Introduction
Pancreatic cancer (PC) is one of most lethal types
of malignancy worldwide, with equal incidence rates in men and
women (~2.5%) (1). PC is a
significant health burden worldwide, affecting >411,600
individuals and exhibiting a 5-year survival rate of <5%
(2,3). Although medical technologies have
improved in recent decades, no reliable diagnostic biomarkers are
currently available for the detection of asymptomatic PC at an
early stage. Consequently, PC is usually diagnosed at a late stage,
when the majority of patients exhibit distant metastasis.
Therefore, >80% of patients with PC are not eligible for
surgical resection, which is currently considered to be the most
effective treatment (4).
In humans, the incidence of PC is complex, and risk
factors include a high-protein diet, high-fat diet, smoking and
susceptibility to genetic mutations (5). However, another risk factor is gastric
mucosal injury following Helicobacter pylori (HP) infection
(6). Previous studies have reported
that the pathophysiological actions of H. pylori
colonization, alongside the gastric acidity, potentially modulate
pancreatic tumorigenesis via N-nitrosamine intake-mediated
hyperchlorhydria (6,7). Due to the crucial role that the tumor
microenvironment serves in cancer progression, increasing attention
has been paid to the extracellular matrix, stromal, immune and stem
cells that predominate the PC microenvironment (8).
According to previous studies, H. pylori has
developed mechanisms to co-exist in the harsh gastric
microenvironment by inducing mucosal inflammation and immune
activation (9–11). H. pylori may induce the
secretion of inflammatory cytokines or neuroendocrine mediators by
the infected gastric mucosa (GM) cells (12). In contrast to the direct carcinogenic
impact of metabolites on GM or esophageal mucosa, H. pylori
infection may promote neoplasia and metastasis of distant organs
such as the pancreas by mediating the cellular microenvironment
(13,14).
The results of the aforementioned studies have
provided initial evidence of the incidence of H. pylori
infection-associated PC; however, the biological mechanisms and key
factors involved in this process remain poorly understood. In order
to improve the current understanding, the present study aimed to
identify the factors associated with the potential pathogenicity of
H. pylori in PC tumorigenesis and metastasis.
Materials and methods
Cell culture
The human PC cell line PANC-1 and the human normal
ductal epithelial cell line HPDE6-C7 were purchased from the Cell
Resource Center of Shanghai Institute of Biochemistry and Cell
Biology, The Chinese Academy of Sciences. The cells were cultured
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and maintained at 37°C with 5% CO2.
Patient studies
Between June and December 2019, a total of 32
inpatient volunteers, among whom 20 patients had chronic gastritis
without PC (10 HP− and 10 HP+) and 12 had PC
(6 HP− and 6 HP+), were recruited at The
Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical
University (Changzhou, China) (Table
I). The patients were diagnosed with H. pylori
infection, PC or both. H. pylori infection was diagnosed by
carbon 13 breath test and gastroscopy. GM injury was determined by
gastroscopy. The diagnosis and tumor stage of PC were systemically
assessed using imaging, surgery and biopsy. The study was approved
by the Ethics Committee of the Affiliated Changzhou No. 2 People's
Hospital of Nanjing Medical University [approval no: (2019) KYO
073-01], and all patients provided informed consent. PC and
adjacent tissue samples (>5 cm from the tumor) were obtained
during surgery. Blood samples were obtained after diagnosis. All
samples (20 GM, 12 tumor, 12 adjacent tissue and 12 blood samples)
were stored at −80°C and preprocessed for further research.
 | Table I.Clinicopathological characteristics
of patients included in the present study. |
Table I.
Clinicopathological characteristics
of patients included in the present study.
|
| Gastritis | PC |
|---|
|
|
|
|
|---|
| Characteristic | HP+ | HP− | HP+ | HP− |
|---|
| Total, n | 10 | 10 | 6 | 6 |
| Sex, n |
|
|
|
|
|
Male | 6 | 5 | 4 | 4 |
|
Female | 4 | 5 | 2 | 2 |
| Age, years | 39.56±4.64 | 40.32±5.03 | 53.87±4.23 | 56.42±8.53 |
| GM injury, n
(%) | 8 (80%) | 7 (70%) | 2 (33%) | 2 (33%) |
| Lymph node
invasion, n (%) | N/A | N/A | 2 (33%) | 2 (33%) |
| Distant metastasis,
n (%) | N/A | N/A | 1 (16%) | 0 (0%) |
Microarrays
Microarray datasets of H. pylori-infected GM
[dataset nos. GSE6143 (15),
GSE47797 (16), GSE70394 (17) and GSE5081 (18)] and PC [dataset nos. GSE85991
(19), GSE27890 and GSE55643
(20)] were downloaded from the Gene
Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). By referring to the
annotation information, the probe IDs were converted into the
corresponding gene symbols. Basic information of the microarray
datasets is displayed in Table
II.
 | Table II.Microarray datasets of patients with
Helicobacter pylori infection and pancreatic cancer. |
Table II.
Microarray datasets of patients with
Helicobacter pylori infection and pancreatic cancer.
| First author,
year | ID | Platform | Sample type | Sample
character | n | (Refs.) |
|---|
| Wen et al,
2007 | GSE6143 | GPL193 | Gastric mucosa | H.
pylori+ | 9 | (15) |
|
|
|
|
| Normal | 8 |
|
| Hanada et
al, 2014 | GSE47797 | GPL13497 | Gastric mucosa | H.
pylori+ | 4 | (57) |
|
|
|
|
| Normal | 4 |
|
| Costa et al,
2016 | GSE70394 | GPL6480 | Gastric mucosa | H.
pylori+ | 3 | (17) |
|
|
|
|
| Normal | 3 |
|
| Galamb et
al, 2008 | GSE5081 | GPL570 | Injured gastric
mucosa | H.
pylori+ | 8 | (18) |
|
|
|
|
| H.
pylori− | 8 |
|
| Koutsioumpa et
al, 2019 | GSE85991 | GPL570 | Pancreatic
cancer | Tumor | 2 | (19) |
|
|
|
|
| Normal | 2 |
|
| Chinaranagari et
al, | GSE27890 | GPL570 | Pancreatic
cancer | Tumor | 4 | (58) |
| 2015 |
|
|
| Normal | 4 |
|
| Lunardi et
al, 2014 | GSE55643 | GPL6480 | Pancreatic
cancer | Tumor | 45 | (20) |
|
|
|
|
| Normal | 8 |
|
Identification and overlap of
differentially expressed genes (DEGs)
The DEGs between the tumor and control groups were
identified using the GEO2R web tool (http://www.ncbi.nlm.nih.gov/geo/geo2r) by comparing
the GEO microarray datasets. Probes without corresponding gene
symbols or genes with redundant probe sets were eliminated or
processed using the DAVID online tool (https://david.ncifcrf.gov/), respectively. A |log
(fold-change)| >1 and P<0.01 were selected to identify
statistically significant differences. The overlap analysis of DEGs
from the H. pylori-infected GM and PC datasets was conducted
and displayed in Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/). In
addition, the mRNA expression levels of the DEGs were presented in
heatmaps.
