Introduction
Hepatocellular carcinomas (HCCs) are one of the
fourth most common cause of cancer-associated death worldwide
(1,2). Globally, there is an increased
incidence of HCC (1,2). The risk factors for HCCs are well
understood, such as chronic hepatitis or cirrhosis due to
persistent infection with hepatitis B and C virus, fatty liver
disease, diabetes and excessive alcohol intake (3); however, the molecular mechanisms in HCC
have not been elucidated.
microRNAs (miRNAs/miRs) are small, endogenous
non-coding RNAs (4,5) that are reported to be involved in gene
regulatory mechanisms. miRNAs are reported to be widely involved in
regulatory networks, such as proliferation, differentiation and
inflammation (5–8). One miRNA is estimated to regulate
>200 target genes (5–8). In animals, single-stranded miRNA binds
to the 3′ untranslated region (3′-UTR) of the target mRNAs through
a homologous sequence, which inhibits translation or degrades the
mRNAs (5–8).
The importance of these regulators has been
recognized; however, the precise function of these regulatory
non-coding RNAs is not completely understood. Various human
malignancies (9–13), including HCC, are associated with
aberrant expression of miRNAs. This indicates that the miRNAs
function as oncogenes or tumor suppressor genes (13). miRNAs have an important role in
carcinogenesis as half of the known aberrantly expressed miRNA
genes are located in the cancer-associated genomic regions or
fragile sites (14,15). Changes in miRNA expression have been
reported in various malignancies, such as Burkitt lymphoma
(16), colorectal cancer (17,18) and
lung cancer (19).
Several studies have reported that the HCC cells or
malignant liver tissues exhibit aberrant expression of specific
miRNAs, such as miR-26a, let-7a, let-7b, let-7c, let-7d, let- f-1
and let-g, when compared with non-malignant hepatocyte tissues in
the previous reports (20–27), which indicates an association between
miRNA and HCC. Previous studies have demonstrated that the
modulation of non-coding RNA expression, especially miRNAs, may
contribute to HCC formation (20–27).
However, there are no studies that have reported the correlation
between miRNA-527 and HCC, to the best of our knowledge.
Additionally, the target genes of the aberrantly expressed miRNAs
in HCC have not been fully elucidated. The present study aimed to
examine these aberrantly expressed miRNAs, which may contribute to
HCC pathogenesis by modulating the expression of gene products.
Glypican (GPC)-3 is an oncofetal glycoprotein
attached to the cell membrane by a glycophosphatidylinositol anchor
(28). GPC-3 is overexpressed in
some tumors, especially HCC (29).
GPC-3 may be involved in the regulation of Wnt, hedgehog, bone
morphogenic protein, and fibroblast growth factor (FGF) signaling
pathways (30). Therefore, GPC-3
contributes to cell proliferation and apoptosis in certain cell
types, including malignant cells (29). Moreover, overexpression of GPC-3 is
reported to be associated with poor prognosis in patients with HCC
(31,32). The present study aimed to investigate
GPC-3 as a target gene of miR-527.
Materials and methods
Chemicals
All chemicals were obtained from Sigma-Aldrich;
Merch KGaA.
Cell lines and culture
All HCC cell lines (Huh7, HLE, HLF, Alexander (code
no. IFO50069), Li-7 and Hep3B (code no. 86062703) and Cos7 cells,
one of the most commonly used cell system for mammalian transient
expression, were obtained from the Japanese Cancer Research
Resources Bank and KAC Co., Ltd. Cos7 cells do not express GPC-3.
The normal human hepatocytes were obtained from DS Pharma
Biomedical (Gibco; Thermo Fisher Scientific, Inc.). The HCC cell
lines were cultured in RPMI-640 medium (Gibco; Thermo Fisher
Scientific, Inc.), while the normal hepatocytes were cultured using
the CS-c COMPLETE Medium kit R (Sumitomo Dainippon Pharma Co.,
Ltd.). Cos-7 cells were cultured in Dulbecco's modified Eagle's
medium (DMEM) (Gibco; Thermo Fisher Scientific, Inc.). All media
were supplemented with 10% fetal bovine serum (cat. no. 533-69545;
FUJIFILM Wako Pure Chemical Corporation) and
penicillin-streptomycin (100 mg/l; Invitrogen; Thermo Fisher
Scientific, Inc.). All the cells were cultured in a humidified
atmosphere at 5% CO2 and 37°C.
Human tissues
HCC tissues were limited to patients who had not
received any treatment, including any chemotherapy, prior to
partial hepatectomy. Partial hepatectomy surgery was performed from
May 2015 to September 2015. Four tissue samples were collected
during partial hepatectomy of patients with HCC in Kagawa
University Hospital (Kita-gun, Japan). Histology of tissues were
pathologically analyzed using a light microscope. These tissues
were analyzed retrospectively. The HCC tissue samples were frozen
on dry ice within 30 min of collection. The samples were preserved
in the freezer at −70°C.
The histological analysis of the surrounding
non-cancerous tissues (N) revealed that the tissues were cirrhotic.
Of the four patients, three were male and one was female (mean age
60.5±6.98 years; range, 51–69). All patients tested positive for
hepatitis C virus and negative for hepatitis B surface antigen. The
HCC tissues from all patients were moderately differentiated. The
study was approved by The Human Subjects Committee of Kagawa
University (approval no. 22-163). Additionally, informed written or
verbal consent was provided by all patients.
Tissue and cell line lysate
The samples were homogenized in TNE buffer (10
mmol/l Tris-HCl [pH 7.4], 10 mmol/l Na3VO4,
50 mmol/l, Na2MoO4, 1% Nonidet P-40 and 100
U/ml aprotinin). The tissue lysate was centrifuged at 2,900 × g and
4°C for 60 min. The protein concentration in the supernatant was
measured by bicinchoninic acid protein assay.
Analysis of miRNA microarrays
miRNAs expression in the cancerous and surrounding N
tissues was evaluated using microarray analysis. Total RNA was
extracted from the liver tissues using the miRNeasy Mini kit
(Qiagen AB) as described previously (33,34). The
OD260 nm/OD280 nm ratio of the total RNA
samples used in this study ranged from 1.8 to 2.0. The samples were
labeled using the miRCURY Hy3 Power Labeling kit (Exiqon; Qiagen
AB). miRNA expression was measured by hybridizing the RNA sample on
a human miRNA Oligo chip, version 14.0 (Toray Industries, Inc.) as
described previously (33,34). The chip was scanned using a 3D-Gene
Scanner 3000 (Toray Industries, Inc.). The raw intensity of the
image was measured using the 3D-Gene extraction software (version
1.2; Toray Industries, Inc.). The raw data were analyzed using
GeneSpringGX (version 10.0; Agilent Technologies, Inc.) and
subjected to quantile normalization (35). The fold changes in the miRNA
expression levels were evaluated between the cancerous and
surrounding N tissues. Hierarchical clustering was performed using
the furthest neighbor method and Pearson's product-moment
correlation coefficient as a metric, with P<0.01 and false
discovery rate (FDR) <0.05.
