Introduction
Colorectal cancer is a common malignant tumor of the
digestive tract. Present statistics show that colorectal cancer
ranks third in incidence and forth in mortality rates among
malignant tumors worldwide (1). In
recent years, the age of onset of colorectal cancer has decreased.
A survey showed that between 2000 and 2013, the incidence of
colorectal cancer among Americans aged <50 increased by 22%
(2).
Although an increasing number of measures for cancer
prevention and treatment are being implemented, the incidence and
mortality of colorectal cancer remain high (3). The development of precision medicine in
recent years is expected to revolutionize the diagnosis and
treatment of colorectal cancer (4).
Identifying molecular markers corresponding to tumors and achieving
individualized treatment to maximize efficacy and minimize side
effects is an important direction for future clinical tumor
treatment (5,6). Colorectal cancer is caused by a variety
of factors, such as a series of genetic events and gene mutations,
and generally involves the activation of oncogenes and inactivation
of tumor suppressor genes. Youth colorectal cancer has unique
molecular characteristics. Mork et al (7) found that the proportion of youth
colorectal cancer secondary to familial adenomatous polyposis with
an APC gene mutation was significantly higher compared with
colorectal cancer in the elderly. Compared with elderly colorectal
cancer, youth colorectal cancer has unique molecular biological
characteristics, including a high proportion of microsatellite high
instability, a high proportion of mismatch repair gene defects and
relatively lower mutation rates of the BRAF and KRAS genes in the
EGFR pathway (8–10). However, the underlying molecular
biological mechanism of young colorectal cancer is still not
completely clear, rendering it necessary to explore specific
molecular markers for colorectal cancer in young people. In the
present study, Gene Expression Omnibus (GEO), The Cancer Genome
Atlas (TCGA) database data mining and reverse
transcription-quantitative (RT-q)PCR were used to explore key
molecular markers for youth colorectal cancer.
Materials and methods
Microarray data
The GSE41657 (analyzed on Agilent-014850 Whole Human
Genome Microarray 4x44K G4112F platform; conducted by
Peking Union Medical College and Chinese Academy of Medical
Sciences in China) and GSE41258 (analyzed on Affymetrix Human
Genome U133A Array platform; conducted by Weizmann Institute of
Science in Israel) gene expression profiles in human colorectal
cancer were downloaded from the GEO database of the National Center
for Biotechnology Information (www.ncbi.nlm.nih.gov/geo). The patients selected for
the cancer and normal groups were all aged <50 years.
Data preprocessing and screening
strategy
Firstly, GEO2R was used to identify the
differentially expressed genes (DEGs) between youth colorectal
cancer and normal tissues. GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r/) is a convenient web
tool for DEG screening by comparing two groups of samples in a GEO
dataset (11). An adjusted P<0.05
and |log2 fold change (FC)|≥1 were set as the cut-off
criteria.
Secondly, these DEGs were classified according to
Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; www.genome.jp/kegg) pathways using Database for
Annotation, Visualization and Integration Discovery (DAVID;
http://david.ncifcrf.gov/) software.
Based on the enriched GO terms and significant KEGG pathways, the
disease-associated pathways and genes were screened.
Thirdly, DEGs were imported into STRING software
(version 3.6.0; http://string-db.org/) to produce protein-protein
interaction networks, and then Cytoscape software (version 3.6.0;
http://cytoscape.org/) was used to construct a
visible network diagram. In this network, cytoHubba plugin was used
to screen the top 10 hub genes using degree algorithm. Combined
with the analysis of signaling pathways, disease-associated genes
were screened out.
TCGA, human protein atlas (HPA)
validation and survival analysis
TCGA (http://cancergenome.nih.gov/) dataset was used to
validate the disease-associated genes screened by GEO datasets.
TCGA dataset was analyzed using Gene Expression Profiling
Interactive Analysis (http://gepia.cancer-pku.cn/), which is a commonly used
interactive website that plots expression profiles of selected
genes. A total of 362 colorectal cancer and 51 normal tissues from
TCGA database were selected for analysis. Survival analysis was
further performed on these genes, based on gene expression levels,
as previously described (12). Key
genes were obtained by overall survival (OS) and disease-free
survival (DFS) analysis, and the association between the expression
of these genes and tumor staging was explored. Patients with high
expression levels (50%) of key genes were assigned to high
expression group. The expression levels of key genes were also
validated using the HPA database (http://www.proteinatlas.org/) (13).