Kyoto encyclopedia genes and genomes
(KEGG) signaling pathway and gene ontology (GO) functional term
enrichment analyses
The KEGG Orthology Based Annotation System (KOBAS
3.0; http://kobas.cbi.pku.edu.cn/kobas3) web server was
used for gene/protein functional annotation (Annotation module) and
functional set enrichment (Enrichment module) in the present study.
GO functional term enrichment analysis bioinformatics annotation
tool was used to determine gene functions and perform biological
analysis (21). KEGG signaling
pathway enrichment analysis was used to illustrate gene functions
and biological pathways (22).
P<0.01 was considered to indicate a statistically significant
difference.
Data acquisition from The Cancer
Genome Atlas (TCGA) database
Gene expression data of PC were acquired from TCGA
(https://portal.gdc.cancer.gov) (23). Overall survival (OS) and disease-free
survival (DFS) analyses of patients grouped into high/low mucin 4
(MUC4) mRNA expression groups based on the median expression levels
were performed using the Gene Expression Profiling Interactive
Analysis (GEPIA) online database (http://gepia.cancer-pku.cn) (24). The associations between the
expression levels and tumor grades and the meta-analysis of the
data from three previous studies (25–27) were
analyzed using the Oncomine database (http://www.oncomine.com) (28).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the PC cell line and
tissues, as well as the corresponding controls, using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The total RNA was reverse-transcribed into cDNA using a
PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.). qPCR
was subsequently performed using a SYBR® Premix Ex Taq
kit (Takara Biotechnology Co., Ltd.) on a 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The reverse
transcription was performed at 42°C for 1 h, followed by 95°C for 5
min. The thermocycling conditions were as follows: Initial
denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C
for 15 sec and at 60°C for 1 min. The expression levels were
calculated using the 2−ΔΔCq method (29). The primer pairs used were as follows:
MUC4 forward, 5′-GACTTGGAGCTCTTTGAGAATGG-3′ and reverse,
5′-TGCAATGGCAGACCACAGTCC-3′; GAPDH forward,
5′-ATCATCCCTGCCTCTACTGG-3′ and reverse,
5′-GTCAGGTCCACCACTGACAC-3′.
Multilevel logistic regression
analysis
Basic information of 22 patients (12
PC+HP+/− and 10 PC−HP−)
was obtained from the medical records. The patients were grouped by
age (>/<50 years), sex (male/female), MUC4 expression, HP
infection (HP+/HP−) and PC diagnosis
(PC+/PC−). Multilevel logistic regression
analysis was made with grouped patients.
Statistical analysis
Data are presented as the mean ± SEM. SPSS 17.0
(SPSS statistics, IBM Inc.) was used for statistical analysis.
GraphPad Prism 6.0 software (GraphPad Software, Inc.) was used to
generate the receiver operating characteristic (ROC) curve and
perform multilevel logistic regression analysis. Statistical
differences were determined using one-way ANOVA with the Bonferroni
post hoc test for multiple comparisons. Correlation analysis was
performed using the Spearman correlation coefficient. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of overlapping DEGs in
H. pylori-infected GM and PC
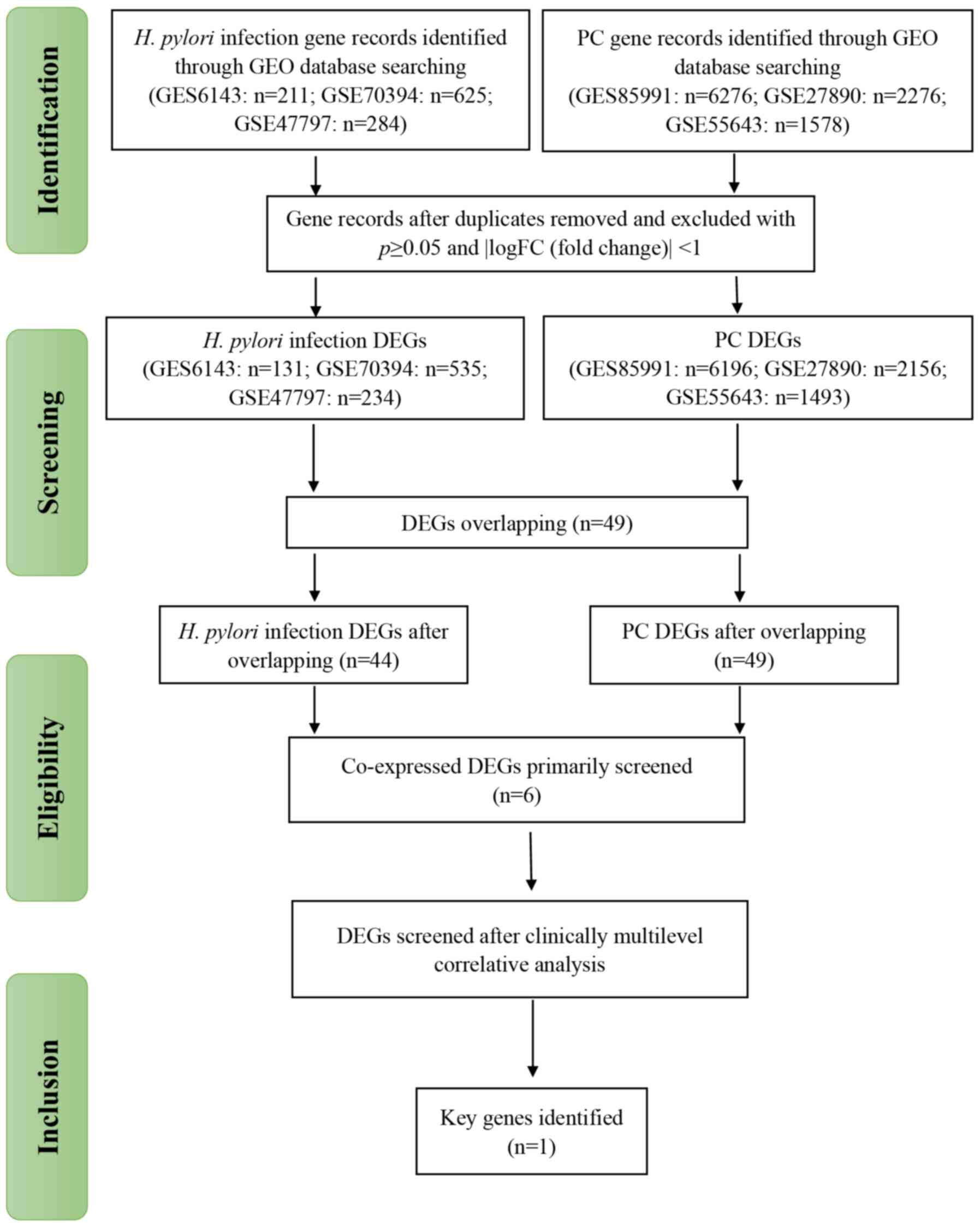
A total of 32 patients and seven microarray datasets
were included in the present study (Fig.
