Introduction
Colorectal cancer (CRC) is the third most common
malignancy among both sexes and the third leading cause of
cancer-associated mortality worldwide, and the rates of incidence
and mortality are 6.1 and 9.2% of the total cases in 2018 (1). Several factors, such as dysregulation
of oncogenes or tumor suppressor genes, are involved in the
occurrence and development of CRC, which affects the therapeutic
efficacy and prognosis of CRC (2).
In addition, tumor recurrence, invasion and metastasis are the main
causes of mortality of patients with CRC (3). However, the underlying molecular
mechanisms remain unknown.
MicroRNAs (miRNAs/miRs) belong to a family of
endogenous non-coding small RNAs that usually consist of 19–24
nucleotides (4). miRNAs perform
multiple functions by targeting the 3′-untranslated region (UTR) of
target genes to induce transcriptional suppression or mRNA
degradation (5–7). Previous studies have reported that
abnormal expression of miRNAs exists in cancer tissues or cancer
cell lines, which is associated with the initiation and progression
of tumors, including breast cancer, bladder cancer and lung cancer
(8–10). miRNAs also serve important roles in
regulating tumor cell proliferation, apoptosis, migration and
invasion by acting as oncogenes or tumor suppressors, or by
targeting oncogenes or tumor suppressor genes (10,11).
miR-206 belongs to the miR-206/133b cluster, which
is responsible for the growth and development of skeletal muscle
(12). Recently, miR-206 expression
has been demonstrated to be downregulated in different types of
human tumors, including CRC (13,14),
prostate (15,16), lung (17) and breast cancer (17,18),
suggesting that miR-206 expression may be closely associated with
the development of tumors. It has been reported that overexpression
of miR-206 can decrease tumor cell viability, proliferation,
migration, invasion and epithelial-to-mesenchymal transition, thus
inhibiting the metastatic potential in breast cancer (8,19), while
miR-206 knockdown promotes CRC promotion by activating C-C motif
chemokine ligand 2 (CCL2) (20) and
upregulating C-X-C chemokine receptor type 4 and VEFG-A (14). However, the underlying molecular
mechanism of how miR-206 regulates the progression of CRC requires
further investigation.
Thus, the present study aimed to investigate whether
miR-206 inhibits CRC progression by regulating the c-Met/AKT/GSK-3
pathway. The effects of miR-206 on proliferation, migration, and
invasion of CRC cells were assessed via gain or loss of function of
miR-206. Taken together, the results of the present study
demonstrated that overexpression of miR-206 suppressed the
proliferation, migration and invasion of CRC cells by targeting the
c-Met/AKT/GSK-3β pathway.
Materials and methods
Cell culture and treatment
The human colonic cancer cell lines, HCT116 and
SW480, were purchased from The Cell Bank of Type Culture Collection
of Chinese Academy of Sciences. Cells were maintained in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Cytiva), at 37°C in 5% CO2.
Reagents and transfection
miR-206 mimic, mimic negative control (mimic NC),
miR-206 inhibitor, inhibitor NC, c-Met small interfering (si)RNA
targeting c-Met (si-c-Met) and si-NC (scramble siRNA) were
purchased from Guangzhou RiboBio Co., Ltd. The sequences of miR-206
mimic and inhibitor were as follows: miR-206 mimic,
5′-UGGAAUGUAAGGAAGUGUGUGG-3′; mimic NC,
5′-UUGUACUACACAAAAGUACUG-3′; miR-206 inhibitor,
5′-CCACACACUUCCUUACAUUCCA-3′; and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′. The sequences of si-c-Met and si-NC
were as follows: si-c-Met sense, 5′-GGUGUUGUCUCAAUAUCAATT-3′, and
antisense, 5′-UUGAUAUUGAGACAACACCTT-3′; si-NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. CRC cells were transfected with
oligonucleotides using Lipofectamine® 2000 reagent
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. Briefly, CRC cells were seeded into 6-well plates at
a density of 1×105 cells/ml and transfected with 50 nM
oligonucleotides or scrambled duplexes using transfection reagent
at 37°C in a humidified incubator with 5% CO2 for 48 h.
Transfection efficiency was assessed via reverse
transcription-quantitative PCR or western blot analyses 48 h
post-transfection.
RT-qPCR
Total RNA was extracted from CRC cells in each group
using TRIzol reagent (Thermo Fisher Scientific, Inc.) and cDNA was
synthesized using the One-Step miRNA RT kit (cat. no. 639503,
Takara Bio, Inc.), at 37°C for 60 min. miR-206 expression was
detected and analysed using qPCR using SYBR PremixEx Taq
(cat. no. RR820A, Takara Bio, Inc.). U6 was used as the internal
control to normalize relative miR-206 expression. The following
primer sequences were used for qPCR: miR-206 RT primer,
5′-CTCAGCGGCTGTCGTGGACTGCGCGCTGCCGCTGAGCCACACAC-3′; miR-206
forward, 5′-GGCGGTGGAATGTAAGGAAG-3′ and reverse,
5′-GGCTGTCGTGGACTGCG-3′; U6 RT primer,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGG-3′; and U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-CTCGCTTCGGCAGCACA-3′. The thermocycling conditions were as
follows: 95°C for 10 min, followed by 40 cycles of amplification
(95°C for 30 sec, 58°C for 60 sec, 72°C for 90 sec), and a final
extension step of 72°C for 5 min. miRNA levels were quantified
using the 2−ΔΔCq method (21).