Gel electrophoresis and western
blotting
The expression level of GPC-3 was determined in the
liver tissue and HCC cell lysates using western blotting. The
lysate samples (10 µg) were subjected to SDS-PAGE following the
methods of Laemmli (36). Next, the
resolved proteins were subjected to western blotting, following the
methods of Towbin et al (37). The membrane was incubated with the
following primary antibodies: Anti-GPC-3 (1:500) and anti-neomycin
phosphotransferase II (1:500) (both Santa Cruz Biotechnology,
Inc.). β-actin was used as the loading control in western blot
analysis. The immunoreactive bands were visualized using an
enhanced chemiluminescence detection system (Funakoshi Co., Ltd.)
on an X-ray film.
Transient transfection
The cDNA of GPC-3 gene was inserted into the
pCMV6-XL5 vector (Funakoshi Co., Ltd.), which contains the target
DNA sequences, poly A signal and CMV promoter. The African green
monkey kidney cells, Cos7 cells, were used for transfection as
these cells do not express GPC-3. Cos7 cells seeded
(0.3×106/ml) in 6-well plates were transfected with 4 µg
of the recombinant (pCMV6-GPC) or empty pCMV6-XL-5 vector
(Funakoshi Co., Ltd.) and 100 nM negative control miRNA
(non-targeting sequence) or mimic miR-527 (Funakoshi Co., Ltd.)
using 10 µl of COSFectin lipid reagent (Bio-Rad Laboratories, Inc.)
at 37°C for 48 h. To confirm transfection efficiency, the cells
were co-transfected with 4 µg of pcDNA3.1(+) plasmid containing the
neomycin phosphotransferase II gene. After transfection, the cells
were harvested and subjected to western blot analysis as
aforementioned.
Bioinformatics
Target sites for miR-527 found in GPC-3 identified
in GPC-3′UTR are aligned among human, mouse and rat using Target
Scan software (http://www.targetscan.org/vert_72/).
Transfection of miRNA
Huh7 cells were transfected with miR-527 mimic
(Funakoshi Co., Ltd.) using Lipofectamine® 2000 (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
HCC cells were transfected with 100 nM miR-527 mimic, or 100 nM
negative control #1 (Funakoshi Co., Ltd.). To study cell
proliferation, the WST-8 assay [3-(4,5-di-methylthiazol-
2-yl)-2,5-diphenyltetrazolium bromide] was conducted after 24 h of
transfection (34).
Statistical analysis
The data were analyzed using the Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference. For each miRNA probe of the microarray analyses,
Student's t-test was performed on the two groups. False discovery
rate (FDR) as determined using the Benjamini-Hochberg method
(38) was used to control the
multiplicity of testing caused by the number of the miRNA probes
used. All statistical analyses were performed in the GraphPad Prism
6 software (GraphPad Software).
Results
Dysregulation of miRNA expression in
the human HCC tissues
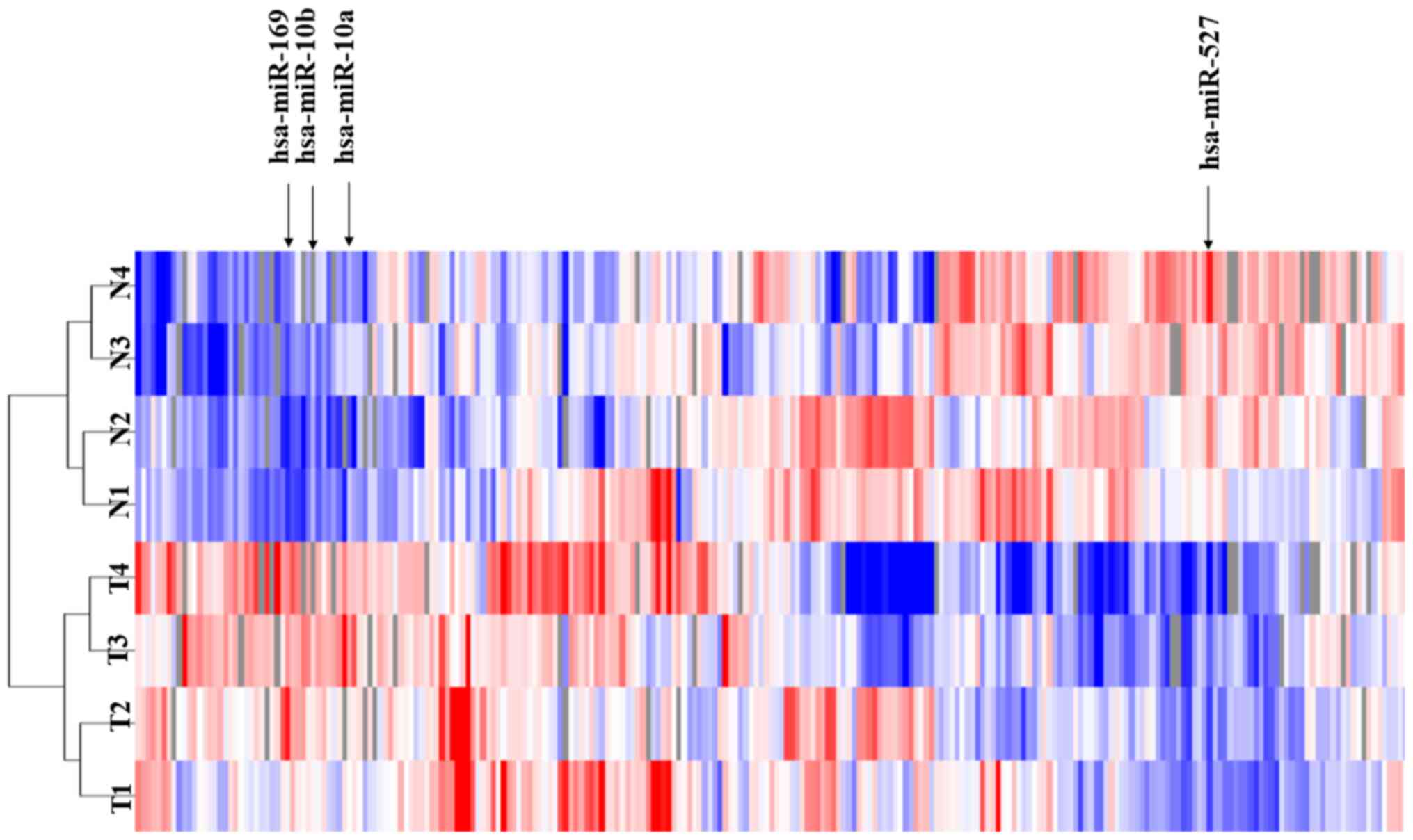
To investigate the roles of miRNAs in HCCs, the
miRNA expression profiles of four paired T tissues and their
corresponding N liver tissues (cirrhotic liver tissues) using 985
miRNAs probes. The human T tissues and their corresponding N liver
tissues exhibited differential miRNA expression profiles. The
unsupervised hierarchical clustering analysis revealed that the T
tissues clustered separately from the N tissues (Fig. 1). Among the tested 985 miRNAs, four
miRNAs with P<0.01 and FDR <0.05 were differentially
expressed (three upregulated and one downregulated) in the T
compared with N cirrhotic tissues (Fig.
