) | 18 (75.0) | 3.760 | 0.0525 |
|
III–IV | 19 | 13 (52.0) | 6 (25.0) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 19 | 13 (52.0) | 6 (25.0) | 3.760 | 0.0525 |
|
Negative | 30 | 12 (48.0) | 18 (75.0) |
|
|
| Location |
|
|
|
|
|
|
Colon | 32 | 17 (68.0) | 15 (62.5) | 0.164 | 0.6860 |
|
Rectum | 17 | 8 (32.0) | 9 (37.5) |
|
|
| Differentiation
grade |
|
|
|
|
|
|
Low | 5 | 4 (17.4) | 1 (4.5) | 1.879 | 0.1705 |
| Medium
and high | 40 | 19 (82.6) | 21 (95.5) |
|
|
 | Table III.Analysis of the association between
TIPE1 expression and patient clinicopathological
characteristics. |
Table III.
Analysis of the association between
TIPE1 expression and patient clinicopathological
characteristics.
|
|
| TIPE1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | n | High, n (%) | Low, n (%) | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
|
Male | 35 | 20 (80.0) | 15 (62.5) | 1.838 | 0.1752 |
|
Female | 14 | 5 (20.0) | 9 (37.5) |
|
|
| Age |
|
|
|
|
|
|
≥60 | 35 | 20 (80.0) | 15 (62.5) | 1.838 | 0.1752 |
|
<60 | 14 | 5 (20.0) | 9 (37.5) |
|
|
| TNM stage |
|
|
|
|
|
|
I–II | 30 | 16 (64.0) | 14 (58.3) | 0.166 | 0.864 |
|
III–IV | 19 | 9 (36.0) | 10 (41.7) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 30 | 16 (64.0) | 14 (58.3) | 0.166 | 0.864 |
|
Negative | 19 | 9 (36.0) | 10 (41.7) |
|
|
| Location |
|
|
|
|
|
|
Colon | 32 | 16 (64.0) | 16 (66.7) | 0.038 | 0.8446 |
|
Rectum | 17 | 9 (36.0) | 8 (33.3) |
|
|
| Differentiation
grade |
|
|
|
|
|
|
Low | 5 | 3 (13.0) | 2 (9.1) | 0.178 | 0.6732 |
| Medium
and high | 40 | 20 (87.0) | 20 (90.9) |
|
|
 | Table IV.Analysis of the association between
TIPE3 expression and patient clinicopathological
characteristics. |
Table IV.
Analysis of the association between
TIPE3 expression and patient clinicopathological
characteristics.
|
|
| TIPE3
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | n | High, n (%) | Low, n (%) | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
