Introduction
Hepatocyte nuclear factor 1β (HNF1B) is a
transcription factor which plays a crucial role during embryonic
development and differentiation of various organs (liver, kidney,
lung, gonads, biliary system, and pancreas) (1). The HNF1B protein regulates the
expression of multiple genes involved in cell cycle modulation,
susceptibility to apoptosis, and response to oxidative stress
(2,3). A growing number of studies have
demonstrated the potential involvement of HNF1B in the
tumorigenesis of some solid tumours. However, the precise mechanism
by which HNF1B influences tumour development has not yet been fully
elucidated. Recent findings suggest that HNF1B may act either as a
tumour suppressor or an oncogene in different cancers, depending on
the type of tissue and the tumour (4). Some authors regard HNF1B as a
pro-differentiation factor with a suppressive influence on
epithelial-to-mesenchymal transition (EMT) in unmethylated healthy
tissues (5). However, others have
described its role as an epithelial-specific oncogene, which
induces a cancerous phenotype, epithelial-to-mesenchymal
transition, and invasive behaviour (6).
To date, the only studies concerned with the
involvement of the HNF1B gene in the carcinogenesis of
tumours of the female genital tract are mostly genome-wide
associated studies (GWAS), exploring the influence of single
nucleotide polymorphisms (SNPs) on ovarian and endometrial cancers.
It has been reported that in endometrial cancer there is an
association between the SNP rs4430796 and the risk and prognosis of
the disease (7–11). In ovarian cancer, several SNPs of
interest have been identified. The SNP rs757210 was found to be
associated with promoter methylation in high-grade serous carcinoma
(HGSC), and its involvement also reached borderline significance in
ovarian clear cell carcinoma (OCCC) (5). There are other SNPs which have also
been implicated in influencing promotor methylation: SNP rs7405776
is associated with increased methylation in HGSC, and SNP
rs11651755 was reported to be associated with unmethylated status
and an increased HNF1B expression in OCCC (10).
The immunohistochemical expression of HNF1B was
initially considered to be a highly specific marker of OCCC
(7). However, recent studies have
also described the positivity of HNF1B in some other malignant and
benign tumours of the female genital tract, and even in
non-neoplastic endometrial lesions and normal endometrium (12–14).
These findings demonstrate that the specificity of HNF1B is less
than was originally assumed. However, if we take into account the
staining intensity, then the characteristic diffuse strong nuclear
expression has been found predominantly in clear cell carcinomas
(15,16).
Concerning the downstream targets, the transcription
factor HNF1B seems to be involved in several key regulatory
pathways including cell cycle regulation, epithelial-mesenchymal
transition, cell migration, adhesion, and proliferation. One of the
possible downstream targets of HNF1B seems to be enoyl-CoA-(Δ)
isomerase 2 (ECI2), which belongs to the acyl-CoA-binding domain
(ACBD) family (17). The ECI2 enzyme
plays a role in glucose and lipid metabolism, allowing for the
re-entry of the enoyl-CoA into the β-oxidation cycle (18,19).
Currently, the exact role of ECI2 in carcinogenesis is still poorly
understood. However, a recent study has shown that the inhibition
of expression of ECI2 in prostate cancer leads to decreased glucose
utilization, the accumulation of fatty acids, and the
down-regulation of the cell cycle-associated genes (19). Furthermore, the regulatory role of
HNF1B in ECI2 expression was investigated on a mouse model of
prostate cancer (17). The authors
found that the increased expression levels of HNF1B in the initial
stages of prostatic cancer play a tumour-protective role and are
associated with ECI2 upregulation. Nevertheless, the precise
mechanism explaining the possible interactions between HNF1B and
ECI2 and their role in tumorigenesis have not yet been fully
elucidated. ECI2 is targeted to mitochondria and peroxisomes, and
probably mediates their mutual interaction (20). The data about the expression of ECI2
in ovarian cancer is currently unknown.
This is the first retrospective analysis of a large
subset of HGSC which includes a complex genetic, epigenetic, and
histochemical analysis of HNF1B, complete with an analysis of the
possible HNF1B's downstream target ECI2. The goals of our study
were: i) To perform a comprehensive molecular and
immunohistochemical analysis of HNF1B in HGSC, including the
protein and mRNA expression, and the epigenetic and genetic changes
of HNF1B, ii) to perform a protein and mRNA expression analysis of
its downstream target ECI2, iii) to correlate HNF1B methylation
with mRNA or protein expression, and to study the association of
HNF1B and ECI2 expression, iv) to analyse the relationship between
the molecular and immunohistochemical patterns with the
clinicopathological variables and clinical outcomes.
Materials and methods
Samples
Formalin-fixed paraffin-embedded (FFPE) tissue
blocks were primarily used for the analyses. Where available, the
corresponding fresh-frozen tissue (FT) extracted from the same
individuals was used for the subsequent molecular DNA/RNA analysis.
The FFPE samples were obtained from the archive files of the
Institute of Pathology, and the FT samples were provided by the
Bank of Biological Material (BBM) of the First Faculty of Medicine,
Charles University in Prague. The FFPE samples were stored in the
archives at 10–14°C and the FT samples were stored in the RNAlater
stabilization solution (Qiagen) at −80°C, according to the
manufacturer's protocol (Stabilization of RNA in Harvested Animal
Tissues; Qiagen).
In total, 122 FFPE samples of HGSC were selected for
immunohistochemical analysis, including 69 cases with an available
FT for subsequent epigenetic, genetic, and expression analysis. All
FFPE and FT tissue samples were reviewed by senior pathologists who
selected the eligible areas for tumour analysis. The mean age of
patients was 59 years (median, 60 years; range, 36–81 years). The
clinicopathological characteristics of the samples analysed are
summarized in Tables I and II.
 | Table I.Association of HNF1B protein
expression (H-score) and clinicopathological characteristics, based
on 122 cases of HGSC. |
Table I.
Association of HNF1B protein
expression (H-score) and clinicopathological characteristics, based
on 122 cases of HGSC.