1). These results indicated that there are differences in
microRNA expression between T and N cirrhotic tissues.
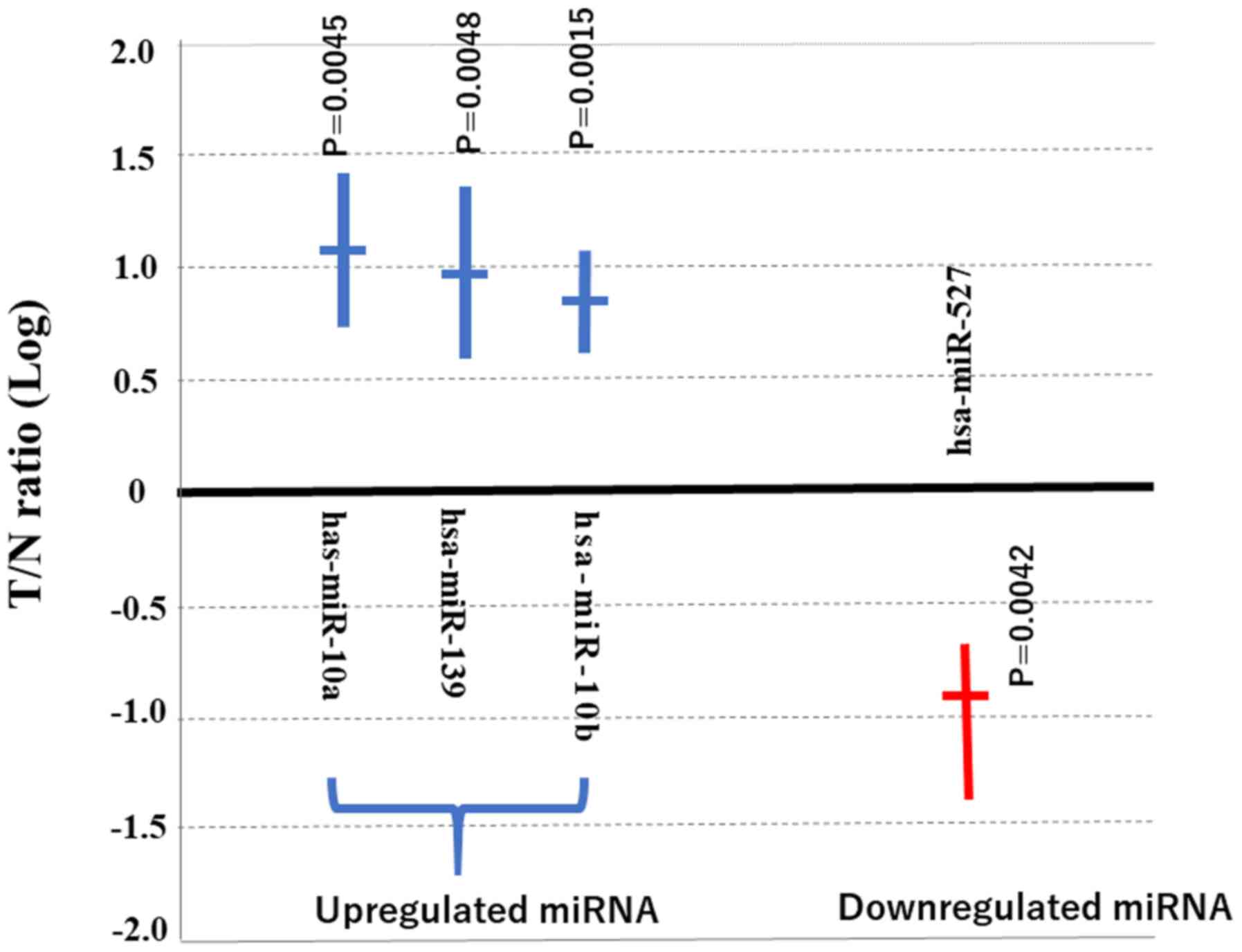
As shown in Fig. 2,
miR-527 was significantly downregulated in the T tissues
(P=0.0042). The data presented in Fig.
2 represent the average of the ratio of miR-527 expression in T
compared with that in N tissues of the four specimens obtained from
surgery. Previous studies have reported the target genes of
miRNA-10a (39) miRNA-10b (40) and miRNA-379 (41). However, the target gene of miR-527 is
not clear, to the best of our knowledge. Therefore, miRNA-527 was
chosen for further investigation. In short, miR-527 was
significantly downregulated in the T tissues compared with N
tissues.
