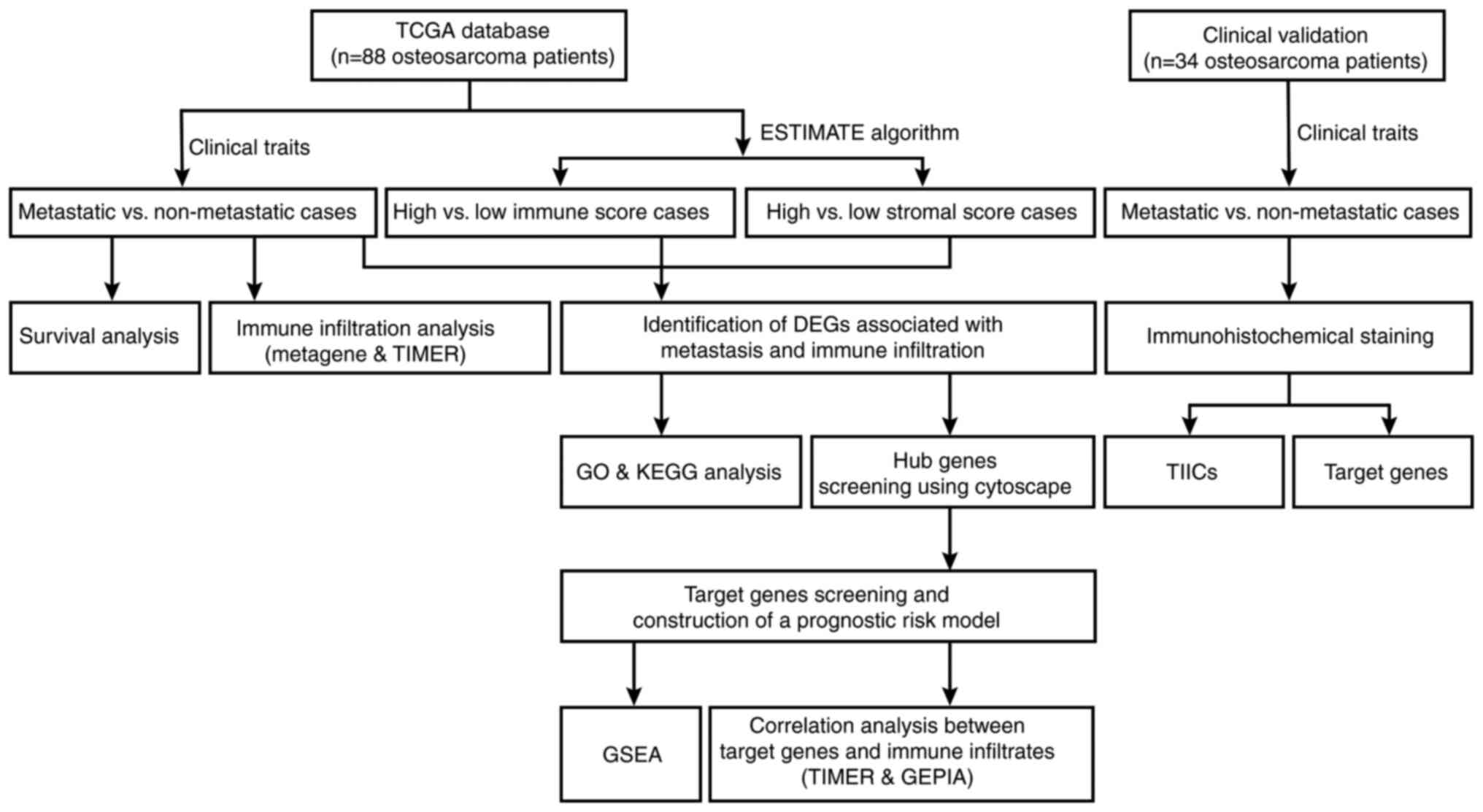
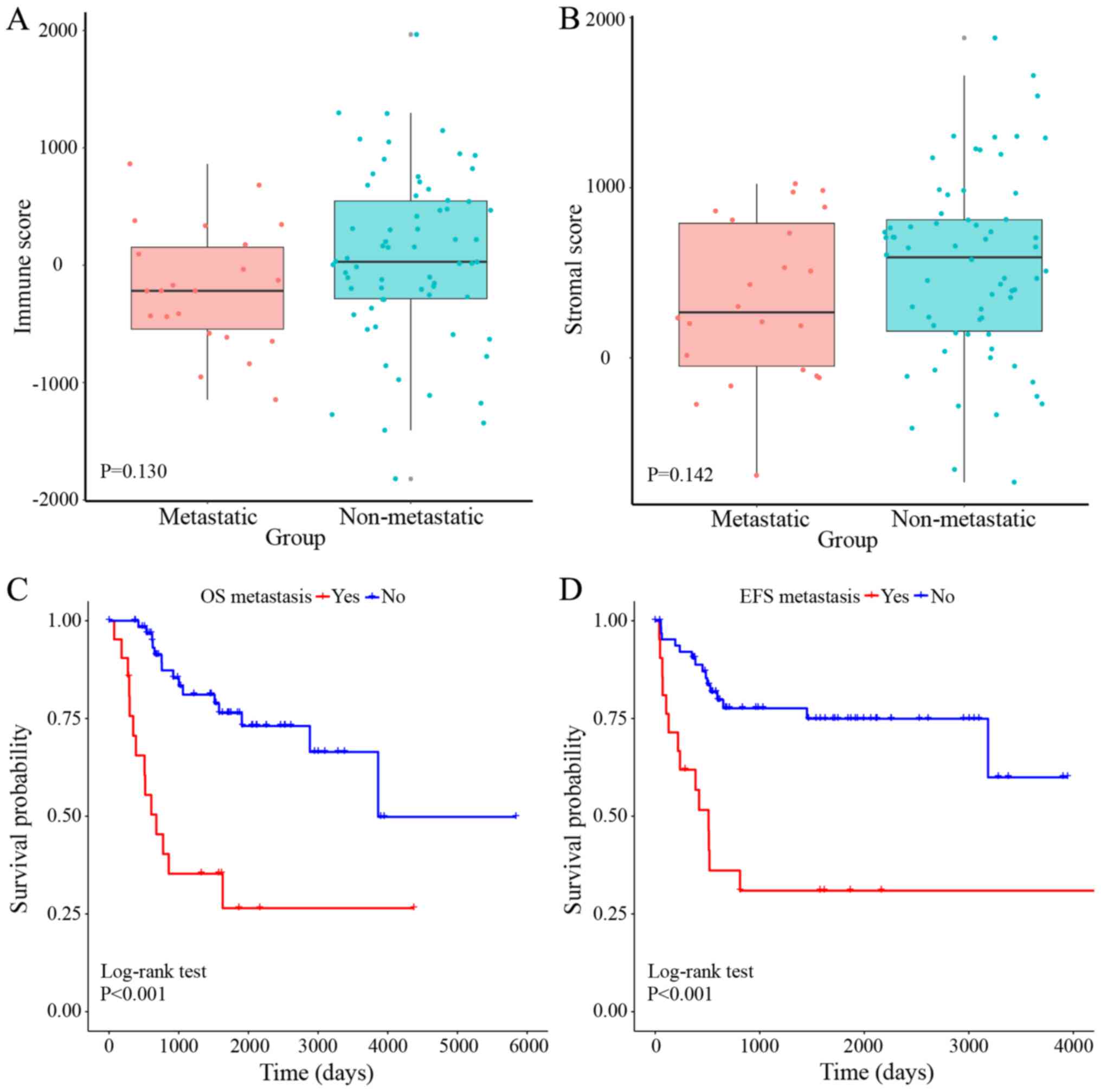
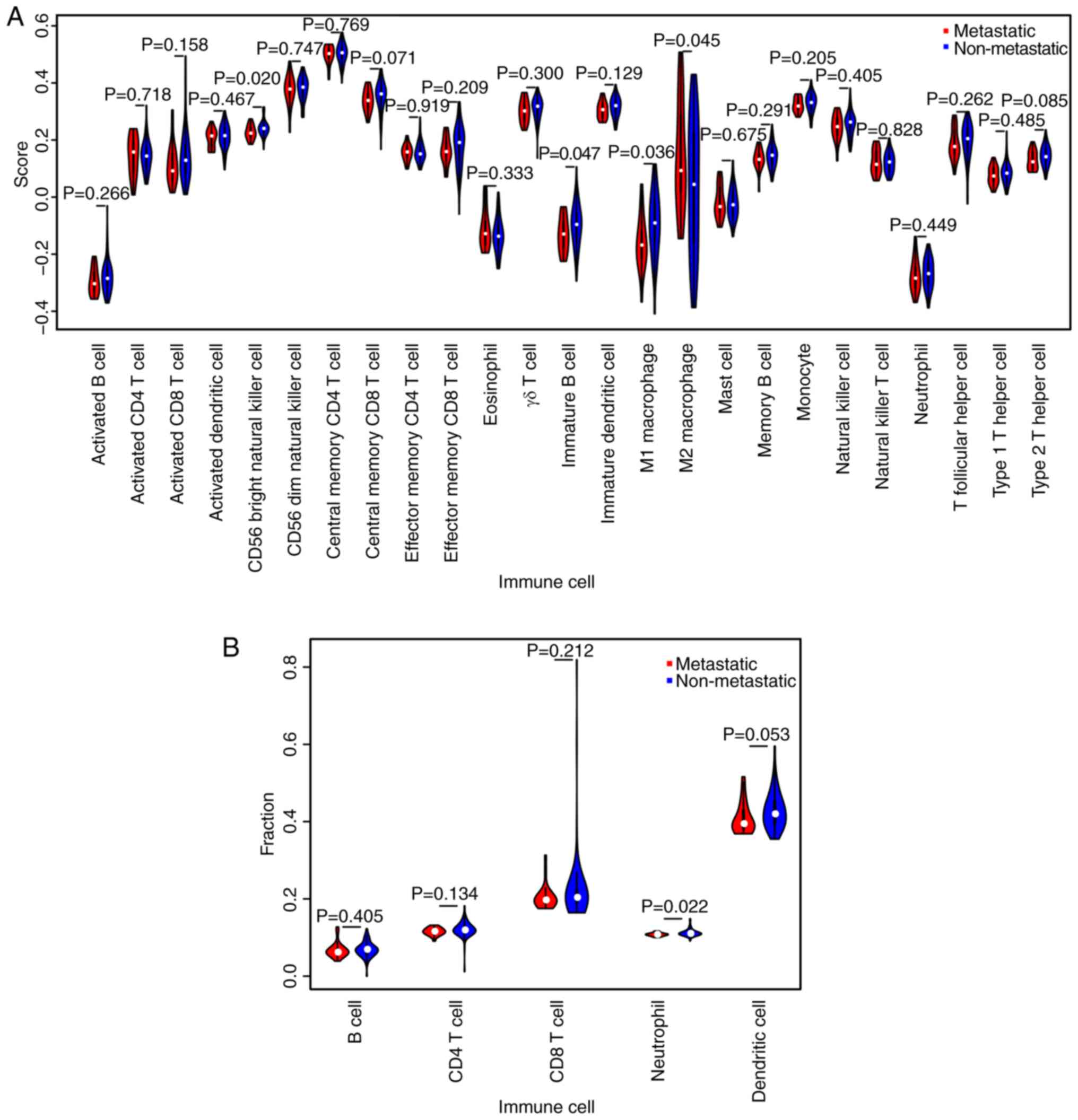
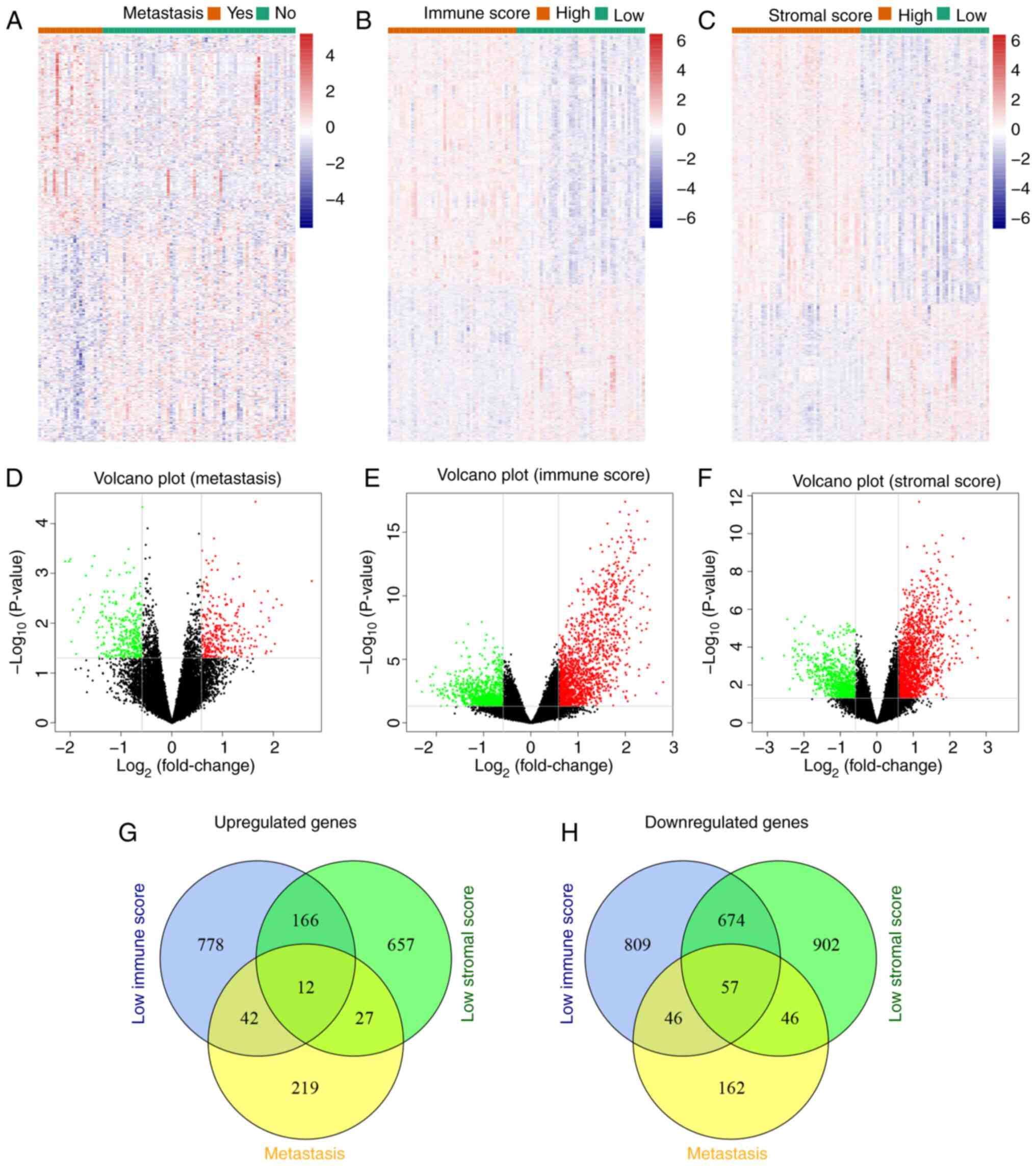
|
1
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 (Suppl 7):vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brown HK, Tellez-Gabriel M and Heymann D:
Cancer stem cells in osteosarcoma. Cancer Lett. 386:189–195. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang L, Huang Z, Lin W, Wang L, Zhu X,
Chen X, Yang S and Lv C: Salidroside suppresses the growth and
invasion of human osteosarcoma cell lines MG63 and U2OS in vitro by
inhibiting the JAK2/STAT3 signaling pathway. Int J Oncol.