|
Male | 35 | 20 (80.0) | 15 (62.5) | 1.838 | 0.1752 |
|
Female | 14 | 5 (20.0) | 9 (37.5) |
|
|
| Age, years |
|
|
|
|
|
|
≥60 | 35 | 18 (72.0) | 17 (70.8) | 0.008 | 0.9280 |
|
<60 | 14 | 7 (28.0) | 7 (29.2) |
|
|
| TNM stage |
|
|
|
|
|
|
I–II | 30 | 13 (52.0) | 17 (70.8) | 1.829 | 0.1762 |
|
III–IV | 19 | 12 (48.0) | 7 (29.2) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 19 | 12 (48.0) | 7 (29.2) | 1.829 | 0.1762 |
|
Negative | 30 | 13 (52.0) | 17 (70.8) |
|
|
| Location |
|
|
|
|
|
|
Colon | 32 | 19 (76.0) | 13 (54.2) | 2.576 | 0.1085 |
|
Rectum | 17 | 6 (24.0) | 11 (45.8) |
|
|
| Differentiation
grade |
|
|
|
|
|
|
Low | 5 | 4 (17.4) | 1 (4.5) | 1.879 | 0.1705 |
| Medium
and high | 40 | 19 (82.6) | 21 (95.5) |
|
|
Linear regression analysis of TIPE
family member expression levels in CRC
To further investigate the relationships between
TIPE family members in CRC, the linear fitting of the relative mRNA
levels of TIPE family members with each other in the tumor tissues
alone or the paired CRC tumor/adjacent tissues was assessed by
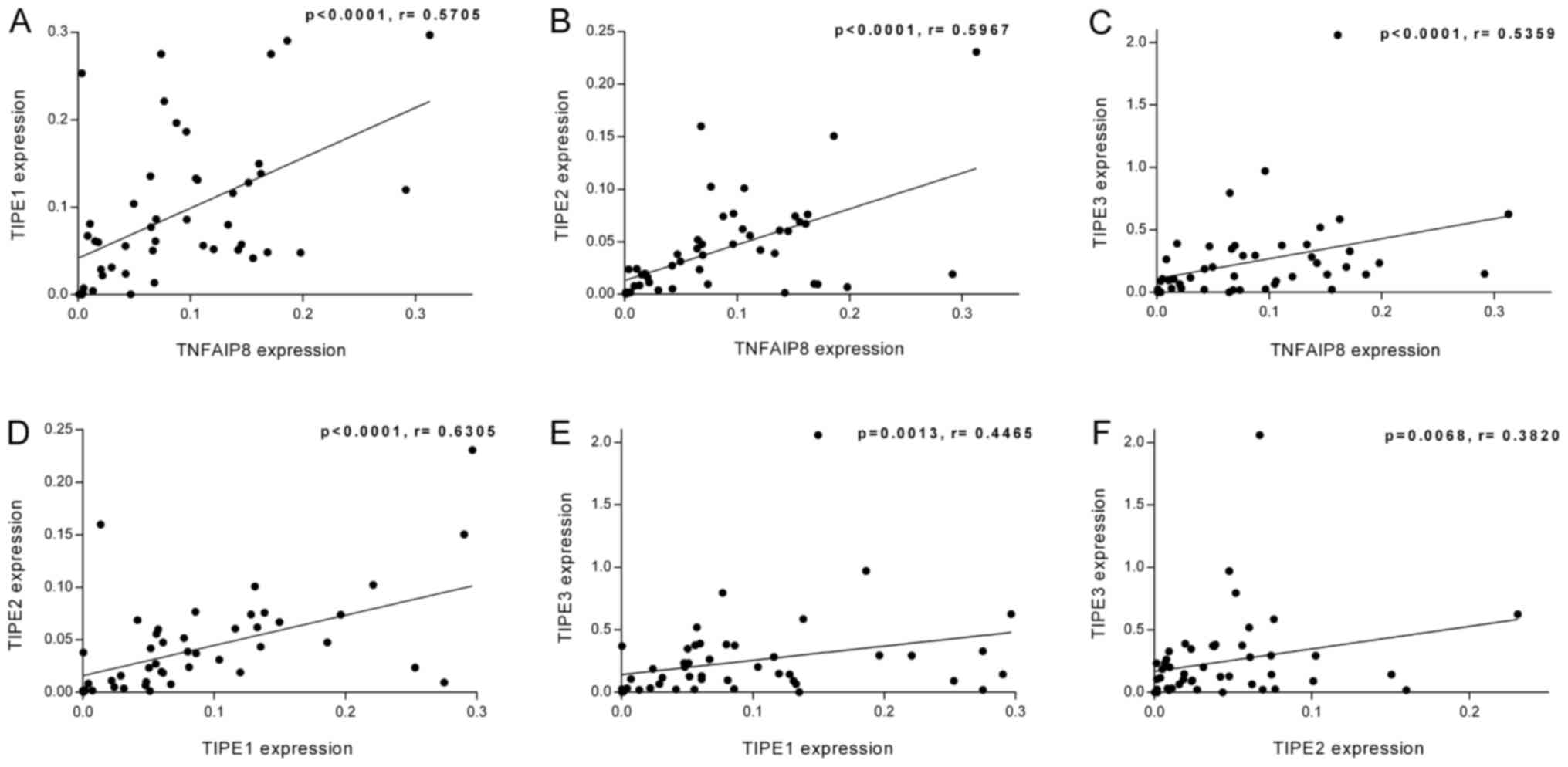
linear regression analysis. As presented in Fig. 2A-F, TNFAIP8 expression levels
exhibited a positive linear relationship with those of TIPE1
(r=0.5705; P<0.0001), TIPE2 (r=0.5967; P<0.0001) and TIPE3
(r=0.5359; P<0.0001) in the tumor tissues. In addition, a
positive linear association was identified between the expression
levels of TIPE1 and TIPE2 (r=0.6305; P<0.0001), TIPE1 and TIPE3
(r=0.4465; P=0.0013), and TIPE2 and TIPE3 (r=0.3820; P=0.0068). The
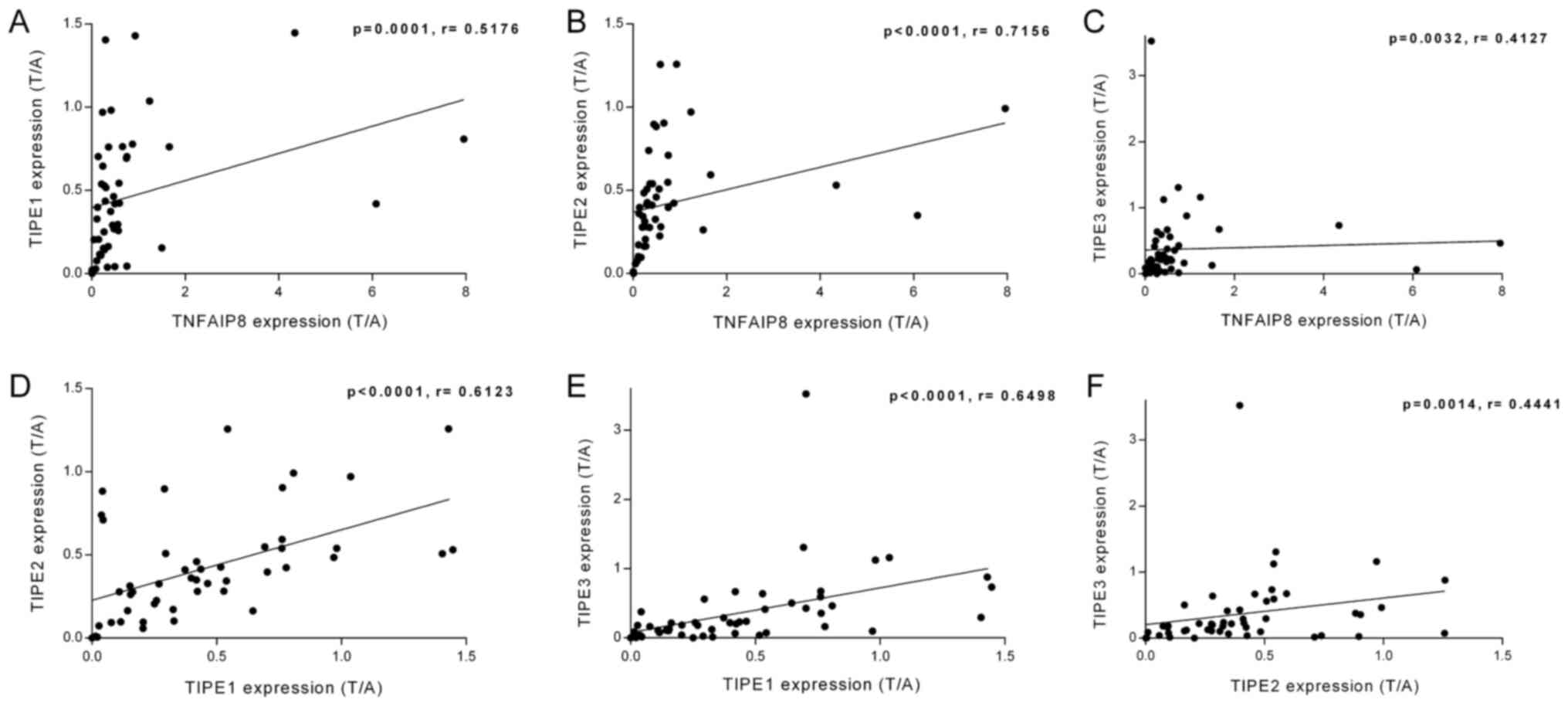
linear regression analysis was also performed on the ratio of the
relative mRNA expression of TIPE family members in the tumor
tissues to those in the adjacent tissues (T/A). As demonstrated in
Fig. 3A-F, the T/A ratio of TNFAIP8
was positively associated with those of TIPE1 (r=0.5176; P=0.0001),