Dual-luciferase reporter assay
The 3′-UTR of the human c-Met gene has a putative
hsa-miR-206 binding sequence using the TargteScan database (release
7.2; http://www.targetscan.org/vert_72/). The luciferase
reporter vector Pgl4Luc-RLuc (Addgene, Inc.) was used to establish
a wild-type (WT) c-Met luciferase reporter plasmid or a mutant
(MUT) c-Met luciferase reporter plasmid (Shanghai GenePharma Co.,
Ltd.). SW480 cells were subsequently co-transfected with miR-206
mimic (100 nM) or mimic NC and 200 ng WT or MUT c-Met 3′-UTR
luciferase reporter plasmid using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Following
incubation for 48 h at 37°C, relative luciferase activities were
detected using a Dual-luciferase reporter assay system (Beijing
Abace Biotech, Co., Ltd.). and normalized by using the ratio of
firefly to Renilla luciferase activity.
MTT assay
HCT116 or SW480 cells were transfected with
oligonucleotides (miR-206 mimic, mimic NC, si-c-MET or si-NC), with
concentrations ranging from 50–100 nM. Following incubation for 48
h at 37°C, cell viability was assessed via the MTT assay. Briefly,
cells were seeded into 96-well microdilution plates at a density of
2×103 cells/well with DMEM containing 10% FBS. MTT
reagent (cat. no. C0009S; Beyotime Biotech, Co., Ltd.) was added on
3 consecutive days at a final concentration of 5 mg/ml. Following
the MTT incubation at 37°C for 4 h, the purple formazan crystals
were dissolved using 150 µl DMSO and viability was subsequently
analyzed at a wavelength of 570 nm, using an Enzyme Immunoassay
Analyzer (Bio-Rad Laboratories, Inc.). Cell viability was
calculated and normalized to the respective NC group.
Cell migration and invasion
assays
Cell migration and invasion were assessed via the
Transwell assays, with or without Matrigel. For the invasion assay,
Transwell membranes were precoated with 50 µl diluted Matrigel (BD
Biosciences) for 2 h at 37°C. Following transfection,
6×104 cells were plated in the upper chambers of
Transwell plates in 200 µl serum-free DMEM medium, while 600 µl
cell-free DMEM medium supplemented with 10% FBS was plated in the
lower chambers. Following incubation for 24 h at 37°C, cells in the
upper chambers were removed using a cotton swab. The invasive cells
in the lower chambers were fixed with 100% pre-cold methanol for 10
min and stained with 0.1% crystal violet for 30 min at room
temperature. For the migration assay, the same process was
performed; however, Matrigel was not added in the upper chambers.
Stained cells were counted in five randomly selected fields using a
light microscope (Olympus Corporation; magnification, ×200).
Western blotting
Transfected cells were collected and lysed using
RIPA lysis buffer (Beyotime Institute of Biotech). Extracted total
protein was quantified using the BCA protein assay kit (cat. no.
P0012; Beyotime Institute of Biotech). A total of 25 µg protein was
separated via 8 or 10% SDS-PAGE and transferred onto PVDF
membranes. The membranes were blocked with 5% skim milk for 1 h at
room temperature and then incubated with primary antibodies
against: AKT (1:1,000; cat. no. 9272; Cell Signaling Technology,
Inc.), phosphorylated (p)-AKT (Ser473) (1:500; cat. no. 4060; Cell
Signaling Technology, Inc.), GSK-3β (1:1,000; cat. no. 12456; Cell
Signaling Technology, Inc.), phosphorylated (p)-GSK-3β (Ser9)
(1:500; cat. no. 5558; Cell Signaling Technology, Inc.), β-actin
(1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.) and c-Met
(1:1,000; cat. no. ab216330; Abcam) overnight at 4°C. Following the
primary incubation, membranes were incubated with HRP-linked
secondary antibodies (anti-mouse IgG, 1:10,000; cat. no. 7076;
anti-rabbit IgG, 1:10,000, cat. no. 7074; Cell Signaling
Technology, Inc.) for 1 h at room temperature. Immunoreactive bands
were detected using Immobilon Forte Western HRP substrate reagents
(EMD Millipore) and imaged using automatic chemiluminescence
analyzer (Tanon Science and Technology Co., Ltd.). The gray value
of target bands in each group was analyzed using ImageJ software
(1.50d; National Institutes of Health).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.). All experiments were performed in triplicate
and data are presented as the mean ± standard deviation. Paired or
unpaired Student's t-test were used to compare differences between
two groups, while one-way ANOVA followed by Tukey's post hoc test
were used to compare differences between multiple groups.