| Characteristic | N | H-score mean | H-score median | P-value | H-score group
1 | H-score group
2 | P-value |
|---|
| Age of diagnosis
(mean=59, median=60), years |
|
|
| 0.094b |
|
| 0.273c |
|
<60 | 59 | 13.8 | 0 |
| 48 | 11 |
|
|
≥60 | 63 | 29.3 | 0 |
| 46 | 17 |
|
| FIGOa |
|
|
| 0.953b |
|
| 0.676c |
| I | 10 | 27.6 | 0 |
| 7 | 3 |
|
| II | 7 | 18.6 | 0 |
| 6 | 1 |
|
|
III | 83 | 22.5 | 0 |
| 62 | 21 |
|
| IV | 20 | 19.2 | 0 |
| 17 | 3 |
|
| Lymphovascular
invasiona |
|
|
| 0.025b |
|
| 0.023c |
|
Yes | 63 | 27.7 | 0 |
| 45 | 18 |
|
| No | 28 |
8.6 | 0 |
| 26 | 2 |
|
| Neoadjuvant
therapy |
|
|
| 0.607b |
|
| 0.768c |
|
Yes | 28 | 24.4 | 0 |
| 21 | 7 |
|
| No | 94 | 20.9 | 0 |
| 73 | 21 |
|
| Local
recurrencea |
|
|
| 0.261b |
|
| 0.734c |
|
Yes | 55 | 16.8 | 0 |
| 42 | 13 |
|
| No | 49 | 29.5 | 0 |
| 36 | 13 |
|
| Distant
recurrencea |
|
|
| 0.087b |
|
| 0.651c |
|
Yes | 52 | 15.8 | 0 |
| 40 | 12 |
|
| No | 52 | 30.1 | 0 |
| 38 | 14 |
|
|
Methylationa |
|
|
| 0.056b |
|
| 0.089c |
|
Yes | 26 | 12.3 | 0 |
| 23 | 3 |
|
| No | 41 | 21.6 | 0 |
| 29 | 12 |
|
|
rs4430796a |
|
|
| 0.993b |
|
| 0.535d |
|
Yes | 44 | 20.4 | 0 |
| 35 | 9 |
|
| No | 21 | 13.9 | 0 |
| 15 | 6 |
|
|
rs757210a |
|
|
| 0.613b |
|
| 0.476d |
|
Yes | 52 | 17.9 | 0 |
| 41 | 11 |
|
| No | 13 | 19.8 | 0 |
| 9 | 4 |
|
|
rs7405776a |
|
|
| 0.071b |
|
| 0.115d |
|
Yes | 44 | 24.8 | 0 |
| 31 | 13 |
|
| No | 21 |
5.0 | 0 |
| 19 | 2 |
|
 | Table II.Association of ECI2 protein
expression (H-score) and clinico-pathological characteristics,
based on 122 cases of HGSC. |
Table II.
Association of ECI2 protein
expression (H-score) and clinico-pathological characteristics,
based on 122 cases of HGSC.
| Characteristic | N | H-score mean | H-score median | P-value | H-score group
1 | H-score group
2 | P-value |
|---|
| Age of diagnosis
(mean=59, median=60), years |
|
|
| 0.673b |
|
| 0.508c |
|
<60 | 59 | 110.1 | 105 |
| 14 | 45 |
|
|
≥60 | 63 | 104.7 | 105 |
| 18 | 45 |
|
| FIGOa |
|
|
| 0.372b |
|
| 0.242d |
| I | 10 | 96.7 | 102 |
| 4 | 6 |
|
| II | 7 | 119.5 | 120 |
| 0 | 7 |
|
|
III | 83 | 106.2 | 105 |
| 21 | 62 |
|
| IV | 20 | 107.6 | 102 |
| 7 | 13 |
|
| Lymphovascular
invasiona |
|
|
| 0.217b |
|
| 0.278b,c |
|
Yes | 63 | 104.3 | 104 |
| 18 | 45 |
|
| No | 28 | 116.1 | 114 |
| 5 | 23 |
|
| Neoadjuvant
therapy |
|
|
| 0.901b |
|
| 0.511c |
|
Yes | 28 | 109.6 | 105 |
| 6 | 22 |
|
| No | 94 | 106.6 | 105 |
| 26 | 68 |
|
| Local
recurrencea |
|
|
| 0.517b |
|
| 0.332c |
|
Yes | 55 | 107.5 | 104 |
| 17 | 38 |
|
| No | 49 | 105.1 | 110 |
| 11 | 38 |
|
| Distant
recurrencec |
|
|
| 0.556b |
|
| 0.658c |
|
Yes | 52 | 111.0 | 107 |
| 13 | 39 |
|
| No | 52 | 101.8 | 105 |
| 15 | 37 |
|
Immunohistochemical analysis
The FFPE blocks were used for the construction of
the tissue microarrays (TMAs). Two cores (each 2.0 mm in diameter)
were taken from a single donor block from each case, using the
tissue microarray instrument TMA Master (3DHISTECH Ltd.). All
samples of FFPE tissue were sectioned at a thickness of 4–5 µm, and
the immunohistochemical (IHC) analysis was performed using an
antibody against HNF1B (polyclonal, dilution 1:500; Sigma-Aldrich;
Merck KGaA), and a rabbit antibody against ECI2 (polyclonal,
dilution 1:100, product no. ab235322; Abcam) in the automated
staining instrument Ventana BenchMark ULTRA (Roche). Heat induced
antigen retrieval, including pre-treatment, was carried out in a
citrate buffer (pH 6.0). For visualization, the OptiView DAB IHC
Detection Kit (Ventana, Roche) was used. For HNF1B, only nuclear
staining was regarded as positive, and for ECI2 only the
cytoplasmic staining was evaluated. The expression of both markers
was double-blindly evaluated by two pathologists.
The immunohistochemical results were assessed
semi-quantitatively, using the H-score method described previously
by others (21). The H-score
combines the percentage of positive cells and the level of staining
intensity (1+ for weak, 2+ for moderate, and 3+ for strong
intensity). The final H-score is then calculated according to the
following formula: [1 × (% of cells 1+) + 2× (% of cells 2+) + 3×
(% of cells 3+)], with the results ranging from 0 to 300.
DNA and RNA isolation, quality control
and cDNA synthesis
Firstly, the FT tissues were thawed and homogenized
(10–30 mg) in the presence of 600 µl of RLT Buffer (Qiagen) with 6
µl of 14.3M 2-mercaptoehthanol (Sigma-Aldrich; Merck KGaA) using
MagNA Lyser Green Beads tubes in MagNA Lyser Instrument (Roche), as
described in Bartu et al (22).
The total DNAs and RNAs were isolated according to
the Simultaneous Purification of Genomic DNA and Total RNA from
Animal Tissues protocol using an AllPrep DNA/RNA Mini kit (Qiagen).
The isolated DNA and RNA samples were quantified by the NanoDrop
2000 (Thermo Fisher Scientific, Inc.).
The RNA Quality Number (RQN) of the isolated total
RNA was determined using the Fragment Analyzer (AATI) capillary
electrophoresis system and Standard RNA kit (AATI). Those RNA
samples with an RQN lower than 7.5 were removed from further
analysis (tissue samples RQN mean=9.3; range 5–10). Otherwise, 3.75
µg of total RNA of each sample (where available) was treated by
DNase I (Thermo Fisher Scientific, Inc.), and cDNA was synthetized
in a 40 µl reaction using SuperScript III Reverse Transcriptase
(Thermo Fisher Scientific, Inc.) with random hexamers (Roche) as
described in Dundr et al (23).
HNF1B mutation analysis
The mutation analysis of the HNF1B gene included the
analysis of all the coding region (exons 1–9, RefSeq NM_000458.2)
with adjacent intronic sequences (±15 bp) and two deep intronic
regions containing the rs7527210 and rs4430796 polymorphisms. The
FT samples were analysed by amplicon next generation sequencing, as
described previously (22).
Primers for the analysis of rs7405776 were designed
(rs7405776_Forward: agccacagactctagatctgg, rs7405776_Reverse:
caaagtgctgggattataagtgtg), and the amplicons were sequenced by
Sanger sequencing on the ABI3500 Genetic Analyser (Thermo Fisher
Scientific, Inc.).
Mutations which are not found in the literature, the
Single Nucleotide Polymorphism Database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/), the ClinVar
Database (https://www.ncbi.nlm.nih.gov/clinvar/), or in the
Catalogue of Somatic Mutations in Cancer (COSMIC, http://www.sanger.ac.uk/cosmic; databases
accessed September 2020) are considered as novel.
