Introduction
As one of the most severe human malignant diseases,
liver cancer ranks as the second leading cause of cancer-associated
mortality worldwide (1). The annual
liver cancer-associated mortalities are estimated at ~700,000
worldwide (2). Liver cancer is often
extremely heterogeneous, making the treatment of the disease even
more difficult (2). Compared with
developed countries, the incidence of liver cancer exhibits an
increasing trend in developing countries (3). The morbidity rate of liver cancer ranks
second among all cancer-associated mortalities (4). Liver cancer is further classified into
primary and secondary liver cancer. Primary liver cancers mainly
include hepatocellular carcinoma (HCC), intrahepatic
cholangiocarcinoma (ICC), and combined HCC and ICC, while secondary
liver cancer mainly refers to metastatic disease (5). Several factors have been associated
with liver cancer, including viral infections such as hepatitis B
virus and hepatitis C viruses, fatty liver disease, smoking,
obesity, diabetes mellitus, iron overload, alcohol abuse and
deregulation of metabolism (6). The
early stage of liver cancer may often be asymptomatic; therefore,
the majority of patients are diagnosed in advanced stages of the
disease, when metastasis has already occurred, thus leading to a
poor prognosis and a high mortality rate (7).
As a heterogeneous malignant disease, liver cancer
is considered one of the most difficult types of cancer to treat
(2). To date, multidisciplinary
approaches are used to treat liver cancer based on the clinical
characteristics of each patient, including the complex interplay of
tumor stage and the extent of underlying liver disease, as well as
the patient's general health (8). A
variety of therapies for liver cancer have been developed, mainly
including cytotoxic chemotherapy, immunotherapy
(immune-checkpoint), oncolytic virus therapy and novel targeted
therapy (8). Other treatment
strategies, including transarterial embolization (TAE), where
embolic particles without chemotherapy are used, or transarterial
chemoembolization (TACE), where embolic particles are combined with
chemotherapeutic drugs, have been reported to be effective against
liver cancer via regulating arterial blood supply to induce tumor
necrosis (1). However, neither
current ablation therapies nor chemotherapy are ideal in improving
the outcomes of patients with liver cancer; therefore, more
effective therapies for treating this devastating disease are
urgently required (9).
The intravenous administration of tumoricidal agents
is limited due to their inability to access the entire tumor mass,
which is mediated by the high interstitial pressure of the solid
tumor (10). Recently, the minimally
invasive interventional drug delivery method for treating human
cancer has received increasing attention (11). In interventional therapy, a needle or
catheter that enters the body through a fine skin incision is
guided by an imaging system (fluoroscopy) to the solid tumor
(11). Compared with conventional
therapies, in interventional therapy, the anticancer drugs are
directly delivered into the tumor, thereby providing several
advantages, including fewer anesthetic administrations, fewer
traumas, less pain and shorter hospitalization time (10).
Chloroquine is a chemically synthesized compound,
which has been widely used as an antimalarial agent for a few
decades (12). Recently, chloroquine
and its derivatives have been reported to exert antiviral effects
against SARS-CoV-2 infection (13).
In addition, chloroquine diphosphate (CQ) was used to treat
Plasmodium falciparum parasite infections (14). Accumulating evidence has indicated
that chloroquine and its derivatives exert anticancer effects. For
example, Wei et al (15)
demonstrated that CQ may suppress pancreatic cancer via modulating
the autophagy process. Furthermore, a study revealed that CQ
exerted antitumor effect on breast cancer in a murine model
(16). Sasaki et al (17) demonstrated that 5-fluorouracil
combined with chloroquine may suppress colon cancer in a colon
cancer cell line and mouse model. However, the effects of
chloroquine and its derivatives on liver cancer remain to be
investigated. Therefore, in the present study, an interventional
therapy was applied to investigate the effects of CQ on liver
cancer in C57BL/6 mice. The findings of the present study may
provide novel insight into the development of novel treatment
strategies against liver cancer.
Materials and methods
Chemicals and reagents
CQ was purchased from Sigma-Aldrich; Merck KGaA
(cat. no. 50-63-5). CellTiter-Glo® Luminescent Cell
Viability assay (cat. no. G7570) and CellTiter-Glo® 3D
Cell Viability assay (cat. no. G9681) were both obtained from
Promega Corporation. The Cell Counting Kit-8 (CCK-8) assay was
purchased from Beyotime Institute of Biotechnology (cat. no. C0037)
and Dulbecco's phosphate-buffered saline (DPBS; cat. no. 14190250)
from Thermo Fisher Scientific, Inc. All other reagents used were of
analytical grade.
Cell culture
Human liver cancer HepG2 cells were purchased from
the American Type Culture Collection (HB-8065™). The cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; cat. no.
11995065), supplemented with 10% fetal bovine serum (FBS; cat. no.
16140071) and 50 U/ml penicillin-streptomycin (cat. no. 15070063;
all Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified
atmosphere of 5% CO2. Upon reaching 80–90% confluence,
the cells were passaged.
3D liver spheroid culture
A 3D liver spheroid culture was performed and
modified as previously described (18,19). In
brief, HepG2 cells were cultured in 75 cm2 cell culture
flasks, trypsinized using the TrypLE™ Express Enzyme kit (cat. no.
12605010; Thermo Fisher Scientific, Inc.) and pelleted by
centrifugation at 100 × g for 5 min at 4°C. Cells were resuspended,
and were then seeded onto a Corning Matrigel Growth Factor Reduced
Basement Membrane Matrix (cat. no. 356231; Corning Incorporated).
The morphology of 3D liver spheroids was observed under a
phase-contrast microscope (Olympus Corporation).
In vitro drug treatment assay
Subsequently, the effect of CQ on HepG2 cell growth
and 3D spheroids was investigated. At least two independent
experiments or four replicates were performed. For the HepG2
monolayer cultures, the cells (1×105 cells/well) were
seeded onto 48 well-plates and, upon reaching 50–60% confluence,
they were treated with the indicated concentrations of CQ (0, 1, 5,
10, 50 and 100 µM). For 3D liver spheroids, when 3D liver spheroids
were formed, they were harvested from Matrigel using cold PBS,
followed by filtration through a 70-µm cell strainer. Next, the
spheroids were centrifuged at 100 × g for 5 min at 4°C, added into
the 48-well plates, and treated with the indicated concentrations
of CQ (0, 1, 5, 10, 50 and 50 µM).
CCK-8 assays
HepG2 cell viability was assessed using a CCK-8
assay (cat. no. C0037; Beyotime Institute of Biotechnology),
according to the manufacturers' protocols. In brief, cells
(2×103 cells/well) or 3D liver spheroids were seeded
onto a 96-well plate, supplemented with 100 µl culture medium
containing the indicated concentrations of CQ and cultured for 48 h
at 37°C in a humidified atmosphere containing 5% CO2.