54:1969–1980. 2019.PubMed/NCBI
|
|
4
|
Sergi C and Zwerschke W: Osteogenic
sarcoma (osteosarcoma) in the elderly: Tumor delineation and
predisposing conditions. Exp Gerontol. 43:1039–1043. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jaffe N: Osteosarcoma: Review of the past,
impact on the future. The American experience. Cancer Treat Res.
152:239–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miwa S, Shirai T, Yamamoto N, Hayashi K,
Takeuchi A, Igarashi K and Tsuchiya H: Current and emerging targets
in immunotherapy for osteosarcoma. J Oncol. 2019:70350452019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reed DR, Hayashi M, Wagner L, Binitie O,
Steppan DA, Brohl AS, Shinohara ET, Bridge JA, Loeb DM, Borinstein
SC and Isakoff MS: Treatment pathway of bone sarcoma in children,
adolescents, and young adults. Cancer. 123:2206–2218. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reina-Campos M, Moscat J and Diaz-Meco M:
Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol.
48:47–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meurette O and Mehlen P: Notch signaling
in the tumor microenvironment. Cancer Cell. 34:536–548. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen JL, Lucas JE, Schroeder T, Mori S, Wu
J, Nevins J, Dewhirst M, West M and Chi JT: The genomic analysis of
lactic acidosis and acidosis response in human cancers. PLoS Genet.
4:e10002932008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar
|
|
12
|
Mori K, Rédini F, Gouin F, Cherrier B and
Heymann D: Osteosarcoma: Current status of immunotherapy and future
trends (Review). Oncol Rep. 15:693–700. 2006.PubMed/NCBI
|
|
13
|
Buddingh EP, Kuijjer ML, Duim RA, Bürger
H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC,
Lankester AC and Cleton-Jansen AM: Tumor-infiltrating macrophages
are associated with metastasis suppression in high-grade
osteosarcoma: A rationale for treatment with macrophage activating
agents. Clin Cancer Res. 17:2110–2119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ratti C, Botti L, Cancila V, Galvan S,
Torselli I, Garofalo C, Manara MC, Bongiovanni L, Valenti CF,
Burocchi A, et al: Trabectedin overrides osteosarcoma
differentiative block and reprograms the tumor immune environment
enabling effective combination with immune checkpoint inhibitors.
Clin Cancer Res. 23:5149–5161. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kawano M, Itonaga I, Iwasaki T and Tsumura
H: Enhancement of antitumor immunity by combining anti-cytotoxic T
lymphocyte antigen-4 antibodies and cryotreated tumor lysate-pulsed
dendritic cells in murine osteosarcoma. Oncol Rep. 29:1001–1006.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saraf AJ, Fenger JM and Roberts RD:
Osteosarcoma: Accelerating progress makes for a hopeful future.
Front Oncol. 8:42018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Li B, Ren Y and Ye Z: T-Cell-based
immunotherapy for osteosarcoma: Challenges and opportunities. Front
Immunol. 7:3532016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miwa S, Yamamoto N, Hayashi K, Takeuchi A,
Igarashi K and Tsuchiya H: Therapeutic targets for bone and
soft-tissue sarcomas. Int J Mol Sci. 20:1702019. View Article : Google Scholar
|
|
19
|
Angelova M, Charoentong P, Hackl H,
Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik
B, Galon J and Trajanoski Z: Characterization of the
immunophenotypes and antigenomes of colorectal cancers reveals
distinct tumor escape mechanisms and novel targets for
immunotherapy. Genome Biol. 16:642015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu Z, Zhang Y, Xu M, Zheng X, Lin M, Pan
J, Ye C, Deng Y, Jiang C, Lin Y, et al: Demethylation and
overexpression of CSF2 are involved in immune response,
chemotherapy resistance, and poor prognosis in colorectal cancer.