Spearman's correlation analysis was performed to assess the
correlation between miR-206 and c-Met. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-206 inhibits the proliferation,
migration and invasion of CRC cells
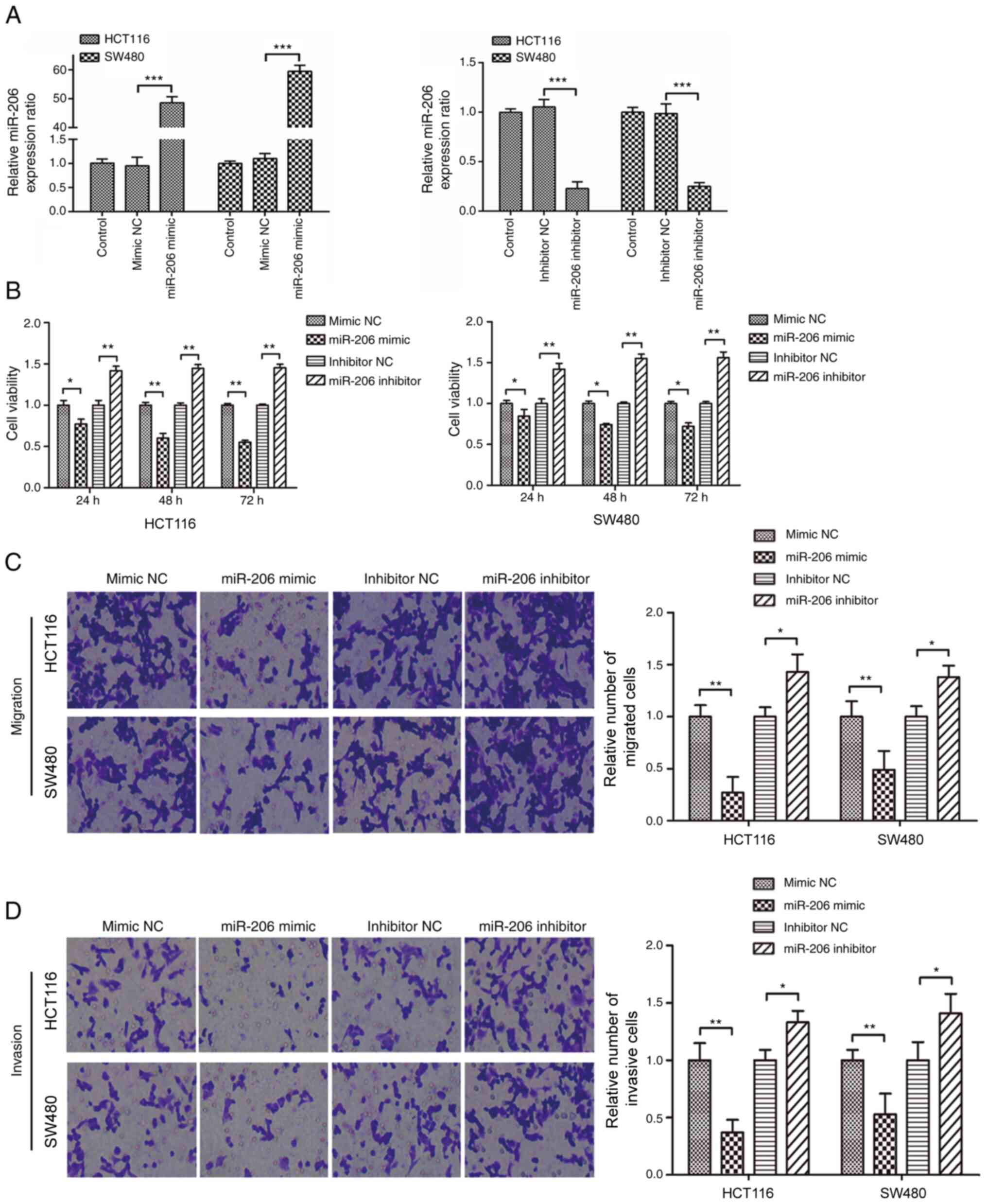
As presented in Fig.
1A, miR-206 expression significantly increased in HCT116 and
SW480 cells following transfection with miR-206 mimic (P<0.001),
while transfection with miR-206 inhibitor significantly decreased
miR-206 expression (P<0.001). In addition, overexpression of
miR-206 significantly suppressed the proliferative, migratory and
invasive abilities of CRC cells (P<0.05); however, transfection
with miR-206 inhibitor promoted the proliferation, migration and
invasion of cells (Fig. 1B-D).