RT-qPCR validation
Six pairs of youth colorectal cancer and adjacent
normal tissues from patients aged <50 years (age range, 35–48; 3
men and 3 women) were collected from the First Department of
Gastrointestinal Surgery of the Affiliated Hospital of Putian
University (Putian, China). All colorectal cancer and normal
tissues were stored in a liquid nitrogen tank within 30 min from
their removal from the patient. For RNA extraction, the sample was
ground into pieces in a mortar filled with liquid nitrogen. Total
RNA was then extracted using RNAiso Plus (Takara Bio, Inc.) and
reverse transcribed into cDNA using PrimeScript™ RT reagent kit
with gDNA Eraser (Takara Bio, Inc.), according to the
manufacturer's instructions. RT-qPCR was performed using the SYBR
Premix Ex Taq™ II (Takara Bio, Inc.), according to the
manufacturer's instructions. The PCR primers were designed by the
National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/) and provided by SangonBiotech
Co., Ltd. (Table SI). The gene
GAPDH was selected as an internal reference, to compare gene
expression in different samples. The RT-qPCR process included the
following steps: Pre-denaturation at 95°C for 30 sec, followed by
40 cycles of denaturation at 95°C for 5 sec and extension at 60°C
for 35 sec (7500 Real Time PCR System; Thermo Fisher Scientific,
Inc.). Each sample was tested in triplicate. Gene expression values
in each sample were calculated using the 2−∆∆Cq method
(14). The present study was
approved by the Ethics Committee of the Affiliated Hospital of
Putian University (approval no. 202016). Written informed consent
was obtained from each patient for sample collection and
analysis.
Statistical analysis
SPSS version 17.0 (SPSS Inc.) was used to analyze
the data. Continuous variables are presented as the mean ± standard
deviation. One-way ANOVA with least significant difference post hoc
test was used to analyze the data of more than two groups.
Student's t-test was used for comparisons between two groups. A
paired t-test was used when comparing the six pairs of youth
colorectal cancer and adjacent normal tissues, and an unpaired
t-test was used for other comparisons. If the variance was not
equal between two groups, Mann-Whitney U test was used for
statistical analysis. The statistical significance of survival time
was determined by the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of DEGs in youth
colorectal cancer
The gene expression levels of genes were downloaded
from the GEO database. Based on GEO2R analysis, 6,322 and 12,585
DEGs in GSE41657 and GSE41258 were screened in youth colorectal
cancer compared with normal colorectal tissues. Screened with the
cut-off criteria of adjusted P<0.05 and |log2
FC|>1, 1,256 upregulated and 1,782 downregulated genes were
identified in the GSE41657 dataset (Fig. S1A), and 311 upregulated and 568
downregulated genes were identified in the GSE41258 dataset
(Fig. S1B). In the integrated
analysis of the two datasets, 443 overlapping genes, including 131
upregulated and 312 downregulated, were obtained (Fig. S1C and D).
GO analysis of selected genes
The functional enrichment of 443 candidate genes was
analyzed using DAVID. Three GO category results are presented in
Table SII, including biological
processes, cellular components and molecular functions. The
biological process results revealed that the selected genes were
mainly enriched in ‘cellular response to zinc ion’, ‘negative
regulation of growth’ and ‘cell proliferation’. The cellular
component results revealed that selected genes were mainly enriched
in ‘extracellular exosome’, ‘extracellular space’ and ‘apical
plasma membrane’. The molecular function analysis revealed that the
selected genes were mainly enriched in ‘protein binding’,
‘carbonate dehydratase activity’ and ‘CXCR chemokine receptor
binding’. These results demonstrated that the majority of the
candidate genes were significantly enriched in ‘binding’ and ‘cell
proliferation’.
Signaling pathway enrichment
analysis
The signaling pathway enrichment of 443 candidate
genes was analyzed using KEGG pathway online databases. A total of
23 genes were significantly enriched in ‘Pathways in cancer’
(Table SIII). Based on previous
reports (15,16), ‘Pathways in cancer’ is an important
signaling pathway associated with the occurrence and development of
cancer.
Protein-protein network
construction
A total of 443 candidate genes were analyzed using
the STRING 11.0 database, and then the protein-protein interaction
(PPI) was imported into Cytoscape 3.6.0 software to build a visible
network diagram. A total of 390 nodes and 1,462 edges are presented
in the network (Fig. S2).
Hub gene selection and analysis with
key signaling pathway
Within the PPI network, the cytoHubba plugin was
used to screen for hub genes. Based on the degree algorithm, the
top 10 hub genes were VEGFA, CXCL8, MYC, CD44, CXCL12, CCND1, IGF1,
CXCL1, KIT and SOX9 (Table I).
Combining signaling pathway analysis, CXCL8, IGF1, KIT, CXCL12,
CCND1, VEGFA and MYC in the ten hub genes were enriched in the
‘pathways in cancer’ (Fig. S3).
Among these genes, CXCL8, CCND1, VEGFA and MYC were upregulated,
and the others downregulated.
 | Table I.10 hub genes ranked by degree. |
Table I.