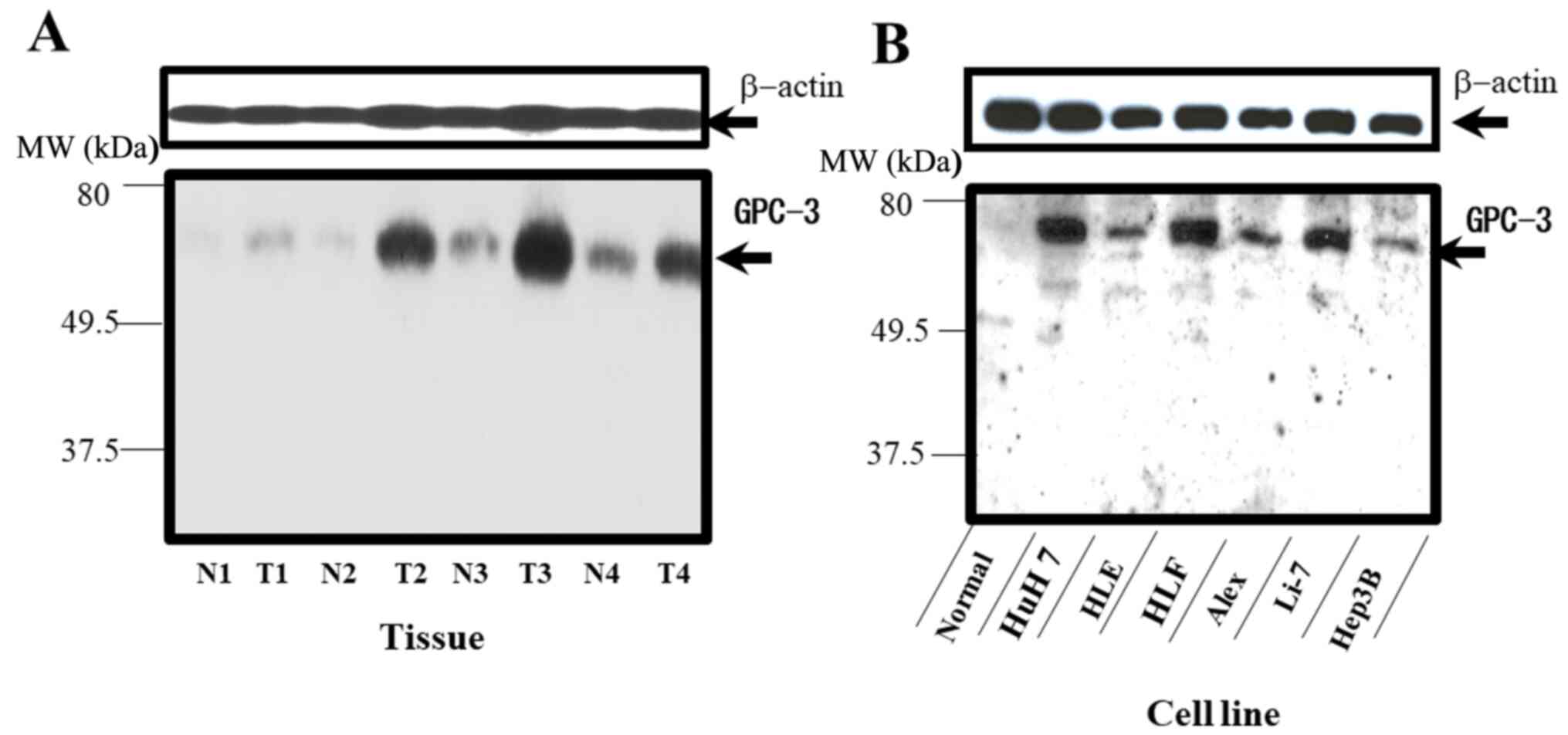
HCC cells exhibit enhanced expression
of GPC-3
GPC-3 is an important carcinogenic factor that is
highly sensitive for HCC (42).
Western blotting revealed that the four HCC (T) tissues exhibited
higher GPC-3 expression levels compared with the N tissues
(Fig. 3A). Consistent with the
observations in human tissues, the expression of GPC-3 was detected
in various HCC cell lines (Huh7, HLE, HLF, Alexander, Li-7 and
Hep3B) but not in normal hepatocyte line (Fig. 3B). These results suggested that the
expression of GPC-3 in the N tissues was lower compared with that
in the normal hepatocyte cell line.
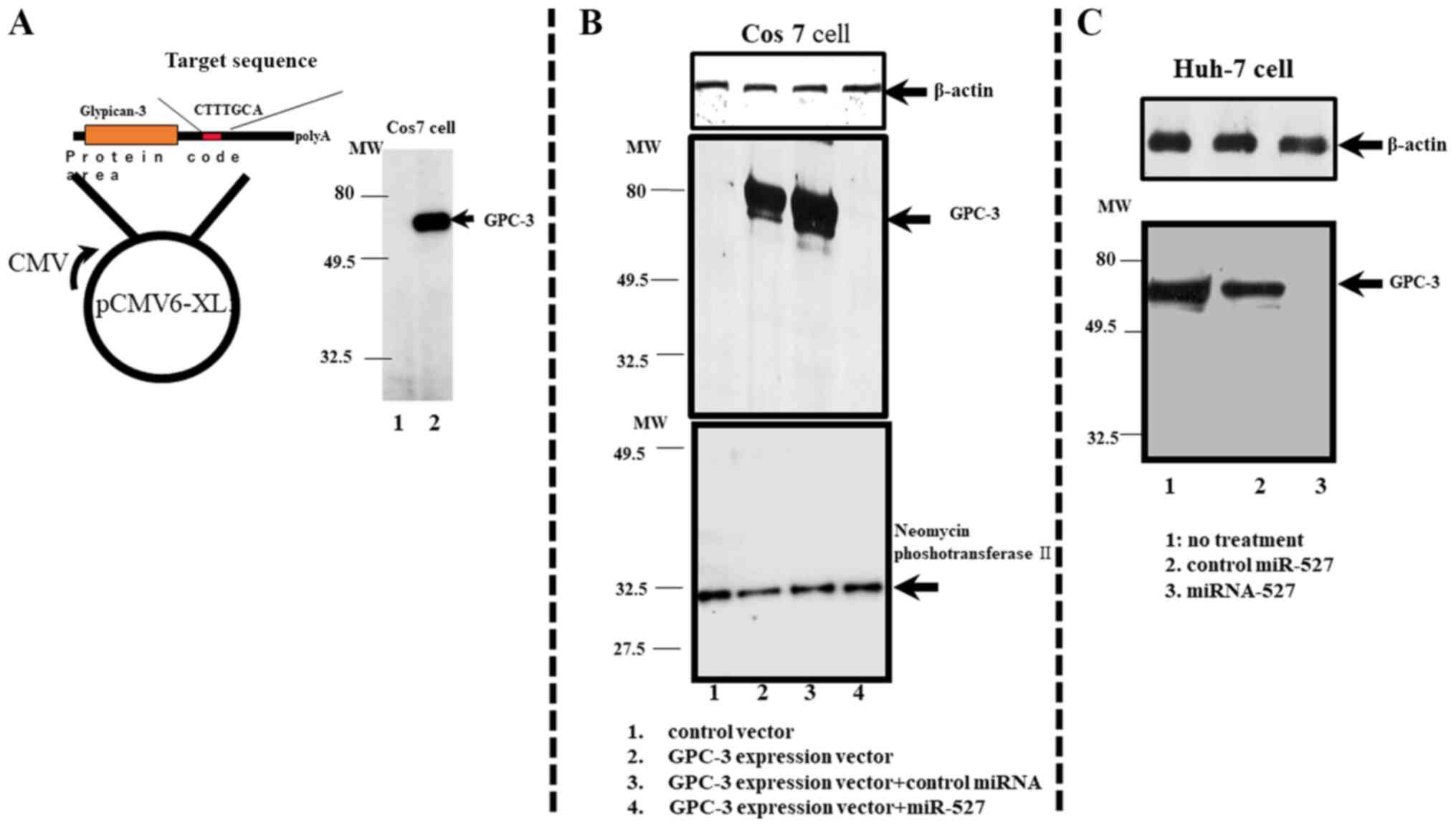
miRNA-527 suppresses GPC-3
expression
To clarify if GPC-3 is the target gene of miRNA-527,
transient gene transfection analysis was performed. The Cos7 cells
transfected with the pCMV6-XL5 vector (control vector) did not
exhibit GPC-3 protein expression (lane 1; Fig. 4A). The Cos7 cells transfected with
the pCMV6-GPC recombinant vector exhibited strong GPC-3 protein
expression (Fig. 4A, lane 2).