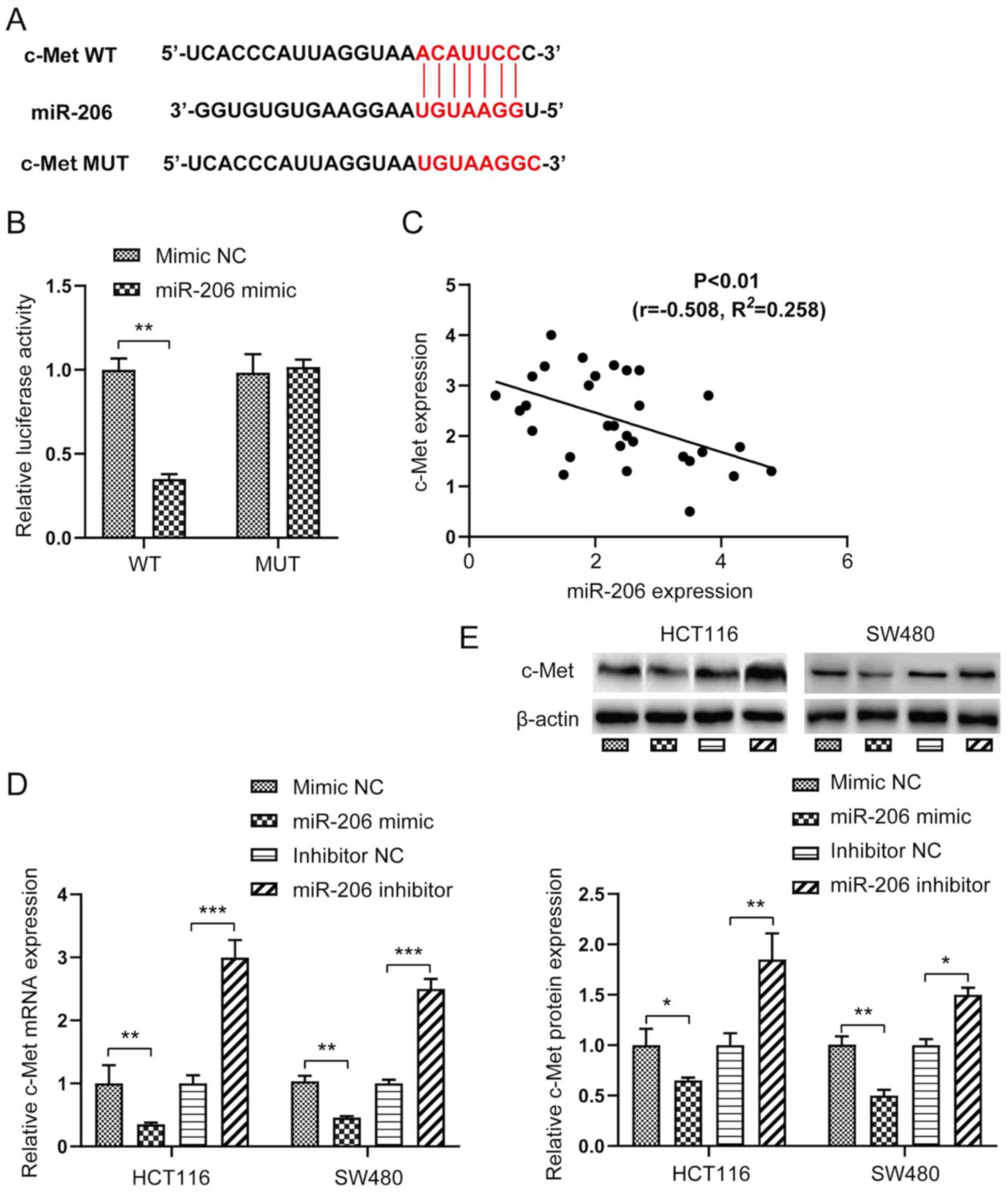
c-Met is a direct target gene of
miR-206
The potential target genes of miR-206 were screened
using the TargetScan Human database. c-Met was identified as one of
the potential targets of miR-206. To further verify that c-Met was
a direct target gene of miR-206, the fluorescent reporter vectors
with WT or MUT 3′-UTR sequences of c-Met were co-transfected with
miR-206 mimic (Fig. 2A). As
presented in Fig. 2B, relative
luciferase activity of the c-Met WT 3′-UTR vector significantly
decreased (P<0.01), whereas relative luciferase activity of the
c-Met MUT 3′-UTR vector was not affected following transfection
with miR-206 mimic, suggesting that c-Met is direct target gene of
miR-206. Furthermore, Spearman's correlation analysis demonstrated
that c-Met expression was negatively correlated with miR-206
expression (Fig. 2C).
The present study investigated whether c-Met was
involved in the miR-206-mediated effects in CRC. As expected,
overexpression of miR-206 significantly decreased the mRNA and
protein expression levels of c-Met in HCT116 and SW480 cells
(P<0.05 or P<0.01), the effects of which were reversed
following transfection with miR-206 inhibitor (Fig. 2D and E).
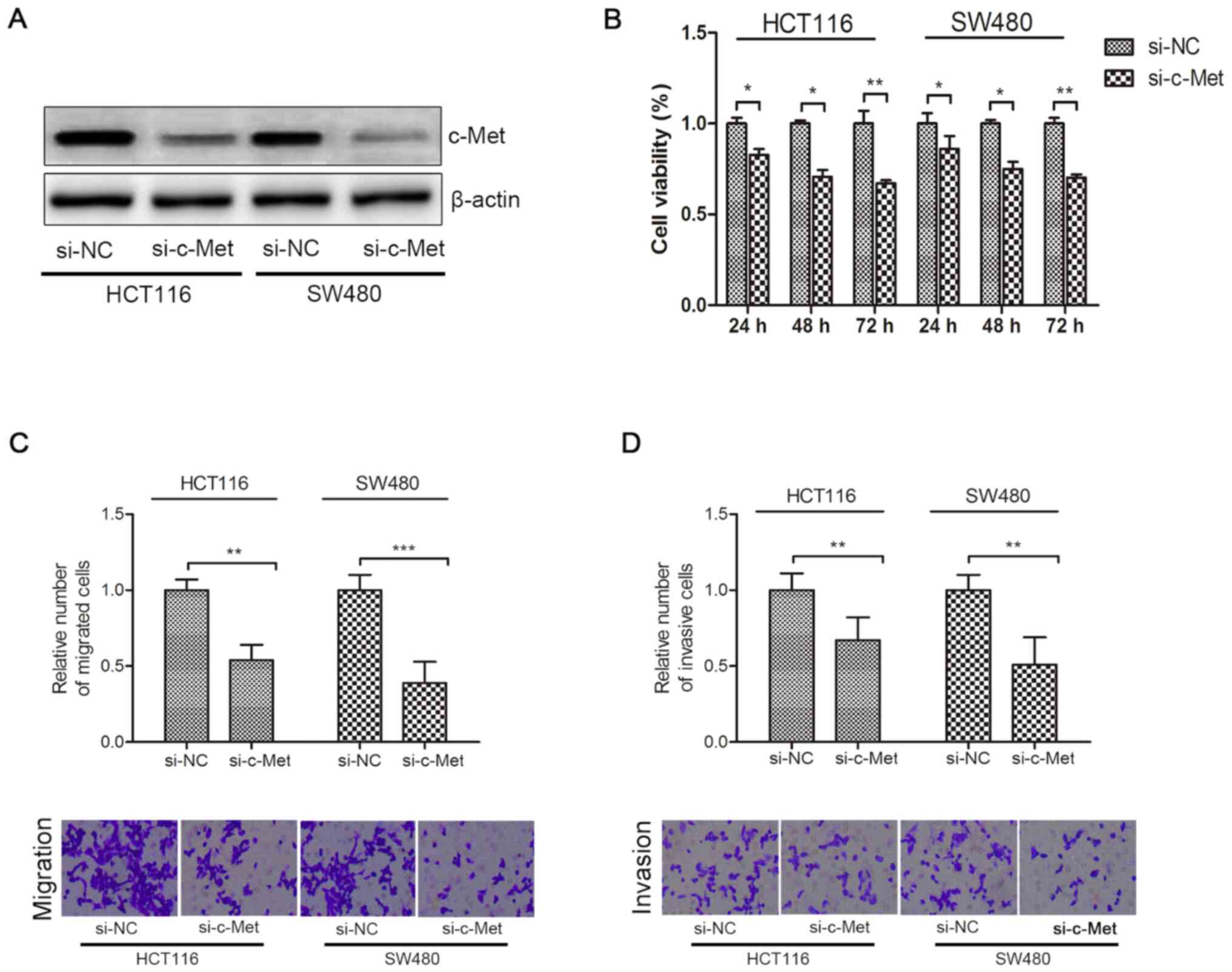
Knockdown of c-Met inhibits the
proliferation, migration and invasion of HCT116 and SW480
cells
As presented in Fig.
3A, c-Met protein expression significantly decreased in HCT116
and SW480 cells following transfection with si-c-Met. Knockdown of
c-Met significantly decreased the viability, migration and invasion
of CRC cells compared with the si-NC group (P<0.05; Fig. 3B-D).
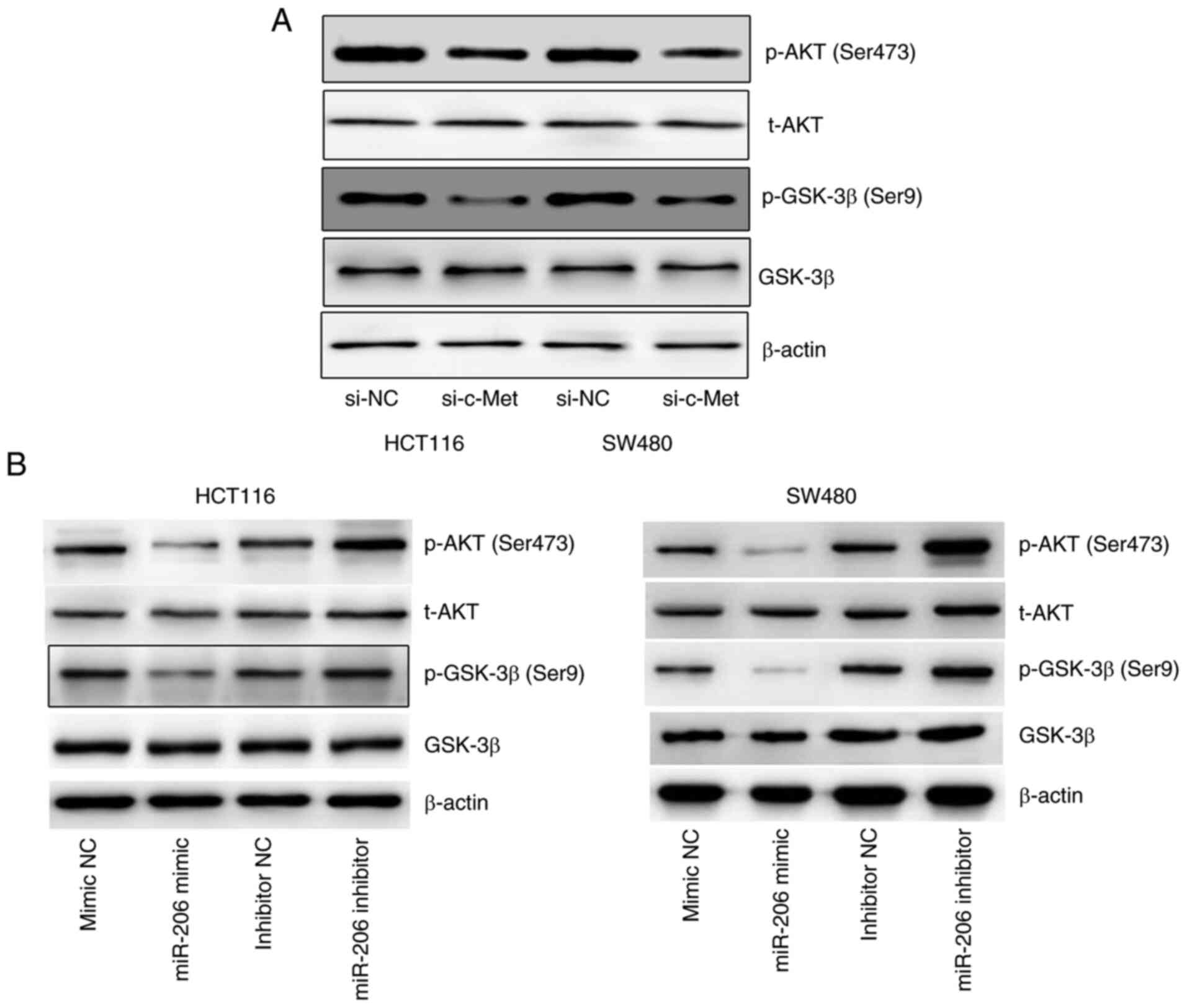
miR-206-mediates the downregulation of
c-Met and inhibits AKT/GSK-3β signaling
The expression levels of p-AKT (Ser473) and p-GSK-3β
(Ser9) markedly decreased following inhibition of c-Met (Fig. 4A), suggesting that c-Met regulates
the activation of AKT/GSK-3β signaling. In addition, overexpression
of miR-206 was associated with decreased expression levels of p-AKT
and p-GSK-3β, the effects of which were reversed following
transfection with miR-206 inhibitor (Fig. 4B). Taken together, these results