Subsequently, 10 µl CCK-8 reagent was added into the culture medium
and cells were incubated for an additional 2 h. The absorbance at
450 nm was measured using a microplate reader (Thermo Fisher
Scientific, Inc.).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissues using
the Beyozol total RNA extraction kit (cat. no. R0011; Thermo Fisher
Scientific, Inc.), according to the manufacturers' protocol. The
RNA was reverse transcribed into complementary DNA (cDNA) using the
PrimeScript™ RT Master mix (cat. no. RR036A; Takara Bio, Inc.),
according to the manufacturers' protocol. Subsequently, RT-qPCR was
performed using the TB Green® Fast qPCR mix (cat. no.
RR036A; Takara Bio, Inc.) on the ABI Prism 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The pre-denaturation
and denaturation temperatures were set at 95°C and that of
annealing/extension at 60°C. Pre-denaturation was performed for 10
min, denaturation for 15 sec and annealing/extension for 60 sec.
The number of cycles was set to 40. The relative expression of the
target genes was analyzed using the 2−ΔΔCq method
(20). GAPDH was used as a
housekeeping reference gene. All primer sequences are listed in
Table I.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Primer | Primer
sequences | Tm |
|---|
| Caspase-3 | Sense |
TGCTATTGTGAGGCGGTTGT | 59.96 |
|
| Antisense |
TCACGGCCTGGGATTTCAAG | 60.32 |
| Caspase-9 | Sense |
AGGCCCCATATGATCGAGGA | 59.88 |
|
| Antisense |
TCGACAACTTTGCTGCTTGC | 59.97 |
| GAPDH | Sense |
ATGTTGCAACCGGGAAGGAA | 60.18 |
|
| Antisense |
GCATCACCCGGAGGAGAAAT | 59.82 |
CellTiter-Glo Luminescent Cell
Viability assay
To measure the viability of cells and 3D liver
spheroids, 2D and 3D CellTiter-Glo Luminescent Cell Viability
assays were performed. In brief, for HepG2 cells, cells were seeded
onto a 96-well culture plate at a density of 3×103
cells/100 µl and treated with different concentrations of CQ for 48
h at 37°C in a humidified atmosphere containing 5% CO2.
For 3D liver spheroids, spheroids were isolated from the Matrigel,
seeded onto a 100× pre-coated Matrigel 96-well culture plate at a
density of ~100 spheroids/100 µl, and treated with different
concentrations of CQ for 48 h at 37°C in a humidified atmosphere
containing 5% CO2. Luminescence signals were measured
using the LMax II kit (Molecular Devices, LLC). The concentration
of ATP was calculated based on an ATP reference calibration curve
(cat. no. 18330019; Thermo Fisher Scientific, Inc.).
Animals, grouping and animal
experiments
All animal experiments were performed according to
the Guide for the Care and Use of Laboratory Animals from the
National Institutes of Health, and the study was approved by the
Animal Experimentation Committee of The Fourth Hospital of Medical
University (Shijiazhuang, China). A total of 30 male Wistar rats
(age, 6–7 weeks old; weight, 200±20 g) and 3 male young Wistar rats
(age, 3–4 weeks; weight, 100±10 g) were obtained from the
Experimental Center of The Fourth Hospital of Medical University.
All rats were housed in plastic cages (dimensions, 500×360×200 mm).
Each cage held 3 rats of the same sex and all cages were placed in
the same specific-pathogen-free animal room. The environment
conditions of the animal room were strictly controlled and
maintained at a temperature of 20.4–23.0°C, a relative humidity of
40.1–68.9%, an air change 8–15 times/h, a 12/12 h-light/dark cycle,
and ad libitum access to food. The rats were sacrificed
using deep anesthesia with thiopental (50 mg/kg). Death was
confirmed using cervical dislocation. HepG2 cell suspensions were
prepared (density, 1×104 cells/ml), and three young rats
were subcutaneously injected with 0.3 ml of the prepared cell
suspension. Following 10 days from the injection, the tumors were
isolated and dissected into 1.5 mm3 pieces.
Subsequently, the dissected tissues were implanted under the
capsule of the left liver lobe of the other 30 Wistar rats. The
rats were intramuscularly injected with 30,000 units long-acting
penicillin. The rats were maintained in a disease-free environment,
had ad libitum access to food and water and the litter was
changed every other day. On the 11th day following implantation,
the rats were divided into three groups, namely the control group
(no treatment) and the 0.5- and 1.5-mg/kg CQ treatment groups.
Following anesthetization, the hepatic artery of the animals was
retrograde intubated into the gastroduodenal artery (Portex PE10
microcatheter; inner diameter, 0.28 mm; outer diameter, 0.61 mm)
and the experimental groups were then perfused with 0.5 and 1.5
mg/kg CQ through the hepatic artery. Animals in the control group
were perfused with 1 ml saline. All animals were treated daily with
CQ or saline for 9 days according to a previous study (21). At the end of the treatment period,
animals were sacrificed, and the tumors were isolated, and their
size was measured. The tumor volume was calculated as previously
described (22). In brief, the tumor
length and width were measured using a precise clipper, and the
tumor volume was estimated using the following formula:
V=(W2 × L)/2, where V, W and L indicate tumor volume,
width, and length, respectively.
Flow cytometry
Cell suspensions were generated from tumor tissues,
followed by rinsing with a total volume of 10 ml of PBS buffer and
washing twice in 10 ml PBS buffer. Following the last wash step,
the supernatant was discarded and the pellet of cells was suspended
in washing buffer (PBS 5% his 0.1% NaN3). Cells were centrifuged
for 5 min at 200 × g and 4°C. The pellet was resuspended with 50 µl
primary anti-cytokeratin 19 antibody (cat. no. ab52625; Abcam) and
anti-Sox9 (cat. no. ab185966; Abcam) diluted 5 times in washing
buffer and incubated overnight at 4°C. Cells were also incubated
with the control isotype corresponding to each primary antibody.
Following incubation, primary antibodies were removed and cells
were washed for three times with washing buffer. Next, cells were
incubated with goat anti-rabbit IgG H&L (Alexa
Fluor® 488; cat. no. ab150077; Abcam) for 1 h at room
temperature. Next, cells were analyzed by flow cytometry using a
FACScalibur (BD Biosciences) with 488 channel. The data was
analyzed using FlowJo software (v10; BD Biosciences).
Statistical analysis
All data are expressed as the mean ± standard error
of the mean. Pairwise comparisons of the analytical data were
performed using an unpaired Student's t-test. For multiple
comparisons, one-way analysis of variance was performed, followed
by Bonferroni's and Dunnett's test. P<0.05 was considered to
indicate a statistically significant difference.
Results
CQ potently inhibits the growth of
HepG2 cells
To investigate the effect of CQ on cell growth,
HepG2 cells were treated with different concentrations (0, 1, 5,
10, 50 and 100 µM) of CQ. The results demonstrated that CQ markedly
attenuated the growth of HepG2 cells (Fig. 1A and B). To further investigate the
effect of CQ on HepG2 cell viability, a CellTiter-Glo Luminescent
Cell Viability assay was performed. Treatment of cells with 1, 5,
10, 50 and 100 µM CQ significantly decreased the ATP concentration
in HepG2 cells (Fig. 1C).
Furthermore, the CCK-8 assay revealed that treatment with 1, 5, 10,
50 or 100 µM CQ significantly inhibited HepG2 viability (Fig. 1D). Taken together, these results
indicated that CQ may potently inhibit the growth of HepG2 cells
in vitro.