Onco Targets Ther. 12:11255–11269. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pan JH, Zhou H, Cooper L, Huang JL, Zhu
SB, Zhao XX, Ding H, Pan YL and Rong L: LAYN is a prognostic
biomarker and correlated with immune infiltrates in gastric and
colon cancers. Front Immunol. 10:62019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang S, Liu T, Nan H, Wang Y, Chen H,
Zhang X, Zhang Y, Shen B, Qian P, Xu S, et al: Comprehensive
analysis of prognostic immune-related genes in the tumor
microenvironment of cutaneous melanoma. J Cell Physiol.
235:1025–1035. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen Y, Peng X and Shen C: Identification
and validation of immune-related lncRNA prognostic signature for
breast cancer. Genomics. 112:2640–2646. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li R, Qu H, Wang S, Wei J, Zhang L, Ma R,
Lu J, Zhu J, Zhong WD and Jia Z: GDCRNATools: An R/Bioconductor
package for integrative analysis of lncRNA, miRNA and mRNA data in
GDC. Bioinformatics. 34:2515–2517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2012, ISBN 3-900051-07-0, URL
http://www.R-project.org/.
|
|
28
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Therneau TM and Grambsch PM: Modeling
Survival Data: Extending the Cox Model. Springer; New York, NY:
2000, ISBN 0-387-98784-3. https://www.springer.com/gp/book/9780387987842#
View Article : Google Scholar
|
|
31
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ye Z, Zeng Z, Wang D, Lei S, Shen Y and
Chen Z: Identification of key genes associated with the progression
of intrahepatic cholangiocarcinoma using weighted gene
co-expression network analysis. Oncol Lett. 20:483–494. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsukahara T, Emori M, Murata K, Mizushima
E, Shibayama Y, Kubo T, Kanaseki T, Hirohashi Y, Yamashita T, Sato
N and Torigoe T: The future of immunotherapy for sarcoma. Expert
Opin Biol Ther. 16:1049–1057. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang SD, Li HY, Li BH, Xie T, Zhu T, Sun
LL, Ren HY and Ye ZM: The role of CTLA-4 and PD-1 in anti-tumor
immune response and their potential efficacy against osteosarcoma.
Int Immunopharmacol. 38:81–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Heymann MF, Lézot F and Heymann D: The
contribution of immune infiltrates and the local microenvironment
in the pathogenesis of osteosarcoma. Cell Immunol. 343:1037112019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ehnman M, Chaabane W, Haglund F and
Tsagkozis P: The tumor microenvironment of pediatric sarcoma:
Mesenchymal mechanisms regulating cell migration and metastasis.
Curr Oncol Rep. 21:902019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng Y, Wang G, Chen R, Hua Y and Cai Z:
Mesenchymal stem cells in the osteosarcoma microenvironment: Their
biological properties, influence on tumor growth, and therapeutic
implications. Stem Cell Res Ther. 9:222018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Scott MC, Temiz NA, Sarver AE, LaRue RS,
Rathe SK, Varshney J, Wolf NK, Moriarity BS, O'Brien TD, Spector
LG, et al: Comparative transcriptome analysis quantifies immune
cell transcript levels, metastatic progression, and survival in
osteosarcoma. Cancer Res. 78:326–337. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Merchant MS, Melchionda F, Sinha M, Khanna
C, Helman L and Mackall CL: Immune reconstitution prevents
metastatic recurrence of murine osteosarcoma. Cancer Immunol
Immunother. 56:1037–1046. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Merchant MS, Bernstein D, Amoako M, Baird
K, Fleisher TA, Morre M, Steinberg SM, Sabatino M, Stroncek DF,
Venkatasan AM, et al: Adjuvant immunotherapy to improve outcome in
high-risk pediatric sarcomas. Clin Cancer Res. 22:3182–3191. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li X, Gao Y, Xu Z, Zhang Z, Zheng Y and Qi
F: Identification of prognostic genes in adrenocortical carcinoma
microenvironment based on bioinformatic methods. Cancer Med.
9:1161–1172. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang S, Zhang B, Fan W, Zhao Q, Yang L,
Xin W and Fu D: Identification of prognostic genes in the acute
myeloid leukemia microenvironment. Aging (Albany NY).