suggest that miR-206 mediates downregulation of c-Met and inhibits
AKT/GSK-3β signaling.
Discussion
The initiation of malignant tumors usually begins
from dysregulation of the normal growth regulatory mechanism, as
well as cells undergoing hyperproliferation, evading apoptosis,
becoming invasive and ultimately acquiring a malignant phenotype
(22). Aberrant expression of miRNAs
is closely associated with the tumorigenesis and progression of
tumor, including CRC. For example, it has been reported that miRNAs
can act as proto-oncogenes, such as miR-346, or tumor suppressors,
such as miR-187-3p, to regulate cell proliferation, apoptosis and
differentiation (23). Previous
studies have demonstrated that miR-206 is dysregulated in different
types of human cancer, including colon (24), lung (10), prostate (15) and breast cancer (19), suggesting that miR-206 serves an
important role in tumorigenesis. In addition, clinical research has
reported that serum miR-206 expression significantly decreases in
patients with CRC compared with healthy controls, and was confirmed
to be an independent prognostic indicator for CRC, as patients with
CRC with low miR-206 expression experience poor overall survival
and disease-free survival than those with high miR-206 expression,
suggesting that miR-206 may be a promising biomarker for the
diagnosis and prognosis of CRC (25). However, the underlying molecular
mechanism of miR-206 in regulating the progression of CRC remains
unknown, although several researchers have suggested the repression
of miR-206 on proliferation, migration, invasion (21) and angiogenesis (14).
Previous studies have reported that overexpression
of miR-206 can inhibit CRC cell migration and invasion (26), as well as prostaglandin E2-induced
CRC cell proliferation, migration and invasion by targeting
transmembrane 4 L six family member 1 (27). In addition, miR-206 knockdown
promotes drug resistance and decreases the apoptosis of HCT116/FR
cells (28). The results of the
present study demonstrated that overexpression of miR-206
suppressed the proliferation, migration and invasion of HCT116 and