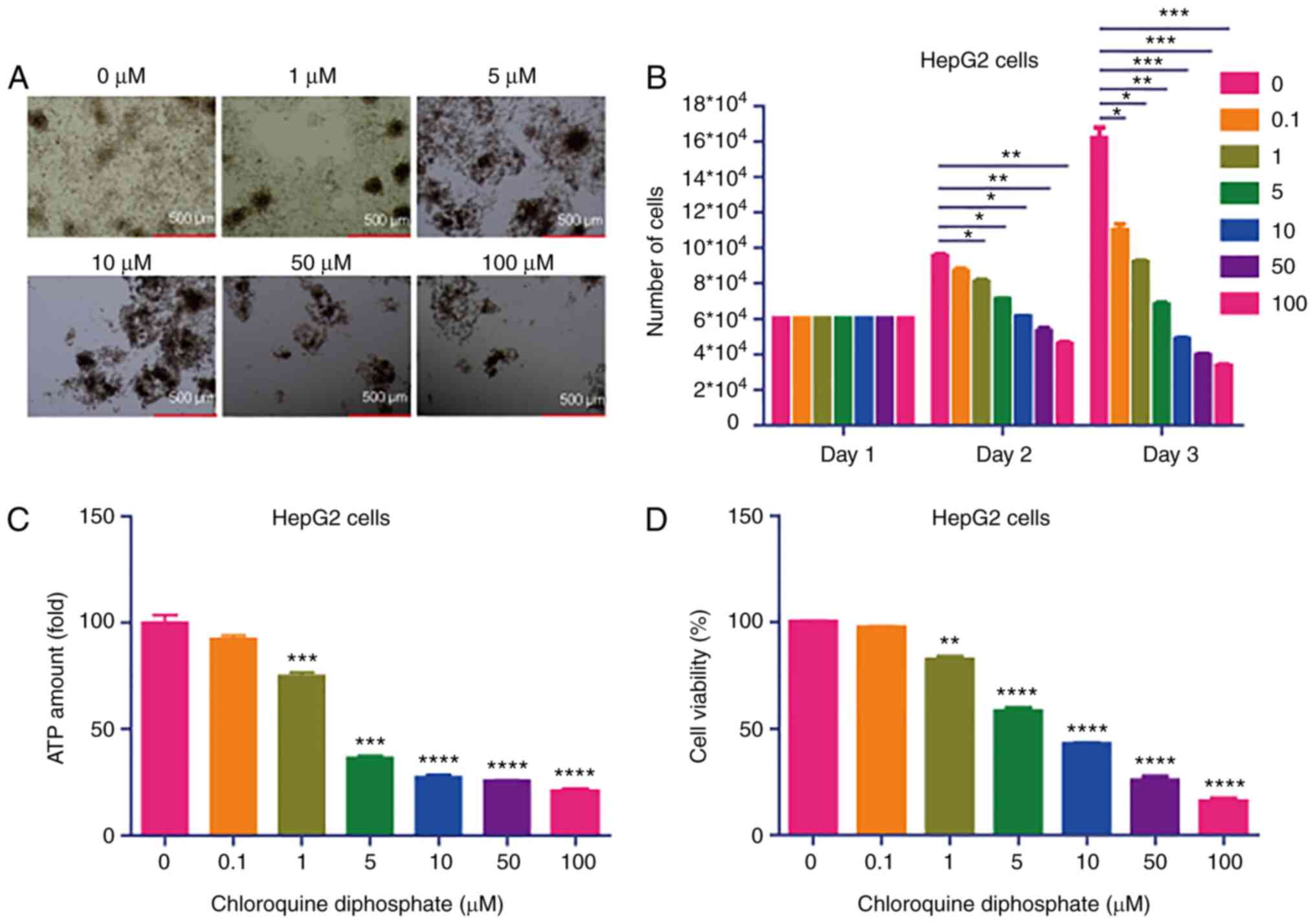 | Figure 1.CQ potently inhibits HepG2 cell
growth. (A) Morphology of HepG2 cells treated with different
concentrations (0, 1, 5, 10, 50 and 100 µM) of CQ under a light
microscope. (B) Quantification of the number of HepG2 cells treated
with different concentrations (0, 1, 5, 10, 50 and 100 µM) of CQ
(n=6). *P<0.05, **P<0.01 and ***P<0.001 vs. the control
group. (C) ATP concentration was measured in HepG2 cells treated
with 1, 5, 10, 50 and 100 µM CQ using a CellTiter-Glo Luminescent
Cell Viability assay (n=10). ***P<0.001 and ****P<0.0001 vs.
the control group. (D) Viability of HepG2 cells treated with 1, 5,
10, 50 and 100 µM of CQ was determined using a Cell Counting kit-8
assay (n=10). **P<0.01 and ****P<0.0001 vs. the control
group. CQ, chloroquine diphosphate. |
CQ potently inhibits the growth of 3D
liver spheroids
It has been reported that the 3D structure more
precisely mimics the in vivo physiology, compared with 2D
monolayer cells (23). Therefore, 3D
liver spheroids were cultured to investigate the effects of CQ on
the 3D model. CQ strongly suppressed the growth of 3D liver
spheroids (Fig. 2A and B). To
further investigate the effects of CQ on the viability of 3D liver
spheroids, a CCK-8 assay was performed, demonstrating that
treatment with 1, 5, 10, 50 or 100 µM CQ significantly inhibited 3D
liver spheroid viability (Fig. 2C).
These results confirmed that CQ may significantly inhibit the
growth of 3D liver spheroids.