HNF1B promoter methylation
analysis
The epigenetic analysis of the HNF1B promoter region
was performed as described previously (22). The isolated DNA was converted using
the EZ DNA Methylation-Lightning Kit (Zymo Research) according to
the manufacturer's instructions. The primers for the PCR
amplification of both the methylated and unmethylated alleles were
designed using Methprimer software (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi;):
HNF1B_met_forward_TTTTTGGATTTGTTAAGTTAGTGTTTT, HNF1B_met_reverse
CCCTTCCTAAATAATCAATTTCTCTT (PCR product chr17:36105251-36105506,
GrCh37). The PCR amplification was carried out using the following
protocol: 95°C_12 min, 40× (95°C_15 sec, 58°C_30 sec, 72°C_30 sec),
72°C_5 min, and followed by melting curve analysis in the
LightCycler 480 II instrument (Roche). The amplification and
melting curves were analysed by the LightCycler 480 II Software,
and then compared to the control mixes.
Analysis of mRNA expression
The expression analysis was performed by using cDNA
samples and the droplet digital PCR system (ddPCR; Bio-Rad). All
the ddPCR steps, including the expression of three potential
reference mRNA targets (POLR2A, HPRT1 and ATP5F1B), two HNF1B mRNA
targets (in 5′ and 3′UTR), and one ECI2 target, as well as
repeatability and reproducibility, were optimized prior to the
general ddPCR analysis.
The ddPCR reactions were prepared according to the
manufacturer's instructions using the QX200 ddPCR EvaGreen Supermix
(Bio-Rad), 1 µl of cDNA template (which corresponds to
approximately 90 ng of total RNA) and 4 pmol of each of the primer
pairs (200 nM final concentration) in 20 µl of reaction volume.
Master mix droplets were generated by the QX200 AutoDG instrument
(Bio-Rad), and the samples were amplified by a 5 min incubation at
95°C, then 40 cycles of 95°C for 30 sec and 58°C for 1 min,
followed by the final signal stabilization steps consisting of 4°C
for 5 min, finishing with 90°C for 5 min. The resulting data was
acquired using the QX200 Droplet Reader instrument (Bio-Rad) with
the standard acquisition protocol for the EvaGreen master mix and
analysed by QuantaSoft software (Bio-Rad). The threshold for
positive droplet signals of each of the three final amplicons
(reference POLR2A; HNF1B 3′UTR and ECI2 targets) was set as the
average value of the thresholds, which were calculated
automatically by the QuantaSoft software during the optimization
steps. The thresholds of all the acquired targets were manually
confirmed. The final data of the targets (HNF1B and ECI2),
expressed as the number of templates in 20 µl of the master mix
(which corresponds to 1 µl of cDNA), were re-calculated to the
number of targets per one thousand reference POLR2A targets, and
analysed as described below in the Statistical analysis section.
Only samples with a positive HNF1B mRNA expression (the reliable
limit for positivity was set to more than 50 copies/µl of cDNA)
were further compared to the mRNA ECI2 expression.
Statistical analysis
All the statistical analyses were performed using
the software Statistica (TIBCO). The Shapiro-Wilk test was used to
control data normality. With respect to the non-normal data
distribution, non-parametric analyses were conducted (Mann-Whitney
U test or Kruskal-Wallis H-test) in order to analyse the
association between HNF1B or ECI2 protein expression (H-score) and
the clinico-pathological variables (age at the time of diagnosis,
FIGO stage, lymphovascular invasion (LVSI), neoadjuvant therapy,
local and distant recurrence, and, in the case of HNF1B, promoter
methylation status and the presence of the three analysed SNPs:
rs4430796, rs757210, rs7405776. For the evaluation of the effect of
independent clinicopathological characteristics on the categorized
H-score or methylation status, the Pearson χ2 test or
the Fisher's exact test was used, depending on the expected
frequencies (24). Correlations
between two continuous variables were analysed using Pearson's
method.
For the purposes of χ2 tests and survival
analyses, the H-score of both HNF1B and ECI2 was categorized into
two groups with respect to their median values (HNF1B: Group 1:
H-score 0–19; group 2: H-score 20–300; ECI2: Group 1: H-score 0–99,
group 2: H-score 100–300).
Survival analyses were plotted using the
Kaplan-Meier model and the differences between curves were tested
for significance using the log-rank test. Disease-free survival
(DFS), local recurrence-free survival (LFS) and metastasis-free
survival (MFS) were defined as the time from the date of the
diagnosis to the date of a specific event: Death as a result of the
diagnosis (DFS), the first local recurrence (LFS) and/or the first
distant metastasis (MFS). If the patient did not show any of the
monitored events, the case was censored in the analysis at the date
of the last follow-up. The probability of survival between the
HNF1B and ECI2 H-score group 1 and group 2 was also compared. All
tests were two-sided and a P-value of <0.05 was considered as
significant.
The Cancer Genome Atlas (TCGA) data
access and analysis of HNF1B promoter methylation and mRNA
expression of HNF1B and ECI2
The data from the TCGA, including the
clinico-pathological findings and mRNA expression (z-score) of
HNF1B and ECI2, was downloaded through the cBioPortal (www.cbioportal.org; (TCGA, Ovarian Serous
Cystadenocarcinoma, Firehose Legacy, access June 2020)).
Additionally, the HNF1B promoter methylation data was downloaded
through the portal https://mexpress.be/ (gene: HNF1B, cancer type:
Ovarian serous cystadenocarcinomas, access April 2020) to compare
our results. The TCGA sample set included 569 samples of the
histological type of serous cystadenocarcinoma with stated stage,
neoplasm histologic grade 2 or 3, and methylation status. The mRNA
expression of HNF1B and ECI2 was available only for 178 of these
samples. The locus cg12788467 (position to the relative
transcription start site −238, GRCh37), which is also included in
our analysed promoter region, was used for the investigation of the
methylation status of the HNF1B promoter region. The β-value
>0.3 of this locus was determined as hypermethylated (25).