10 hub genes ranked by degree.
| Gene name | Category | Score | Rank |
|---|
| VEGFA | Upregulated | 65 | 1 |
| CXCL8 | Upregulated | 53 | 2 |
| MYC | Upregulated | 53 | 3 |
| CD44 | Upregulated | 43 | 4 |
| CXCL12 | Downregulated | 39 | 5 |
| CCND1 | Upregulated | 39 | 6 |
| IGF1 | Downregulated | 38 | 7 |
| CXCL1 | Upregulated | 32 | 8 |
| KIT | Downregulated | 30 | 9 |
| SOX9 | Upregulated | 29 | 10 |
TCGA, HPA validation and survival
analysis
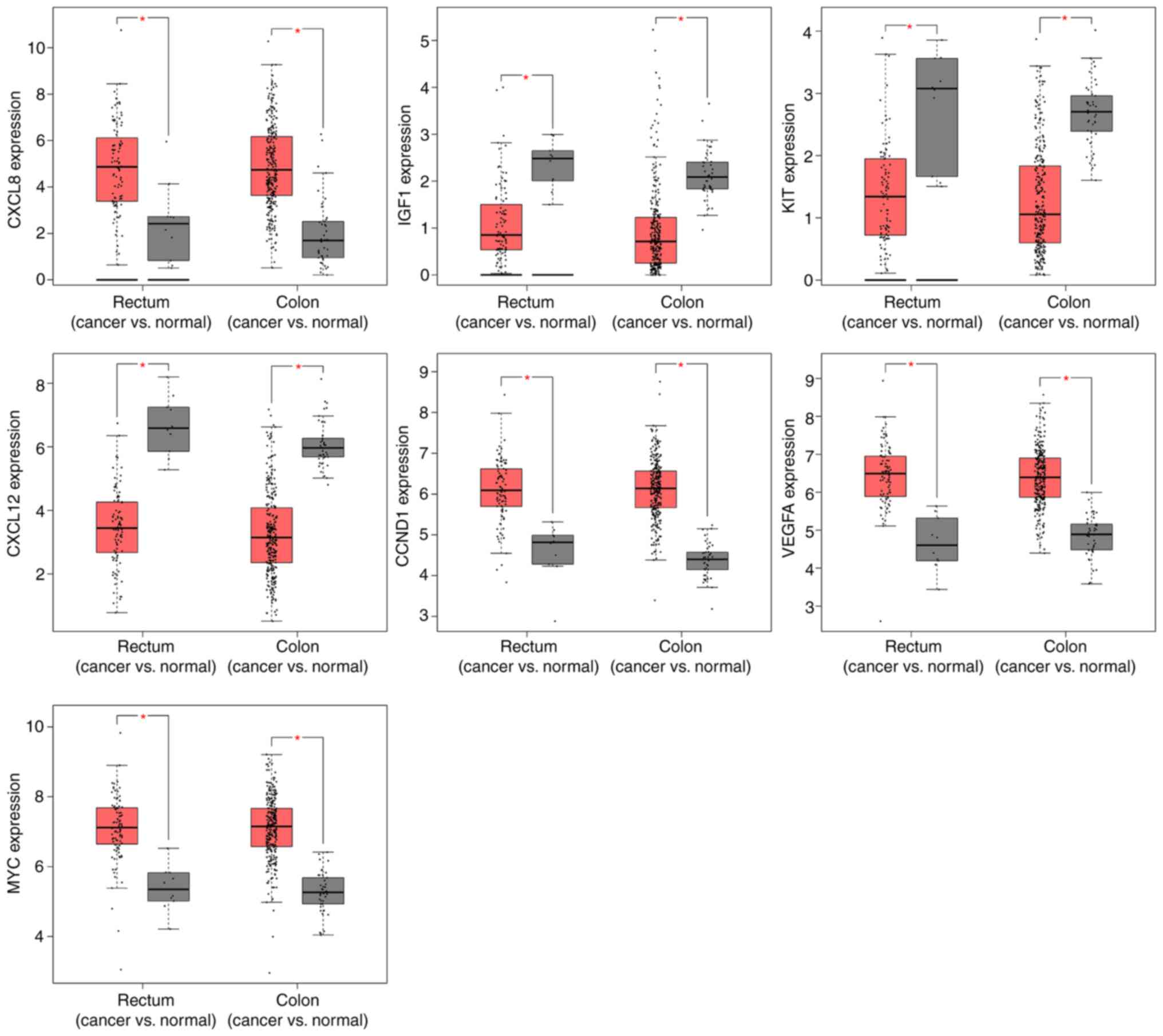
Differential expression of CXCL8, IGF1, KIT, CXCL12,
CCND1, VEGFA and MYC in colorectal cancer compared with normal
colorectal tissues, was screened in TCGA database. The expression
levels of these eight genes were significantly higher in both colon
and rectal cancer tissues, as compared with normal colorectal
tissues (P<0.05; Fig. 1).