1). Following standardization and processing on the GEO
database, DEGs were selected from three H. pylori-infected
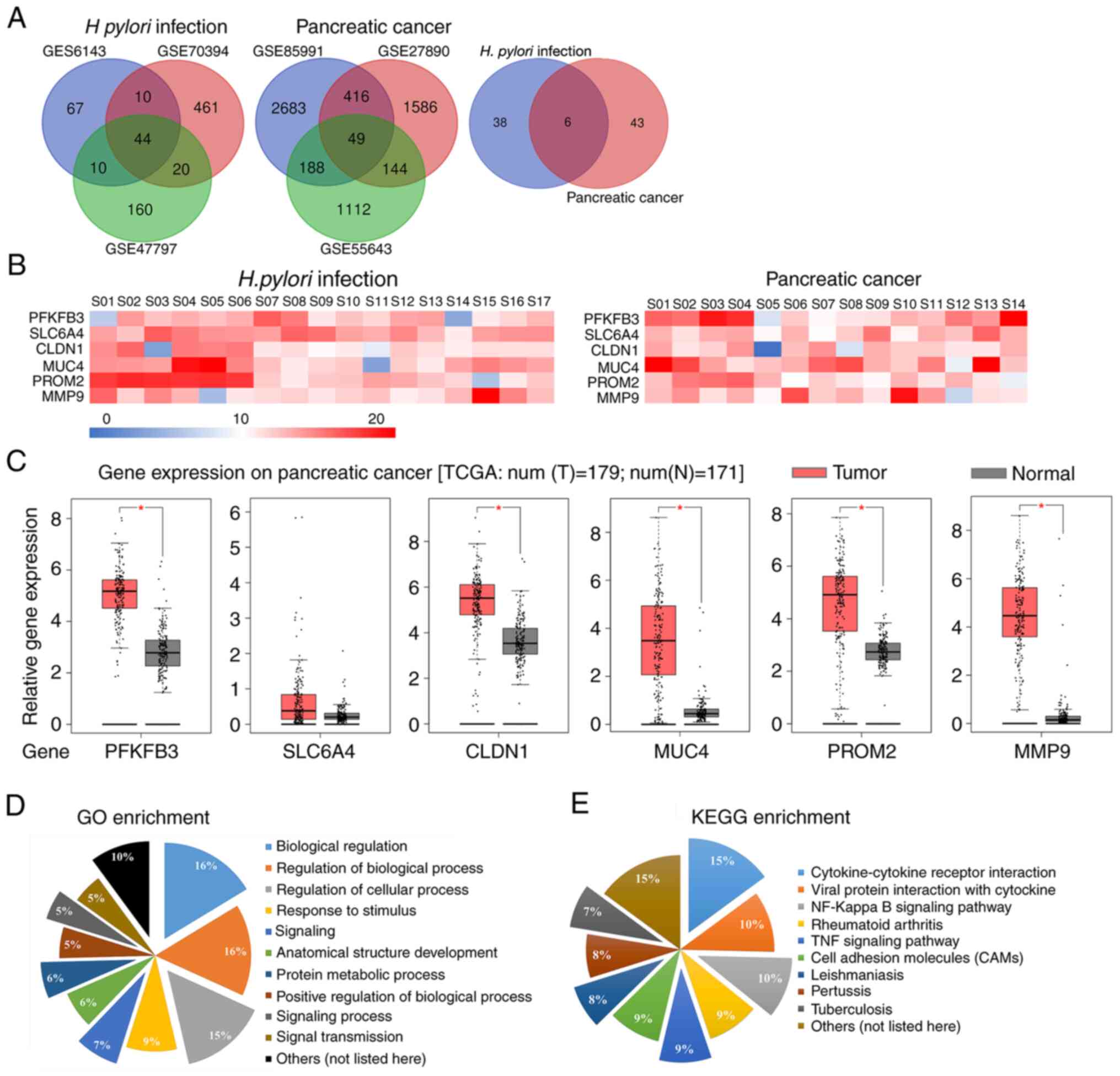
GM and three PC datasets. Using overlap analysis, six genes were
selected among the 44 and 49 DEGs from H. pylori-infected GM
and PC datasets, respectively, which were 6-phosphofructo-2-kinase
(PFKFB3), sodium-dependent serotonin transporter (SLC6A4),
claudin-1 (CLDN1), MUC4, prominin-2 (PROM2) and matrix
metalloproteinase 9 (MMP9) (Fig.
2A). The functional annotation of these six genes is presented
in Table III. The relative mRNA
expression levels of the genes that were significantly upregulated
are presented in heat maps in Fig.
2B. For further verification, gene expression data from
patients with PC were downloaded from TCGA database. As presented
in Fig. 2C, TCGA data analysis
revealed that the expression levels PFKFB3, CLDN1, MUC4, PROM2 and
MMP9 were significantly upregulated in the tumor tissues compared
with those in the normal tissues (P<0.05). However, the
expression levels of SLC6A4 were not significantly different
between the tumor and normal tissues (P>0.05). These enriched
genes were associated with a number of GO functional terms, such as
‘biological regulation’, ‘regulation of biological process’ and
‘regulation of cellular process’, and KEGG signaling pathways,
including ‘cytokine-cytokine receptor interaction’, ‘viral protein
interaction with cytokine’ and ‘NF-kappa B signaling pathway’
(Fig. 2D and E).
 | Table III.Full names and functions of the six
key differentially expressed genes identified in the present
study. |
Table III.
Full names and functions of the six
key differentially expressed genes identified in the present
study.