Results
Immunohistochemical findings and
clinicopathological associations
The immunohistochemical analysis of both markers
(HNF1B and ECI2) was performed in all 122 cases of HGSC. The
nuclear protein expression of HNF1B was generally very low
(mean=21.8, median=0). The HNF1B positivity was observed in 28
cases (H-score ranging from 20 to 99 in 15 cases, H-score ranging
from 100 to 200 in 13 cases). The rest of the samples were negative
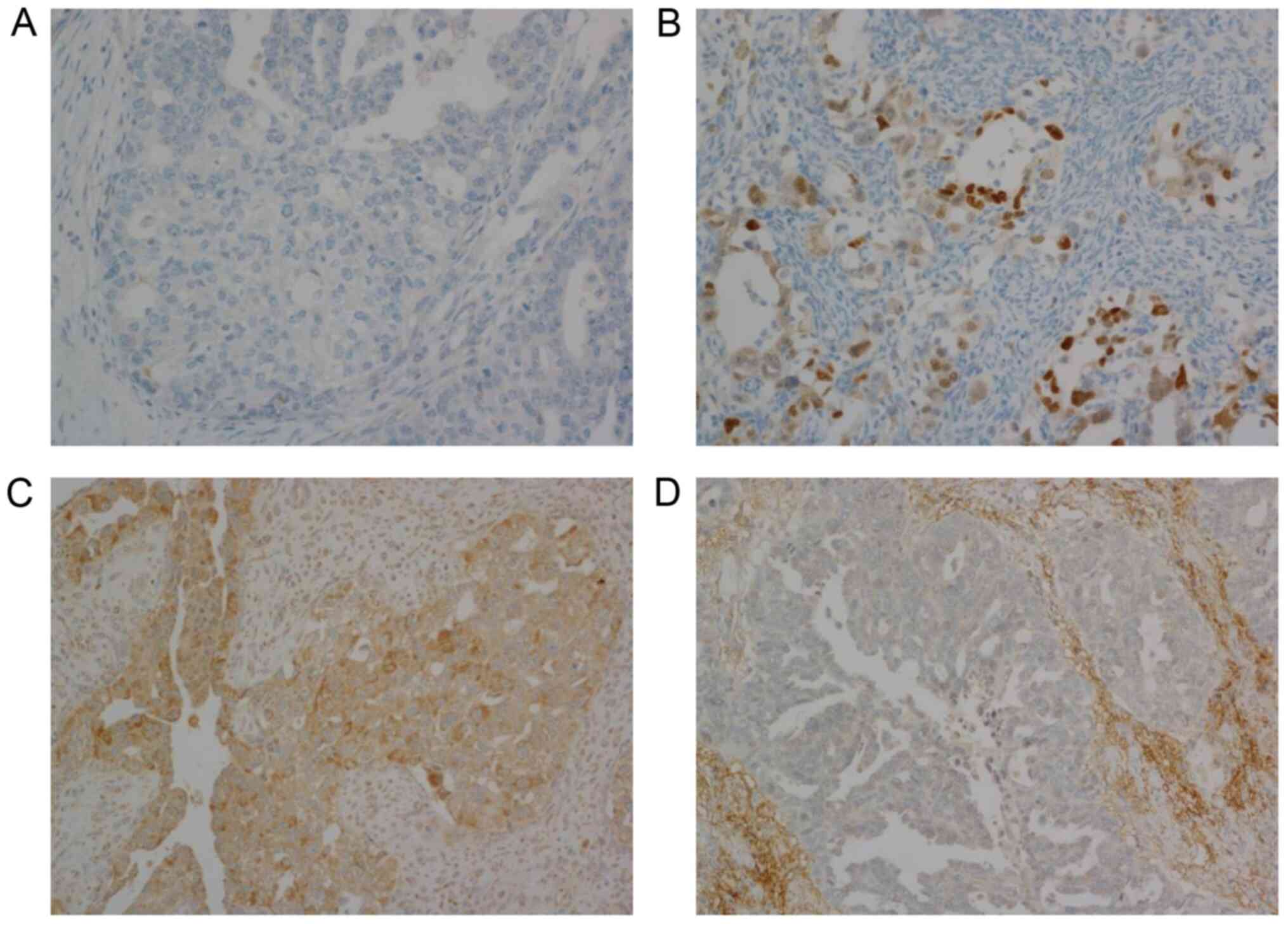
(H-score=0, n=112; Fig. 1A). The
intensity of staining was mostly mild to moderate (Fig. 1B). Focal strong nuclear staining was
observed in 17 cases, but the portion of positive cells did not
exceed 30% of all the tumour cells. None of our cases exceeded an
H-score value of 200. The cytoplasmic expression of ECI2 ranged
from 0 to 190 (mean=107.3, median=105; Fig. 1C). Seven cases did not express ECI2
at all (H-score=0; Fig. 1D).
Adjacent non neoplastic ovarian tissue showed positive cytoplasmic
staining of ECI2 and nuclear negativity of HNF1B staining in all
samples.
A higher protein HNF1B expression was associated
with lymphovascular invasion (Z=−2.23, N=91, P=0.025; Table I). No other significant associations
of HNF1B or ECI2 protein expression with clinico-pathological
characteristics were observed (Tables
I and II).
The relationship between HNF1B or ECI2 expression
and disease outcome, including disease-free survival (DFS), local
recurrence-free survival (LFS), and metastasis-free survival (MFS)
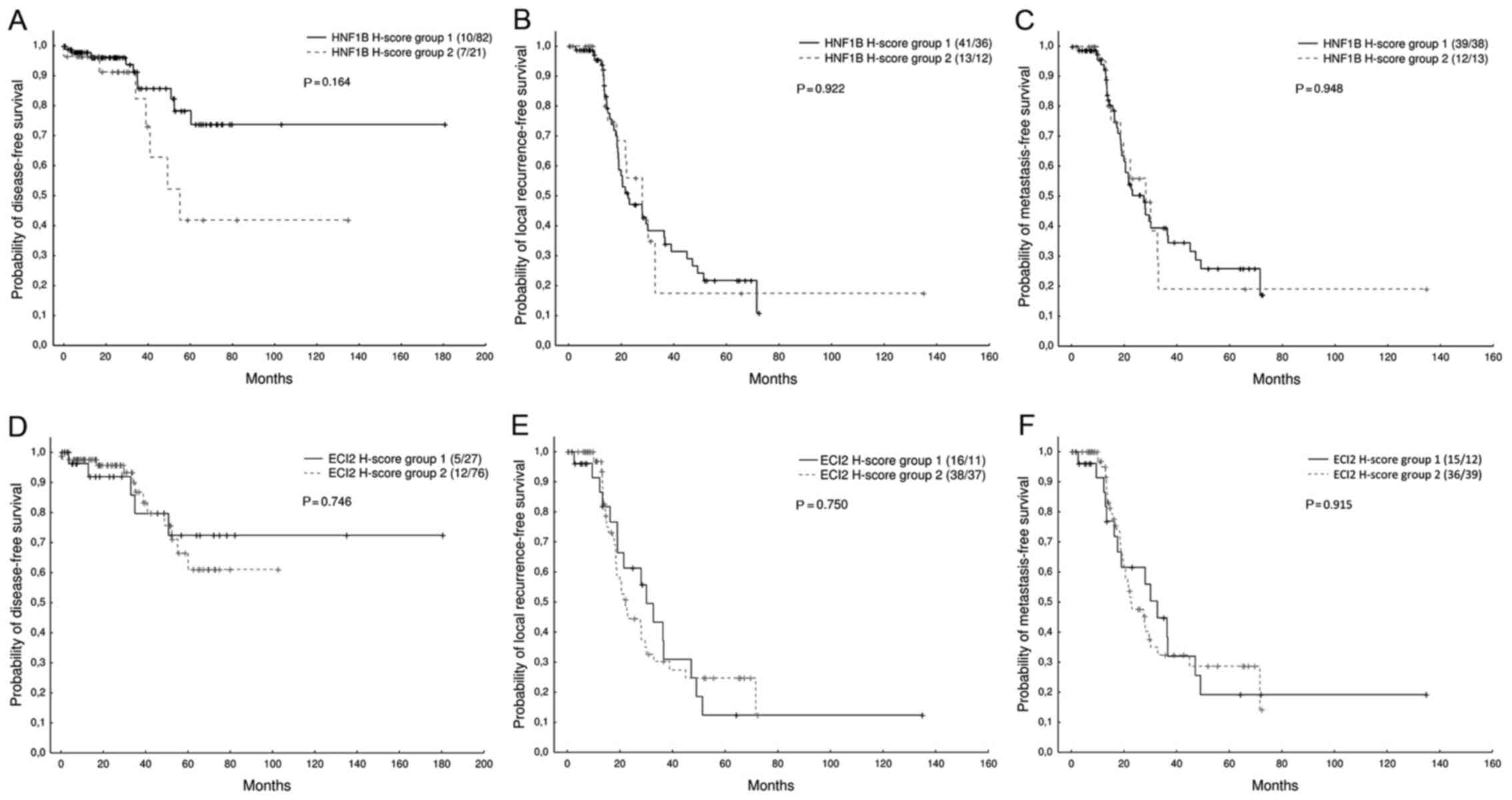
was analysed. No significant association between the HNF1B or ECI2
expression (H-score categorized into two groups, as described in
Materials and methods) and the probability of DFS (Z=1.39, N=120,
P=0.164/Z=−0.32, N=120, P=0.746, respectively), LFS (Z=−0.09,
N=102, P=0.922/Z=0.32, N=102, P=0.750, respectively), or MFS
(Z=−0.06, N=102, P=0.948/Z=−0.11, N=102, P=0.915, respectively) was
found. However, we observed a non-significant trend suggesting an
association between a lower HNF1B expression and a higher
probability of disease-free survival (Fig. 2).