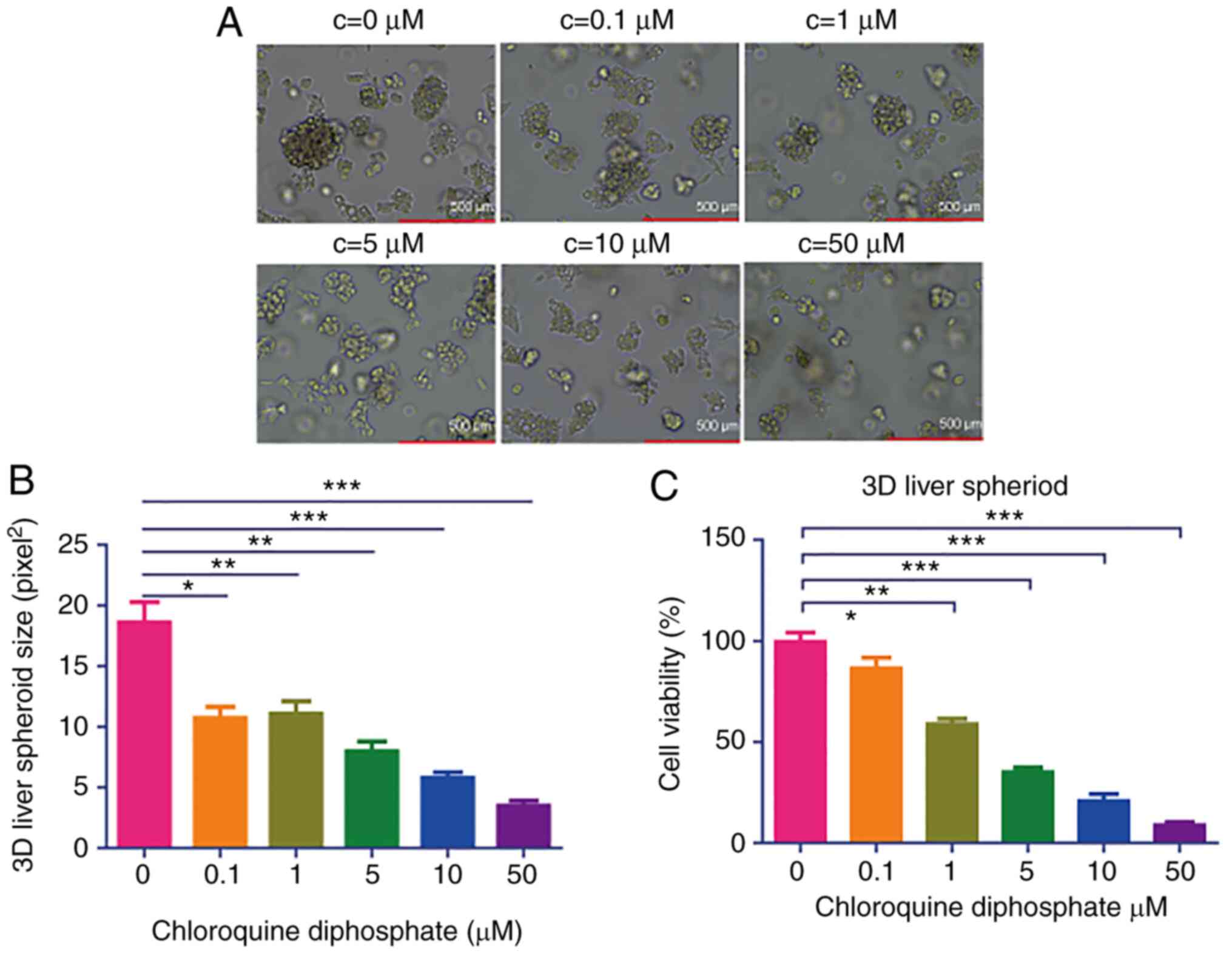 | Figure 2.CQ potently inhibits the growth of 3D
HepG2 spheroids. (A) Morphology of 3D liver spheroids treated with
different concentrations (0, 1, 5, 10 and 50 µM) of CQ under a
light microscope. (B) The concentration of ATP in 3D liver
spheroids treated with 1, 5, 10 and 50 µM CQ was measured using a
CellTiter-Glo Luminescent Cell Viability assay (n=10). *P<0.05,
**P<0.01 and ***P<0.001 vs. the control group. (C) The
viability of 3D liver spheroids treated with 1, 5, 10 and 50 µM of
CQ was evaluated using a Cell Counting kit-8 assay (n=10).
*P<0.05, **P<0.01 and ***P<0.001 vs. the control group..
CQ, chloroquine diphosphate. |
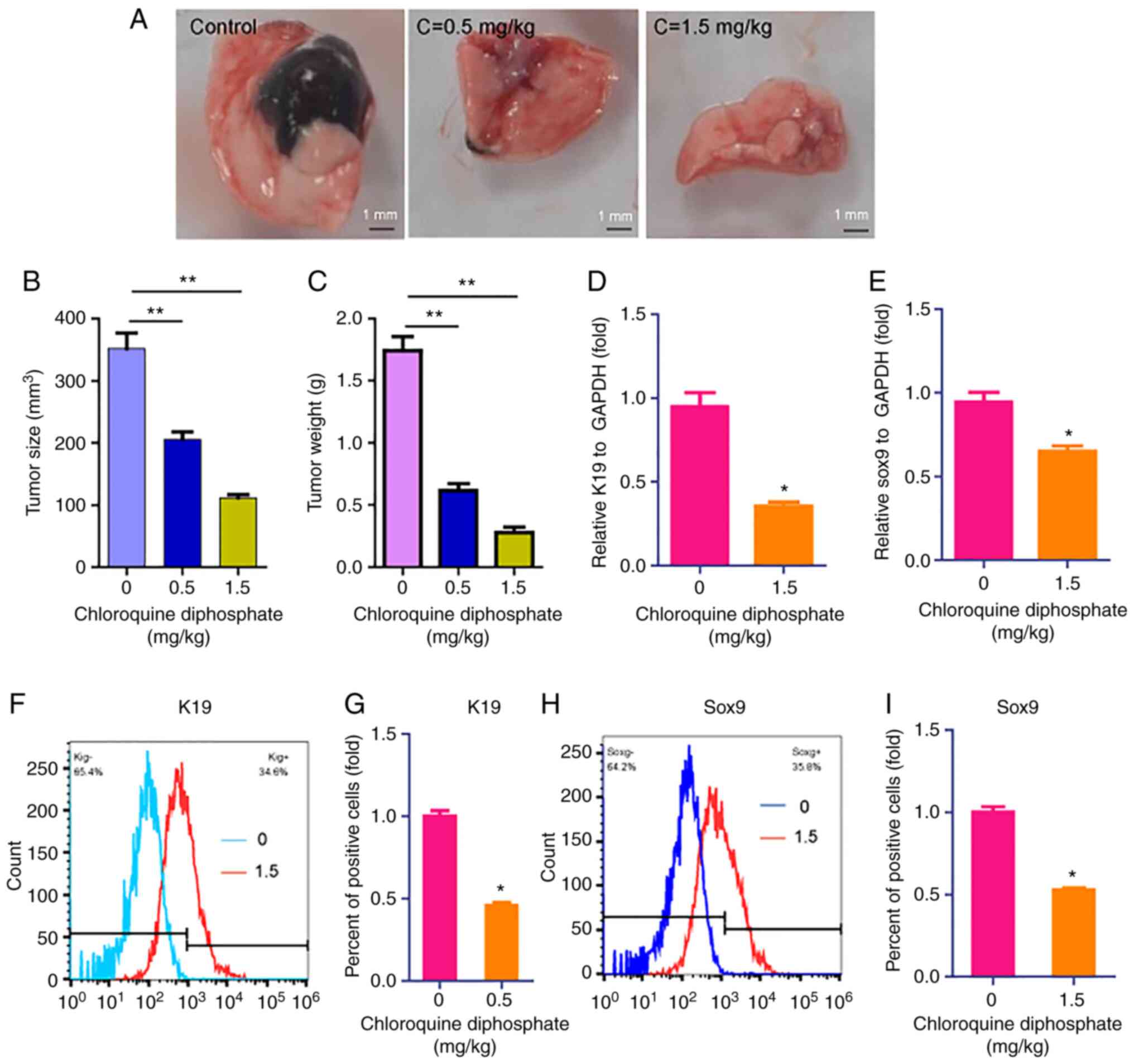
CQ-based interventional therapy
attenuates tumor growth in vivo
Interventional therapy is considered a less invasive
therapy approach to treat cancer, compared with conventional
therapies. Therefore, invasive therapy was applied to investigate
the effect of CQ on liver cancer. The results demonstrated that the
two drug doses (0.5 and 1.5 mg/kg) used inhibited tumor size
(Fig. 3A and B) and weight (Fig. 3C), compared with results in control
rats. Subsequently, to further investigate the effect of CQ on the
expression of liver cancer markers, the expression levels of the
liver cancer stem cell markers, including keratin 19 (K19) and
sox9, were determined (24). RT-qPCR
results revealed that CQ (1.5 mg/kg) downregulated the expression
of K19 and sox9 (Fig. 3D and E).