SW480 cells, the effects of which were reversed following miR-206
knockdown, suggesting that miR-206 may be involved in the
development of CRC by suppressing the proliferation, migration and
invasion of CRC cells.
c-Met, also known as MET, acts as an oncogene and
has been identified as a direct target of miR-206 (29), and has also been demonstrated to
regulate proliferation, invasion and metastasis of tumor cells
(10,30). It has been reported that miR-206
suppresses the proliferation, migration and invasion of lung cancer
cells by downregulating c-Met signaling (31,32).
c-Met upregulation has been observed in different types of human
cancer, including CRC and exhibits oncogenic activity (33,34). It
has been demonstrated that specific inhibition of the STAT3/c-Met
pathway can potentially neutralize the intrahepatic metastatic
implantation and increase growth over a short-lived temporal window
caused by radiofrequency ablation of healthy liver in an
intrahepatic CRC mice model (35). A
positive association between c-Met expression and tumor stages of
CRC liver metastasis and lymph node metastasis has been suggested
(36), indicating that c-Met
expression may be a significant prognostic factor in CRC
metastasis. In addition, a previous study revealed that
overexpression of miR-206 and inhibition of the
c-Met/ERK/Elk-1/HIF-1α/VEGF-A pathway by CCL19 suppresses
angiogenesis in CRC (37).
The results of the present study demonstrated that
miR-206 expression was negatively correlated with c-Met expression
in CRC cells, and identified that c-Met knockdown significantly
inhibited the proliferation, migration and invasion of CRC cells.
Taken together, these results suggest that miR-206 may regulate the
progression of CRC cells by targeting c-Met. It has been reported
that c-Met can mediate the activation of several intracellular
signaling pathways, including the AKT/GSK-3β, STAT3, SRC/FAK and
MAPK/ERK pathways (38–40). Activation of these pathways results
in the emergence of diverse cellular hallmarks of cancer, including
cell proliferation, survival, inhibition of inflammation and
apoptosis, migration, invasion and metastasis (41–43).