| Gene symbol | Gene name | Functions |
|---|
| PFKFB3 |
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase
3 | Required for cell
cycle progression and prevention of apoptosis; functions as a
regulator of cyclin-dependent kinase 1, linking glucose metabolism
to tumor cell proliferation and survival. |
| SLC6A4 | Solute carrier
family 6 member 4 | A target of
psychomotor stimulants, such as amphetamines and cocaine; a member
of the sodium: neurotransmitter symporter family; a repeat length
polymorphism in the promoter of this gene has been demonstrated to
affect the rate of serotonin uptake. |
| CLDN1 | Claudin 1 | A member of the
claudin family; an integral membrane protein; a component of tight
junction strands, serving as a physical barrier to prevent solutes
and water from passing freely through the paracellular space. |
| MUC4 | Mucin 4, cell
surface associated | A major constituent
of mucus; serves important roles in the protection of the
epithelial cells; has been implicated in epithelial renewal and
differentiation. |
| PROM2 | Prominin 2 | A member of the
prominin family of pentaspan membrane glycoproteins; may be
involved in the organization of plasma membrane microdomains. |
| MMP9 | Matrix
metallopeptidase 9 | Involved in the
breakdown of extracellular matrix in normal physiological
processes, such as embryonic development, reproduction and tissue
remodeling, as well as in disease processes, such as arthritis and
tumor metastasis. |
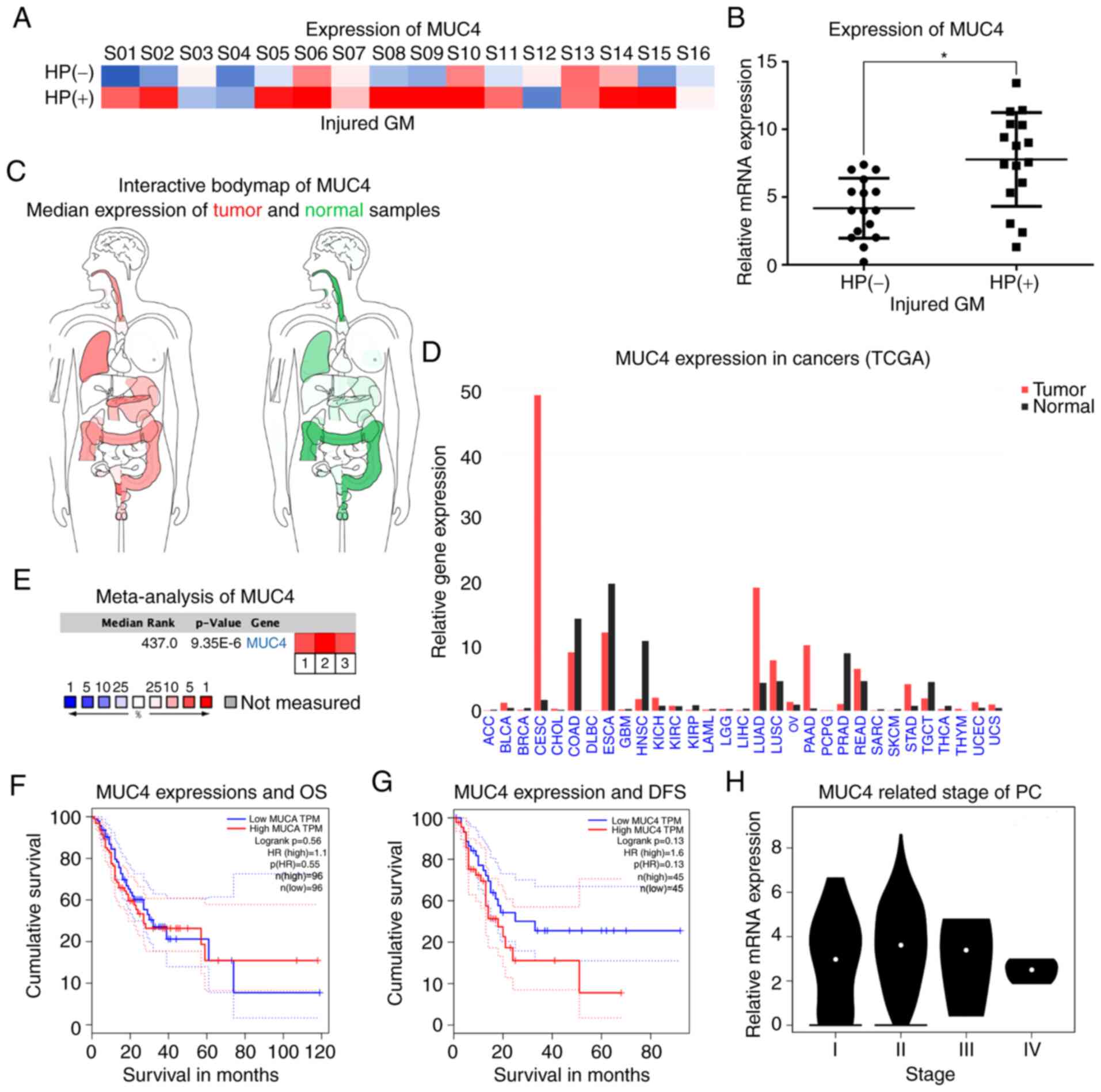
MUC4 contributes to H. pylori
infection-associated PC
The GSE5081 dataset was used to compare the whole
genome gene expression profile between patients with HP+
and HP− GM injury. Among the six selected genes, MUC4,
MMP9 and CLDN1 were differentially expressed in the two groups.
Notably, compared with normal GM tissues, MUC4 expression levels
were upregulated in HP+ and downregulated in
HP− GM injured tissues (Fig.
3A and B). These results indicated that MUC4 may be a potential
factor involved in the biological processes of the H.
pylori-infected GM. For further analysis, the expression
profile of MUC4 in human tissues and the associated clinical
information were produced using the GEPIA webtool. The human body
maps illustrated that MUC4 expression levels were upregulated in
pancreatic adenocarcinoma, cervical squamous cell carcinoma and
endocervical adenocarcinoma, lung adenocarcinoma (LUAD) and stomach
adenocarcinoma (STAD), but downregulated in prostate adenocarcinoma
and head & neck squamous cell carcinoma compared with those the
corresponding normal tissues (Fig. 3C
and D). In addition, Oncomine-based meta-analysis indicated
that the expression levels of MUC4 were significantly upregulated
in cancer tissues compared with those in normal tissues in the
three PC datasets (Fig. 3E).
However, as presented in Fig. 3F,
MUC4 was not associated with patient OS rates in PC. A weak
association was identified between low MUC4 levels and DFS rates;
however, this result was not significant (P>0.05; Fig. 3G). In addition, although MUC4
expression levels were upregulated at each tumor stage, no
significant differences were identified among the four stages
(P>0.05; Fig. 3H).
The human PC cell line PANC-1, 20 patients with
chronic gastritis without PC and 12 patients with PC with or
without H. pylori infection were used to verify the trends
observed in MUC4 expression. Blood and tissue samples (GM, tumor
and adjacent tissues) were collected and analyzed by RT-qPCR. The
expression levels of MUC4 were upregulated in HP+ GM
compared with those in HP− GM (P<0.05; Fig. 4A). In addition, compared with the
normal tissues and cell lines, the expression levels of MUC4 were
upregulated in both PC tissues and PANC-1 cells (P<0.05;
Fig. 4B). The results of the blood
tests revealed that patients in the HP+PC−
group exhibited markedly upregulated MUC4 expression levels
compared with those in the other groups. (P<0.05; Fig. 4C). These results indicated that the
expression levels of MUC4 were significantly upregulated in H.
pylori-infected GM and PC.