The transfection efficiency was evaluated by
co-transfecting the Cos7 cells with pCDNA3.1(+) vector, which
contains the neomycin phosphotransferase II gene, and pCMV6-GPC or
pCMV6-XL5 vector. The expression of neomycin phosphotransferase II
in the co-transfected cells was evaluated using western blotting
(Fig. 4B, lane 1).
As shown in Fig. 4B,
transfection with non-specific control miRNA did not suppress GPC-3
protein expression in the Cos7 cells at 48 h post-transfection
(Fig. 4B, lane 3). Meanwhile,
transfection with miRNA-527 suppressed GPC-3 protein expression in
the Cos7 cells (Fig. 4B, lane 4).
Cos-7 cells transfected with only pCMV6-GPC vector exhibited GPC-3
protein expression at 48 h post-transfection (Fig. 4B, lane 2), whereas those transfected
with only pCMV6-XL5 control vector did not exhibit GPC-3 protein
expression (Fig. 4B, lane 1). These
results suggested that miRNA-527 suppresses protein expression of
GPC-3.
miRNA-527 inhibits expression of
intrinsic GPC-3 in the Huh-7 cells
Similar to the HCC tissues, several HCC cell lines
are reported to express GPC-3 (43).
The expression of GPC-3 was confirmed in several HCC cell lines,
including the Huh-7 cell line (Fig.
4C). The effect of miRNA-527 transfection on the intrinsic
expression of GPC-3 in the Huh-7 cells was examined. As shown in
Fig. 4C, GPC-3 is expressed in the
Huh-7 cells (lane 1). Transfection with non-specific miRNAs did not
suppress the expression of GPC-3 in the Huh-7 cells (lane 2).
However, transfection with miRNA-527 completely inhibited the
intrinsic expression of GPC-3 in the Huh-7 cells (lane 3). This
indicated that miRNA-527 can also suppress the intrinsic expression
of GPC-3 in the Huh-7 cell line. The expression of neomycin
phosphotransferase II was similar in all groups, which suggested
that the transfection efficiency was similar in all groups
(Fig. 4B).
Target genes of miRNA-527
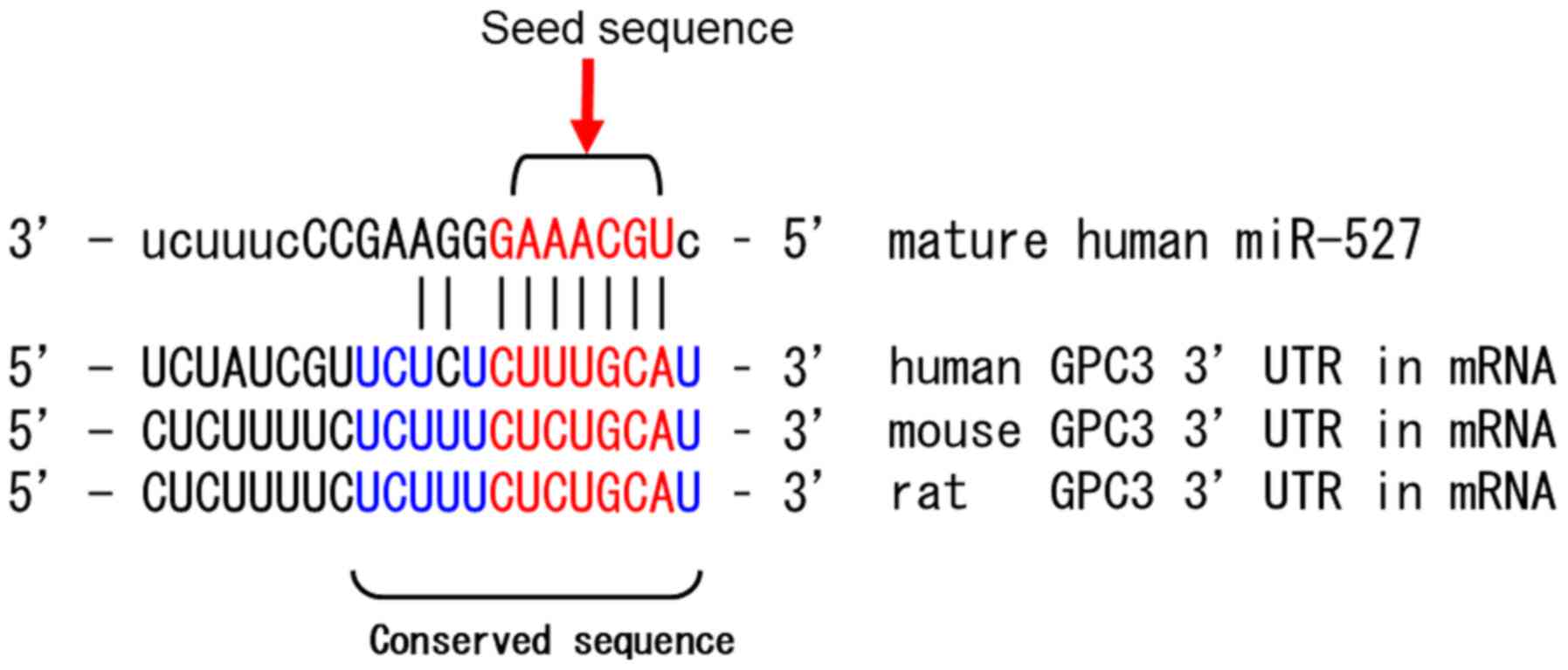
miRNA-527 is encoded on the 19th human chromosome
using the data base of miRNA, miRbase (http://www.mirbase.org/) and its target genes were
identified using Target Scan. The sequences at the 3′-UTR of the
GPC-3 gene, which are potentially involved in the development of
HCC, contained the target sites for miRNA-527. The GAAACGU
sequences (from second to the eight base) at the 5′ end of
miRNA-527, which is referred to as seed sequence, was complementary
to the sequences at the 3′-UTR in the GPC-3 gene among human, rat
and mouse (Fig. 5).