The present study investigated the expression levels
of p-Akt and p-GSK-3β and demonstrated that c-Met knockdown
significantly decreased the expression levels of these factors.
Further investigation revealed that overexpression of miR-206
decreased the expression levels of p-Akt and p-GSK-3β, the effects
of which were reversed following miR-206 knockdown. Collectively,
these results suggest that miR-206 may downregulate activity of the
Akt/GSK-3β pathway by targeting c-Met, thus suppressing the
proliferation, migration and invasion of CRC cells. Further
research is required to confirm whether the effect of miR-206 on
CRC cells can be reversed by specifically activating the Akt/GSK-3β
pathway, thus providing further evidence to clarify the molecular
mechanism of miR-206 in regulating the progression of CRC cells. In
addition, whether miR-206 can regulate tumor growth and metastasis
via the Akt/GSK-3β pathway requires further investigation.
In conclusion, the results of the present study
demonstrated that miR-206 suppressed the proliferation, migration
and invasion of CRC cells by targeting the c-Met/AKT/GSK-3β
signaling pathway. These findings demonstrate the antitumor effects
of miR-206 and its underlying molecular mechanism, which may aid in
the development of more effective and promising therapies against
CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JL performed the experiments and drafted the initial
manuscript. YS performed the statistical analysis. HM and XL
conceived and designed the experiments. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng M, Zhu L, Li L and Kang C: miR-378
suppresses the proliferation, migration and invasion of colon
cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 22:122017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang B, Liu G, Ding L, Zhao J and Lu Y:
FOXA2 promotes the proliferation, migration and invasion, and
epithelial mesenchymal transition in colon cancer. Exp Ther Med.
16:133–140. 2018.PubMed/NCBI
|
|
4
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma W, Liu B, Li J, Jiang J, Zhou R, Huang
L, Li X, He X and Zhou Q: MicroRNA-302c represses
epithelial-mesenchymal transition and metastasis by targeting
transcription factor AP-4 in colorectal cancer. Biomed
Pharmacother. 105:670–676. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q,
Yang W, Zheng X, Tu K and Liu Q: miR-187-3p inhibits the metastasis
and epithelial-mesenchymal transition of hepatocellular carcinoma
by targeting S100A4. Cancer Lett. 381:380–390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tu K, Liu Z, Yao B, Han S and Yang W:
MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT
signaling in hepatocellular carcinoma. Int J Oncol. 48:965–974.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang
G, Wang J and Lu J: MiR-206 suppresses epithelial mesenchymal
transition by targeting TGF-β signaling in estrogen receptor
positive breast cancer cells. Oncotarget. 7:24537–24548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu X, Zhu Y, Liang Z, Li S, Xu X, Wang X,
Wu J, Hu Z, Meng S, Liu B, et al: c-Met and CREB1 are involved in
miR-433-mediated inhibition of the epithelial-mesenchymal
transition in bladder cancer by regulating Akt/GSK-3beta/Snail
signaling. Cell Death Dis. 7:e20882016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ,
Song J, Yan J, Wu LJ, Xu LQ and Shi JG: Comprehensive gene and
microRNA expression profiling reveals miR-206 inhibits MET in lung
cancer metastasis. Mol Biosyst. 11:2290–2302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
12
|
Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B
and Li K: MiR-206, a key modulator of skeletal muscle development
and disease. Int J Biol Sci. 11:345–352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu H, He G and Wang Y, Hu Y, Zhang Z,
Qian X and Wang Y: Long intergenic noncoding RNA 00707 promotes
colorectal cancer cell proliferation and metastasis by sponging
miR-206. Onco Targets Ther. 12:4331–4340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH
and Han SG: Circ_0056618 promoted cell proliferation, migration and
angiogenesis through sponging with miR-206 and upregulating CXCR4
and VEGF-A in colorectal cancer. Eur Rev Med Pharmacol Sci.
24:4190–4202. 2020.PubMed/NCBI
|
|
15
|
Wang Y, Xu H, Si L, Li Q, Zhu X, Yu T and
Gang X: MiR-206 inhibits proliferation and migration of prostate
cancer cells by targeting CXCL11. Prostate. 78:479–490. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang N, Wang L, Liu J, Liu L, Huang J,
Chen X and Luo Z: MicroRNA-206 regulates the epithelial-mesenchymal
transition and inhibits the invasion and metastasis of prostate
cancer cells by targeting Annexin A2. Oncol Lett. 15:8295–8302.
2018.PubMed/NCBI
|
|
17
|
Liao M and Peng L: MiR-206 may suppress
non-small lung cancer metastasis by targeting CORO1C. Cell Mol Biol
Lett. 25:222020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan C, Liu N, Zheng D, Du J and Wang K:
MicroRNA-206 inhibits metastasis of triple-negative breast cancer
by targeting transmembrane 4 L6 family member 1. Cancer Manag Res.
11:6755–6764. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu Y, Shao ZM, He QZ, Jiang BQ, Wu Y and
Zhuang ZG: Hsa-miR-206 represses the proliferation and invasion of
breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci.