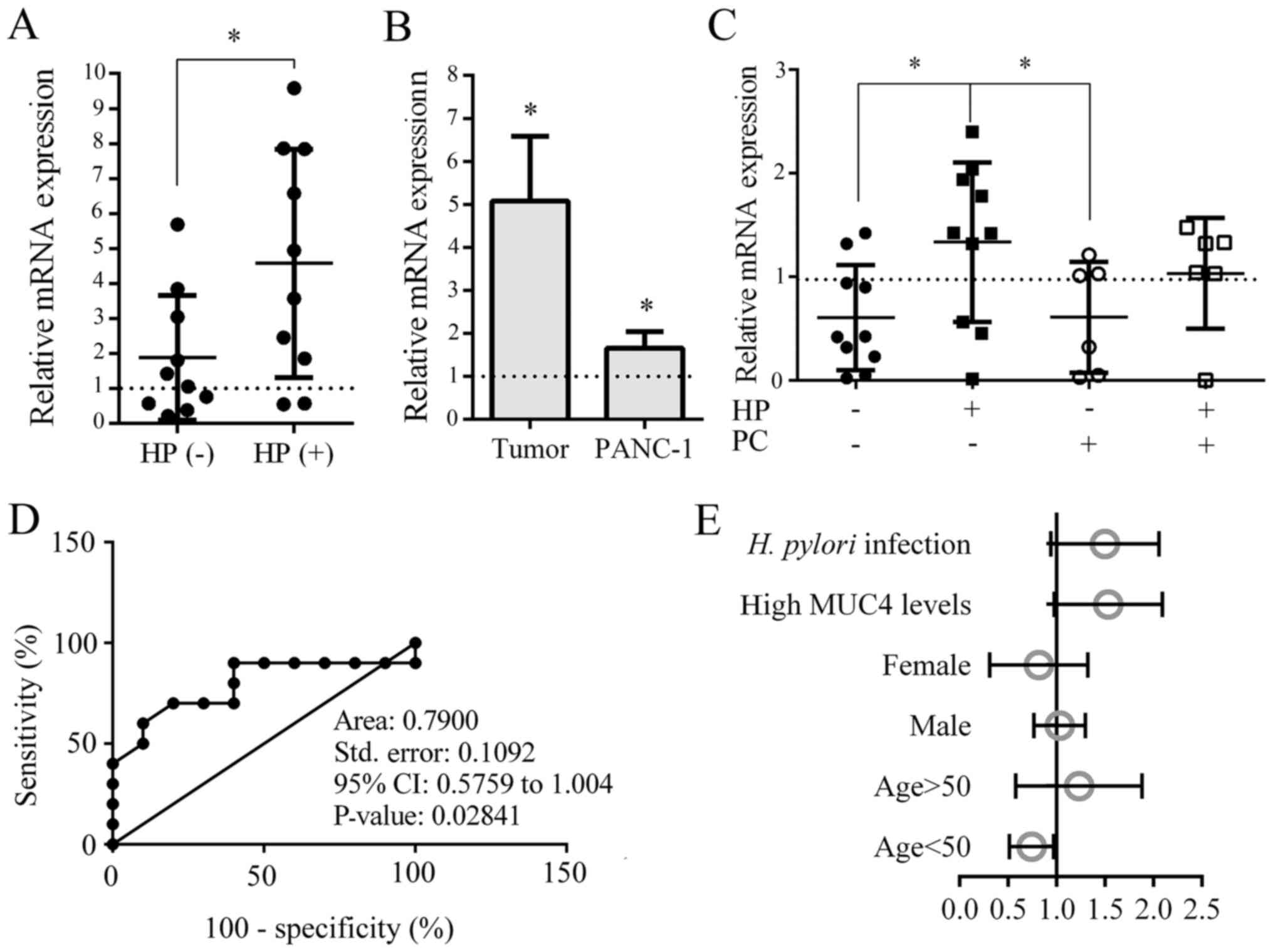 | Figure 4.Verification of MUC4 expression
levels in the GM, PC and blood. (A-C) mRNA expression levels of
MUC4 in (A) injured GMs with or without H. pylori infection,
(B) tumor tissues (relative to normal tissues) and PC cell lines
(relative to a human normal ductal epithelial cell line), and (C)
blood samples from patients with H. pylori infection or PC.
(D) In a total of 26 patients, a ROC curve was generated to assess
the sensitivity and specificity in HP-infection related PC of MUC4.
(E) Multilevel logistic regression analysis of age, sex, high MUC4
levels and HP infection vs. PC status was conducted. *P<0.01.
MUC4, mucin 4; GM, gastric mucosa; PC, pancreatic cancer; HP/H.
pylori, Helicobacter pylori; ROC, receiver operating
characteristic. |
To assess the sensitivity of MUC4, 26 patients were
divided into very low-risk (HP−PC−), low-risk
(HP+PC−) and high-risk
(HP+PC+) groups. A ROC curve was generated,
and the results revealed that MUC4 exhibited moderate sensitivity
for the identification of H. pylori-related PC (Fig. 4D). Multilevel logistic regression
analysis of the effects of patient age, sex, MUC4 expression levels
and H. pylori infection in PC was conducted, which revealed
that high MUC4 expression levels and the presence of an H.
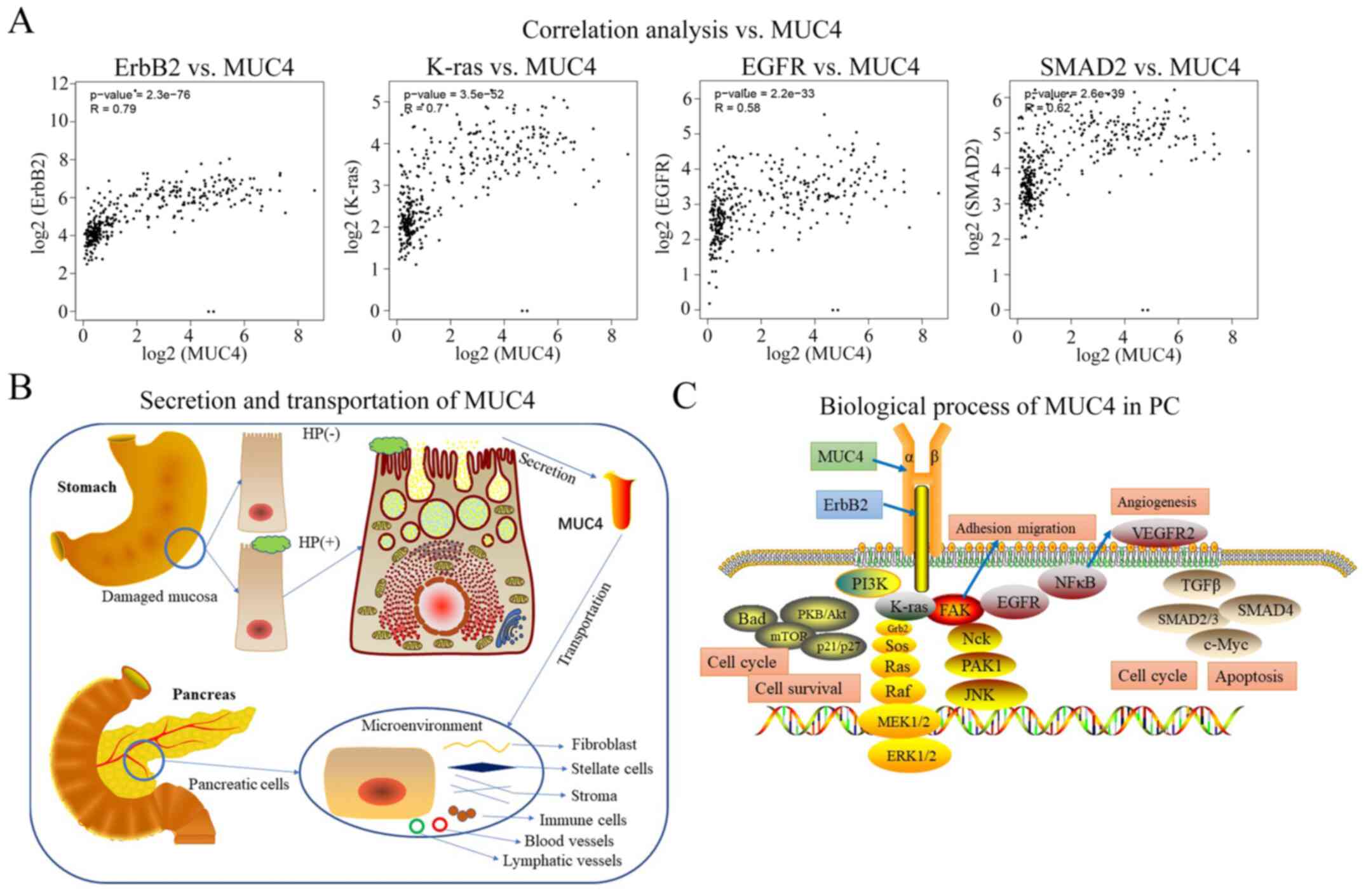
pylori infection were positively associated with PC (Fig. 4E). Correlation analyses of multiple
genes co-expressed with MUC4 were conducted using TCGA data, and
four tumorigenic factors of PC, including receptor tyrosine-protein
kinase erbB-2 (ErbB2), K-Ras, EGFR and SMAD2, were identified to be
moderately positively correlated with MUC4 (Fig. 5A).