These findings were further verified using flow cytometric analysis
(Fig. 3F-I).
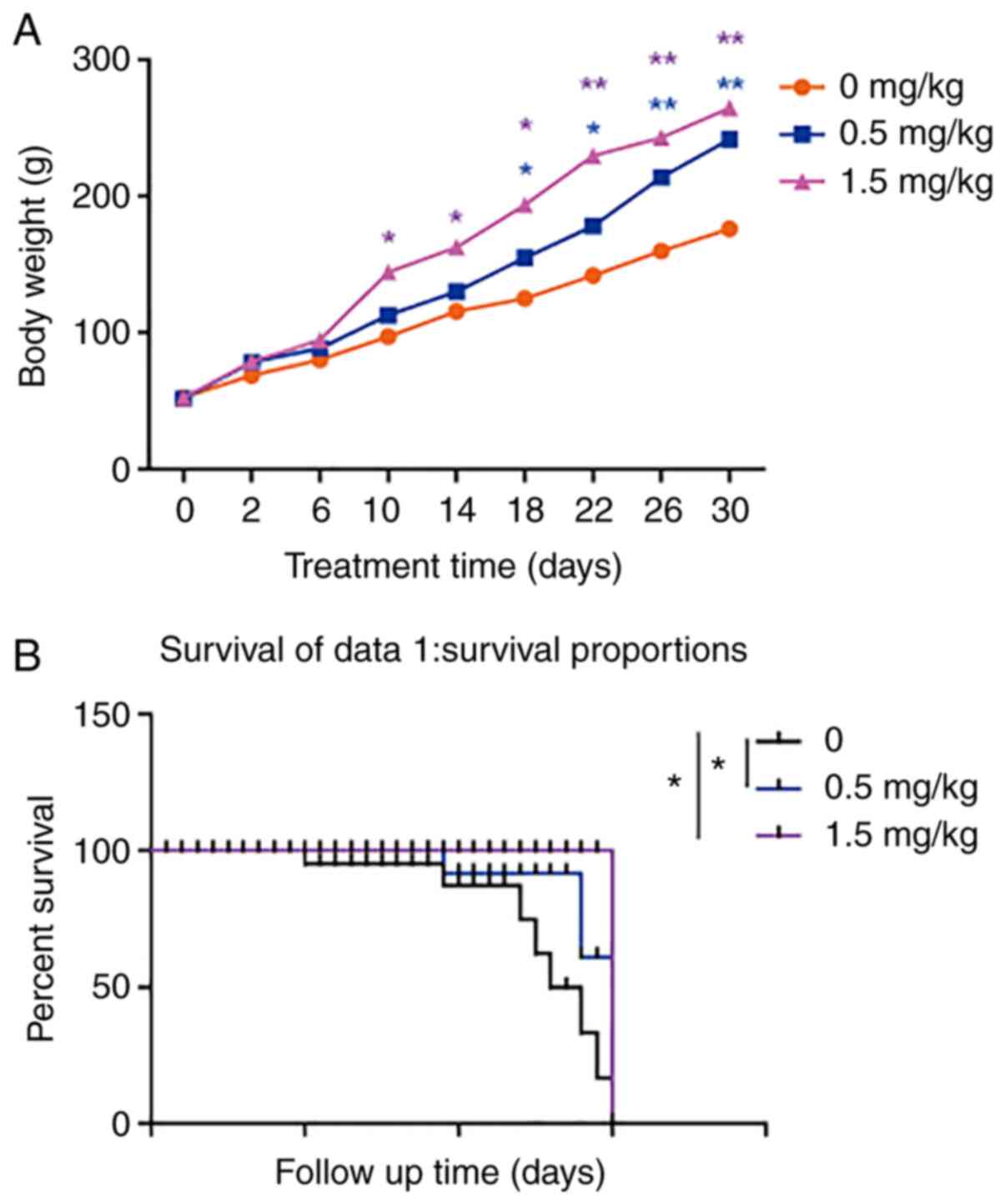
To monitor the growth and survival of rats that had
undergone interventional therapy, their body weight was recorded
daily. Therefore, the body weight of rats treated with CQ-based
interventional therapy was increased, compared with that of the
control group (Fig. 4A). Concerning
the survival rate of rats, 7/10, 3/10 and 1/10 rats died in the
control group, 0.5 (low-dose) and 1.5 mg/kg (high-dose) CQ
treatment groups, respectively (Fig.
4B). Notably, food and drinking water intake were increased in
the CQ treatment groups, compared with the control group (Table II). Taken together, the
aforementioned findings suggested that CQ-based interventional
therapy may effectively treat liver cancer.
 | Table II.Daily feed and water intake for rats
in different groups. |
Table II.
Daily feed and water intake for rats
in different groups.