Relationship between genetic,
epigenetic, and expression characteristics
The mutation analysis of HNF1B was successfully
performed on 61 FT tumour samples. In one of the 61 (1.6%) analysed
tumour samples, a nonsense somatic mutation NM_000458.2:
c.1063C>T, p.(Q355X) with a variant allele frequency 68% was
found. This case showed complete negativity of the HNF1B protein
expression (H-score=0), together with moderate positivity of ECI2
(H-score=100).
The HNF1B promoter methylation was detected
in 26 (38.8%) of the 67 analysed samples. The methylation status
did not correlate with any of the tested clinicopathological
characteristics (Table III).
Moreover, none of the three investigated SNPs (rs7405776,
rs4430796, and rs7527210) showed any association with HNF1B protein
expression (Table I) or HNF1B
promoter methylation (Table III).
Samples without HNF1B promoter methylation had a
significantly higher HNF1B mRNA expression (Z=2.91, N=46, P=0.003).
Association of HNF1B promoter methylation with protein expression
was marginally non-significant (Z=1.91, N=67, P=0.056).
 | Table III.Association of HNF1B promoter
methylation and selected clinicopathological characteristics, based
on 67 cases of HGSC. P-values are based on Pearson χ2
test. |
Table III.
Association of HNF1B promoter
methylation and selected clinicopathological characteristics, based
on 67 cases of HGSC. P-values are based on Pearson χ2
test.
|
Characteristics | Methylation
yes | Methylation no | P-value |
|---|
| Age of diagnosis,
years |
|
| 0.307 |
|
<60 | 16 | 20 |
|
|
≥60 | 10 | 21 |
|
| FIGOa |
|
| 0.714 |
| I | 1 | 4 |
|
| II | 2 | 3 |
|
|
III | 17 | 27 |
|
| IV | 6 | 6 |
|
| Lymphovascular
invasiona |
|
| 0.778 |
|
Yes | 12 | 26 |
|
| No | 5 | 9 |
|
| Neoadjuvant
therapy |
|
| 0.478 |
|
Yes | 5 | 11 |
|
| No | 21 | 30 |
|
| Local
recurrencea |
|
| 0.252 |
|
Yes | 14 | 19 |
|
| No | 6 | 16 |
|
| Distant
recurrencea |
|
| 0.878 |
|
Yes | 11 | 20 |
|
| No | 9 | 15 |
|
|
rs4430796a |
|
| 0.615 |
|
Yes | 16 | 28 |
|
| No | 9 | 12 |
|
|
rs757210a |
|
| 0.524 |
|
Yes | 19 | 33 |
|
| No | 6 | 7 |
|
|
rs7405776a |
|
| 0.614 |
|
Yes | 16 | 28 |
|
| No | 9 | 12 |
|
Correlations of HNF1B and ECI2
expression on mRNA and protein level
HNF1B expression was successfully analysed in all of
the 122 HGSC FFPE samples (protein) and corresponding 47 FT samples
(mRNA). ECI2 mRNA expression was analysed in a limited subset of
14/47 (29.8%) FT samples, where the HNF1B mRNA expression was
positive. ECI2 protein expression was successfully analysed and
compared in all 122 HGSC samples.
Increased mRNA expression of HNF1B significantly
correlated with increased protein expression (F=12.18,
R2=0.213, N=47, P=0.001; Fig.
3). A positive correlation was not observed for ECI2 mRNA and
protein expression (F=0.51, R2=0.041, N=14, P=0.488,
Fig. 3). A significant positive
association was observed when comparing HNF1B with ECI2 protein
expression (F=8.03, R2=0.063, N=122, P=0.005; Fig. 3), however, no association was
observed for mRNA expression (F=0.05, R2=0.005, N=14,
P=0.816; Fig. 3). However, these
results may be influenced by the limited number of samples.
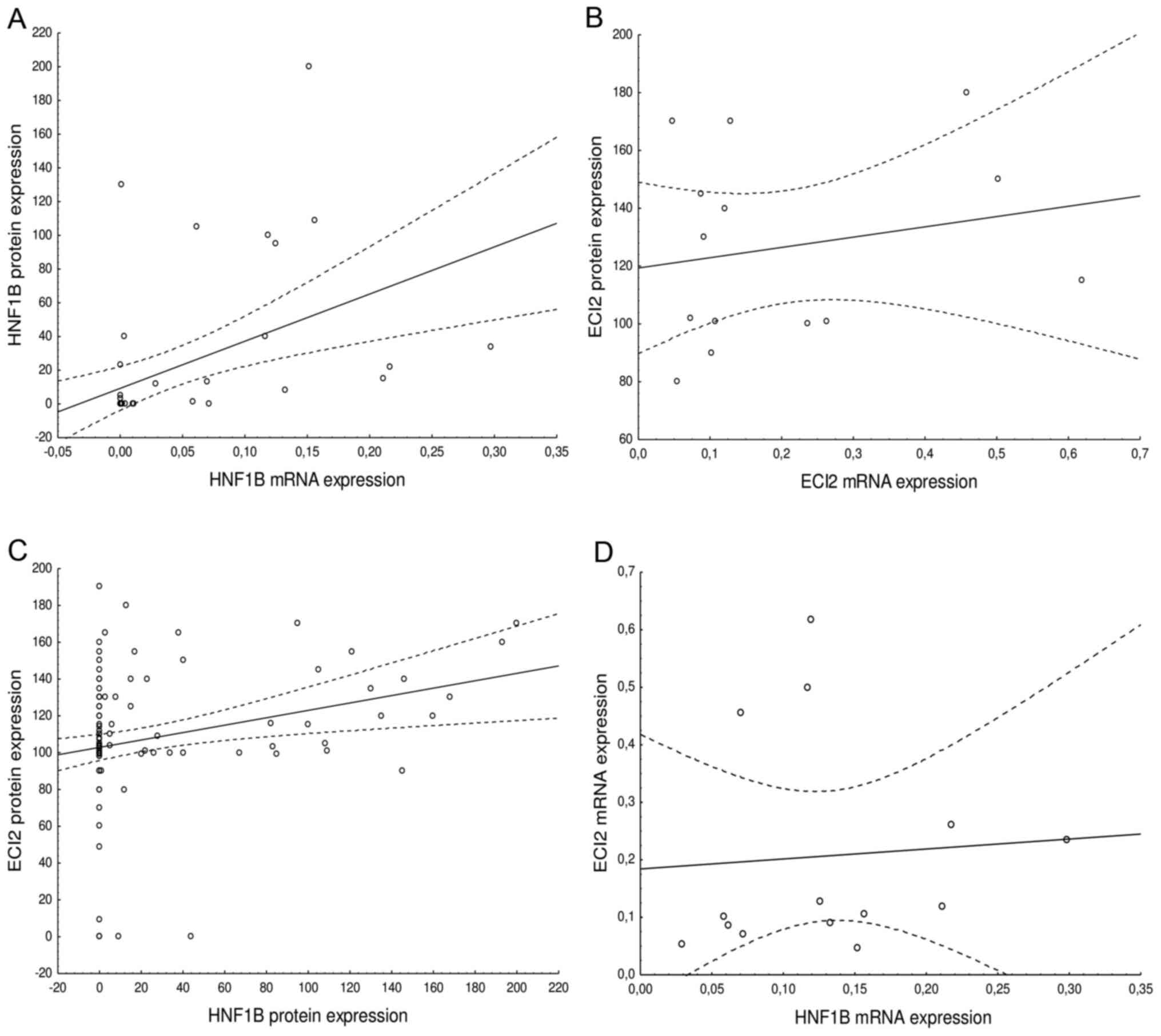 | Figure 3.Association of mRNA expression and
protein expression of (A) HNF1B. (F=12.18, R2=0.213,
N=47, P=0.001) and (B) ECI2 (F=0.51, R2=0.041, N=14,
P=0.488), and the association of expression of HNF1B and ECI2 on a
(C) protein level (F=8.03, R2=0.063, N=122, P=0.005) and
(D) mRNA level (F=0.05, R2=0.005, N=14, P=0.816). Dashed
lines represent 0.95 confidence interval. HNF1B, hepatocyte nuclear
factor 1β; ECI2, enoyl-CoA (Δ) isomerase 2 |
Re-analysis of mRNA and epigenetic
status of TCGA dataset
Based on TCGA data, the methylation of the HNF1B
promoter was detected in 315/569 (55%) samples of HGSC. A
significantly higher expression of HNF1B mRNA was detected in the
non-methylated cases, in comparison to the methylated cases
(Z=3.32, N=71, P<0.001). No statistically significant
correlation between the mRNA expression of HNF1B and ECI2 was found
(F=0.25, R2=0.001, N=71, P=0.614).
Discussion
HNF1B is a transcriptional factor implicated in the
carcinogenesis of solid tumours, and its role in the tumorigenesis
of the female genital tract has so far mostly focused on clear cell
carcinomas, especially OCCC (7,26,27).
However, the expression of this marker was also found in HGSC and
in other types of ovarian tumours (28,29).