Discussion
The pancreas is an important retroperitoneal organ
with exocrine and endocrine functions (30). Non-endocrine pancreatic tumors are
usually malignant and have different histological features that can
be classified as a ductal adenocarcinoma, cystadenocarcinoma or
another type of malignant tumor, such as sarcoma and small cell
carcinoma (31). In recent decades,
the prognosis of PC has remained dismal, with limited improvements
achieved for the diagnosis and treatment of the disease. Despite
advances in surgery and comprehensive treatment regimens, the poor
outcome of patients with PC remains unchanged (32). The results of epidemiological studies
have indicated that age, obesity, smoking, long-term alcohol use,
chronic pancreatitis and family history are all risk factors for PC
(31). With the development of
genomics and bioinformatics tools, several genetic mutations have
been identified to be involved in PC development; for example,
BRCA2 and K-Ras mutations, and loss of p16, SMAD4 or p53 function
occur in the epithelium of precursor pancreatic diseases, which
markedly accelerate the progression of tumorigenesis (33–35).
The stomach serves several exocrine functions that
are similar to the pancreas in the gastrointestinal system, such as
producing and secreting various polypeptides and proteins (36–38).
H. pylori, recognized as the most common gastroduodenal
infection, typically colonizes the human stomach (15). Chronic H. pylori infection has
been associated with an increased risk of several types of disease
or pathological lesions, including gastritis, peptic ulcers,
dysplasia, neoplasia, mucosa-associated lymphoid tissue lymphoma
and invasive gastric adenocarcinoma (39–41). In
addition, a potential role of H. pylori infection in several
extragastric diseases, such as neurodegenerative, cardiovascular,
hepatobiliary, pancreatic and colorectal diseases, has been
reported (7). Epidemiological and
cohort studies in large numbers of PC cases have verified that
H. pylori infection serves a role in the pathogenesis of
chronic and autoimmune pancreatitis, diabetes and PC. Subsequent
studies have suggested that H. pylori infection may affect
the pancreatic physiology and contribute to the tumorigenesis of PC
(17). A number of factors produced
in response to infection, including ammonia, lipopolysaccharides
and inflammatory cytokines, have been demonstrated to induce
progressive damage in the pancreas (8). Maisonneuve and Lowenfels and
Yadav amd Lowenfels (12,42) have reported that similar pathological
manifestations to H. pylori-infected gastric tissues are
also observed in PC. Other previous studies have indicated that
increases in gastrin levels and decreases in somatostatin levels
are involved in the mechanisms associated with H. pylori
infection and PC (43). In addition,
tissue inflammation, genomic DNA damage and various cytokines,
including NF-κB, activator protein-1 and serum response elements,
have been reported to contribute to the malignant transformation of
pancreatic cells (5,44).
MUC4 has been identified to be associated with H.
pylori infection. MUC4 is a major constituent of the mucus, the
viscous secretion that covers epithelial surfaces, such as those in
the trachea, colon and cervix, composed of highly glycosylated
proteins called mucins (45).
Multiple previous studies have reported a role of MUC4 in the
aggressiveness of PC through its ability to enhance tumor growth
and metastasis (46–49). The results of the present study
demonstrated that the expression levels of MUC4 were also
upregulated in LUAD and STAD compared with those in normal tissues,
suggesting that the role of MUC4 may depend on the type of
cancer.
Although MUC4 levels were upregulated in H.
pylori infection-associated GM and PC samples compared with
those in normal tissues, there is currently lack of evidence of an
association between the two in existing studies. Following the
analysis of MUC4 expression levels in patient blood and tissues,
cytokine transportation and biological networks were hypothetically
constructed and visualized in the present study (Fig. 5B and C). The results of the present
study revealed that MUC4 expression levels were upregulated in
patients with H. pylori infection regardless of GM damage
compared with those in normal tissues. Thus, H. pylori
infection potentially promotes the intracellular transcription and
the extracellular secretion of MUC4 in GM cells. The circulatory
and immune systems and the digestive tract appear to be main routes
for MUC4 transportation (46,50).
Generally, the microenvironment of PC cells consists of vessels,
immune cells, stroma, stem cells, stellate cells and fibroblasts
(8,51,52).
MUC4 is transported to the surrounding microenvironment, where it
potentially participates in the cellular biological processes of
PC. MUC4 has been reported to serve as an intramembrane ligand and
co-mediate with ErbB2 (53,54). Correlation analyses in the present
study revealed that the expression levels of PC tumorigenic factors
(ErbB2, K-Ras, EGFR and SMAD2) were associated with those of MUC4
in PC tissues. A previous study has concluded that the MUC4/ErbB2
complex contributes to processes such as differentiation,
proliferation, cell cycle, angiogenesis and migration by regulating
the EGF, K-Ras, PI3K/AKT and TGF-β signaling pathways (48,55).
Therefore, MUC4 produced by the H. pylori-infected GM may be
involved in mediating cell survival and migration by regulating the
PC cell microenvironment.
MUC4 is considered to serve a crucial role in the
tumorigenesis and metastasis of PC (47,56). In
the present study, MUC4 was found to be upregulated in H.
pylori-infected GM. Nevertheless, the underlying mechanisms of
MUC4 between H. pylori infection and PC have received
limited attention, especially regarding the cellular transport
mechanism. Thus, further studies are required. However, limited
samples of H. pylori-infected GM are currently available in
public databases, which is a limitation of the present study.
Additional representative datasets with large samples are needed in
future studies.
In conclusion, the results of the present study
revealed that MUC4 may be a cytokine involved in the pathogenesis
of PC and may represent a novel treatment target. Based on these
results, it may be speculated that silencing MUC4 may reduce the
tumorigenic risk of H. pylori infection in PC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Jiangsu Natural
Science Foundation (grant no. BK20181155), the Nanjing Medical
University (grant no. 2017NJMU043), the Changzhou Department of
Health (grant no. QN201711) and the Changzhou No. 2 People's
Hospital (grant no. 2018K003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YG, LT and HL designed the study. HY and YF
collected tissue samples and basic information from the patients
and carried out statistical analyses. YL and YW carried out the
bioinformatics analysis. SC performed the experiments. HL wrote and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical
University [No: (2019) KYO 073-01]. All blood samples and tissues
involved in this study were collected with informed consent from
the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PC
|
pancreatic cancer
|
|
GM
|
gastric mucosa
|
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
differentially expressed genes
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GO
|
Gene Ontology
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
LUAD
|
lung adenocarcinoma
|
|
STAD
|
stomach adenocarcinoma
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar
|
|
3
|
Ilic M and Ilic I: Epidemiology of
pancreatic cancer. World J Gastroenterol. 22:9694–9705. 2016.