TIPE2 (r=0.7156; P<0.0001) and TIPE3 (r=0.4127; P=0.0032). There
were also positive linear relationships between the T/A ratio
values of TIPE1 and TIPE2 (r=0.6123; P<0.0001), TIPE1 and TIPE3
(r=0.6498; P<0.0001), and TIPE2 and TIPE3 (r=0.4441; P=0.0014).
Therefore, the mRNA levels of the TIPE family members exhibited
significant linear relationships with each other.
TIPE family member expression in TCGA
CRC data
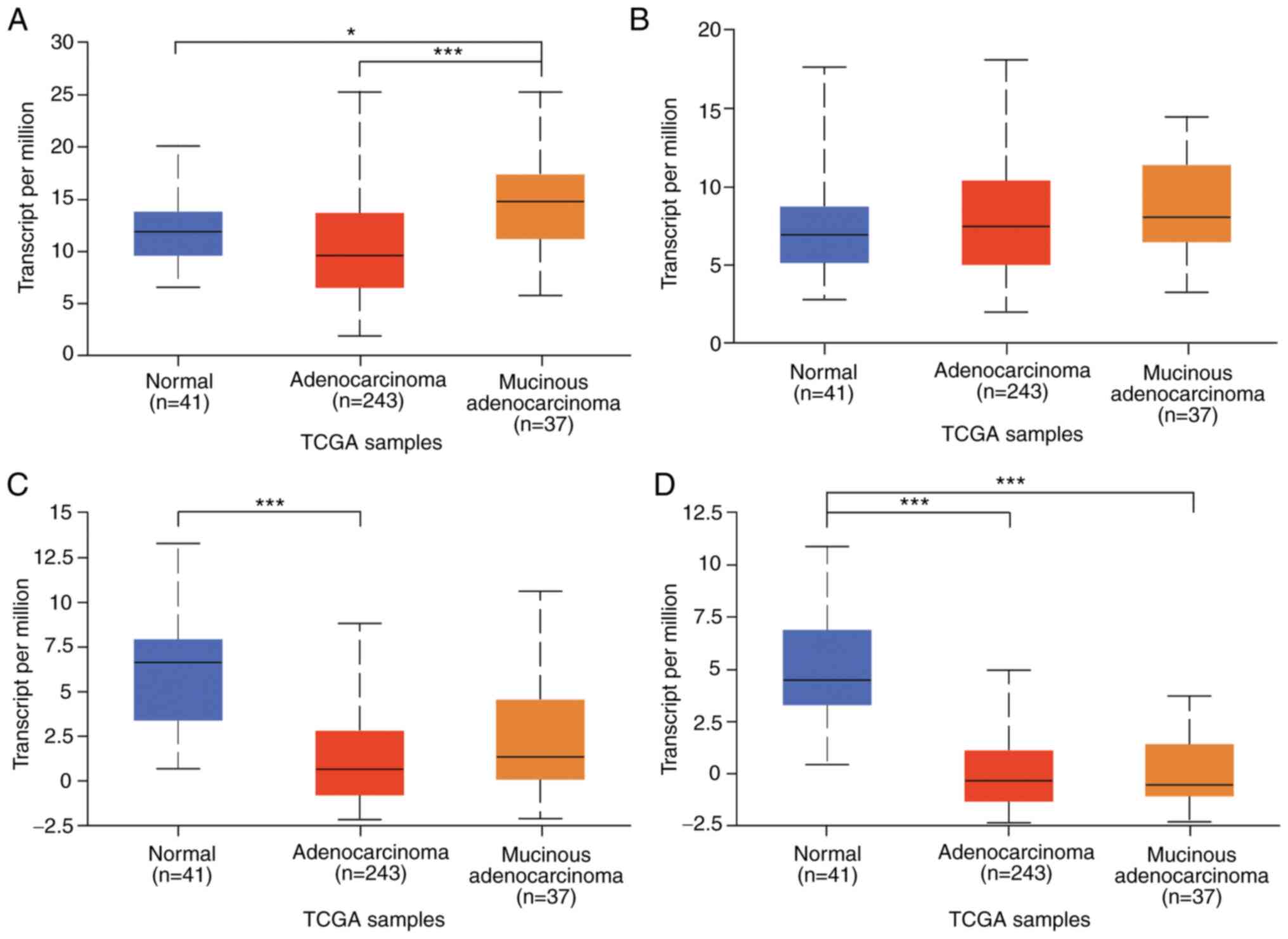
To further investigate TIPE family expression in
CRC, normal colon and colon adenocarcinoma data were obtained from
TCGA database. As presented in Fig.
4, the mucinous adenocarcinoma tissues expressed the highest
levels of TNFAIP8 and TIPE1, and TNFAIP8 expression levels in the
normal tissues were slightly higher compared with those in the
adenocarcinoma tissues, whereas TIPE1 levels were slightly lower in
the normal tissues compared with those in the adenocarcinoma
tissues (both P>0.05; Fig. 4A and
B). The expression levels of TIPE2 and TIPE3 were the highest
in the normal colon tissues (Fig. 4C and
D). The expression levels of TIPE2 in the mucinous
adenocarcinoma tissues were slightly higher compared with those in
the adenocarcinoma tissues, whereas the expression levels of TIPE3
in the adenocarcinoma tissues were slightly higher compared with
those in the mucinous adenocarcinoma tissues (both P>0.05). The
results of TCGA data analysis demonstrated that the expression
trends of TIPE, TIPE2 and TIPE3 were consistent with the
experimental results of the present study, and that their
expression levels in the normal colon tissues were higher compared
with those in adenocarcinoma.
Discussion
The present study evaluated the mRNA expression
levels of all TIPE family members in CRC and adjacent tissues. The
results demonstrated that the mRNA levels of TNFAIP8, TIPE1, TIPE2