11:10557–10580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Schell TD, Knowles BB and Tevethia SS:
Sequential loss of cytotoxic T lymphocyte responses to simian virus
40 large T antigen epitopes in T antigen transgenic mice developing
osteosarcomas. Cancer Res. 60:3002–3012. 2000.PubMed/NCBI
|
|
51
|
Shen JK, Cote GM, Choy E, Yang P, Harmon
D, Schwab J, Nielsen GP, Chebib I, Ferrone S, Wang X, et al:
Programmed cell death ligand 1 expression in osteosarcoma. Cancer
Immunol Res. 2:690–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lussier DM, O'Neill L, Nieves LM, McAfee
MS, Holechek SA, Collins AW, Dickman P, Jacobsen J, Hingorani P and
Blattman JN: Enhanced T-cell immunity to osteosarcoma through
antibody blockade of PD-1/PD-L1 interactions. J Immunother.
38:96–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Steinman RM and Idoyaga J: Features of the
dendritic cell lineage. Immunol Rev. 234:5–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yu Z, Qian J, Wu J, Gao J and Zhang M:
Allogeneic mRNA-based electrotransfection of autologous dendritic
cells and specific antitumor effects against osteosarcoma in rats.
Med Oncol. 29:3440–3448. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kawano M, Tanaka K, Itonaga I, Iwasaki T,
Miyazaki M, Ikeda S and Tsumura H: Dendritic cells combined with
anti-GITR antibody produce antitumor effects in osteosarcoma. Oncol
Rep. 34:1995–2001. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Böttcher JP, Bonavita E, Chakravarty P,
Blees H, Cabeza-Cabrerizo M, Sammicheli S, Rogers NC, Sahai E,
Zelenay S and Reis e Sousa C: NK cells stimulate recruitment of
cDC1 into the tumor microenvironment promoting cancer immune
control. Cell. 172:1022–1037.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chang YH, Connolly J, Shimasaki N, Mimura
K, Kono K and Campana D: A chimeric receptor with NKG2D specificity
enhances natural killer cell activation and killing of tumor cells.
Cancer Res. 73:1777–1786. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kiany S, Huang G and Kleinerman ES: Effect
of entinostat on NK cell-mediated cytotoxicity against osteosarcoma
cells and osteosarcoma lung metastasis. OncoImmunology.
6:e13332142017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Beauvillain C, Delneste Y, Scotet M, Peres
A, Gascan H, Guermonprez P, Barnaba V and Jeannin P: Neutrophils
efficiently cross-prime naive T cells in vivo. Blood.
110:2965–2973. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jablonska J, Lang S, Sionov RV and Granot
Z: The regulation of pre-metastatic niche formation by neutrophils.
Oncotarget. 8:112132–112144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao SJ, Jiang YQ, Xu NW, Li Q, Zhang Q,
Wang SY, Li J, Wang YH, Zhang YL, Jiang SH, et al: SPARCL1
suppresses osteosarcoma metastasis and recruits macrophages by
activation of canonical WNT/β-catenin signaling through
stabilization of the WNT-receptor complex. Oncogene. 37:1049–1061.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Noy R and Pollard JW: Tumor-associated
macrophages: From mechanisms to therapy. Immunity. 41:49–61. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li R, Shi Y, Zhao S, Shi T and Zhang G:
NF-κB signaling and integrin-β1 inhibition attenuates osteosarcoma
metastasis via increased cell apoptosis. Int J Biol Macromol.
123:1035–1043. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shahi P, Wang CY, Chou J, Hagerling C,
Gonzalez Velozo H, Ruderisch A, Yu Y, Lai MD and Werb Z: GATA3
targets semaphorin 3B in mammary epithelial cells to suppress
breast cancer progression and metastasis. Oncogene. 36:5567–5575.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Si W, Huang W, Zheng Y, Yang Y, Liu X,
Shan L, Zhou X, Wang Y, Su D, Gao J, et al: Dysfunction of the
reciprocal feedback loop between GATA3- and ZEB2-nucleated
repression programs contributes to breast cancer metastasis. Cancer
Cell. 27:822–836. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ma L, Xue W and Ma X: GATA3 is
downregulated in osteosarcoma and facilitates EMT as well as
migration through regulation of slug. Onco Targets Ther.