HGSC is the most common and most aggressive subtype of ovarian
cancer with poor prognosis, accounting for up to 80% of all deaths
from ovarian carcinoma (30). The
possible involvement of HNF1B in the carcinogenesis of HGSC has not
yet been fully elucidated. In HGSC, the expression of HNF1B is
predominantly low, and the current hypothesis is that in this
context HNF1B acts as a tumour suppressor gene (5,7,26).
Some knowledge about this issue can be found in the
GWAS, which were mostly focused on the involvement of various SNPs
in ovarian cancer. Ross-Adams et al examined the
relationship between SNPs and epigenetic changes in ovarian and
prostatic cancers, also exploring their potential influence on EMT.
They found a significant association between the SNP rs757210 and
tumour methylation of HNF1B in HGSC, but there was no significant
association with HNF1B expression levels (5). Other authors, such as Shen et
al, investigated selected SNPs in HGSC (rs7405776) and OCCC
(rs11651755), and they did not prove any significant, but only
borderline association between rs7405776 and increased promoter
methylation in HGSC (P=0.07) (10).
In accordance with those findings our results also showed no
significant relationship between promoter methylation and
rs7405776, or the other SNPs (rs4430796, and rs757210) which are
most commonly implicated in carcinomas of the female genital system
(Table III). Therefore, it is not
surprising that we and others have also observed no association
between the abovementioned SNPs and HNF1B protein expression or
mRNA expression (5,10).
We observed methylation of the HNF1B promoter
region in 38.8% (26/67) HGSC, which is similar to the observations
of other authors: 41.3% (12/29) (31), 42% (120/286) (10), 45% (18/40) (32) and 50.8% (31/61) (33). However, the TCGA data shows a higher
frequency of methylated HGSCs (55.4%; 315/569). Our data showed no
statistically significant correlation between the methylation
status and any other tested characteristics, including the tumour
stage (Table III), which is in
concordance with other studies (31). Data from studies published by
Bubancova et al, Baranova et al, and the TCGA data
showed a more frequent HNF1B promoter methylation in tumours of a
later stage (stage I/II 36.2 vs. stage III/IV 60.1%; 28.6 vs.
57.5%; 34.0 vs. 57.3%). We observed the same trend (stage I/II 30
vs. stage III/IV 41.7%), but this finding was not statistically
significant (P>0.05) in our cohort.
The mutation analysis revealed that in one case of
the 61 (1.6%) there was a novel somatic HNF1B truncating
mutation in exon 5 (p.Q355X). This pathogenic variant is predicted
to encode a truncated non-functional protein, which is in
accordance with immunohistochemical analysis where complete
negativity of HNF1B staining (H-score=0) was observed. Our findings
support our previous assumption that the expression of HNF1B in
tumour tissue is downregulated by either hypermethylation of the
promoter, or by other potential regulatory mechanism which affects
transcriptional activation, rather than mutations in the HNF1B
coding sequence or posttranscriptional or posttranslational
modifications of gene expression (22).
We also analysed the immunohistochemical and mRNA
expression of HNF1B, with the aim of exploring whether there is a
correlation between the expression and clinico-pathological
characteristics. In total, we found immunohistochemical positivity
of HNF1B in 23% (28/122) of cases and discovered a significant
association between higher values of the H-score and lymphovascular
invasion (P=0.025). This finding supports the contention of some
authors that HNF1B induces transformation and
epithelial-to-mesenchymal transition, which can contribute to the
acquisition of the invasive properties which are essential for the
metastasis's invasive behaviour (6,34). No
associations between HNF1B or ECI2 protein expression and disease
outcome (Fig. 2) were observed.
So far, studies concerning the expression of HNF1B
in HGSC have been focused on the diagnosis of OCCC and the
differentiation of tumours with clear cell morphological features
from clear cell carcinomas (15,35–37). The
discriminative threshold in these studies was adjusted to
differentiate OCCC from other tumour types and, as such, HNF1B
expression was confirmed as a specific marker for the diagnosis of
OCCC. Huang et al found positive expression of HNF1B in only
2.9% (1/35 cases) of HGSCs (35).