View Article : Google Scholar
|
|
4
|
Chu LC, Goggins MG and Fishman EK:
Diagnosis and detection of pancreatic cancer. Cancer J. 23:333–342.
2017. View Article : Google Scholar
|
|
5
|
Ansari D, Tingstedt B, Andersson B,
Holmquist F, Sturesson C, Williamsson C, Sasor A, Borg D, Bauden M
and Andersson R: Pancreatic cancer: Yesterday, today and tomorrow.
Future Oncol. 12:1929–1946. 2016. View Article : Google Scholar
|
|
6
|
Risch HA, Lu L, Kidd MS, Wang J, Zhang W,
Ni Q, Gao YT and Yu H: Helicobacter pylori seropositivities
and risk of pancreatic carcinoma. Cancer Epidemiol Biomarkers Prev.
23:172–178. 2014. View Article : Google Scholar
|
|
7
|
Rabelo-Goncalves EM, Roesler BM and
Zeitune JM: Extragastric manifestations of Helicobacter
pylori infection: Possible role of bacterium in liver and
pancreas diseases. World J Hepatol. 7:2968–2979. 2015. View Article : Google Scholar
|
|
8
|
Dougan SK: The pancreatic cancer
microenvironment. Cancer J. 23:321–325. 2017. View Article : Google Scholar
|
|
9
|
Rojas A, Araya P, Gonzalez I and Morales
E: Gastric tumor microenvironment. Adv Exp Med Biol. 1226:23–35.
2020. View Article : Google Scholar
|
|
10
|
Sethi V, Vitiello GA, Saxena D, Miller G
and Dudeja V: The role of the microbiome in immunologic development
and its implication for pancreatic cancer immunotherapy.
Gastroenterology. 156:2097–2115.e2. 2019. View Article : Google Scholar
|
|
11
|
Wessler S, Krisch LM, Elmer DP and Aberger
F: From inflammation to gastric cancer-the importance of
Hedgehog/GLI signaling in Helicobacter pylori-induced
chronic inflammatory and neoplastic diseases. Cell Commun Signal.
15:152017. View Article : Google Scholar
|
|
12
|
Maisonneuve P and Lowenfels AB: Risk
factors for pancreatic cancer: A summary review of meta-analytical
studies. Int J Epidemiol. 44:186–198. 2015. View Article : Google Scholar
|
|
13
|
Chen L, Xu W, Lee A, He J, Huang B, Zheng
W, Su T, Lai S, Long Y, Chu H, et al: The impact of Helicobacter
pylori infection, eradication therapy and probiotic
supplementation on gut microenvironment homeostasis: An open-label,
randomized clinical trial. EBioMedicine. 35:87–96. 2018. View Article : Google Scholar
|
|
14
|
Noto JM and Peek RM Jr: The gastric
microbiome, its interaction with Helicobacter pylori, and
its potential role in the progression to stomach cancer. PLoS
Pathog. 13:e10065732017. View Article : Google Scholar
|
|
15
|
Wen S, Velin D, Felley CP, Du L, Michetti
P and Pan-Hammarstrom Q: Expression of Helicobacter pylori
virulence factors and associated expression profiles of
inflammatory genes in the human gastric mucosa. Infect Immun.
75:5118–5126. 2007. View Article : Google Scholar
|
|
16
|
Echizen K, Horiuchi K, Aoki Y, Yamada Y,
Minamoto T, Oshima H and Oshima M: NF-κB-induced NOX1 activation
promotes gastric tumorigenesis through the expansion of
SOX2-positive epithelial cells. Oncogene. 38:4250–4263. 2019.
View Article : Google Scholar
|
|
17
|
Costa AM, Ferreira RM, Pinto-Ribeiro I,
Sougleri IS, Oliveira MJ, Carreto L, Santos MA, Sgouras DN,
Carneiro F, Leite M and Figueiredo C: Helicobacter pylori
activates matrix metalloproteinase 10 in gastric epithelial cells
via EGFR and ERK-mediated pathways. J Infect Dis. 213:1767–1776.
2016. View Article : Google Scholar
|
|
18
|
Galamb O, Gyorffy B, Sipos F, Dinya E,
Krenács T, Berczi L, Szõke D, Spisák S, Solymosi N, Németh AM, et
al: Helicobacter pylori and antrum erosion-specific gene
expression patterns: The discriminative role of CXCL13 and VCAM1
transcripts. Helicobacter. 13:112–126. 2008. View Article : Google Scholar
|
|
19
|
Koutsioumpa M, Hatziapostolou M,
Polytarchou C, Tolosa EJ, Almada LL, Mahurkar-Joshi S, Williams J,
Tirado-Rodriguez AB, Huerta-Yepez S, Karavias D, et al: Lysine
methyltransferase 2D regulates pancreatic carcinogenesis through
metabolic reprogramming. Gut. 68:1271–1286. 2019. View Article : Google Scholar
|
|
20
|
Lunardi S, Jamieson NB, Lim SY, Griffiths
KL, Carvalho-Gaspar M, Al-Assar O, Yameen S, Carter RC, McKay CJ,
Spoletini G, et al: IP-10/CXCL10 induction in human pancreatic
cancer stroma influences lymphocytes recruitment and correlates
with poor survival. Oncotarget. 5:11064–11080. 2014. View Article : Google Scholar
|
|
21
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar
|
|
22
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–103, 119–128, 244–252. 2002. View Article : Google Scholar
|
|
23
|
Tomczak K, Czerwinska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. (45W):
W98–W102. 2017. View Article : Google Scholar
|
|
25
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar
|
|
26
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar
|
|
27
|
Wagner KW, Punnoose EA, Januario T,
Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF,
Totpal K, et al: Death-receptor O-glycosylation controls tumor-cell
sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med.
13:1070–1077. 2007. View
Article : Google Scholar
|
|
28
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Bastidas-Ponce A, Scheibner K, Lickert H
and Bakhti M: Cellular and molecular mechanisms coordinating
pancreas development. Development. 144:2873–2888. 2017. View Article : Google Scholar
|
|
31
|
Storz P and Crawford HC: Carcinogenesis of
pancreatic ductal adenocarcinoma. Gastroenterology. 158:2072–2081.
2020. View Article : Google Scholar
|
|
32
|
The Lancet Gastroenterology Hepatology:
Pancreatic cancer: How can we tackle the lack of progress? Lancet
Gastroenterol Hepatol. 2:732017. View Article : Google Scholar
|
|
33
|
Jin G, Hong W, Guo Y, Bai Y and Chen B:
Molecular mechanism of pancreatic stellate cells activation in
chronic pancreatitis and pancreatic cancer. J Cancer. 11:1505–1515.