11:7579–7589. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Choi JW, Herr DR, Noguchi K, Yung YC, Lee
CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN and Chun J: LPA
receptors: Subtypes and biological actions. Annu Rev Pharmacol
Toxicol. 50:157–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tsujiuchi T, Araki M, Hirane M, Dong Y and
Fukushima N: Lysophosphatidic acid receptors in cancer
pathobiology. Histol Histopathol. 29:313–321. 2014.PubMed/NCBI
|
|
69
|
Araki M, Kitayoshi M, Dong Y, Hirane M,
Ozaki S, Mori S, Fukushima N, Honoki K and Tsujiuchi T: Inhibitory
effects of lysophosphatidic acid receptor-5 on cellular functions
of sarcoma cells. Growth Factors. 32:117–122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Dong Y, Hirane M, Araki M, Fukushima N,
Honoki K and Tsujiuchi T: Lysophosphatidic acid receptor-5
negatively regulates cell motile and invasive activities of human
sarcoma cell lines. Mol Cell Biochem. 393:17–22. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Minami K, Ueda N, Ishimoto K and Tsujiuchi
T: LPA5-mediated signaling induced by endothelial cells and
anticancer drug regulates cellular functions of osteosarcoma cells.
Exp Cell Res. 388:1118132020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zjablovskaja P, Kardosova M, Danek P,
Angelisova P, Benoukraf T, Wurm AA, Kalina T, Sian S, Balastik M,
Delwel R, et al: EVI2B is a C/EBPα target gene required for
granulocytic differentiation and functionality of hematopoietic
progenitors. Cell Death Differ. 24:705–716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Matesanz-Isabel J, Sintes J, Llinàs L, de
Salort J, Lázaro A and Engel P: New B-cell CD molecules. Immunol
Lett. 134:104–112. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lafuente EM, van Puijenbroek AA, Krause M,
Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA,
Gertler FB and Boussiotis VA: RIAM, an Ena/VASP and Profilin
ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion.
Dev Cell. 7:585–595. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Patsoukis N, Bardhan K, Weaver JD, Sari D,
Torres-Gomez A, Li L, Strauss L, Lafuente EM and Boussiotis VA: The
adaptor molecule RIAM integrates signaling events critical for
integrin-mediated control of immune function and cancer
progression. Sci Signal. 10:eaam82982017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mace EM, Monkley SJ, Critchley DR and
Takei F: A dual role for talin in NK cell cytotoxicity: Activation
of LFA-1-mediated cell adhesion and polarization of NK cells. J
Immunol. 182:948–956. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Meri T, Amdahl H, Lehtinen MJ, Hyvärinen
S, McDowell JV, Bhattacharjee A, Meri S, Marconi R, Goldman A and
Jokiranta TS: Microbes bind complement inhibitor factor H via a
common site. PLoS Pathog. 9:e10033082013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bonavita E, Gentile S, Rubino M, Maina V,
Papait R, Kunderfranco P, Greco C, Feruglio F, Molgora M, Laface I,
et al: PTX3 is an extrinsic oncosuppressor regulating
complement-dependent inflammation in cancer. Cell. 160:700–714.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Corrales L, Ajona D, Rafail S, Lasarte JJ,
Riezu-Boj JI, Lambris JD, Rouzaut A, Pajares MJ, Montuenga LM and
Pio R: Anaphylatoxin C5a creates a favorable microenvironment for
lung cancer progression. J Immunol. 189:4674–4683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu J and Hoh J: Loss of complement factor
H in plasma increases endothelial cell migration. J Cancer.
8:2184–2190. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Martin M, Leffler J, Smoląg KI, Mytych J,
Björk A, Chaves LD, Alexander JJ, Quigg RJ and Blom AM: Factor H
uptake regulates intracellular C3 activation during apoptosis and
decreases the inflammatory potential of nucleosomes. Cell Death
Differ. 23:903–911. 2016. View Article : Google Scholar : PubMed/NCBI
|