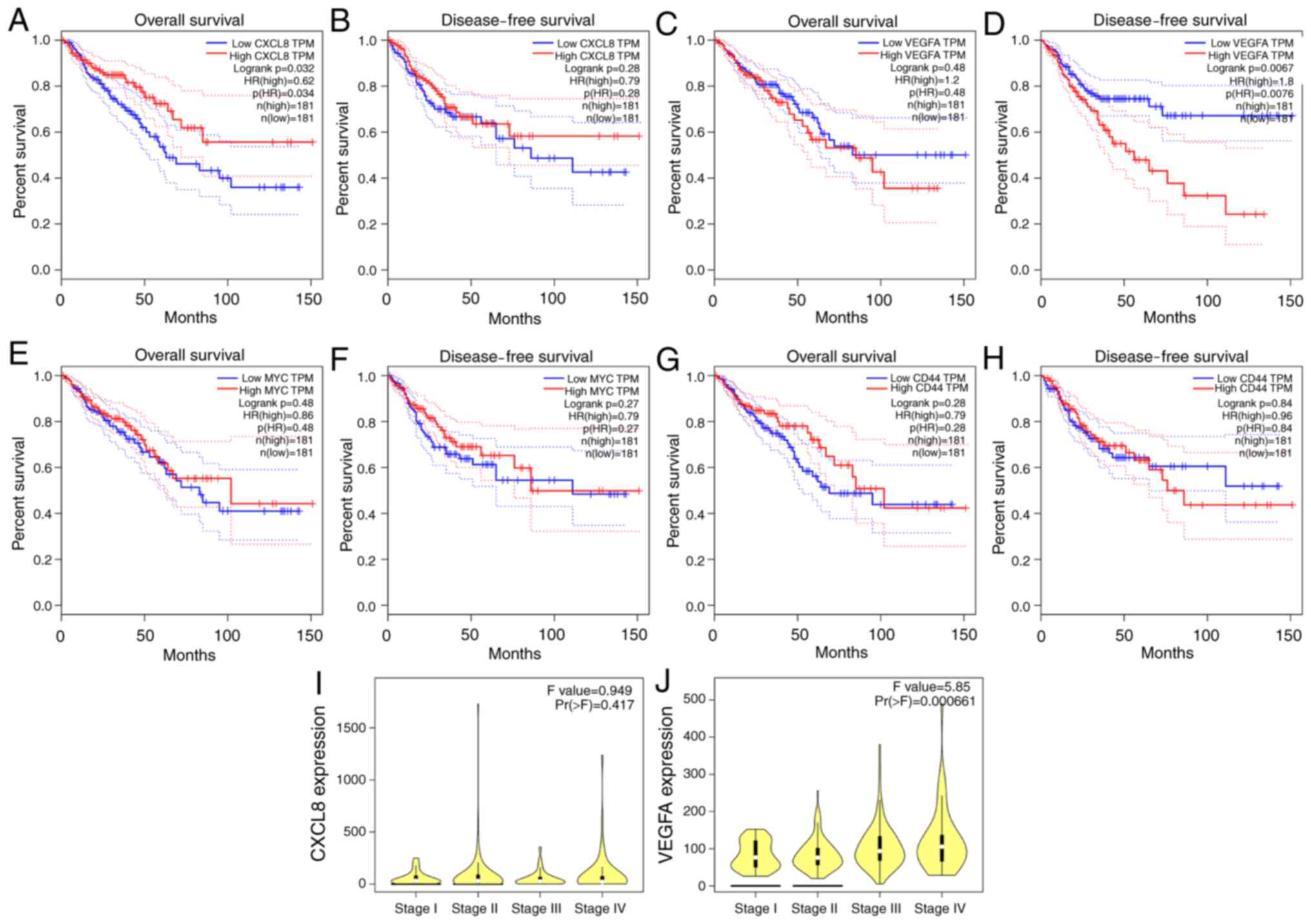
These genes were further subjected to survival
analysis to screen out genes associated with colorectal cancer
prognosis. The expression level of CXCL8 was found to impact
overall survival (OS; P<0.05; Fig.
2A), and that of VEGFA to impact disease-free survival (DFS;
P<0.05; Fig. 2D). The expression
level of MYC and CD44 was found to have no impact on OS or DFS
(P>0.05; Fig. 2E-H). Thus, CXCL8
and VEGFA were identified as key genes.
Compared with the expression at different
Tumor-Node-Metastasis (TNM) stages of colorectal cancer, it was
found that the expression of the CXCL8 gene in high TNM-stage
colorectal cancer was increased, but there was no statistical
significance (P>0.05; Fig. 2I),
while the gene expression of VEGFA in high TNM stages was
significantly higher (P=0.000661; Fig.
2J).