and TIPE3 in the CRC tissues were significantly lower compared with
those in the paired adjacent tissues.
Previous studies have demonstrated that the
expression of TNFAIP8 protein is upregulated in ~50% of cancer
tissues compared with adjacent non-tumor tissue in patients with
CRC and is associated with the degree of malignancy of the tumor
(34,36). However, it is unclear whether TNFAIP8
isoform a functions in cancer and whether the levels of TNFAIP8 v2
mRNA increases alongside those of TNFAIP8 isoform b in CRC tissues.
In the present study, the primers used to amplify TNFAIP8 targeted
TNFAIP8 v1 and v3, which encode isoform a. Notably, the expression
pattern of TNFAIP8 mRNA was consistent with that in the previous
study by Lowe et al (4). A
number of previous studies have confirmed that TNFAIP8 serves a
signal transmission role in tumorigenesis. For example, TNFAIP8
promotes the proliferation and invasion of MDA-MB-435 breast cancer
cells by increasing the expression of VEGFR-2, MMP1 and MMP9
(37). TNFAIP8 interacts with large
tumor suppressor kinase 1, regulates Hippo signaling in lung and
liver cancer cells, and induces cell proliferation, migration and
invasion (38). In addition,
depletion of TNFAIP8 in HeLa cervical cancer cells activates
caspase-3/8, induces p38 phosphorylation and promotes
cisplatin-induced apoptosis and cell death (39). As for upstream regulators, the
expression level of TNFAIP8 can be upregulated by NF-κB and TNF-α
in diversified cell lines (40). Gao
et al, experimentally proved microRNA-9 to negatively
regulate the expression of TIPE, and induction of microRNA-9
resulted in reduced TIPE levels and reduced GC cell proliferation
in vitro and tumor growth in vivo (41). The results of the present study also
demonstrated that in samples from five patients with CRC, the
expression levels of TNFAIP8 protein were upregulated in three
tumor tissues compared with those in the adjacent tissues.