2020. View Article : Google Scholar
|
|
34
|
Kenner BJ: Early detection of pancreatic
cancer: The role of depression and anxiety as a precursor for
disease. Pancreas. 47:363–367. 2018. View Article : Google Scholar
|
|
35
|
Malats N, Molina-Montes E and La Vecchia
C: Genomics in primary and secondary prevention of pancreatic
cancer. Public Health Genomics. 20:92–99. 2017. View Article : Google Scholar
|
|
36
|
Hunt RH, Camilleri M, Crowe SE, El-Omar
EM, Fox JG, Kuipers EJ, Malfertheiner P, McColl KE, Pritchard DM,
Rugge M, et al: The stomach in health and disease. Gut.
64:1650–1668. 2015. View Article : Google Scholar
|
|
37
|
Norris AW and Uc A: A novel
stomach-pancreas connection: More than physical. EBioMedicine.
37:25–26. 2018. View Article : Google Scholar
|
|
38
|
Holst JJ, Knuhtsen S, Jensen SL,
Fahrenkrug J, Larsson LI and Nielsen OV: Interrelation of nerves
and hormones in stomach and pancreas. Scand J Gastroenterol Suppl.
82:85–99. 1983.
|
|
39
|
Wang F, Meng W, Wang B and Qiao L:
Helicobacter pylori-induced gastric inflammation and gastric
cancer. Cancer Lett. 345:196–202. 2014. View Article : Google Scholar
|
|
40
|
Wroblewski LE and Peek RM Jr:
Helicobacter pylori, cancer, and the gastric microbiota. Adv
Exp Med Biol. 908:393–408. 2016. View Article : Google Scholar
|
|
41
|
Bravo D, Hoare A, Soto C, Valenzuela MA
and Quest AF: Helicobacter pylori in human health and
disease: Mechanisms for local gastric and systemic effects. World J
Gastroenterol. 24:3071–3089. 2018. View Article : Google Scholar
|
|
42
|
Yadav D and Lowenfels AB: The epidemiology
of pancreatitis and pancreatic cancer. Gastroenterology.
144:1252–1261. 2013. View Article : Google Scholar
|
|
43
|
Venerito M, Vasapolli R, Rokkas T,
Delchier JC and Malfertheiner P: Helicobacter pylori,
gastric cancer and other gastrointestinal malignancies.
Helicobacter. Sep 22–2017.(Epub ahead of print): doi:
10.1111/hel.12413. 2017. View Article : Google Scholar
|
|
44
|
Goral V: Pancreatic cancer: Pathogenesis
and diagnosis. Asian Pac J Cancer Prev. 16:5619–5624. 2015.
View Article : Google Scholar
|
|
45
|
Carraway KL, Theodoropoulos G, Kozloski GA
and Carothers Carraway CA: Muc4/MUC4 functions and regulation in
cancer. Future Oncol. 5:1631–1640. 2009. View Article : Google Scholar
|
|
46
|
Gautam SK, Kumar S, Dam V, Ghersi D, Jain
M and Batra SK: MUCIN-4 (MUC4) is a novel tumor antigen in
pancreatic cancer immunotherapy. Semin Immunol. 47:1013912020.
View Article : Google Scholar
|
|
47
|
Jahan R, Macha MA, Rachagani S, Das S,
Smith LM, Kaur S and Batra SK: Axed MUC4 (MUC4/X) aggravates
pancreatic malignant phenotype by activating integrin-β1/FAK/ERK
pathway. Biochim Biophys Acta Mol Basis Dis. 1864:2538–2549. 2018.
View Article : Google Scholar
|
|
48
|
Gautam SK, Kumar S, Cannon A, Hall B,
Bhatia R, Nasser MW, Mahapatra S, Batra SK and Jain M: MUC4 mucin-a
therapeutic target for pancreatic ductal adenocarcinoma. Expert
Opin Ther Targets. 21:657–669. 2017. View Article : Google Scholar
|
|
49
|
Vasseur R, Skrypek N, Duchêne B, Renaud F,
Martínez-Maqueda D, Vincent A, Porchet N, Van Seuningen I and
Jonckheere N: The mucin MUC4 is a transcriptional and
post-transcriptional target of K-ras oncogene in pancreatic cancer.
Implication of MAPK/AP-1, NF-κB and RalB signaling pathways.
Biochim Biophys Acta. 1849:1375–1384. 2015. View Article : Google Scholar
|
|
50
|
Cho JS, Park MH, Lee JS and Yoon JH:
Reduced MUC4 expression is a late event in breast carcinogenesis
and is correlated with increased infiltration of immune cells as
well as promoter hypermethylation in invasive breast carcinoma.
Appl Immunohistochem Mol Morphol. 23:44–53. 2015. View Article : Google Scholar
|
|
51
|
Karamitopoulou E: Tumour microenvironment
of pancreatic cancer: Immune landscape is dictated by molecular and
histopathological features. Br J Cancer. 121:5–14. 2019. View Article : Google Scholar
|
|
52
|
Ren B, Cui M, Yang G, Wang H, Feng M, You
L and Zhao Y: Tumor microenvironment participates in metastasis of
pancreatic cancer. Mol Cancer. 17:1082018. View Article : Google Scholar
|
|
53
|
Carraway KL, Perez A, Idris N, Jepson S,
Arango M, Komatsu M, Haq B, Price-Schiavi SA, Zhang J and Carraway
CA: Muc4/sialomucin complex, the intramembrane ErbB2 ligand, in
cancer and epithelia: To protect and to survive. Prog Nucleic Acid
Res Mol Biol. 71:149–185. 2002. View Article : Google Scholar
|
|
54
|
Miyahara N, Shoda J, Kawamoto T, Ishida H,
Ueda T, Akimoto Y, Kawakami H and Irimura T: Interaction of Muc4
and ErbB2 in a transgenic mouse model of gallbladder carcinoma:
Potential pathobiological implications. Oncol Rep. 32:1796–1802.
2014. View Article : Google Scholar
|
|
55
|
Liberelle M, Magnez R, Thuru X, Bencheikh
Y, Ravez S, Quenon C, Drucbert AS, Foulon C, Melnyk P, Van
Seuningen I and Lebègue N: MUC4-ErbB2 oncogenic complex: Binding
studies using microscale thermophoresis. Sci Rep. 9:166782019.
View Article : Google Scholar
|
|
56
|
Seshacharyulu P, Ponnusamy MP, Rachagani
S, Lakshmanan I, Haridas D, Yan Y, Ganti AK and Batra SK: Targeting
EGF-receptor(s)-STAT1 axis attenuates tumor growth and metastasis
through downregulation of MUC4 mucin in human pancreatic cancer.
Oncotarget. 6:5164–5181. 2015. View Article : Google Scholar
|
|
57
|
Hanada K, Uchida T, Tsukamoto Y, Watada M,
Yamaguchi N, Yamamoto K, Shiota S, Moriyama M, Graham DY and
Yamaoka Y: Helicobacter pylori infection introduces DNA
double-strand breaks in host cells. Infect Immun. 82:4182–4189.
2014. View Article : Google Scholar
|
|
58
|
Chinaranagari S, Sharma P, Bowen NJ and
Chaudhary J: Prostate cancer epigenome. Methods Mol Biol.
1238:125–140. 2015. View Article : Google Scholar
|