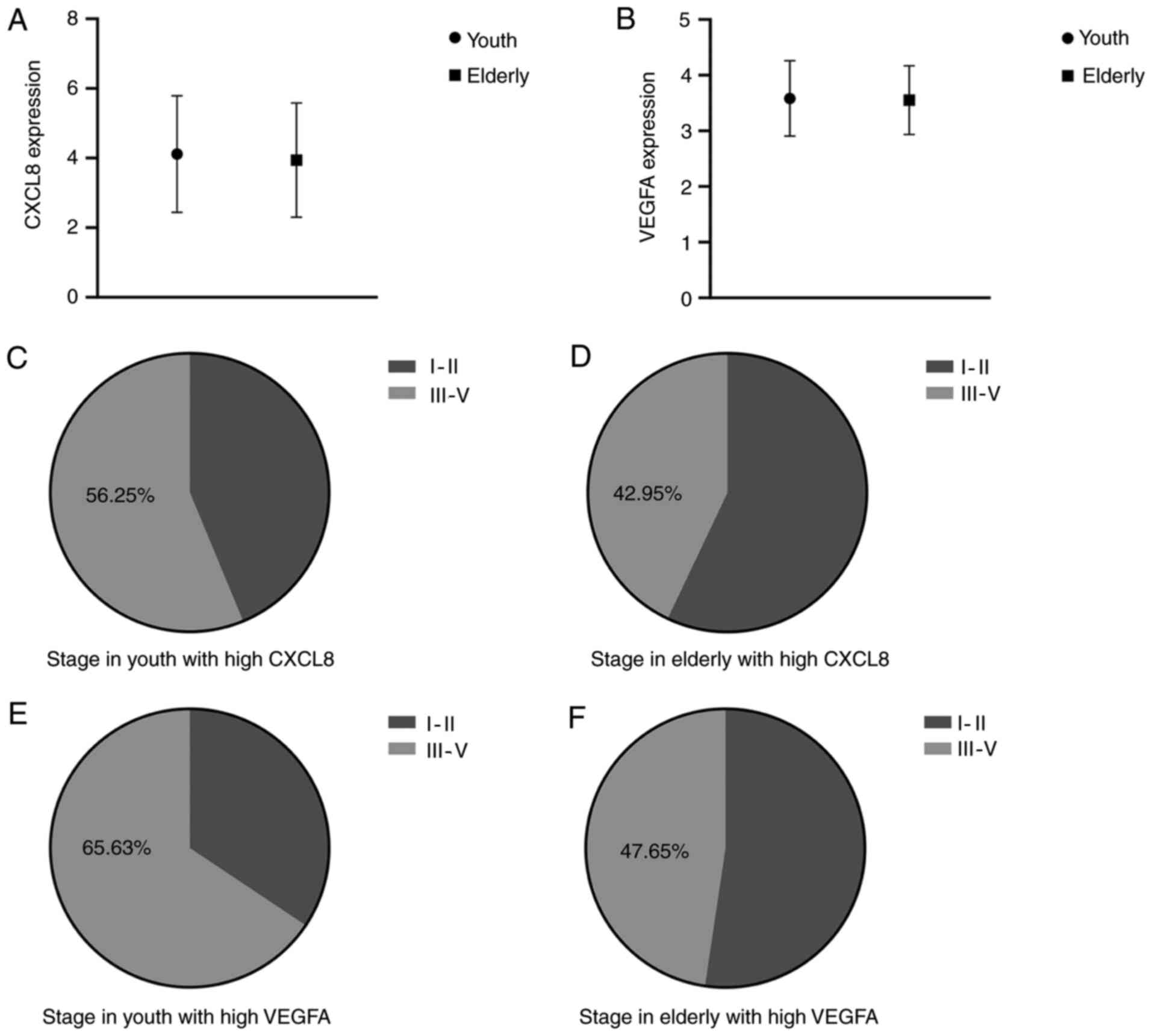
The comparison of CXCL8 and VEGFA expression between
youth and elderly colorectal cancer, revealed no significant
differences (Fig. 3A and B). Further
analysis indicated that high CXCL8 and VEGFA expression was
associated with tumor stage. The proportion of stages III–IV was
56.25% in youth colorectal cancer, 42.95% in elderly colorectal
cancer with high CXCL8 expression, and 65.63% in youth colorectal
cancer, 47.65% in elderly colorectal cancer with high VEGFA
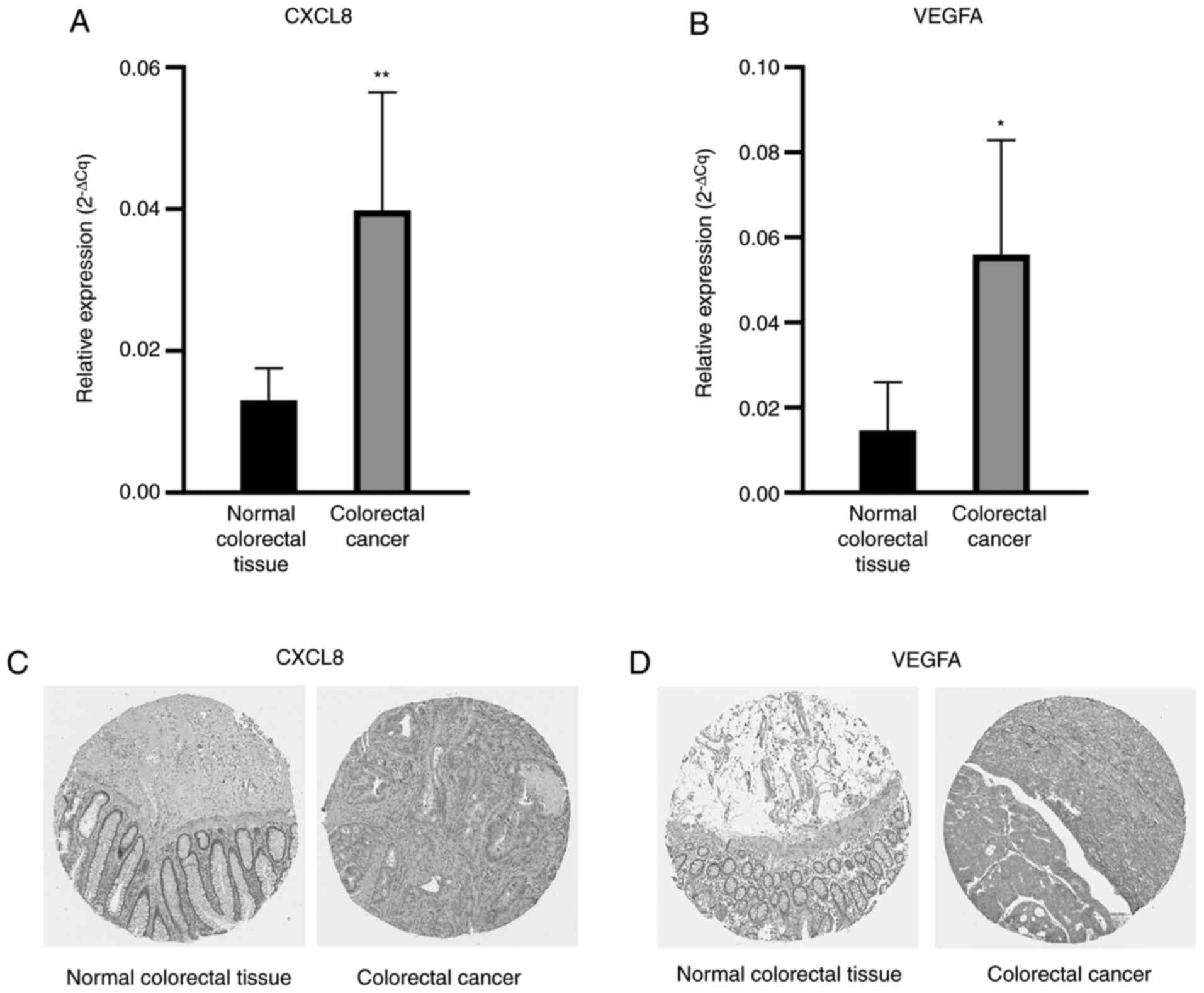
expression (Fig. 3F). The HPA
database was also used to verify the protein expression levels of
key genes. The expression of CXCL8 and VEGFA in colorectal cancer
from the HPA database was also consistent with the aforementioned
bioinformatics analysis (Fig. 4C and
D).
RT-qPCR validation
To confirm the reliability of the findings from data
mining, the two genes, CXCL8 and VEGFA, were selected for
validation by RT-qPCR in six pairs of youth colorectal cancer and
adjacent normal tissues. According to the experimental results, the
difference in expression of these two genes is consistent with the
results of data mining. In youth colorectal cancer, the expression
level of CXCL8 and VEGFA were significantly upregulated (P<0.05;
Fig. 4A and B).
Discussion
Colorectal cancer is a common malignant tumor of the
digestive system, which seriously threatens human health. According
to the 2018 Global Cancer Data Statistics Report, among malignant
tumors, colorectal cancer ranks third in morbidity and second in
mortality (17). In recent years,
the incidence and mortality of colorectal cancer have been on the
rise, particularly in young people. The onset of colorectal cancer
in young people is typically undetected, and most patients are