Therefore, the protein level of TNFAIP8 may be affected by the
crosstalk of a number of signaling pathways.
Although TCGA data analysis in the present study
demonstrated that TIPE1 mRNA levels were not significantly
different between the normal colon and CRC tissues, recent studies
have stated that TIPE1 is downregulated in CRC compared with
adjacent non-tumor tissues, and that TIPE1 overexpression inhibits
cell proliferation by suppressing the Wnt/β-catenin signaling
pathway in CRC (16,32). TIPE1 primers used in the present
study targeted both transcript variants. The results of the present
study demonstrated that TIPE1 mRNA expression levels were
significantly downregulated in the CRC tissues compared with those
in the adjacent tissues, and that TIPE1 protein levels were
consistent with the mRNA levels in five randomly selected pairs of
samples. These results were consistent with previous findings in
HCC, gastric and lung cancer (15,17,18). As
aforementioned, TIPE1 interacts with Rac1 and inhibits p65 and
c-Jun N-terminal kinase in liver cancer cells (15). TIPE1 promotes autophagy by decreasing
mTOR phosphorylation in dopaminergic neuronal cells (42). In addition, TIPE1 is required for
both zVAD and TNF-α-induced necroptosis (43). Therefore, we hypothesize that the
downregulation of TIPE1 may lead to decreased cell death and
enhanced colorectal tumorigenesis.
As a negative regulator of immunity and
inflammation, TIPE2 expression levels are downregulated in most
types of cancer tissues compared with those in adjacent normal
tissues (5,20–23).
However, a previous study has reported higher levels of TIPE2
protein in tissues from patients with CRC compared with those in
healthy human colon tissues collected by colonoscopy (35). In contrast to this, the results of
the present study demonstrated that TIPE2 mRNA expression levels
were downregulated in the CRC tissues compared with those in the
paired adjacent tissues, and TIPE2 protein and mRNA expression
levels were consistent in five random samples. TCGA data also
supported these results. TIPE2 reduces the phosphorylation of
protein kinase B/AKT and ERK1/2 and enhances the activation of
caspase-9 and caspase-3 to promote apoptosis in gastric cancer
cells (44). In HCC cells,
overexpression of TIPE2 decreases the expression levels of MMP-9
and urokinase plasminogen activator by inhibiting the Rac1 pathway,
thus suppressing tumor invasion and metastasis, and abrogates the
effects of TNF-α-induced cell migration by reducing MMP-13/-3,
inhibiting the activation of ERK1/2 and NF-κB (45,46). On
the other hand, a number of inflammatory diseases are associated
with downregulated levels of TIPE2 (47–49).
Therefore, low expression levels of TIPE2 may attenuate apoptosis
and promote invasion and metastasis in CRC.
TIPE3 may function differently in different cancers.
A previous study has reported that TIPE3 promotes tumorigenesis by
increasing the levels of phosphatidylinositol 4,5-bisphosphate and
phosphatidylinositol 3,4,5-trisphosphate in the cell plasma
membrane and is expressed at higher levels in lung, esophageal and
cervical cancer, as well as colorectal adenocarcinoma tissues
compared with the adjacent tissues (27). The results of the present study
demonstrated a decrease in TIPE3 mRNA levels (v1 and v2) in the CRC
samples compared with those in the adjacent tissues, and TIPE3
protein (isoform 1) and mRNA expression patterns were not
consistent in five randomly selected pairs of CRC and adjacent
tissue samples. Thus, the mRNA and protein expression patterns and
functions of TIPE3 in CRC remain to be fully elucidated.
As aforementioned, with the exception of TIPE2, TIPE
family members have a range of transcript variants (4,12,28,30).
The results of the present study confirmed that the mRNA levels of
TIPE2 and the targeted transcript variants of TNFAIP8, TIPE1 and
TIPE3 were downregulated in the CRC tissues compared with those in
the adjacent tissues. Considering the structural similarity of the
TIPE family members, we hypothesize that during the development of
CRC, the downregulation of TIPE family member expression levels may
be interrelated. In TCGA data analysis results, with the exception
of TIPE3, TIPE family member mRNA expression levels were all
upregulated in colon mucinous adenocarcinoma compared with those in
colon adenocarcinoma, which suggested that the high malignancy
histological subtypes of colon cancer may affect the transcription
of the TIPE family members. In addition, TNFAIP8 and TIPE3 protein
levels were not consistent with their mRNA expression levels, which
may be explained by one of the following phenomena: i) Different
transcript variants may have different contributions to the overall
protein levels; ii) posttranscriptional regulation may be involved
in the expression of related proteins; and iii) posttranslational
regulation, such as delayed protein degradation, may participate in
maintaining protein stability, which increases TNFAIP8 and TIPE3
protein levels in CRC.