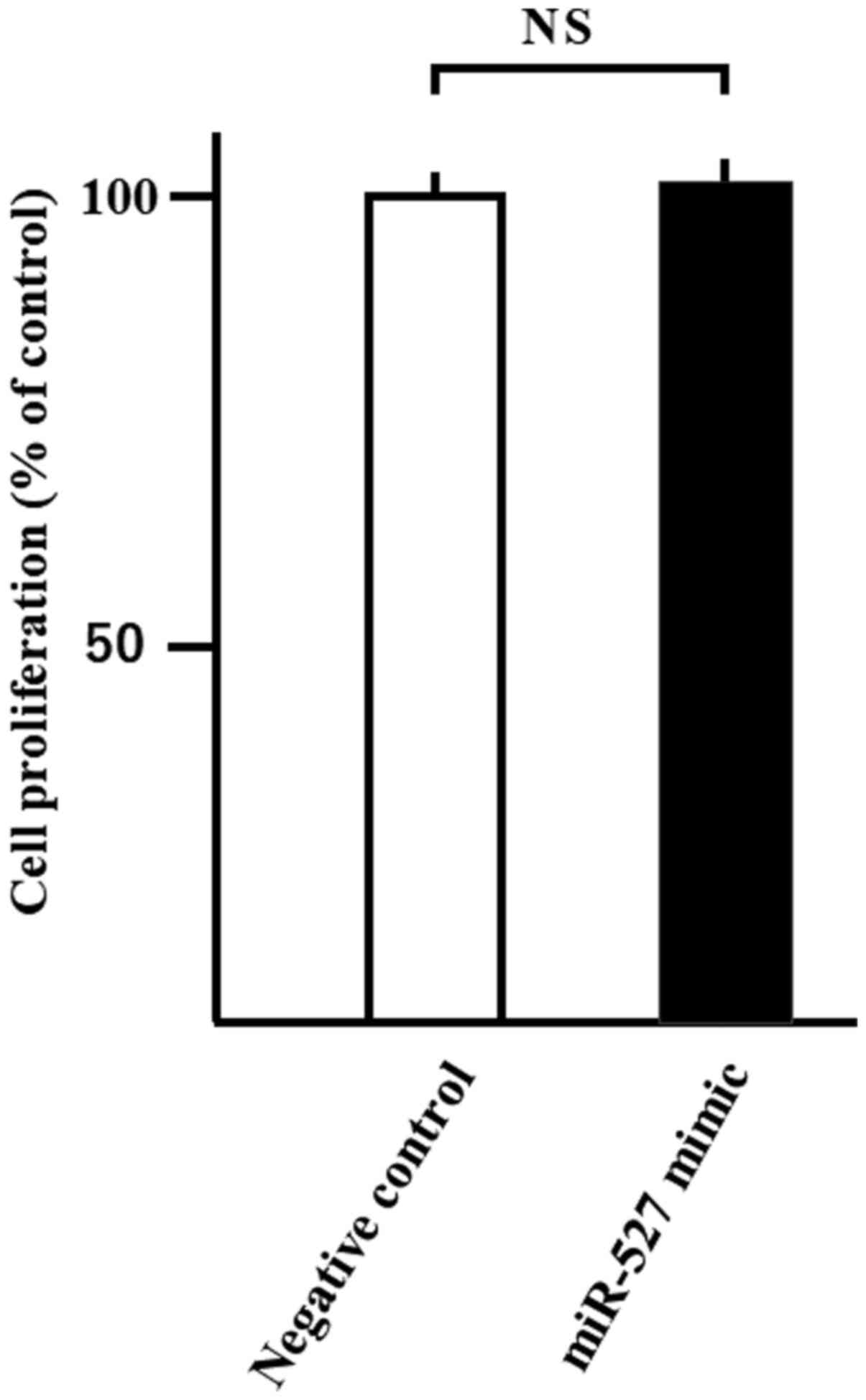
Transfection of miRNA
To study cell proliferation, the WST-8 assay was
conducted after 24 h of miR-527 transfection into Huh-7 cells.
Transfection with miR-527 did not affect the proliferation of Huh-7
cells compared with negative control miRNA (Fig. 6).
Discussion
Globally, HCC is one of the most common cancer types
(1). HCC is associated with complex
and heterogenous carcinogenesis and molecular mechanisms (44). Hence, the pathophysiology of HCCs is
not well understood. Previous studies have reported the molecular
mechanisms of miRNA in the carcinogenic process (42,43).
miRNAs regulate gene expression, and one miRNA is estimated to
regulate >200 target genes (5,6).
Therefore, miRNAs are potential therapeutic targets for various
cancer types, including HCC (7,9–13). The present study analyzed the miRNA
expression profiles of four paired HCCs and their corresponding
non-cancerous liver tissues using 985 miRNA probes to identify the
miRNAs that contribute to HCC pathogenesis.
The microarray analysis revealed one downregulated
and three upregulated miRNAs in the HCC tissues when compared with
the corresponding non-cancerous liver tissues. Generally,
downregulated miRNAs are considered to regulate the carcinogenic or
progressive factors of carcinomas (23–26).
miRNA-527 was downregulated, which may function as a tumor
suppressor miRNA in HCC. The association between miRNA-527 and HCC
has not been previously reported, therefore miRNA-527 was selected
for further investigation.
In addition, the current study identified GPC-3 as a
target gene of miRNA-527 using the Human micro RNA Target database.