However, only tumours with diffuse, and moderate or strong nuclear
positivity were regarded as positive by these authors, which is
quite a rare occurrence in HGSC. In their study, Kao et al
investigated HGSCs with clear cell changes and found that only 5%
(3/60 cases) reached an H-score of >10 (this H-score threshold
was determined as the cut-off to differentiate HGSC and endometroid
ovarian cancer from OCCC in their study) (15). Kobel et al investigated the
expression of HNF1B in 133 OCCCs and 200 HGSCs, and found focally
distributed nuclear positivity in 4.8% of HGSCs (36). In another study, Li et al
reported HNF1B positivity in 13.3% (4/30 cases) of HGSCs (37), and none of the HGSC cases showed
strong nuclear positivity. The rather lower reported positivity
when compared to our results can most likely be explained by
differences in methodology, such as the use of a different antibody
with different pretreatment, or different visualization methods, as
well as different criteria for the evaluation of HNF1B
expression.
Our data showed a statistically significant positive
correlation between mRNA and protein expression of HNF1B (P=0.001).
This finding implies that the regulation of HNF1B expression on the
translational or posttranslational level probably does not play a
significant role.
The data concerning the significance of ECI2 in
ovarian carcinomas, including HGSC, is currently unknown. According
to studies on prostate cancer, the inhibition of ECI2 can trigger
acute metabolic stress of the tumour cells, and thus targeting ECI2
could be potentially used as a therapeutic approach (19). According to some studies, a higher
expression of the ECI2 protein is associated with a poor outcome in
prostate cancer, but the data concerning the expression of ECI2 in
ovarian cancer is lacking (18). In
our study, we found cytoplasmic expression of ECI2 in all but 7
cases (H-score up to 190). Our data showed no correlation between
any of the clinico-pathological characteristics and survival on a
statistically significant level (Table
II). Regarding the connection between the expression of HNF1B
and ECI2, our data showed a significant positive relationship on a
protein level (P=0.005), but we did not observe correlation at an
mRNA level. However, this is in concordance with the re-analysis of
the TGCA dataset, where no correlation between the mRNA expression
of HNF1B/ECI2 has been found.
In conclusion, the comprehensive analysis of
molecular and immunohistochemical characteristics of HNF1B and its
possible downstream target ECI2 brought about several important
findings. Our data from the expression analysis confirmed a
generally low HNF1B expression on a protein level, which positively
correlated with lymphovascular invasion, but with no other
clinico-pathological characteristics. Nevertheless, despite the
observed low levels, the expression of HNF1B was noted in 28/122
cases (23%), which supports the fact that the specificity of HNF1B
as a marker of clear cell carcinoma is relatively low. However, no
case of HGSC in our study reached a H-score higher than 200. Taking
the extent of expression into account, the sensitivity of HNF1B is
significantly higher, given that in CCC the expression of HNF1B is
commonly strong and diffuse. Furthermore, we observed a
statistically significant correlation between the mRNA and protein
expression of HNF1B, which suggests that posttranscriptional and
posttranslational mechanisms are probably not involved in the
regulation of the HNF1B gene expression in HGSC.
This is the first time that protein and mRNA
expression of ECI2, one of the possible HNF1B downstream targets,
has been analysed in HGSC. Our data shows that a low level of
immunohistochemical expression was present in the majority of
HGSCs. We have found a significant positive relationship between
HNF1B and ECI2 on a protein level, which is in accordance with the
previous findings in prostate cancer (19). Interestingly, the expression of HNF1B
and ECI2 on an mRNA level did not show a positive association,
which is supported by the re-analysis of the TCGA dataset. However,
no correlation between any of the clinico-pathological
characteristics or survival analyses and ECI2 was found. Therefore,
ECI2 does not seem to be an eligible prognostic marker for
HGSC.
Acknowledgements
The authors would like to thank Assistant Professor
Zachary H.K. Kendall, B.A. (Institute for History of Medicine and
Foreign Languages, First Faculty of Medicine, Charles University in
Prague) for the English language editing.
Funding
This work was supported by Ministry of Health, Czech
Republic (research project AZV 17-28404A and conceptual development
of research organization 64165; General University Hospital in
Prague), by Charles University (Project Progress Q28/LF1 and SVV
260367), and by European Regional Development Fund (project
EF16_013/0001674, BBMRI_CZ LM2018125, and OPPK-Research Laboratory
of Tumor Diseases, CZ.2.16/3.1.00/24509).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Study concept and design: KN and PD. Samples and
clinical data collection: DC, KJ, MB, HQB. Preparation and analysis
of samples: KN, MB, NH, JH and EK. Statistical analyses and data
interpretation: RM, IS, KN. Drafting of the manuscript: KN, MB, JH,
NH, IS, PD, RM, DC and KJ. Proofread the manuscript: IS, PD and KN.
KN and PD confirmed the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the General University Hospital in Prague in
compliance with the Helsinki Declaration (ethical approval number
41/16 Grant VES 2017 AZV VFN). The requirement for written informed
consent was waived due to the retrospective nature of the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu Y, Kanyomse Q and Xie Y:
Tumor-suppressive activity of Hnf1β in Wilms' tumor. Biosci
Biotechnol Biochem. 83:2008–2015. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suzuki E, Kajita S, Takahashi H, Matsumoto
T, Tsuruta T and Saegusa M: Transcriptional upregulation of HNF-1β
by NF-KB in ovarian clear cell carcinoma modulates susceptibility
to apoptosis through alteration in bcl-2 expression. Lab Invest.
95:962–972. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsuchiya A, Sakamoto M, Yasuda J, Chuma M,
Ohta T, Ohki M, Yasugi T, Taketani Y and Hirohashi S: Expression
profiling in ovarian clear cell carcinoma: Identification of
hepatocyte nuclear factor-1 beta as a molecular marker and a
possible molecular target for therapy of ovarian clear cell
carcinoma. Am J Pathol. 163:2503–2512. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartu M, Dundr P, Nemejcova K, Ticha I,
Hojny H and Hajkova N: The role of HNF1B in tumorigenesis of solid
tumours: A review of current knowledge. Folia Biol (Praha).
64:71–83. 2018.PubMed/NCBI
|
|
5
|
Ross-Adams H, Ball S, Lawrenson K, Halim
S, Russell R, Wells C, Strand SH, Ørntoft TF, Larson M, Armasu S,
et al: HNF1B variants associate with promoter methylation and
regulate gene networks activated in prostate and ovarian cancer.
Oncotarget. 7:74734–74746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsui A, Fujimoto J, Ishikawa K, Ito E,
Goshima N, Watanabe S and Semba K: Hepatocyte nuclear factor 1 beta
induces transformation and epithelial-to-mesenchymal transition.
FEBS Lett. 590:1211–1221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kato N, Sasou S and Motoyama T: Expression
of hepatocyte nuclear factor-1beta (HNF-1beta) in clear cell tumors
and endometriosis of the ovary. Mod Pathol. 19:83–89. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mandato VD, Farnetti E, Torricelli F,
Abrate M, Casali B, Ciarlini G, Pirillo D, Gelli MC, Nicoli D,
Grassi M, et al: HNF1B polymorphism influences the prognosis of
endometrial cancer patients: A cohort study. BMC Cancer.