The results of the present study identified a
significant positive linear relationship among the expression
levels of TIPE family members and among the T/A ratios of mRNA
expression for each pair of genes. TIPE1 and TIPE2 exhibited the
highest regression coefficient. For the T/A ratio of mRNA
expression, the strongest linear relationship was between the T/A
ratios of TNFAIP8 (v1 and v3) and TIPE2. The results of TCGA data
analysis also revealed that the levels of TNFAIP8, TIPE1 and TIPE2,
especially those of TNFAIP8 and TIPE2, were positively associated
with each other. As the linear relationship between TIPE2 and the
rest members of the TIPE family was statistically significant,
whether it is the linear relationship between the tumor or the T/A
ratio, it was hypothesized that low TIPE2 expression levels may
cause the downregulation of the mRNA levels of the other three TIPE
family members and serve a predominant role in CRC
tumorigenesis.
A higher degree of tumor differentiation was
associated with lower expression levels of TNFAIP8 mRNA. In male
patients with CRC, TIPE2 expression levels were significantly
higher compared with those in female patients. No significant
differences were observed between the expression levels of TIPE
family members and other clinicopathological features in the CRC
samples. However, the number of samples was limited in the present
study, suggesting that additional samples should be collected for
further research on the associations between the TIPE family
members and patient clinicopathological characteristics. The
present study demonstrated that 4 members of the TIPE family are
closely related to the tumorigenesis and development of CRC. In
addition, these 4 members influenced each other and TIPE2 may serve
a significant role in CRC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mrs. Xiaoli Liu,
Miss Jiarong Chen and Dr Guoliang Yan (School of Medicine, Xiamen
University, Xiamen, China) for advice on statistical analysis.
Funding
This study was supported by the Natural Science
Foundation of Fujian Province (grant nos. 2015J01530 and
2018J01138), Xiamen Science and Technology Project (grant no.
3502Z20209039), The Joint Research Project of Health and Education
of Fujian Province (grant no. WKJ2016-2-17) and XMU Undergraduate
Innovation and Entrepreneurship Training Programs (grant no.
201810384226).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YW, SZ and GZ designed the study. MZ, ZC, YY, AB,
XC, HC, HR, KQ and ZH performed the experiments and analyzed the
data. MZ, ZC and YY contributed to the drafting of the manuscript..
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Committee on Medical
Ethics of Zhongshan Hospital, Xiamen University (Xiamen, China)
[approval no. xmzsyyky (2020-137)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuipers EJ, Grady WM, Lieberman D,
Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ and Watanabe T:
Colorectal cancer. Nat Rev Dis Primers. 1:150652015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schreuders EH, Ruco A, Rabeneck L, Schoen
RE, Sung JJ, Young GP and Kuipers EJ: Colorectal cancer screening:
A global overview of existing programmes. Gut. 64:1637–1649. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lowe JM, Nguyen TA, Grimm SA, Gabor KA,
Peddada SD, Li L, Anderson CW, Resnick MA, Menendez D and Fessler
MB: The novel p53 target TNFAIP8 variant 2 is increased in cancer
and offsets p53-dependent tumor suppression. Cell Death Differ.
24:181–191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Padmavathi G, Banik K, Monisha J, Bordoloi
D, Shabnam B, Arfuso F, Sethi G, Fan L and Kunnumakkara AB: Novel
tumor necrosis factor-α induced protein eight (TNFAIP8/TIPE)
family: Functions and downstream targets involved in cancer
progression. Cancer Lett. 432:260–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bordoloi D, Banik K, Shabnam B, Padmavathi
G, Monisha J, Arfuso F, Dharmarajan A, Mao X, Lim LHK, Wang L, et
al: TIPE family of proteins and its implications in different
chronic diseases. Int J Mol Sci. 19:29742018. View Article : Google Scholar
|
|
7
|
Sun H, Gong S, Carmody RJ, Hilliard A, Li
L, Sun J, Kong L, Xu L, Hilliard B, Hu S, et al: TIPE2, a negative
regulator of innate and adaptive immunity that maintains immune
homeostasis. Cell. 133:415–426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang S, Zhang Y, Wei X, Zhen J, Wang Z,
Li M, Miao W, Ding H, Du P, Zhang W, et al: Expression and
regulation of a novel identified TNFAIP8 family is associated with
diabetic nephropathy. Biochim Biophys Acta. 1802:1078–1086. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
You Z, Ouyang H, Lopatin D, Polver PJ and
Wang CY: Nuclear factor-kappa B-inducible death effector
domain-containing protein suppresses tumor necrosis factor-mediated
apoptosis by inhibiting caspase-8 activity. J Biol Chem.