The database analysis revealed that the GPC-3 gene had the target
site for miRNA-527. The 5′ end of miRNA-527 contains the seed
sequence, which binds to the 3′-UTR of the GPC-3 gene. GPC-3
belongs to the heparin sulfate proteoglycan (HSPG) family, which
are localized and bound to the cell membrane surface by
glycosylphosphatidylinositol anchors (6,7). GPC-3
is overexpressed in the HCC tissues and is reported to be a
therapeutic target for HCC (44).
GPC-3 expression was also observed in the fetal liver with levels
of proliferation but not in the normal adult liver or cirrhotic
liver (45). The results of the
present study showed that non-cancerous tissues exhibited lower
GPC-3 expression compared with HCC tissues. GPC-3 expression was
not detected in the normal hepatocyte cell line; however, enhanced
GPC-3 expression was observed in numerous hepatoma cell lines. The
current data suggested that upregulated GPC-3 may contribute to HCC
carcinogenesis.
Wang et al (46) reported that out of 111 HCC cases,
GPC-3-overexpression was detected in the cytoplasm, cell membrane
and canaliculus in 84 (75.7%) cases. Of these 84 cases, 61 (72.6%)
cases exhibited diffuse immunoreactivity. Contrastingly, none of
the 110 cases of hepatocellular adenoma, focal nodular hyperplasia
and large regenerative nodule exhibited detectable GPC-3
expression. Additionally, the expression of GPC-3 in HCC tissue is
not correlated with the size, differentiation or advanced stage
based on Union for International Cancer Control Tumor Node
Metastasis classification of the tumor (32). Moreover, patients with HCC exhibit
higher serum GPC-3 levels compared with patients without HCC
(46). Overall, these results and
those of the present study indicate that GPC-3 can be a novel tumor
marker for HCC.
GPC-3 is also reported to be a carcinogenic factor
in HCC (47). GPC-3 may induce HCC
tissue growth by stimulating the canonical Wnt/β-catenin pathway
(46). Additionally, GPC-3 is
closely associated with the function of sulfatase 2 (SULF2), an
enzyme with 6-O-desulfatase activity on HSPGs. SULF2, which is
upregulated in 60% of HCC cases, is closely associated with poor
prognosis (32). SULF2 is also
reported to upregulate FGF signaling in HCC cells via heparin
sulfate-dependent and GPC-3-dependent mechanisms (47). Furthermore, overexpression of SULF2
increases the proliferation and migration of HCC cells (47). Knockdown of GPC-3 attenuates FGF2
binding in the SULF2-expressing HCC cells. The role of SULF2 in
upregulating GPC-3 expression and promoting tumor growth was also
confirmed in a nude mouse xenograft model (47). These studies indicate that GPC-3 may
have an important role in the development of HCC.
Next, it was investigated whether miRNA-527 can
suppress the expression of GPC-3. The pCMV6-GPC vector was
constructed and transfected into the Cos7 cells to overexpress
GPC-3, as Cos7 cells do not usually express GPC-3. miRNA-527
completely suppressed the expression of GPC-3 in the
GPC3-overexpressing Cos7 cells. In addition, whether miRNA-527
could suppress intrinsically expressed GPC-3 in human HCC cells was
investigated. miRNA-527 was transfected into the Huh-7 cells, which
express intrinsic GPC-3. GPC-3 expression was inhibited in the
miR-527-transfected Huh-7 cells. These data suggested that GPC-3 is
a target gene of miRNA-527 is. To the best of our knowledge, the
present study is the first to report the association between GPC-3
and miRNA-527 in HCC. Only one previous study has reported the
correlation between miRNA-527 and cancer; Huo et al
(48) demonstrated that lung tumor
tissues and non-small cell lung cancer cells exhibit decreased
miRNA-527 expression. Overall, the present results indicated that
miR-527 has potential as a diagnostic biomarker for HCC in the
future.
A previous study demonstrated that siRNA-mediated
downregulation of GPC-3 expression in GPC-3-overexpressing HCC cell
lines markedly decreased the expression levels of growth signaling
molecules, matrix metalloproteinases [(MMP)2 and MMP14], FGF
receptor 1 and insulin-like growth factor 1 receptor in the cells
(47). Thus, GPC-3 may be a
potential therapeutic target for HCC (46).
To study the effect of miR-527 transfection on
cancer cell proliferation, miR-527 mimic was transfected into the
Huh-7 cells. Transfection with miR-527 did not affect the
proliferation of Huh-7 cells (data not shown). These results
suggested that gene transfection into HCC cells of with miR-527,
which targets GPC-3, did not inhibit of cancer cells
proliferation.
Further studies are needed to evaluate the
suppression of HCC growth through GPC-3 expression modulation by
miRNA-527. However, the results of the present study and previous
studies indicate that miRNA-527 could have potential as a
therapeutic target for HCCs through regulation of GPC-3
expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KN, AK, HI, JT, TN and MN designed the research and
analyzed the data. KN, TM, JT, HY, HK, AM, KT and TH drafted this
manuscript. KeO and YS collected the sample of patients. KN, KyO,
TT, KF, SM analyzed western blotting and transfection data. KN, AK,
HI, JT, TN, MN, KyO, TT, KF, SM, HY, HK, AM, KeO, YS, KT, TH and TM
were involved in data interpretation and have read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The study was approved by The Human Subjects
Committee of Kagawa University (approval no. 22–163). Additionally,
written informed consent was obtained from all patients to use the
tissue for this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
T
|
cancerous tissue
|
|
N
|
non-cancerous tissue
|
|
GPC-3
|
glypican 3
|
References
|
1
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang CH, Wey KC, Mo LR, Chang KK, Lin RC
and Kuo JJ: Current trends and recent advances in diagnosis,
therapy, and prevention of hepatocellular carcinoma. Asian Pac J
Cancer Prev. 16:3595–3604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sherman M: Hepatocellular carcinoma:
Epidemiology, surveillance, and diagnosis. Semin Liver Dis.
30:3–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iwama H, Masaki T and Kuriyama S:
Abundance of microRNA target motifs in the 3′-UTRs of 20527 human
genes. FEBS Lett. 581:1805–1810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomasello L, Cluts L and Croce CM:
Experimental validation of microRNA targets: Mutagenesis of binding
regions. Methods Mol Biol. 1970:331–339. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Visone R and Croce CM: miRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morishita A and Masaki T: miRNA in
hepatocellular carcinoma. Hepatol Res. 45:128–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA, Liu CG, Sevignani C, Ferracin M,
Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sevignani C, Calin GA, Siracusa LD and
Croce CM: Mammalian microRNAs: A small world for fine-tuning gene
expression. Mamm Genome. 17:189–202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Metzler M, Wilda M, Busch K, Viehmann S
and Borkhardt A: High expression of precursor microRNA-155/BIC RNA
in children with Burkitt lymphoma. Genes Chromosomes Cancer.