diagnosed during the middle and late stages of the disease, at
which point, the best treatment opportunities are no longer
suitable (18). Therefore, early
diagnosis of young patients with colorectal cancer to ensure early
treatment is particularly important for improving survival time and
prognosis. With the advent of precision medicine, it is necessary
to explore biological markers that are closely associated with the
diagnosis, prognosis and treatment of colorectal cancer in young
people.
In the present study, through bioinformatics data
mining combined with gene annotation and signaling pathway
analysis, two genes closely associated with young colorectal
cancer, CXCL8 and VEGFA, were screened out. The difference in the
expression of these two genes was confirmed by RT-qPCR. According
to the data from TCGA database, the expression of these two genes
was closely associated with the survival time of patients with
colorectal cancer.
Both CXCL8 and VEGFA are cytokines that promote
angiogenesis, and play an important role in tumor growth and
metastasis (19). CXCL8, also known
as interleukin 8, is a member of the chemokine family. CXCL8 is
mainly produced by macrophages, and endothelial, epithelial and
other cells (20). The sequence
encoding CXCL8 is located on chromosome 4q13-21 (21). CXCL8 has been found to be highly
expressed in various types of cancer, such as non-small cell lung
(22), gastric (23), breast (24) and colorectal cancer (25). Studies have reported that, in
colorectal cancer, CXCL8 mainly induces epithelial-mesenchymal
transition through the PI3K/Akt/NF-κB signaling pathway (25). Certain studies have reported that the
low expression of CXCL8 is associated with poor prognosis of
patients with colorectal cancer (26,27).