| Treatment | 0 mg/kg | 0.5 mg/kg | 1.5 mg/kg |
|---|
| Feed intake, g | 12.40±0.67 | 14.90±0.64 | 16.2±0.59 |
| Drinking water,
g |
5.60±0.75 |
6.70±0.80 | 9.90±0.85 |
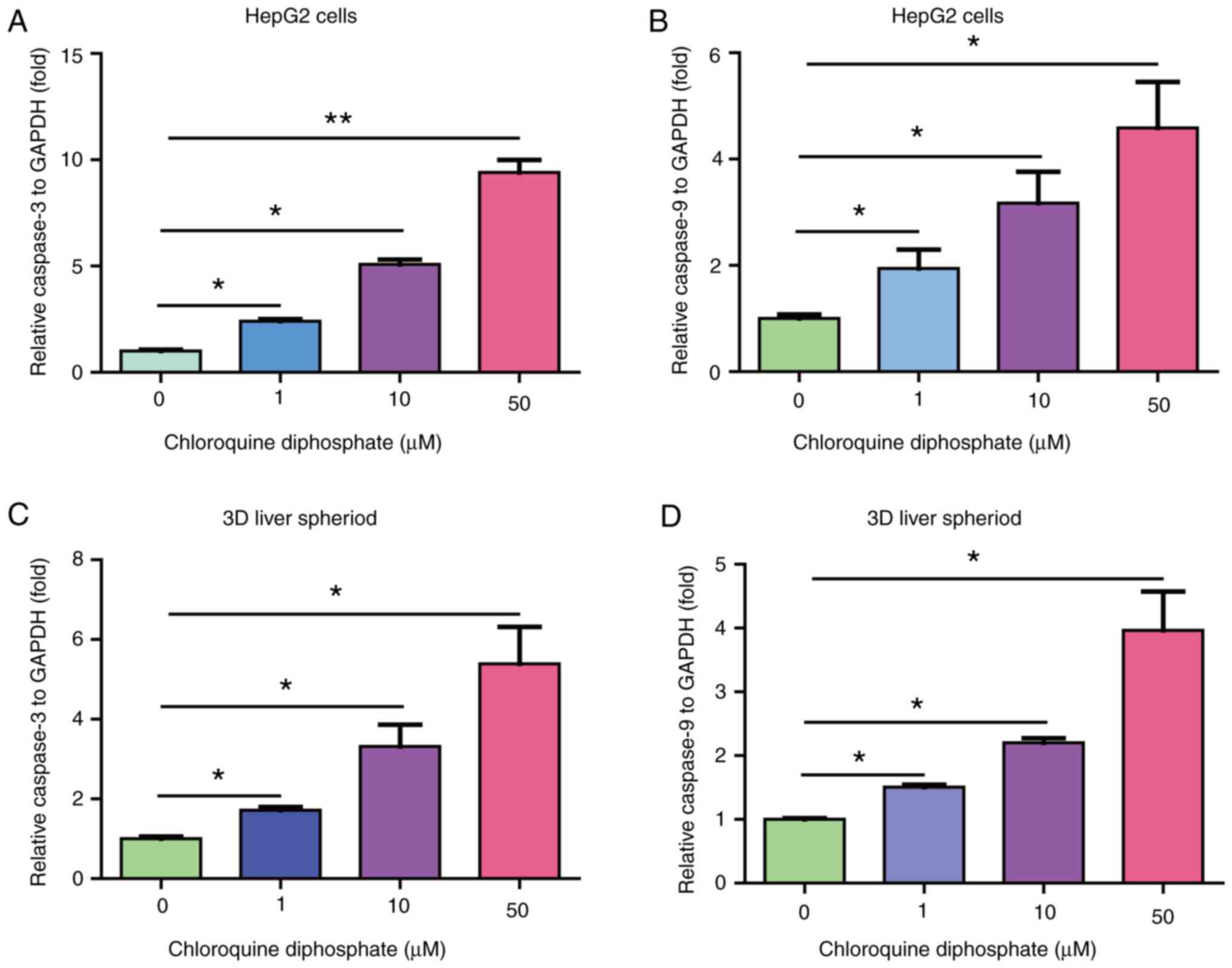
CQ induces apoptosis in HepG2 cells
and 3D liver spheroids
To reveal the mechanism underlying the effect of CQ
on suppressing liver cancer, the expression of the
apoptosis-related genes, caspase-3 and caspase-9, in cells treated
with CQ was determined. Therefore, treatment with 1, 10 or 50 µM CQ
notably increased the mRNA expression levels of caspase-3 and
caspase-9 (Fig. 5A and B).
Consistent with previous results, treatment of 3D liver spheroids
with 1, 10 or 50 µM CQ significantly upregulated the mRNA
expression of caspase-3 and caspase-9 (Fig. 5C and D; n=4; *P<0.05). Taken
together, these results suggested that the anticancer effects of CQ
may be mediated by promoting liver cancer cell apoptosis.
Discussion
As liver cancer is considered one of the most
serious types of cancer, great efforts have been made regarding the
development of optimal treatment approaches. CQ has been used for a
few decades as an antimalarial drug (25). It has been recently reported that CQ
exerts antiviral effects against COVID-19 infection (25). The present study demonstrated that CQ
may significantly inhibit the growth of HepG2 cells and 2D liver
spheroids. Furthermore, CQ-based interventional therapy notably
attenuated tumor growth, and increased the body weight and survival
of Wistar rats. Notably, it was revealed that CQ markedly increased
the expression of the apoptosis-related genes, caspase-3 and
caspase-9, indicating that the two molecules may underly the
anticancer effects of CQ.
CQ is an old drug, which was developed in the last
century (26). The drug is primarily
used to treat infections caused by Plasmodium falciparum and
is orally or parenterally administrated at a dose of 500 mg or 10
mg/kg, respectively (26). However,
CQ has also been widely used to treat multiple diseases. CQ was
found to render direct antiglobulin test-positive red blood cells
(RBCs) free from membrane-bound IgG and remove human leukocyte
antigens (HLAs) from RBCs to aid in identifying or excluding the
presence of antibodies against HLAs expressed on RBCs (27). In addition, treatment with 250 mg/day
CQ may prevent the exacerbation of systemic lupus erythematosus
(28). Notably, accumulating
evidence has indicated that CQ exerts antiviral effects against
SARS-CoV-2 infection. A European group reported that CQ may inhibit
the exacerbation of pneumonia, improve pulmonary imaging findings,
promote a negative conversion of the virus and shorten the disease
course in SARS-CoV-2-infected patients (13). It has also been reported that CQ
exerts anticancer effects on several types of cancer. Wang et
al (29) confirmed that CQ may
enhance gefitinib-mediated apoptosis of cutaneous squamous cell
carcinoma cells via inducing autophagy. Additionally, Wei et
al (15) demonstrated that CQ
was involved in the suppression of pancreatic cancer via regulating
the expression profile of circular RNAs, long non-coding RNAs,
microRNAs and mRNAs. Notably, Hu et al (30) confirmed that chloroquine triggered
G0/G1 cell cycle arrest and promoted DNA damage and apoptosis in
liver cancer cells in a dose- and time-dependent manner. The same
study revealed that chloroquine attenuated tumor growth in an
orthotopic xenograft model of liver cancer (30). The present study demonstrated that CQ
may suppress the growth and viability of liver cancer cells in 2D
and 3D models (Figs. 1 and 2). Furthermore, it was confirmed that the
drug enhanced the expression of apoptosis-related genes, including
caspase-3 and caspase-9 in the two models (Fig. 5). Furthermore, CQ has been found to
exert anticancer effects against other types of cancer, including
colon cancer (17), breast cancer
(16) and lung cancer (31). The aforementioned reports and the
present study suggested that CQ may serve as a broad anticancer
drug.
To date, the goal remains the treatment of liver
cancer patients using non-surgical minimally invasive therapies.
Interventional therapy strategies have been increasingly applied in
cancer due to their low invasiveness and relative safety (32). Different strategies, including
transarterial chemoembolization, radiofrequency ablation,
percutaneous ethanol injection, cryoablation, laser ablation and
upcoming promising procedures such as focused ultrasound and gene
therapy, have been applied as interventional therapy (33). In the present study, interventional
therapy with hepatic artery catheterization was utilized, which
allowed the transfer of CQ directly to the liver tumors, thereby
providing increased treatment efficacy. It was revealed that CQ
could remarkably decrease tumor size and weight (Fig. 3) and increase food intake, water
consumption (Table II), body weight
and survival rate (Fig. 4).
Therefore, these results indicated that the combination of
intervention therapy with CQ may display promising effects on liver
cancer in rats. Additionally, this treatment approach could
possibly confer a health benefit for liver cancer patients infected
with plasmodium or SARS-CoV-2. Nevertheless, more efforts should be
directed toward clarifying the effects of CQ on liver cancer in
mammalian in vivo models and clinical practice.