19:2091–2104. 2015.PubMed/NCBI
|
|
20
|
Shengnan J, Dafei X, Hua J, Sunfu F,
Xiaowei W and Liang X: Long non-coding RNA HOTAIR as a competitive
endogenous RNA to sponge miR-206 to promote colorectal cancer
progression by activating CCL2. J Cancer. 11:4431–4441. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pan JY, Sun CC, Bi ZY, Chen ZL, Li SJ, Li
QQ, Wang YX, Bi YY and Li DJ: miR-206/133b cluster: A weapon
against lung cancer? Mol Ther Nucleic Acids. 8:442–449. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang XW, Xi XQ, Wu J, Wan YY, Hui HX and
Cao XF: MicroRNA-206 attenuates tumor proliferation and migration
involving the downregulation of NOTCH3 in colorectal cancer. Oncol
Rep. 33:1402–1410. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Zheng W, Zhang X, Dong M and Sun G:
The diagnostic and prognostic value of serum miR-206 in colorectal
cancer. Int J Clin Exp Pathol. 10:7528–7533. 2017.PubMed/NCBI
|
|
26
|
Sun P, Sun D, Wang X, Liu T, Ma Z and Duan
L: miR-206 is an independent prognostic factor and inhibits tumor
invasion and migration in colorectal cancer. Cancer Biomark.
15:391–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park YR, Seo SY, Kim SL, Zhu SM, Chun S,
Oh JM, Lee MR, Kim SH, Kim IH, Lee SO, et al: MiRNA-206 suppresses
PGE2-induced colorectal cancer cell proliferation, migration, and
invasion by targetting TM4SF1. Biosci Rep. 38:BSR201806642018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng X and Fu R: miR-206 regulates 5-FU
resistance by targeting Bcl-2 in colon cancer cells. Onco Targets
Ther. 11:1757–1765. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren XL, He GY, Li XM, Men H, Yi LZ, Lu GF,
Xin SN, Wu PX, Li YL, Liao WT, et al: MicroRNA-206 functions as a
tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer
Res Clin Oncol. 142:581–592. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dai C, Xie Y, Zhuang X and Yuan Z: MiR-206
inhibits epithelial ovarian cancer cells growth and invasion via
blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother.
104:763–770. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mataki H, Seki N, Chiyomaru T, Enokida H,
Goto Y, Kumamoto T, Machida K, Mizuno K, Nakagawa M and Inoue H:
Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR
oncogenic signaling in lung squamous cell carcinoma. Int J Oncol.
46:1039–1050. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen X, Tong ZK and Zhou JY, Yao YK, Zhang
SM and Zhou JY: MicroRNA-206 inhibits the viability and migration
of human lung adenocarcinoma cells partly by targeting MET. Oncol
Lett. 12:1171–1177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Raghav K, Bailey AM, Loree JM, Kopetz S,
Holla V, Yap TA, Wang F, Chen K, Salgia R and Hong D: Untying the
gordion knot of targeting MET in cancer. Cancer Treat Rev.
66:95–103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Y, Tao Q, Li S, Zheng P, Liu J and
Liang X: Both endogenous and exogenous miR-139-5p inhibit
Fusobacterium nucleatum-related colorectal cancer
development. Eur J Pharmacol. 888:1734592020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao H, Ahmed M, Markezana A, Zeng G,
Stechele M, Galun E and Goldberg SN: Thermal ablation induces
transitory metastatic growth by means of the STAT3/c-Met molecular
pathway in an intrahepatic colorectal cancer mouse model.
Radiology. 294:464–472. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao JF, Li XJ, Yan LK, He S, Zheng JB,
Wang XR, Zhou PH, Zhang L, Wei GB and Sun XJ: Role of HGF/c-Met in
the treatment of colorectal cancer with liver metastasis. J Biochem
Mol Toxicol. 33:e223162019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu Z, Zhu C, Chen C, Zong Y, Feng H, Liu
D, Feng W, Zhao J and Lu A: CCL19 suppresses angiogenesis through
promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1α/VEGF-A
pathway in colorectal cancer. Cell Death Dis. 9:9742018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gherardi E, Birchmeier W, Birchmeier C and
Vande Woude G: Targeting MET in cancer: Rationale and progress. Nat
Rev Cancer. 12:89–103. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Viticchiè G and Muller PAJ: c-Met and
other cell surface molecules: Interaction, activation and
functional consequences. Biomedicines. 3:46–70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamaoka T, Kusumoto S, Ando K, Ohba M and
Ohmori T: Receptor tyrosine kinase-targeted cancer therapy. Int J
Mol Sci. 19:34912018. View Article : Google Scholar
|
|
41
|
Organ SL and Tsao MS: An overview of the
c-MET signaling pathway. Ther Adv Med Oncol. 3 (Suppl 1):S7–S19.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Comoglio PM, Trusolino L and Boccaccio C:
Known and novel roles of the MET oncogene in cancer: A coherent
approach to targeted therapy. Nat Rev Cancer. 18:341–358. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu Z, Song N, Shen B, Xu X, Fang Y, Shi Y,
Ning Y, Hu J, Dai Y, Ding X, et al: Syndecan-1 shedding inhibition
to protect against ischemic acute kidney injury through HGF target
signaling pathway. Transplantation. 102:e331–e344. 2018. View Article : Google Scholar : PubMed/NCBI
|