276:26398–26404. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar D, Whiteside TL and Kasid U:
Identification of a novel tumor necrosis factor-alpha-inducible
gene, SCC-S2, containing the consensus sequence of a death effector
domain of fas-associated death domain-like
interleukin-1beta-converting enzyme-inhibitory protein. J Biol
Chem. 275:2973–2978. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Liu R, Luan YY and Yao YM: Tumor
necrosis factor-α induced protein 8: Pathophysiology, clinical
significance, and regulatory mechanism. Int J Biol Sci. 14:398–405.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niture S, Dong X, Arthur E, Chimeh U,
Niture SS, Zheng W and Kumar D: Oncogenic role of tumor necrosis
factor α-induced protein 8 (TNFAIP8). Cells. 8:92018. View Article : Google Scholar
|
|
13
|
Laliberte B, Wilson AM, Nafisi H, Mao H,
Zhou YY, Daigle M and Albert PR: TNFAIP8: A new effector for
Galpha(i) coupling to reduce cell death and induce cell
transformation. J Cell Physiol. 225:865–874. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Day TF, Mewani RR, Starr J, Li X,
Chakravarty D, Ressom H, Zou X, Eidelman O, Pollard HB, Srivastava
M and Kasid UN: Transcriptome and proteome analyses of TNFAIP8
knockdown cancer cells reveal new insights into molecular
determinants of cell survival and tumor progression. Methods Mol
Biol. 1513:83–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Z, Liang X, Gao L, Ma H, Liu X, Pan
Y, Yan W, Shan H, Wang Z, Chen YH and Ma C: TIPE1 induces apoptosis
by negatively regulating Rac1 activation in hepatocellular
carcinoma cells. Oncogene. 34:2566–2574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye T, Yang B, Wang C, Su C, Luo J, Yang X,
Yu H, Yuan Z, Meng Z and Xia J: TIPE1 impairs stemness maintenance
in colorectal cancer through directly targeting β-catenin.
Carcinogenesis. 41:25–35. 2020.PubMed/NCBI
|
|
17
|
Wu X, Ma Y, Cheng J, Li X, Zheng H, Jiang
L and Zhou R: TIPE1 function as a prognosis predictor and negative
regulator of lung cancer. Oncotarget. 8:78496–78506. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu W, Chen Y, Xie H, Guo Y, Ren D, Li Y,
Jing X, Li D, Wang X, Zhao M, et al: TIPE1 suppresses invasion and
migration through down-regulating Wnt/β-catenin pathway in gastric
cancer. J Cell Mol Med. 22:1103–1117. 2018.PubMed/NCBI
|
|
19
|
Cui J, Zhang G, Hao C, Wang Y, Lou Y,
Zhang W, Wang J and Liu S: The expression of TIPE1 in murine
tissues and human cell lines. Mol Immunol. 48:1548–1555. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen P, Zhang H, Su Z, Wang S and Xu H: In
silico analysis of tumor necrosis factor α-induced protein 8-like-1
(TIPE1) protein. PLoS One. 10:e01341142015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao Q, Zhao M, Dong T, Zhou C, Peng Y,
Zhou X, Fan B, Ma W, Han M and Liu S: Tumor necrosis
factor-α-induced protein-8 like-2 (TIPE2) upregulates p27 to
decrease gastic cancer cell proliferation. J Cell Biochem.
116:1121–1129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang K, Ren Y, Liu Y, Zhang J and He JJ:
Tumor necrosis factor (TNF)-α-induced protein 8-like-2 (TIPE2)
inhibits proliferation and tumorigenesis in breast cancer cells.
Oncol Res. 25:55–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Li X, Liu G, Sun R, Wang L, Wang J
and Wang H: Downregulated TIPE2 is associated with poor prognosis
and promotes cell proliferation in non-small cell lung cancer.
Biochem Biophys Res Commun. 457:43–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gus-Brautbar Y, Johnson D, Zhang L, Sun H,
Wang P, Zhang S, Zhang L and Chen YH: The anti-inflammatory TIPE2
is an inhibitor of the oncogenic Ras. Mol Cell. 45:610–618. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu MW, Liu R, Wu HY, Zhang W, Xia J, Dong
MN, Yu W, Wang Q, Xie FM, Wang R, et al: Protective effect of
Xuebijing injection on D-galactosamine- and
lipopolysaccharide-induced acute liver injury in rats through the
regulation of p38 MAPK, MMP-9 and HO-1 expression by increasing
TIPE2 expression. Int J Mol Med. 38:1419–1432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Zhu T, Liu W, Qu X, Chen Y, Ren
P, Wang Z, Wei X, Zhang Y and Yi F: TIPE2 acts as a negative
regulator linking NOD2 and inflammatory responses in myocardial
ischemia/reperfusion injury. J Mol Med (Berl). 93:1033–1043. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fayngerts SA, Wu J, Oxley CL, Liu X,
Vourekas A, Cathopoulis T, Wang Z, Cui J, Liu S, Sun H, et al:
TIPE3 is the transfer protein of lipid second messengers that
promote cancer. Cancer Cell. 26:465–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuan F, Liu B, Xu Y, Li Y, Sun Q, Xu P,
Geng R, Den G, Yang J, Zhang S, et al: TIPE3 is a regulator of cell
apoptosis in glioblastoma. Cancer Lett. 446:1–14. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang G, Guo C, Zhao H, Pan Z, Zhu F, Zhang
L and Wang Q: TIPE3 differentially modulates proliferation and
migration of human non-small-cell lung cancer cells via distinct
subcellular location. BMC Cancer. 18:2602018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ren XY, Wen X, Li YQ, Zhang J, He QM, Yang
XJ, Tang XR, Wang YQ, Zhang PP, Chen XZ, et al: TIPE3
hypermethylation correlates with worse prognosis and promotes tumor
progression in nasopharyngeal carcinoma. J Exp Clin Cancer Res.