Although the present study has demonstrated that CQ
may suppress liver cancer via induction of apoptosis genes, there
are several limitations to the present study. To begin with, it was
confirmed that CQ increased expression of caspase-3 and caspase-9;
however, apoptosis is a complicated physiological process, and
multiple signaling pathways are involved in the process, including
TNF signaling (34), the intrinsic
mitochondrial pathway (35), the
intrinsic endoplasmic reticulum pathway (35) and the microRNA pathway (36). As one of inhibitors of autophagy, CQ
was reported to inhibit the growth of several tumors via
mTOR-autophagy-induced apoptosis (37,38),
reactive oxygen species-dependent apoptosis (39) and the mitochondrial pathway (40). Therefore, it is plausible that the
aforementioned apoptosis pathways require further investigation in
order to identify the inhibitory effects of CQ on liver cancer.
Another limitation to the present study is that only HepG2 cells
were used as a model. A more advanced in vitro model, named
organoid, has been developed by Prof. Dr. Hans Clevers of the
Hubrecht Institute in Netherlands (41), and has been used in infectious
diseases (42,43), immunity (44), nutrition (45) and cancer (46,47).
Therefore, the liver cancer organoid may be a promising model for
investigating the effects of CQ on liver cancer in future
studies.
In conclusion, CQ demonstrated effective anticancer
effects on HepG2 cells and 3D liver spheroids. The drug
significantly inhibited cancer cell growth and viability in 2D and
3D in vitro models. Furthermore, the CQ-based intervention
therapy effectively attenuated tumor size and weight, increased
food intake and drinking water consumption, and improved body
weight and survival rate. In addition, treatment of cells with CQ
potently upregulated the expression of apoptosis-related genes.
Therefore, the findings of the present study may provide novel
insight into the development of safe and effective therapies for
liver cancer.
Acknowledgements
Not applicable.
Funding
The study was supported by Hebei Provincial Health
Department (grant no. 20160623).
Availability of data and materials
All data were generated at The Fourth Hospital of
Hebei Medicine University and are available from the corresponding
author on reasonable request.
Authors' contributions
XH and WL had full access to all the data in the
present study and take responsibility for the integrity of the data
and the accuracy of the data analysis. WL conceived and designed
the present study; XH and WL acquired, analyzed and interpreted the
data; XH drafted the manuscript; WL critically revised the
manuscript for important intellectual content; XH and WL performed
the statistical analysis; XH and WL provided administrative,
technical and material support; WL supervised the study. All
authors read and approved the final manuscript.
Ethics statement and consent to
participate
The present study was approved by the Animal
Experimentation Committee of The Fourth Hospital of Medical
University (Shijiazhuang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yamashita T and Kaneko S: Liver cancer.
Rinsho Byori. 64:787–796. 2016.(In Japanese). PubMed/NCBI
|
|
2
|
Li L and Wang H: Heterogeneity of liver
cancer and personalized therapy. Cancer Lett. 379:191–197. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anwanwan D, Singh SK, Singh S, Saikam V
and Singh R: Challenges in liver cancer and possible treatment
approaches. Biochim Biophys Acta Rev Cancer. 1873:1883142020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu
X, Xia C, Yang Z, Li H, Wei W, et al: Liver cancer incidence and
mortality in China: Temporal trends and projections to 2030. Chin J
Cancer Res. 30:571–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiraha H, Iwamuro M and Okada H: Hepatic
stellate cells in liver tumor. Adv Exp Med Biol. 1234:43–56. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Losic B, Craig AJ, Villacorta-Martin C,
Martins-Filho SN, Akers N, Chen X, Ahsen ME, von Felden J, Labgaa
I, D'Avola D, et al: Intratumoral heterogeneity and clonal
evolution in liver cancer. Nat Commun. 11:2912020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo Z, Chen W, Dai G and Huang Y:
Cordycepin suppresses the migration and invasion of human liver
cancer cells by downregulating the expression of CXCR4. Int J Mol
Med. 45:141–150. 2020.PubMed/NCBI
|
|
8
|
Liu CY, Chen KF and Chen PJ: Treatment of
Liver Cancer. Cold Spring Harb Perspect Med. 5:a0215352015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan LK and Ng IO: Joining the dots for
better liver cancer treatment. Nat Rev Gastroenterol Hepatol.
17:74–75. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Phillips WT, Bao A, Brenner AJ and Goins
BA: Image-guided interventional therapy for cancer with
radiotherapeutic nanoparticles. Adv Drug Deliv Rev. 76:39–59. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
French JT, Goins B, Saenz M, Li S,
Garcia-Rojas X, Phillips WT, Otto RA and Bao A: Interventional
therapy of head and neck cancer with lipid nanoparticle-carried
rhenium 186 radionuclide. J Vasc Interv Radiol. 21:1271–1279. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weyerhäuser P, Kantelhardt SR and Kim EL:
Re-purposing chloroquine for glioblastoma: Potential merits and
confounding variables. Front Oncol. 8:3352018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Borba MGS, Val FFA, Sampaio VS, Alexandre
MAA, Melo GC, Brito M, Mourao MPG, Brito-Sousa JD, Baia-da-Silva D,
Guerra MVF, et al: Effect of high vs low doses of chloroquine
diphosphate as adjunctive therapy for patients hospitalized with
severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
infection: A randomized clinical trial. JAMA Netw Open.
3:e2088572020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kashyap A, Kaur R, Baldi A, Jain UK,
Chandra R and Madan J: Chloroquine diphosphate bearing dextran
nanoparticles augmented drug delivery and overwhelmed drug
resistance in Plasmodium falciparum parasites. Int J Biol
Macromol. 114:161–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei DM, Jiang MT, Lin P, Yang H, Dang YW,
Yu Q, Liao DY, Luo DZ and Chen G: Potential ceRNA networks involved
in autophagy suppression of pancreatic cancer caused by chloroquine
diphosphate: A study based on differentially-expressed circRNAs,
lncRNAs, miRNAs and mRNAs. Int J Oncol. 54:600–626. 2019.PubMed/NCBI
|
|
16
|
Jiang PD, Zhao YL, Deng XQ, Mao YQ, Shi W,
Tang QQ, Li ZG, Zheng YZ, Yang SY and Wei YQ: Antitumor and
antimetastatic activities of chloroquine diphosphate in a murine
model of breast cancer. Biomed Pharmacother. 64:609–614. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasaki K, Tsuno NH, Sunami E, Kawai K,
Hongo K, Hiyoshi M, Kaneko M, Murono K, Tada N, Nirei T, et al:
Resistance of colon cancer to 5-fluorouracil may be overcome by
combination with chloroquine, an in vivo study. Anticancer Drugs.