15:2292015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Setiawan VW, Haessler J, Schumacher F,
Cote ML, Deelman E, Fesinmeyer MD, Henderson BE, Jackson RD,
Vöckler JS, Wilkens LR, et al: HNF1B and endometrial cancer risk:
Results from the PAGE study. PLoS One. 7:e303902012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen H, Fridley BL, Song H, Lawrenson K,
Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, et
al: Epigenetic analysis leads to identification of HNF1B as a
subtype-specific susceptibility gene for ovarian cancer. Nat
Commun. 4:16282013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spurdle AB, Thompson DJ, Ahmed S, Ferguson
K, Healey CS, O'Mara T, Walker LC, Montgomery SB, Dermitzakis ET;
Australian National Endometrial Cancer Study Group, ; et al:
Genome-wide association study identifies a common variant
associated with risk of endometrial cancer. Nat Genet. 43:451–454.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kato N and Motoyama T: Hepatocyte nuclear
factor-1beta (HNF-1beta) in human urogenital organs: Its expression
and role in embryogenesis and tumorigenesis. Histol Histopathol.
24:1479–1486. 2009.PubMed/NCBI
|
|
13
|
Yamamoto S, Tsuda H, Aida S, Shimazaki H,
Tamai S and Matsubara O: Immunohistochemical detection of
hepatocyte nuclear factor 1beta in ovarian and endometrial
clear-cell adenocarcinomas and nonneoplastic endometrium. Hum
Pathol. 38:1074–1080. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fadare O and Liang SX: Diagnostic utility
of hepatocyte nuclear factor 1-beta immunoreactivity in endometrial
carcinomas: Lack of specificity for endometrial clear cell
carcinoma. Appl Immunohistochem Mol Morphol. 20:580–587. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kao YC, Lin MC, Lin WC, Jeng YM and Mao
TL: Utility of hepatocyte nuclear factor-1β as a diagnostic marker
in ovarian carcinomas with clear cells. Histopathology. 61:760–768.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nemejcova K, Ticha I, Kleiblova P, Bártů
M, Cibula D, Jirsová K and Dundr P: Expression, epigenetic and
genetic changes of HNF1B in endometrial lesions. Pathol Oncol Res.
22:523–530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dan C, Zhang H, Zeng W, Huang L, Gong X,
Li H, Yang E, Wang L and Yao Q: HNF1B expression regulates ECI2
gene expression, potentially serving a role in prostate cancer
progression. Oncol Lett. 17:1094–1100. 2019.PubMed/NCBI
|
|
18
|
Houten SM, Violante S, Ventura FV and
Wanders RJ: The biochemistry and physiology of mitochondrial fatty
acid β-oxidation and its genetic disorders. Annu Rev Physiol.
78:23–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Itkonen HM, Brown M, Urbanucci A, Tredwell
G, Ho Lau C, Barfeld S, Hart C, Guldvik IJ, Takhar M, Heemers HV,
et al: Lipid degradation promotes prostate cancer cell survival.
Oncotarget. 8:38264–38275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan J, Li X, Issop L, Culty M and
Papadopoulos V: ACBD2/ECI2-mediated peroxisome-mitochondria
interactions in leydig cell steroid biosynthesis. Mol Endocrinol.
30:763–782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirsch FR, Dziadziuszko R, Thatcher N,
Mann H, Watkins C, Parums DV, Speake G, Holloway B, Bunn PA Jr and
Franklin WA: Epidermal growth factor receptor immunohistochemistry:
Comparison of antibodies and cutoff points to predict benefit from
gefitinib in a phase 3 placebo-controlled study in advanced
nonsmall-cell lung cancer. Cancer. 112:1114–1121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartu M, Hojny J, Hajkova N, Michálková R,
Krkavcová E, Simon K, Frýba V, Stružinská I, Němejcová K and Dundr
P: Expression, epigenetic, and genetic changes of HNF1B in
colorectal lesions: An analysis of 145 cases. Pathol Oncol Res.
26:2337–2350. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dundr P, Bartu M, Hojny J, Michálková R,
Hájková N, Stružinská I, Krkavcová E, Hadravský L, Kleissnerová L,
Kopejsková J, et al: HNF1B, EZH2 and ECI2 in prostate carcinoma.
Molecular, immunohistochemical and clinico-pathological study. Sci
Rep. 10:143652020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim HY: Statistical notes for clinical
researchers: Chi-squared test and Fisher's exact test. Restor Dent
Endod. 42:152–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cancer Genome Atlas Research Network, .
The molecular taxonomy of primary prostate cancer. Cell.
163:1011–1025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kato N, Tamura G and Motoyama T:
Hypomethylation of hepatocyte nuclear factor-1beta (HNF-1beta) CpG
island in clear cell carcinoma of the ovary. Virchows Arch.
452:175–180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lau HH, Ng NHJ, Loo LSW, Jasmen JB and Teo
AKK: The molecular functions of hepatocyte nuclear factors-In and
beyond the liver. J Hepatol. 68:1033–1048. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kalloger SE, Kobel M, Leung S, Mehl E, Gao
D, Marcon KM, Chow C, Clarke BA, Huntsman DG and Gilks CB:
Calculator for ovarian carcinoma subtype prediction. Mod Pathol.
24:512–521. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tomassetti A, De Santis G, Castellano G,
Miotti S, Mazzi M, Tomasoni D, Van Roy F, Carcangiu ML and Canevari
S: Variant HNF1 modulates epithelial plasticity of normal and
transformed ovary cells. Neoplasia. 10:1481–1492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ,
Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Terasawa K, Toyota M, Sagae S, Ogi K,
Suzuki H, Sonoda T, Akino K, Maruyama R, Nishikawa N, Imai K, et
al: Epigenetic inactivation of TCF2 in ovarian cancer and various
cancer cell lines. Br J Cancer. 94:914–921. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bubancova I, Kovarikova H, Laco J, Ruszova
E, Dvorak O, Palicka V and Chmelarova M: Next-generation sequencing
approach in methylation analysis of HNF1B and GATA4 genes:
Searching for biomarkers in ovarian cancer. Int J Mol Sci.
18:4742017. View Article : Google Scholar
|
|
33
|
Baranova I, Kovarikova H, Laco J,
Sedlakova I, Vrbacky F, Kovarik D, Hejna P, Palicka V and
Chmelarova M: Identification of a four-gene methylation biomarker
panel in high-grade serous ovarian carcinoma. Clin Chem Lab Med.
58:1332–1340. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chaffer CL, San Juan BP, Lim E and
Weinberg RA: EMT, cell plasticity and metastasis. Cancer Metastasis
Rev. 35:645–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang W, Cheng X, Ji J, Zhang J and Li Q:
The application value of HNF-1β transcription factor in the
diagnosis of ovarian clear cell carcinoma. Int J Gynecol Pathol.
35:66–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kobel M, Kalloger SE, Carrick J, Huntsman
D, Asad H, Oliva E, Ewanowich CA, Soslow RA and Gilks CB: A limited
panel of immunomarkers can reliably distinguish between clear cell
and high-grade serous carcinoma of the ovary. Am J Surg Pathol.
33:14–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Q, Zeng X, Cheng X, Zhang J, Ji J, Wang
J, Xiong K, Qi Q and Huang W: Diagnostic value of dual detection of
hepatocyte nuclear factor 1 beta (HNF-1β) and napsin A for
diagnosing ovarian clear cell carcinoma. Int J Clin Exp Pathol.
8:8305–8310. 2015.PubMed/NCBI
|