37:2272018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM classification of malignant tumours. 8th edition.
Wiley-Blackwell; Hoboken: 2016
|
|
34
|
Miao Z, Zhao T, Wang Z, Xu Y, Song Y, Wu J
and Xu H: SCC-S2 is overexpressed in colon cancers and regulates
cell proliferation. Tumour Biol. 33:2099–2106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li XM, Su JR, Yan SP, Cheng ZL, Yang TT
and Zhu Q: A novel inflammatory regulator TIPE2 inhibits
TLR4-mediated development of colon cancer via caspase-8. Cancer
Biomark. 14:233–240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang C, Xu W, Meng X, Zhou S, Zhang M and
Cui D: SCC-S2 facilitates tumor proliferation and invasion via
activating Wnt signaling and depressing hippo signaling in
colorectal cancer cells and predicts poor prognosis of patients. J
Histochem Cytochem. 67:65–75. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang C, Chakravarty D, Sakabe I, Mewani
RR, Boudreau HE, Kumar D, Ahmad I and Kasid UN: Role of SCC-S2 in
experimental metastasis and modulation of VEGFR-2, MMP-1, and MMP-9
expression. Mol Ther. 13:947–955. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han Y, Tang Z, Zhao Y, Li Q and Wang E:
TNFAIP8 regulates Hippo pathway through interacting with LATS1 to
promote cell proliferation and invasion in lung cancer. Mol
Carcinog. 57:159–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu S, Li W, Wu Z, Cheng T, Wang P, Li N,
Liang X, Chi M, Zhang S, Ma Y, et al: TNFAIP8 promotes cisplatin
resistance in cervical carcinoma cells by inhibiting cellular
apoptosis. Oncol Lett. 17:4667–4674. 2019.PubMed/NCBI
|
|
40
|
Lou Y and Liu S: The tipe (tnfaip8) family
in inflammation, immunity, and cancer. Mol Immunol. 49:4–7. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao HY, Huo FC, Wang HY and Pei DS:
MicroRNA-9 inhibits the gastric cancer cell proliferation by
targeting TNFAIP8. Cell Prolif. 50:e123312017. View Article : Google Scholar
|
|
42
|
Ha JY, Kim JS, Kang YH, Bok E, Kim YS and
Son JH: Tnfaip8 l1/Oxi-β binds to FBXW5, increasing autophagy
through activation of TSC2 in a Parkinson's disease model. J
Neurochem. 129:527–538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hitomi J, Christofferson DE, Ng A, Yao J,
Degterev A, Xavier RJ and Yuan J: Identification of a molecular
signaling network that regulates a cellular necrotic cell death
pathway. Cell. 135:1311–1323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu Y, Tao M, Wu J, Meng Y, Xu C, Tian Y,
Zhou X, Xiang J, Zhang H and Xie Y: Adenovirus-directed expression
of TIPE2 suppresses gastric cancer growth via induction of
apoptosis and inhibition of AKT and ERK1/2 signaling. Cancer Gene
Ther. 23:98–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang YH, Yan HQ, Wang F, Wang YY, Jiang
YN, Wang YN and Gao FG: TIPE2 inhibits TNF-α-induced hepatocellular
carcinoma cell metastasis via Erk1/2 downregulation and NF-κB
activation. Int J Oncol. 46:254–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cao X, Zhang L, Shi Y, Sun Y, Dai S, Guo
C, Zhu F, Wang Q, Wang J, Wang X, et al: Human tumor necrosis
factor (TNF)-alpha-induced protein 8-like 2 suppresses
hepatocellular carcinoma metastasis through inhibiting Rac1. Mol
Cancer. 12:1492013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang W, Zhang J, Zhao L, Shao J, Cui J,
Guo C, Zhu F, Chen YH and Liu S: TIPE2 protein negatively regulates
HBV-specific CD8+ T lymphocyte functions in humans. Mol Immunol.
64:204–209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Jiang Y, Zhou J, Song W, Li J,
Wang M, Chen J, Xu R, Zhang J, Ma F, et al: Hepatitis C virus
promotes hepatocellular carcinogenesis by targeting TIPE2, a new
regulator of DNA damage response. Tumour Biol. 37:15265–15274.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shi G, Zhao JW, Sun XX, Ma JF, Wang P, He
FC and Ming L: TIPE2 is negatively correlated with tissue factor
and thrombospondin-1 expression in patients with bronchial asthma.
Exp Ther Med. 15:3449–3454. 2018.PubMed/NCBI
|