23:675–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Godoy P, Hewitt NJ, Albrecht U, Andersen
ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger
J, et al: Recent advances in 2D and 3D in vitro systems using
primary hepatocytes, alternative hepatocyte sources and
non-parenchymal liver cells and their use in investigating
mechanisms of hepatotoxicity, cell signaling and ADME. Arch
Toxicol. 87:1315–1530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyamoto Y, Ikeuchi M, Noguchi H, Yagi T
and Hayashi S: Spheroid formation and evaluation of hepatic cells
in a three-dimensional culture device. Cell Med. 8:47–56. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mazzei M, Vascellari M, Zanardello C,
Melchiotti E, Vannini S, Forzan M, Marchetti V, Albanese F and
Abramo F: Quantitative real time polymerase chain reaction
(qRT-PCR) and RNAscope in situ hybridization (RNA-ISH) as effective
tools to diagnose feline herpesvirus-1-associated dermatitis. Vet
Dermatol. 30:491–e147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Browning DJ: Pharmacology of chloroquine
and hydroxychloroquine. Hydroxychloroquine and Chloroquine
Retinopathy. Browning DJ: Springer; New York, NY: pp. 35–63. 2014,
View Article : Google Scholar
|
|
22
|
Faustino-Rocha A, Oliveira PA,
Pinho-Oliveira J, Teixeira-Guedes C, Soares-Maia R, da Costa RG,
Colaco B, Pires MJ, Colaco J, Ferreira R and Ginja M: Estimation of
rat mammary tumor volume using caliper and ultrasonography
measurements. Lab Anim (NY). 42:217–224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yin YB, de Jonge HR, Wu X and Yin YL:
Mini-gut: A promising model for drug development. Drug Discov
Today. 24:1784–1794. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawai T, Yasuchika K, Ishii T, Miyauchi Y,
Kojima H, Yamaoka R, Katayama H, Yoshitoshi EY, Ogiso S, Kita S, et
al: SOX9 is a novel cancer stem cell marker surrogated by
osteopontin in human hepatocellular carcinoma. Sci Rep.
6:304892016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ferner RE and Aronson JK: Chloroquine and
hydroxychloroquine in covid-19. BMJ. 369:m14322020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Multicenter collaboration group of
Department of Science and Technology of Guangdong Province and
Health Commission of Guangdong Province for chloroquine in the
treatment of novel coronavirus pneumonia, . Expert consensus on
chloroquine phosphate for the treatment of novel coronavirus
pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 43:185–188. 2020.(In
Chinese). PubMed/NCBI
|
|
27
|
Aye T and Arndt PA: Utility of chloroquine
diphosphate in the blood bank laboratory. Immunohematology.
34:98–102. 2018.PubMed/NCBI
|
|
28
|
Meinao IM, Sato EI, Andrade LE, Ferraz MB
and Atra E: Controlled trial with chloroquine diphosphate in
systemic lupus erythematosus. Lupus. 5:237–241. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Wang C, Hu X, Yu C, Zhou L, Ding Z
and Zhou M: Gefitinib-mediated apoptosis is enhanced via inhibition
of autophagy by chloroquine diphosphate in cutaneous squamous cell
carcinoma cells. Oncol Lett. 18:368–374. 2019.PubMed/NCBI
|
|
30
|
Hu T, Li P, Luo Z, Chen X, Zhang J, Wang
C, Chen P and Dong Z: Chloroquine inhibits hepatocellular carcinoma
cell growth in vitro and in vivo. Oncol Rep.
35:43–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan C, Wang W, Zhao B, Zhang S and Miao J:
Chloroquine inhibits cell growth and induces cell death in A549
lung cancer cells. Bioorg Med Chem. 14:3218–3222. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brooks R and Bannigan K: Occupational
therapy interventions in child and adolescent mental health: A
mixed methods systematic review protocol. JBI Database System Rev
Implement Rep. 16:1764–1771. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guan YS and Liu Y: Interventional
treatments for hepatocellular carcinoma. Hepatobiliary Pancreat Dis
Int. 5:495–500. 2006.PubMed/NCBI
|
|
34
|
Perez-Garijo A, Fuchs Y and Steller H:
Apoptotic cells can induce non-autonomous apoptosis through the TNF
pathway. Elife. 2:e010042013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wong RS: Apoptosis in cancer: From
pathogenesis to treatment. J Exp Clin Cancer Res. 30:872011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pistritto G, Trisciuoglio D, Ceci C,
Garufi A and D'Orazi G: Apoptosis as anticancer mechanism: Function
and dysfunction of its modulators and targeted therapeutic
strategies. Aging (Albany NY). 8:603–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang Q, Xiao Y, Liu K, Zhong C, Zeng M
and Xiao F: Cr(VI)-induced autophagy protects L-02 hepatocytes from
apoptosis through the ROS-AKT-mTOR pathway. Cell Physiol Biochem.
51:1863–1878. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan L, Guo N, Cao Y, Zeng S, Wang J, Lv F,
Wang Y and Cao X: miRNA-145 inhibits myocardial infarction-induced
apoptosis through autophagy via Akt3/mTOR signaling pathway in
vitro and in vivo. Int J Mol Med. 42:1537–1547.
2018.PubMed/NCBI
|
|
39
|
Tong Y, Zhang G, Li Y, Xu J, Yuan J, Zhang
B, Hu T and Song G: Corilagin inhibits breast cancer growth via
reactive oxygen species-dependent apoptosis and autophagy. J Cell
Mol Med. 22:3795–3807. 2018.(Epub ahead of print). View Article : Google Scholar
|
|
40
|
Zeng Y, Li S, Wu J, Chen W, Sun H, Peng W,
Yu X and Yang X: Autophagy inhibitors promoted aristolochic acid I
induced renal tubular epithelial cell apoptosis via mitochondrial
pathway but alleviated nonapoptotic cell death in mouse acute
aritolochic acid nephropathy model. Apoptosis. 19:1215–1224. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu H, Gehart H, Artegiani B,
LÖpez-Iglesias C, Dekkers F, Basak O, van Es J, Chuva de Sousa
Lopes SM, Begthel H, Korving J, et al: Long-term expansion of
functional mouse and human hepatocytes as 3D organoids. Cell.
175:1591–1606 e19. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dutta D and Clevers H: Organoid culture
systems to study host-pathogen interactions. Curr Opin Immunol.
48:15–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yin Y, Chen S, Hakim MS, Wang W, Xu L,
Dang W, Qu C, Verhaar AP, Su J, Fuhler GM, et al: 6-Thioguanine
inhibits rotavirus replication through suppression of Rac1 GDP/GTP
cycling. Antiviral Res. 156:92–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Angelini F, Corbetti F, Nassuato G,
Okolicsanyi L and Zacchi C: Echography with portable equipment. Its
possibilities and limits in the hepatobiliary area. Radiol Med.
76:337–339. 1988.(In Italian). PubMed/NCBI
|
|
45
|
Yin YB, de Jonge HR, Wu X and Yin YL:
Enteroids for nutritional studies. Mol Nutr Food Res.
63:e18011432019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nuciforo S, Fofana I, Matter MS, Blumer T,
Calabrese D, Boldanova T, Piscuoglio S, Wieland S, Ringnalda F,
Schwank G, et al: Organoid models of human liver cancers derived
from tumor needle biopsies. Cell Rep. 24:1363–1376. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sun L, Wang Y, Cen J, Ma X, Cui L, Qiu Z,
Zhang Z, Li H, Yang RZ, Wang C, et al: Modelling liver cancer
initiation with organoids derived from directly reprogrammed human
hepatocytes. Nat Cell Biol. 21:1015–1026. 2019. View Article : Google Scholar : PubMed/NCBI
|