This view is consistent with the results of the present study. As
CXCL8 is a cancer-promoting factor. The reason for the association
between its high expression and favorable prognosis seems to be
contradictory, and the underlying mechanism remains unclear. This
favorable prognosis may be associated with other functions of
CXCL8. For example, CXCL8 as a chemokine has the ability to guide
neutrophils to infectious diseases and control infection (28).
VEGFA is a member of the vascular endothelial growth
factor (VEGF) protein family (29).
VEGFA is the main regulator of angiogenesis, and its combination
with VEGFR2 promotes the proliferation of endothelial cells through
the RAS-RAF-MAPK-ERK signaling pathway (30). VEGFA is widely distributed in the
body, and the high expression of VEGFA can be seen in a variety of
cancer types, such as lung (31),
gastric (32), breast (33), ovarian (34) and colorectal cancer (35). VEGFA can promote angiogenesis, thus
it is useful to understand that the high expression of VEGFA
promotes tumor growth, which in turn leads to a worse prognosis. In
the present study, it was found that compared with the high VEGFA
expression, the low VEGFA expression prolongs the survival time of
patients with colorectal cancer, particularly the DFS. Moreover,
colorectal cancer with a high VEGFA expression is associated with a
higher disease stage. In recent years, various therapeutic targets
for VEGFA have been manufactured. Bevacizumab was the first VEGFA
inhibitor approved by the US Food and Drug Administration in 2004
for the treatment of metastatic colorectal cancer; since then, this
drug has been widely used in clinical treatment and is known to
significantly prolong the survival time of multiple patients
(36).
The present study was not, however, without its
limitations. Firstly, the number of specimens used to verify the
results of the study was small. Secondly, no prospective
experiments have been conducted to verify the association between
candidate genes and patient prognosis.
In conclusion, through GEO datasets, TCGA database
analysis and RT-qPCR validation, two key genes associated with
prognosis, CXCL8 and VEGFA, were identified in youth colorectal
cancer. These key genes were enriched in the ‘pathways in cancer’,
and may be ideal prognostic indicators or therapeutic targets for
youth colorectal cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Open
Foundation of Engineering Research Center of Big Data Application
in Private Health Medicine, The Fujian Province University (grant
no. KF2020009) and The National Natural Science Foundation of China
(grant no. 81860334).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
JC performed the experiments and was a major
contributor in writing the manuscript. ML and YC downloaded the
datasets and performed statistical analysis. JC and JG analyzed the
obtained data. WL designed the study. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Affiliated Hospital of Putian University (approval
no. 202016). Written informed consent was obtained from each
patient for sample collection and analysis.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DEGs
|
differentially expressed genes
|
|
DAVID
|
Database for Annotation, Visualization
and Integration Discovery
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
FC
|
fold change
|
|
TCGA
|
The Cancer Genome Atlas
|
|
HPA
|
Human Protein Atlas
|
References
|
1
|
Naeem A, Tun AM and Guevara E: Molecular
genetics and the role of molecularly targeted agents in metastatic
colorectal carcinoma. J Gastrointest Cancer. 51:387–400. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weinberg BA, Marshall JL and Salem ME: The
growing challenge of young adults with colorectal cancer. Oncology
(Williston Park). 31:381–389. 2017.PubMed/NCBI
|
|
3
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ravegnini G and Angelini S: Toward
precision medicine: How far is the goal? Int J Mol Sci. 17:2452016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson TM: Perspective on precision
medicine in oncology. Pharmacotherapy. 37:988–989. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei Q: Personalized medicine at a prime
time for cancer medicine-introducing cancer medicine. Cancer Med.
1:12012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mork ME, You YN, Ying J, Bannon SA, Lynch
PM, Rodriguez-Bigas MA and Vilar E: High prevalence of hereditary
cancer syndromes in adolescents and young adults with colorectal
cancer. J Clin Oncol. 33:3544–3549. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khan SA, Morris M, Idrees K, Gimbel MI,
Rosenberg S, Zeng Z, Li F, Gan G, Shia J, LaQuaglia MP and Paty PB:
Colorectal cancer in the very young: A comparative study of tumor
markers, pathology and survival in early onset and adult onset
patients. J Pediatr Surg. 51:1812–1817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Magnani G, Furlan D, Sahnane N, Reggiani
Bonetti L, Domati F and Pedroni M: Molecular features and
methylation status in early onset (≤40 years) colorectal cancer: A
population based, case-control study. Gastroenterol Res Pract.