39:167–169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
18
|
Tokarz P and Blasiak J: The role of
microRNA in metastatic colorectal cancer and its significance in
cancer prognosis and treatment. Acta Biochim Pol. 59:467–474. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Del Vescovo V and Denti MA: microRNA and
lung cancer. Adv Exp Med Biol. 889:153–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang X, Liang L, Zhang XF, Jia HL, Qin Y,
Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, et al: MicroRNA-26a
suppresses tumor growth and metastasis of human hepatocellular
carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology.
58:158–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang
JR, Zeng C and Zhuang SM: MicroRNA-26a/b and their host genes
cooperate to inhibit the G1/S transition by activating the pRb
protein. Nucleic Acids Res. 40:4615–4625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AYF, Jooi LLP and Lee CG: Lethal-7 is down-regulated by
the hepatitis B virus × protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X,
Xie D, Shen Z, Sze J, Li K, et al: MYC protein inhibits
transcription of the microRNA cluster MC-let-7a-1~let-7d via
noncanonical E-box. J Biol Chem. 286:39703–39714. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsang WP and Kwok TT: Let-7a microRNA
suppresses therapeutics-induced cancer cell death by targeting
caspase-3. Apoptosis. 13:1215–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Di Fazio P, Montalbano R, Neureiter D,
Alinger B, Schmidt A, Merkel AL, Quint K and Ocker M:
Downregulation of HMGA2 by the pan-deacetylase inhibitor
panobinostat is dependent on hsa-let-7b expression in liver cancer
cell lines. Exp Cell Res. 318:1832–1843. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu XM, Wu LJ, Xu J, Yang R and Wu FS:
Let-7c microRNA expression and clinical significance in
hepatocellular carcinoma. J Int Med Res. 39:2323–2329. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lan FF, Wang H, Chen YC, Chan CY, Ng SS,
Li K, Xie D, He ML, Lin MC and Kung HF: Hsa-let-7g inhibits
proliferation of hepatocellular carcinoma cells by downregulation
of c-Myc and upregulation of p16(INK4A). Int J Cancer. 128:319–331.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Traister A, Shi W and Filmus J: Mammalian
Notum induces the release of glypicans and other GPI-anchored
proteins from the cell surface. Biochem J. 410:503–511. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li N, Gao W, Zhang YF and Ho M: Glypicans
as Cancer Therapeutic Targets. Trends Cancer. 4:741–754. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Capurro MI, Xu P, Shi W, Li F, Jia A and
Filmus J: Glypican-3 inhibits Hedgehog signaling during development
by competing with patched for Hedgehog binding. Dev Cell.
14:700–711. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shirakawa H, Suzuki H, Shimomura M, Kojima
M, Gotohda N, Takahashi S, Nakagohri T, Konishi M, Kobayashi N,
Kinoshita T, et al: Glypican-3 expression is correlated with poor
prognosis in hepatocellular carcinoma. Cancer Sci. 100:1403–1407.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang J, Zhang M, Ma H, Song X, He L, Ye X
and Li X: Overexpression of glypican-3 is a predictor of poor
prognosis in hepatocellular carcinoma: An updated meta-analysis.
Medicine (Baltimore). 97:e111302018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fujihara S, Kato K, Morishita A, Iwama H,
Nishioka T, Chiyo T, Nishiyama N, Miyoshi H, Kobayashi M, Kobara H,
et al: Antidiabetic drug metformin inhibits esophageal
adenocarcinoma cell proliferation in vitro and in
vivo. Int J Oncol. 46:2172–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fujita K, Iwama H, Sakamoto T, Okura R,
Kobayashi K, Takano J, Katsura A, Tatsuta M, Maeda E, Mimura S, et
al: Galectin-9 suppresses the growth of hepatocellular carcinoma
via apoptosis in vitro and in vivo. Int J Oncol.
46:2419–2430. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: Procedure and some applications. Proc Natl
Acad Sci USA. 76:4350–4354. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
39
|
Chen Y-H, Song Y, Yu YL, Cheng W and Tong
X: miRNA-10a promotes cancer cell proliferation in oral squamous
cell carcinoma by upregulating GLUT1 and promoting glucose
metabolism. Oncol Lett. 17:5441–5446. 2019.PubMed/NCBI
|
|
40
|
Pal R and Greene S: microRNA-10b is
overexpressed and critical for cell survival and proliferation in
medulloblastoma. PLoS One. 10:e01378452015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Werk AN, Bruckmueller H, Haenisch S and
Cascorbi I: Genetic variants may play an important role in
mRNA-miRNA interaction: Evidence for haplotype-dependent
downregulation of ABCC2 (MRP2) by miRNA-379. Pharmacogenet
Genomics. 24:283–291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nishida T and Kataoka H: Glypican
3-targeted therapy in hepatocellular carcinoma. Cancers (Basel).
11:E13392019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vongchan P and Linhardt RJ:
Characterization of a new monoclonal anti-glypican-3 antibody
specific to the hepatocellular carcinoma cell line, HepG2. World J
Hepatol. 9:368–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Singh AK, Kumar R and Pandey AK:
Hepatocellular Carcinoma: Causes, Mechanism of Progression and
Biomarkers. Curr Chem Genomics Transl Med. 12:9–26. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou F, Shang W, Yu X and Tian J:
Glypican-3: A promising biomarker for hepatocellular carcinoma
diagnosis and treatment. Med Res Rev. 38:741–767. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang L, Yao M, Pan LH, Qian Q and Yao DF:
Glypican-3 is a biomarker and a therapeutic target of
hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int.
14:361–366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lai JP, Sandhu DS, Yu C, Han T, Moser CD,
Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, et
al: Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth
factor signaling, and decreases survival in hepatocellular
carcinoma. Hepatology. 47:1211–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huo W, Zhu XM, Pan XY, Du M, Sun Z and Li
ZM: MicroRNA-527 inhibits TGF-β/SMAD induced epithelial-mesenchymal
transition via downregulating SULF2 expression in non-small-cell
lung cancer. Math Biosci Eng. 16:4607–4621. 2019. View Article : Google Scholar : PubMed/NCBI
|