2015:1321902015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pearlman R, Frankel WL, Swanson B, Zhao W,
Yilmaz A, Miller K, Bacher J, Bigley C, Nelsen L, Goodfellow PJ, et
al: Prevalence and spectrum of germline cancer susceptibility gene
mutations among patients with early-onset colorectal cancer. JAMA
Oncol. 3:464–471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database Issue)):
D991–D995. 2013.PubMed/NCBI
|
|
12
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang L, Wang J, Sun X, Cao Y, Ning S,
Zhang H, Chen L, Li R, Tian Q, Wang L, et al: Identifying a
polymorphic ‘switch’ that influences miRNAs' regulation of a
myasthenia gravis risk pathway. PLoS One. 9:e1048272014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X, Zheng R, Zhang H and He T: Pathway
crosstalk analysis of microarray gene expression profile in human
hepatocellular carcinoma. Pathol oncol Res. 21:563–569. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marjon PL, Bobrovnikova-Marjon EV and
Abcouwer SF: Expression of the pro-angiogenic factors vascular
endothelial growth factor and interleukin-8/CXCL8 by human breast
carcinomas is responsive to nutrient deprivation and endoplasmic
reticulum stress. Mol Cancer. 3:42004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matsushima K, Baldwin ET and Mukaida N:
Interleukin-8 and MCAF: Novel leukocyte recruitment and activating
cytokines. Chem Immunol. 51:236–265. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Masuya D, Huang C, Liu D, Kameyama K,
Hayashi E, Yamauchi A, Kobayashi S, Haba R and Yokomise H: The
intratumoral expression of vascular endothelial growth factor and
interleukin-8 associated with angiogenesis in nonsmall cell lung
carcinoma patients. Cancer. 92:2628–2638. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yamamoto Y, Kuroda K, Sera T, Sugimoto A,
Kushiyama S, Nishimura S, Togano S, Okuno T, Yoshii M, Tamura T, et
al: The clinicopathological significance of the CXCR2 ligands,
CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8 in gastric
cancer. Anticancer Res. 39:6645–6652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liubomirski Y, Lerrer S, Meshel T, Morein
D, Rubinstein-Achiasaf L, Sprinzak D, Wiemann S, Körner C, Ehrlich
M and Ben-Baruch A: Notch-mediated tumor-stroma-inflammation
networks promote invasive properties and CXCL8 expression in
triple-negative breast cancer. Front Immunol. 10:8042019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen T, Yang Z, Cheng X, Xiao Y, Yu K, Cai
X, Xia C and Li Y: CXCL8 induces epithelial-mesenchymal transition
in colon cancer cells via the PI3K/Akt/NF-κB signaling pathway.
Oncol Rep. 37:2095–2100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Doll D, Keller L, Maak M, Boulesteix AL,
Siewert JR, Holzmann B and Janssen KP: Differential expression of
the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and
their impact on metastatic disease and survival. Int J Colorectal
Dis. 25:573–581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao QQ, Jiang C, Gao Q, Zhang YY, Wang G,
Chen XP, Wu SB and Tang J: Gene expression and methylation profiles
identified CXCL3 and CXCL8 as key genes for diagnosis and prognosis
of colon adenocarcinoma. J Cell Physiol. 235:4902–4912. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Das ST, Rajagopalan L, Guerrero-Plata A,
Sai J, Richmond A, Garofalo RP and Rajarathnam K: Monomeric and
dimeric CXCL8 are both essential for in vivo neutrophil
recruitment. PLoS One. 5:e117542010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ferrara N: VEGF and the quest for tumour
angiogenesis factors. Nat Rev Cancer. 2:795–803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Herbert SP and Stainier DY: Molecular
control of endothelial cell behaviour during blood vessel
morphogenesis. Nat Rev Mol Cell Biol. 12:551–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang SD, McCrudden CM and Kwok HF:
Prognostic significance of combining VEGFA, FLT1 and KDR mRNA
expression in lung cancer. Oncol Lett. 10:1893–1901. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Korobeinikova E, Ugenskiene R, Insodaite
R, Rudzianskas V, Jaselske E, Poskiene L and Juozaityte E:
Association of angiogenesis and inflammation-related gene
functional polymorphisms with early-stage breast cancer prognosis.
Oncol Lett. 19:3687–3700. 2020.PubMed/NCBI
|
|
34
|
Zhou P, Xiong T, Chen J, Li F, Qi T and
Yuan J: Clinical significance of melanoma cell adhesion molecule
CD146 and VEGFA expression in epithelial ovarian cancer. Oncol
Lett. 17:2418–2424. 2019.PubMed/NCBI
|
|
35
|
Wang R, Ma Y, Zhan S, Zhang G, Cao L,
Zhang X, Shi T and Chen W: B7-H3 promotes colorectal cancer
angiogenesis through activating the NF-κB pathway to induce VEGFA
expression. Cell Death Dis. 11:552020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ferrara N and Adamis AP: Ten years of
anti-vascular endothelial growth factor therapy. Nat Rev Drug
Discov. 15:385–403. 2016. View Article : Google Scholar : PubMed/NCBI
|